ABSTRACT
Long noncoding RNAs (lncRNAs) regulate the expression of mRNA and can affect various biological processes and phenotypes. Currently, studies of lncRNAs in cattle are under way, but their exact function for several tissues has not yet been established. Hanwoo cattle (Bos taurus coreanae) have inhabited the Korean peninsula for about 6000 years and are one of the representative domesticated animals in Korea. As a result of intensive breeding, the meat of Hanwoo cattle is high in marbling content and is preferred by Koreans and other East Asian people. In this study, the expression of lncRNAs was identified in 36 samples from skeletal muscle and three adipose tissues (intramuscular, subcutaneous, and omental) of nine Hanwoo individuals. We identified 76 tissue-specific lncRNAs for each of the four tissues using the differences in expression levels. Through QTL information, we could identify 12 lncRNAs associated with shear force and six lncRNAs associated with body weight, which are two important traits in the Hanwoo population breeding strategy. By the physical position comparison of lncRNA and Bovine transcripts information, we could identify 11 lncRNAs that were in bovine transcripts, and four of the 11 genes related to transcripts of lncRNAs were biologically associated with muscle function. We believe this Hanwoo lncRNAs study will help reveal the lncRNA role in the physiological mechanisms of these four tissues.
KEYWORDS: Hanwoo, beef cattle, LncRNA, muscle, adipose tissues
Introduction
Korean native Hanwoo cattle (Bos taurus coreanae) are a hybrid of Bos Taurus and Bos zebu. Hanwoo cattle migrated into and settled on the Korean peninsula in about 4000 BC (Han 1996). In the late 1970s, the Korean government launched a Hanwoo gene-breeding program to improve the quantity and quality of meat (Lee et al. 2013). Tenderness, taste, and aroma are usually considered to be important criteria for consumers’ purchase of beef (Savell et al. 1987). Hanwoo beef is famous for its high marbling content and relatively thin muscle fibers (Kim et al. 1994). Korean consumers are accustomed to the taste of Hanwoo beef, with a high content of oleic acid, and prefer it because of the high intramuscular fat content (Kim et al. 1999; Jung and Choi 2003). Thus, Hanwoo beef is regarded as the most expensive and delicious beef in Korea (Kim and Lee 2003) and is an important subject of study by many researchers. Hanwoo cattle are known to be prolific, but are not good at producing meat and milk because of their slow growth rate and milking capacity. Thus, the Korean cattle industry has aimed to improve their meat-production ability and to increase the number of cattle in order to meet the demand of the growing beef market in Korea (Kim and Lee 2000). In recent years, research has been conducted on muscle fibers and fatty tissues that affect the quality of beef in order to improve the meat quality of Hanwoo cattle (Moon et al. 2006; Hwang et al. 2010).
The bovine genome was one of the first decoded mammalian genomes. Because cattle are important farm animals as a major source of nutrients for humans (Tellam et al. 2009). RNA-seq technology was used to identify transcript expression patterns in bovine muscle and various adipose tissues (He and Liu 2013; Sheng et al. 2014).
Transcripts longer than 200 nucleotides that are not translated into protein are defined as long noncoding RNAs (lncRNAs). In previous studies, an lncRNA was thought to be a transcriptional noise like other ncRNAs (Wang et al. 2004; Struhl 2007). However, several studies have shown that the number of lncRNAs found in eukaryotes is increasing (Ulitsky 2016) in recent years. LncRNA is involved in post-transcriptional gene regulation through processes such as RNA maturation, transport, protein synthesis, and transcriptional gene silencing through chromosome regulation (Bernstein and Allis 2005; Whitehead et al. 2009; Geisler and Coller 2013). LncRNAs have a large effect on biological processes, such as cell differentiation, development, immune response, and tumorigenesis, by regulation of mRNA expression (Consortium et al. 2002; Ota et al. 2004; Wilusz et al. 2009; Fan et al. 2015; Gong et al. 2016).
LncRNAs have been studied in many species, including cattle. In cattle, 449 total lncRNAs are located in 405 intergenic regions. This provides a catalog of bovine lncRNAs for gene expression and confer systematic characterization of genomic features (Huang et al. 2012). Deep-transcriptome sequencing studies have identified many lncRNAs in bovine skin specimens (Weikard et al. 2013). The large intergenic noncoding RNAs (lincRNAs) in cattle have been studied, and 584 skeletal-muscle lincRNAs in nine Limousin bull calves were identified (Billerey et al. 2014). Previous studies looked at the metabolic differences between muscle and intramuscular adipose tissues of Hanwoo cattle using the RNA-seq technology and a systems biology approach (Lee et al. 2014). Therefore, we can carry out a Hanwoo lncRNA study using RNA seq data and other study results.
In this study, we compared the expression patterns of lncRNA between muscle and adipose tissues of Hanwoo cattle to better understand the physiological characteristic in Hanwoo meat production. We identified 76 tissue-specific lncRNAs for each of the four types of tissue and found 12 lncRNAs associated with shear force and six lncRNAs associated with the body weight, which are important traits in Hanwoo population breeding strategy. Additionally, thorough the physical position comparison of lncRNA and Bovine transcript information, we could identify 11 lncRNAs in Bovine transcripts, and two of these 11 genes related to transcripts of lncRNAs were associated with cow muscle function. This study will clarify the bovine biological characteristics and contribute to the production of high-quality Hanwoo cattle. In addition, we want to provide the basis for further research on the molecular biological characteristics in the energy storage and usage of Hanwoo cattle.
Materials and method
Sample preparation and RNA-seq analysis
All analysis was conducted with data reprocessed from a previous study (Lee et al. 2014). Animal and sample preparation are as follows.: A total of nine (three each of cows, steers, and bulls) Hanwoo cattle were used in this study. They were fed the same diet and managed at the same location, the Hanwoo Experimental Station in the National Institute of Animal Science, throughout the experiment. The average (± standard deviation) carcass weight of the cattle at slaughter was 430.2 (± 40.66) kg. Immediately after slaughter, muscle, intramuscular adipose tissues, and subcutaneous adipose tissues were separated and sampled. The omental adipose tissue was taken within the lesser curvature of the abomasum. All of the tissue samples were immediately frozen using liquid nitrogen and stored at −80°C until the analysis. Animal use, care, and experimental protocols for this experiment were reviewed and preapproved by the Institutional Animal Care and Use Committee of the National Institute of Animal Science (number 2010-042). Total RNA of tissues was isolated using TRIzol (Invitrogen) and an RNeasy RNA purification kit with DNase treatment (Qiagen). The mRNA was isolated from the total RNA using oligo-dT beads and was reverse transcribed into double-strand cDNA fragments. Constructing and sequencing the RNA-seq library for each sample were carried out based on the protocols of Illumina HiSeq2000 to generate 101 pair-end reads. The quality of RNA-seq reads from all of the tissues was checked using FastQC.
Analysis of lncRNA discovery
Filtering was conducted to remove the low-quality sequences. The filtered sequences were mapped to a Bovine Taurus genome (bosTau6) using STAR v2.4.0b (Dobin et al. 2013). Expression levels were calculated using Cufflinks v2.2.1. Bovine gene information was used to measure expression levels (Trapnell et al. 2010). The bovine lncRNA analysis was used to conduct mapping with reference to annotated bos taurus ensemble ID results (Koufariotis et al. 2015).
Statistical analysis
Differences in the expression level of each tissue were expressed by Heatmap using R package gplots (v3.0.1) (Warnes et al. 2016). An lncRNA principal component analysis (PCA) plot was used to identify differences between muscle and adipose tissues using Mev (http://mev.tm4.org/) (Howe et al. 2011). The DEGseq R package was used to identify differential expression of lncRNA between muscle and adipose samples (Wang et al. 2009). Significant lncRNAs were identified using cut-off of: log2(fold-change)| ≥ 1 and p-value ≤ 0.001. Tissue-specific differentially expressed lncRNAs statistics analysis used the Prism 5 program (San Diego, CA, USA) (Motulsky 2007). The Venn diagram was used to identify the lncRNA assemblages that were extracted from the four tissues. The Venn diagram was displayed using InteractiVenn (http://www.interactivenn.net/) (Heberle et al. 2015).
Quantitative trait locus (QTL) analysis
QTL regions for comparative analysis with lncRNAs were identified from the Cattle QTL Database (http://www.animalgenome.org/cgi-bin/QTLdb/BT/index). In the cattle, the QTL associated with the quality and productivity of the meat was selected. The position of the selected QTL was compared with the lncRNA expressed in muscle and adipose tissues.
Overlapped gene analysis
The location of lncRNA was identified by ensembl biomart (ensembl.org/biomart) using transcript ID. The databases used ensemble Genes 92 Cow genes (UMD3.1). We found for by overlapped gene predicted to be affected by lncRNA. The position information of lncRNA in the genome and the gene position information of the bovine were matched using Python script. Transcription directions of lncRNA and overlapped genes were used as transcript information by ensemble. Identification of pseudogenes was also confirmed through ensemble data base. (ensembl.org).
Results and discussion
RNA sequencing information
All analysis was conducted with data reprocessed from a previous study (Lee et al. 2014). RNA-seq raw data information is as follows. Totals of 34.2, 35.8, 35.1, and 38.1 Mb of raw reads were obtained on average from muscle, intramuscular adipose, subcutaneous adipose, and omental adipose tissues, respectively. More than 99.5% of the reads remained after being filtered by the quality control, and more than 95.9% of these were mapped to the reference genome. The average length of lncRNA was 866 bp, the minimum length was 209 bp, and the maximum was 3748 bp.
Expression patterns of lncRNA in Hanwoo cattle
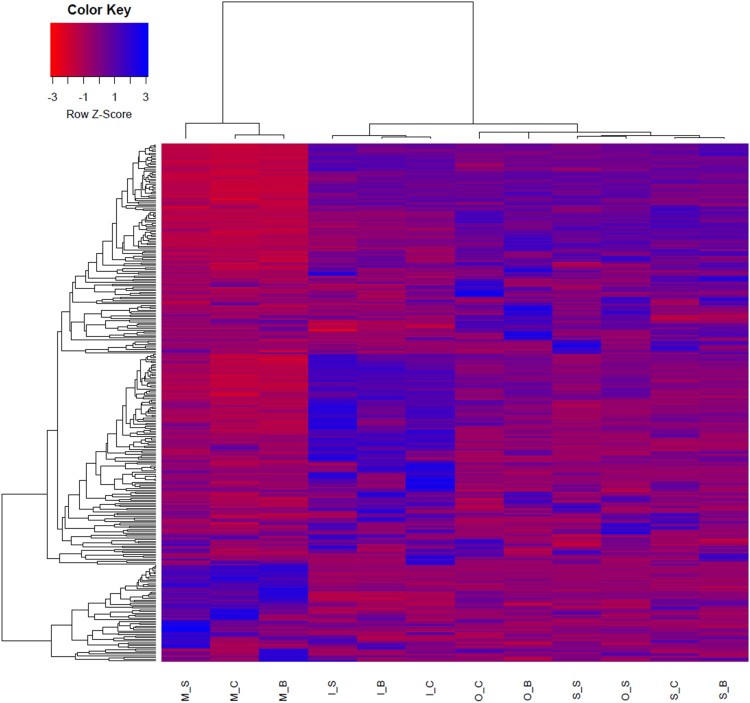
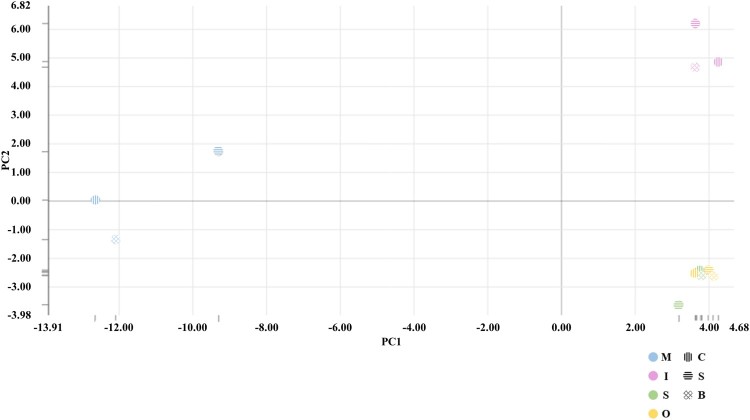
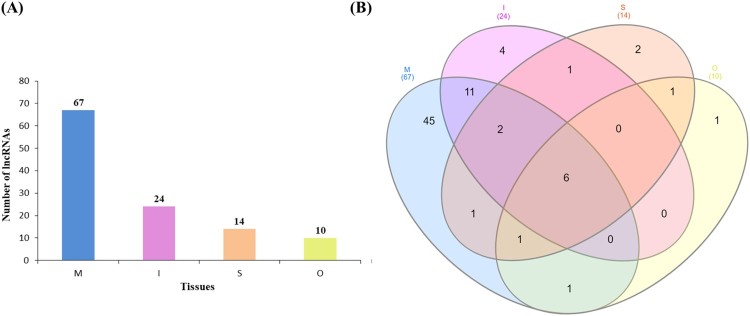
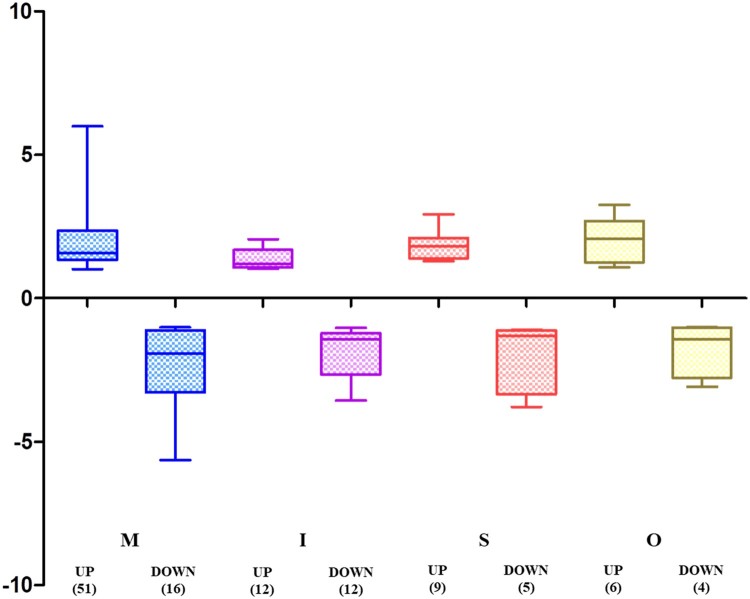
Clustering analysis using PCA analysis showed different patterns for muscle and adipose tissues. The expression of intramuscular tissue was also different from that of the other two adipose tissues (Figure 1). Hierarchical clustering analysis was performed on transcripts expression of muscle and adipose tissues in Hanwoo cattle. This study was similar to the muscle and adipose tissues DEG expression pattern in previous Hanwoo RNA-seq. Clustering analysis of the expression of transcripts showed a distinct transcript expression profile in muscle tissue expression patterns different from those of adipose tissues. Subcutaneous adipose and omental adipose tissues showed expression patterns similar, but intramuscular adipose showed an expression pattern different from those of two adipose tissues (Figure 2). We identified an expression pattern of lncRNA with muscle and adipose tissues.. The lncRNAs isolated from each tissue were 10 for omental adipose, 24 for intramuscular adipose, 14 for subcutaneous adipose, and 67 for muscle (Supplemental Tables 1 and 2). Significantly more lncRNAs were expressed in muscle tissues than in adipose tissues. We conducted PCA analysis to assess the relationship between muscle and adipose tissues. PCA analysis showed that distinguished between muscle and adipose tissues. Intramuscular adipose tissues were also distinguished from other adipose tissues (Figure 3). Expression of lncRNA differently expressed in muscle and adipose tissues were identified (Figure 4). Previous studies have examined lncRNA in several breeds of cattle. Huang et al. (2012) identified 449 putative lncRNAs using Bos taurus expressed sequence tags (ESTs) data (Huang et al. 2012). Weikard et al. (2013) identified 4,849 potential lncRNAs in the F2 of Charolais and German Holstein (Weikard et al. 2013). The expression patterns of lncRNAs were identified in skin with different pigmentation patterns. Billerey et al. (2014) identified 584 lincRNAs in the muscles of nine Limousin bulls (Billerey et al. 2014).
Figure 1.
Transcripts expressed by Hanwoo cattle in muscle and adipose tissues. The abbreviations under the bar mean tissues (I, Intramuscular adipose; M, Muscles; S, Subcutaneous adipose; O, Omental adipose) and sex (C, Cow; S, Steer; B, Bull).
Figure 2.
PCA plot of muscle and adipose tissues transcripts. The abbreviations in the colored circles mean tissues (I, Intramuscular adipose; M, Muscles; S, Subcutaneous adipose; O, Omental adipose) and the stripes in the circles mean sex (C, Cow; S, Steer; B, Bull).
Figure 3.
(A) Number of lncRNAs in muscle and adipose tissues. (B) Venn diagram showing the co-expression of lncRNAs muscle and three adipose tissues. The abbreviations mean tissues (I, Intramuscular adipose; M, Muscles; S, Subcutaneous adipose; O, Omental adipose).
Figure 4.
Number of tissue-specific differentially expressed lncRNAs. The abbreviations mean tissues (I, Intramuscular adipose; M, Muscles; S, Subcutaneous adipose; O, Omental adipose).
The specificity of the muscle tissues is presumed to be due to the difference from the adipose tissues. Energy is consumed mainly in muscle, and adipose tissue is the main energy-storage organization (Joe et al. 2009). The characteristics of muscle fibers affect meat quality characteristics, such as color, moisture retention, marbling, and texture (Totland et al. 1988). Adipose tissue is an important characteristic of meat and nutrition in livestock and affects animal productivity (Basu et al. 2009; Hausman et al. 2009). The amount and distribution of fatty acids is an important factor affecting meat quality in the beef industry (Brooks et al. 2011). Adipose tissue functions as an active metabolism and endocrine organ, and these are different functions that depend on the location (Kirkland and Dax 1984; Arner 1997; Kirkland et al. 1997).
Recent studies have shown that lncRNA plays an important role in the growth and differentiation of skeletal muscle. Lnc-MD1 is the lncRNA that plays an important role in myogenesis. It is specifically expressed during myoblast differentiation and leads to a transition from early- to late-stage muscle differentiation (Cesana et al. 2011). Many lncRNAs are similar to coding RNAs and are capped, spliced, and polyadenylated (Rose et al. 2011; Mercer et al. 2013). LncRNA evolved more rapidly than protein-coding genes and did not exhibit strict species conservation similar to protein-coding RNAs (Pang et al. 2006; Ulitsky et al. 2011).
lncRNA-related bovine economic traits
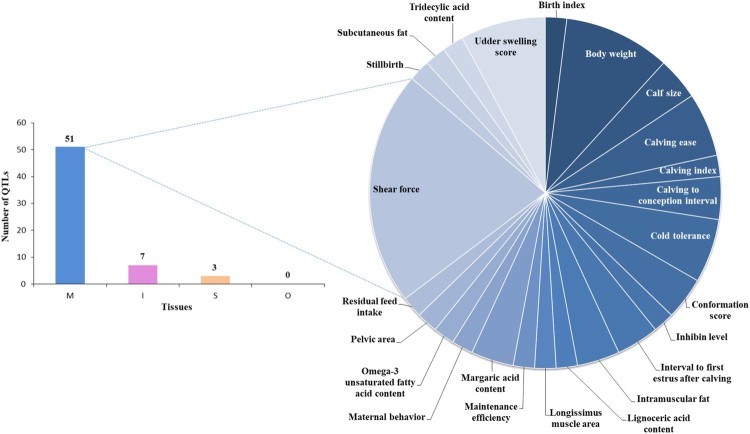
Identification of genomic loci that glow up the complex traits was facilitated by the development of quantitative trait locus (QTL) mapping (Sonah et al. 2014). QTL is important for analyzing the putative functions of genes. Integrating QTL with gene expression or location information may enable identification of candidate genes involved in the development of a specific phenotype in cattle. 507 lincRNAs within 550 QTLs relating to meat quality and muscle related traits (Billerey et al. 2014). The Korean government introduced the beef classification standard in 1992 to assess the quality of beef (National Federation of Animal Cooperatives (NLCF) 1998). The meat-quality grading system in Korea is mainly based on the marbling score (Moon et al. 2003; Park et al. 2002). Hanwoo cattle have been bred and selected to improve meat quality by emphasis on greater marbling to improve taste (Han et al. 2009; Choi et al. 2013). In this study, we conducted lncRNA and QTL analysis in muscle and adipose tissues of Hanwoo cattle. The QTL loci associated with the meat quality and productivity of the cattle were selected by referring to the Cattle QTL Database. LncRNA extracted from muscle and intramuscular adipose overlaps with the QTL domain in terms of meat quality and muscle development characteristics. We identified 61 lncRNA overlaps with QTL, and 51 of them were in the muscle. Seven lncRNAs were identified in the intramuscular adiposes, and three lncRNAs were identified in the subcutaneous adiposes, mainly in the Shear force QTL, body weight (yearling) QTL, and udder-swelling score QTL. These results suggest that more lncRNAs were extracted from muscles than from adipose tissues (Figure 5, Table 1). The lncRNAs associated with the meat quality and productivity of Hanwoo cattle were identified. These results are expected to be based on various studies of Hanwoo cattle.
Figure 5.
Number of QTLs associated with Hanwoo cattle economic traits in muscle and adipose tissues. The abbreviations mean tissues (I, Intramuscular adipose; M, Muscles; S, Subcutaneous adipose; O, Omental adipose).
Table 1. List of main QTLs associated with Hanwoo cattle economic traits.
| QTL | ID | Chr | QTL Peak | Reference | Transcript ID |
|---|---|---|---|---|---|
| Muscle | |||||
| Body weight (weaning) | 24711 | 3 | 28.72 | Mahdi Saatchi | ENSBTAT00000033843 |
| 24749 | 3 | 20.34 | ENSBTAT00000033843 | ||
| Body weight (yearling) | 22770 | 1 | 109.62 | Peters SO | ENSBTAT00000009029 |
| 24790 | 6 | 25.08 | Mahdi Saatchi | ENSBTAT00000034217 | |
| ENSBTAT00000065813 | |||||
| Shear force | 20762 | 5 | 115.33 | McClure MC | ENSBTAT00000056426 |
| 20764 | 6 | 71.21 | ENSBTAT00000034217 | ||
| ENSBTAT00000039582 | |||||
| ENSBTAT00000045907 | |||||
| ENSBTAT00000066034 | |||||
| 20770 | 8 | 27.55 | ENSBTAT00000017165 | ||
| 20773 | 8 | 119 | ENSBTAT00000015430 | ||
| 20817 | 25 | 2.61 | ENSBTAT00000049975 | ||
| 20824 | 26 | 40.66 | ENSBTAT00000054173 | ||
| 20826 | 27 | 23.61 | ENSBTAT00000007942 | ||
| 20833 | 29 | 56.05 | ENSBTAT00000044622 | ||
| Intramuscular adipose | |||||
| Shear force | 20770 | 8 | 27.55 | McClure MC | ENSBTAT00000017165 |
| Subcutaneous adipose | |||||
| Body weight (birth) | 24555 | 21 | 25.52 | Mahdi Saatchi | ENSBTAT00000063594 |
| Body weight (yearling) | 22770 | 1 | 109.62 | Peters SO | ENSBTAT00000045699 |
Analysis of lncRNA-related genes
A pseudogene is a gene that has developed from genes that encode proteins. However, pseudogenes have lost the ability to produce proteins and have long been regarded as nonfunctional genomes of evolution (Poliseno 2012). The pseudogene transcript is a subclass of lncRNA. It exhibits tissue specificity and is involved in various biological processes. Previous studies have shown that lncRNA can regulate the expression of nearby genes (Rinn et al. 2007; Mercer et al. 2009).
Some of the lncRNAs differentially expressed in the muscle and adipose tissues were confirmed to be pseudogenes of overlapped genes. In addition, we classified the transcription direction of the lncRNA and the overlapped gene (Table 2). Mutation in the Coenzyme Q2 (COQ2) gene is associated with Primary Coenzyme Q10 Deficiency. It causes defects of bioenergetics and myopathy with central nervous-system involvement (Quinzii et al. 2006; López-Martín et al. 2007). The regulation of the transcriptional coactivator megakaryoblastic leukemia 1 (MKL1) by actin cytoskeleton dynamics induces mouse adipocyte differentiation mediated by the peroxisome proliferator-activated receptor γ (PPARγ), a transcriptional regulator of adipogenesis (Nobusue et al. 2014).
Table 2. LncRNAs information with overlapped genes.
| LncRNA transcript ID | Gene ID | Gene Symbol | Loci | LncRNA strand | Gene strand |
|---|---|---|---|---|---|
| ENSBTAT00000033843 | ENSBTAG00000017566 | TUFT1 | 3:19,238,265-19,289,315 | Forward | Reverse |
| ENSBTAT00000026486 | ENSBTAG00000012307 | DTNA | 24:22,445,691-22,767,026 | Forward | Reverse |
| ENSBTAT00000064565 | ENSBTAG00000010542 | SPIRE1 | 24:43,323,645-43,488,851 | Forward | Reverse |
| ENSBTAT00000045699 | ENSBTAG00000010394 | MCF2L2 | 1:84,324,970-84,525,526 | Reverse | Forward |
| ENSBTAT00000047753 | ENSBTAG00000000687 | POC1B | 5:19,357,951-19,462,573 | Reverse | Reverse |
| ENSBTAT00000065010 | ENSBTAG00000002630 | MKL1 | 5:112,261,806-112,372,282 | Reverse | Reverse |
| ENSBTAT00000066034 | ENSBTAG00000005744 | COQ2 | 6:99,812,238-99,839,387 | Reverse | Reverse |
| ENSBTAT00000015430 | ENSBTAG00000046602 | PALM2 | 8:101,018,440-101,182,488 | Reverse | Forward |
| ENSBTAT00000065436 | ENSBTAG00000009427 | PPM1D | 19:12,602,357-12,662,178 | Reverse | Reverse |
| ENSBTAT00000049295 | ENSBTAG00000024801 | RANBP17 | 20:2,680,574-3,054,892 | Reverse | Forward |
| ENSBTAT00000065849 | ENSBTAG00000035705 | MTMR8 | X:101,228,033-101,491,280 | Reverse | Forward |
We also classified lncRNAs that are opposite to the transcription direction of the overlapped genes. The pairing of sense and antisense transcription leads to the formation of double-stranded RNA (dsRNA), which can trigger activation of the RNA interference (RNAi) pathway. The protein component of the RNAi pathway, Dicer, splits the dsRNA into smaller pieces known as small interfering RNAs (siRNAs). This siRNA is integrated into the RNA-induced silencing complex (RISC), which degrades and inhibits the mRNA of the parent coding gene (Tam et al. 2008; Watanabe et al. 2008). Two lncRNAs were identified as pseudogenes of muscle-associated genes. β-karyopherin genes include the RAN Binding Protein 17 (RANBP17) gene. In eukaryotes, β karyopherin protein mediates the nuclear cytoplasmic transport of macromolecules. In addition, Mouse Ranbp17 mRNA has high expression in skeletal muscle (Quan et al. 2008). Dystrobrevin-α, encoded by Dtna, belongs to a family of dystrophin-related proteins. The Dystrobrevin Alpha (DTNA) gene is highly expressed in skeletal muscle and is related to muscle diseases (Rees et al. 2007). Therefore, the lncRNAs identified here may have a crucial role for expression of genes at the specific locus that can functionally affect skeletal muscle, which could help clarify the function of the corresponding lncRNA.
Conclusion
This study is the first paper to profile tissue-specific lncRNAs by using comparative analysis of muscle and adipose tissue in Hanwoo cattle. We compared the bovine lncRNA found in the previous study using the transcript data of Hanwoo cattle and identified 76 lncRNAs. Expression patterns of lncRNAs in each tissue were identified. Many lncRNAs were identified in muscles, which have biological metabolism characteristics different from those of adipose tissue. We found the lncRNAs located in the QTL locus. QTL loci related to the meat quality and productivity of the cattle were selected for analysis. These lncRNAs were associated with cattle economic characteristics, such as shear force and body weight. In addition, we found the pseudogenes that are predicted to affect function in the muscles of Hanwoo cattle. In conclusion, we conducted a basic study on the characteristics of lncRNA that is expressed specifically in the skeletal muscle and adipose tissues of Hanwoo cattle. We also identified the candidate lncRNA for the economic traits of Hanwoo cattle. This study will contribute to metabolic function studies by cattle organizations.
Supplementary Material
Funding Statement
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology [grant number 2015R1C1A1A02037582] and a grant from the Next-Generation BioGreen 21 Program [grant number PJ01315101], Rural Development Administration, Republic of Korea. It is the result of Open-type agricultural SW convergence R&BD technology support project of Jeonju IT&CT industry promotion agency.
Acknowledgements
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2015R1C1A1A02037582) and a grant from the Next-Generation BioGreen 21 Program (PJ01315101), Rural Development Administration, Republic of Korea. It is the result of Open-type agricultural SW convergence R&BD technology support project of Jeonju IT&CT industry promotion agency.
Disclosure statement
No potential conflict of interest was reported by the authors.
ORCID
Jae-Young Choihttp://orcid.org/0000-0001-8264-1482
References
- Arner P.1997. Regional adipocity in man. J Endocrinol. 155:191–192. doi: 10.1677/joe.0.1550191 [DOI] [PubMed] [Google Scholar]
- Basu U, Guan L, Taniguchi M, Zhao Y, Dodson M.. 2009. Application of ‘omics’ technologies on improvement of meat quality. In Nutritional biochemistry: genomics, metabolomics and food supply. New York: Nova Science Publishers; p. 165–194. [Google Scholar]
- Bernstein E, Allis CD.. 2005. RNA meets chromatin. Genes Dev. 19:1635–1655. doi: 10.1101/gad.1324305 [DOI] [PubMed] [Google Scholar]
- Billerey C, Boussaha M, Esquerré D, Rebours E, Djari A, Meersseman C, Klopp C, Gautheret D, Rocha D.. 2014. Identification of large intergenic non-coding RNAs in bovine muscle using next-generation transcriptomic sequencing. BMC Genom. 15:499. doi: 10.1186/1471-2164-15-499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks M, Choi C, Lunt D, Kawachi H, Smith S.. 2011. Subcutaneous and intramuscular adipose tissue stearoyl-coenzyme a desaturase gene expression and fatty acid composition in calf-and yearling-fed angus steers1. J Anim Sci. 89:2556–2570. doi: 10.2527/jas.2010-3369 [DOI] [PubMed] [Google Scholar]
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I.. 2011. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell. 147:358–369. doi: 10.1016/j.cell.2011.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J-W, Lee K-T, Liao X, Stothard P, An H-S, Ahn S, Lee S, Lee S-Y, Moore SS, Kim T-H.. 2013. Genome-wide copy number variation in Hanwoo, black angus, and Holstein cattle. Mamm Genome. 24:151–163. doi: 10.1007/s00335-013-9449-z [DOI] [PubMed] [Google Scholar]
- Consortium F, I RGERGP, Team I 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nat. 420:563–573. doi: 10.1038/nature01266 [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15-21. doi: 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan C, Hao Z, Yan J, Li G.. 2015. Genome-wide identification and functional analysis of lincRNAs acting as miRNA targets or decoys in maize. BMC Genom. 16:793. doi: 10.1186/s12864-015-2024-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler S, Coller J.. 2013. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. 14:699–712. doi: 10.1038/nrm3679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Yang Q, Zeng Z, Zhang W, Li X, Zu X, Deng H, Chen P, Liao Q, Xiang B.. 2016. An integrative transcriptomic analysis reveals p53 regulated miRNA, mRNA, and lncRNA networks in nasopharyngeal carcinoma. Tumour Biol. 37:3683–3695. doi: 10.1007/s13277-015-4156-x [DOI] [PubMed] [Google Scholar]
- Han S-Y.1996. Animal breeds. In: Han S-Y, editor. Breeds of cattle. Seoul: Sunjin Publishing Company; p. 53–55. [Google Scholar]
- Han S-H, Cho I-C, Kim J-H, Ko M-S, Jeong H-Y, Oh H-S, Lee S-S.. 2009. AGHR polymorphism and its associations with carcass traits in Harrwoo cattle. Genes Genom. 31:35–41. doi: 10.1007/BF03191136 [DOI] [Google Scholar]
- Hausman G, Dodson M, Ajuwon K, Azain M, Barnes K, Guan L, Jiang Z, Poulos S, Sainz R, Smith S.. 2009. Board-invited review: the biology and regulation of preadipocytes and adipocytes in meat animals. J Anim Sci. 87:1218–1246. doi: 10.2527/jas.2008-1427 [DOI] [PubMed] [Google Scholar]
- He H, Liu X.. 2013. Characterization of transcriptional complexity during longissimus muscle development in bovines using high-throughput sequencing. PLoS One. 8:e64356. doi: 10.1371/journal.pone.0064356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heberle H, Meirelles GV, da Silva FR, Telles GP, Minghim R.. 2015. Interactivenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 16:169. doi: 10.1186/s12859-015-0611-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe EA, Sinha R, Schlauch D, Quackenbush J.. 2011. RNA-Seq analysis in MeV. Bioinformatics. 27:3209–3210. doi: 10.1093/bioinformatics/btr490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Long N, Khatib H.. 2012. Genome-wide identification and initial characterization of bovine long non-coding RNAs from EST data. Anim Genet. 43:674–682. doi: 10.1111/j.1365-2052.2012.02325.x [DOI] [PubMed] [Google Scholar]
- Hwang Y-H, Kim G-D, Jeong J-Y, Hur S-J, Joo S-T.. 2010. The relationship between muscle fiber characteristics and meat quality traits of highly marbled Hanwoo (Korean native cattle) steers. Meat Sci. 86:456–461. doi: 10.1016/j.meatsci.2010.05.034 [DOI] [PubMed] [Google Scholar]
- Joe AW, Yi L, Even Y, Vogl AW, Rossi F.. 2009. Depot-specific differences in adipogenic progenitor abundance and proliferative response to high-Fat diet. Stem Cells. 27:2563–2570. doi: 10.1002/stem.190 [DOI] [PubMed] [Google Scholar]
- Jung K., Choi C.. 2003. Development of technologies to improve competitiveness of Hanwoo. Seoul: Report to the Ministry of Agriculture, p; 85–98. [Google Scholar]
- Kim C, Lee E.. 2003. Effects of quality grade on the chemical, physical and sensory characteristics of Hanwoo (Korean native cattle) beef. Meat Sci. 63:397–405. doi: 10.1016/S0309-1740(02)00099-2 [DOI] [PubMed] [Google Scholar]
- Kim J, Lee C.. 2000. Historical look at the genetic improvement in Korean cattle. review. Asian-Australas J Anim Sci. 13:1467–1481. doi: 10.5713/ajas.2000.1467 [DOI] [Google Scholar]
- Kim I, Lee S, Kang S, Byun J, Lee M.. 1999. The physiochemical, microbial and sensory characteristics of Hanwoo and imported chilled beef. Korean J. Food Sci. An. 4:331–338. [Google Scholar]
- Kim C, Suck J, Ko W, Lee E.. 1994. Studies on the cold and frozen storage for the production of high quality meat of Korean native cattle ii: effects of cold and frozen storage on the drip, storage loss and cooking loss in Korean native cattle. J Food Sci. 14:151–154. [Google Scholar]
- Kirkland J, Cummins P, Steppan C, Dobson D, Cladaras M.. 1997. Decreasing preadipocyte differentiation capacity with aging is associated with blunted expression of the transcription factor, CCAAT enhancer binding protein α. Obes Res. 5:22. [Google Scholar]
- Kirkland JL, Dax EM.. 1984. Adipocyte hormone responsiveness and aging in the rat: problems in the interpretation of aging research. J Am Geriatr Soc. 32:219–228. doi: 10.1111/j.1532-5415.1984.tb02006.x [DOI] [PubMed] [Google Scholar]
- Koufariotis LT, Chen Y-PP, Chamberlain A, Vander Jagt C, Hayes BJ.. 2015. A catalogue of novel bovine long noncoding RNA across 18 tissues. PloS one. 10:e0141225. doi: 10.1371/journal.pone.0141225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K-T, Chung W-H, Lee S-Y, Choi J-W, Kim J, Lim D, Lee S, Jang G-W, Kim B, Choy YH.. 2013. Whole-genome resequencing of hanwoo (Korean cattle) and insight into regions of homozygosity. BMC Genom. 14:519. doi: 10.1186/1471-2164-14-519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-J, Park H-S, Kim W, Yoon D, Seo S.. 2014. Comparison of metabolic network between muscle and intramuscular adipose tissues in Hanwoo beef cattle using a systems biology approach. Int J Genom. 2014:679437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Martín JM, Salviati L, Trevisson E, Montini G, DiMauro S, Quinzii C, Hirano M, Rodriguez-Hernandez A, Cordero MD, Sánchez-Alcázar JA.. 2007. Missense mutation of the COQ2 gene causes defects of bioenergetics and de novo pyrimidine synthesis. Hum Mol Genet. 16:1091–1097. doi: 10.1093/hmg/ddm058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS.. 2009. Long non-coding RNAs: insights into functions. Nat Rev Genet. 10:155–159. doi: 10.1038/nrg2521 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Mattick JS.. 2013. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 20:300–307. doi: 10.1038/nsmb.2480 [DOI] [PubMed] [Google Scholar]
- Moon S, Hwang I, Jin S, Lee J, Joo S, Park G.. 2003. Carcass traits determining quality and yield grades of hanwoo steers. Asian-Australas J Anim Sci. 16:1049–1054. doi: 10.5713/ajas.2003.1049 [DOI] [Google Scholar]
- Moon S, Yang H, Park G, Joo S.. 2006. The relationship of physiological maturity and marbling judged according to Korean grading system to meat quality traits of hanwoo beef females. Meat Sci. 74:516–521. doi: 10.1016/j.meatsci.2006.04.027 [DOI] [PubMed] [Google Scholar]
- Motulsky H.2007. Prism 5 statistics guide. 2007. GraphPad Software. 31:39–42. [Google Scholar]
- National Livestock Cooperatives Federation (NLCF) 1998. Korean carcass grading standard. Seoul: National Livestock Cooperatives Federation. [Google Scholar]
- Nobusue H, Onishi N, Shimizu T, Sugihara E, Oki Y, Sumikawa Y, Chiyoda T, Akashi K, Saya H, Kano K.. 2014. Regulation of MKL1 via actin cytoskeleton dynamics drives adipocyte differentiation. NComms. 5:3368. [DOI] [PubMed] [Google Scholar]
- Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K.. 2004. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 36:40–45. doi: 10.1038/ng1285 [DOI] [PubMed] [Google Scholar]
- Pang KC, Frith MC, Mattick JS.. 2006. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 22:1–5. doi: 10.1016/j.tig.2005.10.003 [DOI] [PubMed] [Google Scholar]
- Park GB, Moon SS, Ko YD, Ha JK, Lee JG, Chang HH, Joo ST.. 2002. Influence of slaughter weight and sex on yield and quality grades of Hanwoo (Korean native cattle) carcasses. J Anim Sci. 80:129–136. doi: 10.2527/2002.801129x [DOI] [PubMed] [Google Scholar]
- Poliseno L.2012. Pseudogenes: newly discovered players in human cancer. Sci Signal. 5:re5–re5. doi: 10.1126/scisignal.2002858 [DOI] [PubMed] [Google Scholar]
- Quan Y, Ji Z-L, Wang X, Tartakoff AM, Tao T.. 2008. Evolutionary and transcriptional analysis of karyopherin β superfamily proteins. Mol Cell Proteomics. 7:1254–1269. doi: 10.1074/mcp.M700511-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinzii C, Naini A, Salviati L, Trevisson E, Navas P, DiMauro S, Hirano M.. 2006. A mutation in para-hydroxybenzoate-polyprenyl transferase (COQ2) causes primary coenzyme Q10 deficiency. Am J Hum Genet. 78:345–349. doi: 10.1086/500092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees ML, Lien C-F, Górecki DC.. 2007. Dystrobrevins in muscle and non-muscle tissues. Neuromuscul Disord. 17:123–134. doi: 10.1016/j.nmd.2006.11.003 [DOI] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E.. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 129:1311–1323. doi: 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose D, Hiller M, Schutt K, Hackermüller J, Backofen R, Stadler PF.. 2011. Computational discovery of human coding and non-coding transcripts with conserved splice sites. Bioinformatics. 27:1894–1900. doi: 10.1093/bioinformatics/btr314 [DOI] [PubMed] [Google Scholar]
- Savell J, Branson R, Cross H, Stiffler D, Wise J, Griffin D, Smith G.. 1987. National consumer retail beef study: palatability evaluations of beef loin steaks that differed in marbling. J Food Sci. 52:517–519. doi: 10.1111/j.1365-2621.1987.tb06664.x [DOI] [Google Scholar]
- Sheng X, Ni H, Liu Y, Li J, Zhang L, Guo Y.. 2014. Rna-seq analysis of bovine intramuscular, subcutaneous and perirenal adipose tissues. Mol Biol Rep. 41:1631–1637. doi: 10.1007/s11033-013-3010-8 [DOI] [PubMed] [Google Scholar]
- Sonah H, O’Donoughue L, Cober E, Rajcan I, Belzile F.. 2014. Combining genome-wide association and qtl analysis: Opportunities and challenges. ISB News Report. [DOI] [PubMed]
- Struhl K.2007. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 14:103–105. doi: 10.1038/nsmb0207-103 [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nat. 453:534–538. doi: 10.1038/nature06904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tellam RL, Lemay DG, Van Tassell CP, Lewin HA, Worley KC, Elsik CG.. 2009. Unlocking the bovine genome. BMC Genomics. 10:193. doi: 10.1186/1471-2164-10-193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totland GK, Kryvi H, Slinde E.. 1988. Composition of muscle fibre types and connective tissue in bovine m. semitendinosus and its relation to tenderness. Meat sci. 23:303–315. doi: 10.1016/0309-1740(88)90014-9 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, Salzberg SL, Wold BJ, Pachter L.. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 28:511–515. doi: 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I.2016. Evolution to the rescue: using comparative genomics to understand long non-coding RNAs. Nat Rev Genet. 17:601–614. doi: 10.1038/nrg.2016.85 [DOI] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP.. 2011. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell. 147:1537–1550. doi: 10.1016/j.cell.2011.11.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Feng Z, Wang X, Wang X, Zhang X.. 2009. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 26:136–138. doi: 10.1093/bioinformatics/btp612 [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang J, Zheng H, Li J, Liu D, Li H, Samudrala R, Yu J, Wong GK-S.. 2004. Mouse transcriptome: neutral evolution of ‘non-coding’complementary DNAs. Nat. 431. [PubMed] [Google Scholar]
- Warnes MGR, Bolker B, Bonebakker L, Gentleman R.. 2016. Package ‘gplots’. Various R Programming Tools for Plotting Data.
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nat. 453:539–543. doi: 10.1038/nature06908 [DOI] [PubMed] [Google Scholar]
- Weikard R, Hadlich F, Kuehn C.. 2013. Identification of novel transcripts and noncoding RNAs in bovine skin by deep next generation sequencing. BMC Genom. 14:789. doi: 10.1186/1471-2164-14-789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead J, Pandey GK, Kanduri C.. 2009. Regulation of the mammalian epigenome by long noncoding RNAs. Biochim Biophys Acta. 1790:936–947. doi: 10.1016/j.bbagen.2008.10.007 [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL.. 2009. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 23:1494–1504. doi: 10.1101/gad.1800909 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.