Abstract
Epigenetic mechanisms traditionally have been studied in the domains of development and disease, but they may also play important roles in ecological and evolutionary processes. In this article, we revisit historical as well as recent studies that indicate significant impacts of epigenetic processes on evolution. Our main focus is DNA methylation, which is a prevalent chemical modification of genomic DNA. First, it has been long known that DNA methylation acts as a major mutational facilitator in animal genomes and influences nucleotide compositions of genomes. More recently, genome-wide analyses have demonstrated that the current levels of DNA methylation can be predicted from the evolutionary signatures of DNA methylation, indicating that these two processes are intimately correlated. Indeed, the recent explosive growth in the knowledge of genomic DNA methylation in wide-ranging taxa has revealed that patterns of DNA methylation are surprisingly conserved across deep phylogenies. Interestingly, comparative analyses of humans and closely related primate species show that genomic regions that do show evolutionary divergence of DNA methylation are enriched for developmental and tissue specializations. A key question is how epigenetic patterns transmit between generations and impact evolutionary dynamics. On the one hand, some studies report direct transmissions of epigenetic features to the next generation. On the other hand, it is becoming clear that genomic sequence variants exist that encode and presumably regulate distinctive epigenetic patterns. For instance, numerous single-nucleotide polymorphisms that affect DNA-methylation patterns have been discovered in human populations. These studies begin to unveil a dynamic interplay between genomic and epigenomic factors across long and short evolutionary timescales.
Introduction
The definition, scope, and temporal scale of the term “epigenetics” are ever-evolving (Burggren and Crews 2014). Broadly speaking, epigenetic modifications, which do not involve direct alteration of underlying DNA sequences, determine how chromatin fibers are packaged in the nucleus, which in turn affect which genomic regions are accessible to cellular regulatory machineries. Therefore, epigenetic modifications may include variation in genome regulation that occurs during the lifespan of an organism (inheritance across mitotic divisions) as well as variation occurring in germ-lines and transmitted to the next generation. Traditionally, epigenetics has been pursued in the areas of development and diseases. However, it is becoming clear that the impact of epigenetics extends far beyond these traditional foci of research. The potential impact of natural epigenetic variation on ecology and evolution, in particular, has recently attracted much attention (Richards 2008; Flores et al. 2013). In this article, we discuss recent studies examining different aspects of epigenetics as they pertain to evolutionary processes.
The main focus of this review is DNA methylation, although other epigenetic mechanisms can also be observed through an evolutionary lens. DNA methylation refers to the addition of a methyl group (–CH3) to DNA nucleotides. In plant and animal genomes, DNA methylation is heavily biased toward cytosine nucleotides. While recent studies have identified additional chemical modifications of genomic DNA (Nabel et al. 2011; Yu et al. 2012), DNA methylation constitutes the major chemical modification of genomic DNA across much of the tree of life. Together with other epigenetic mechanisms such as histone-tail modifications in nucleosomes and non-coding RNA, DNA methylation affects how genomic DNA is packaged within each nucleus, and as such plays an essential role in critical cellular processes such as regulation of transposons, transcriptional silencing, genomic imprinting, and X-chromosome inactivation (Yoder et al. 1997; Jones and Takai 2001; Henderson and Jacobsen 2007).
Recent technological advances to characterize genomic DNA methylation without using pre-defined arrays (Bock et al. 2010; Harris et al. 2010) have enabled researchers to decipher whole-genome DNA-methylation maps from many organisms. Consequently, information on genomic DNA methylation from diverse species has rapidly accumulated during the past few years, providing unique and exciting insights into how DNA methylation evolves as well as and if, and how, it affects evolutionary processes.
Evolutionary signatures of DNA methylation correlate with current methylation patterns
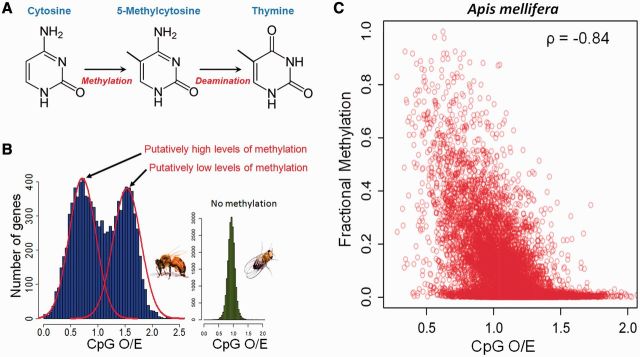
DNA methylation has undoubtedly had a significant impact on the evolution of genomic sequences. The best-known example occurs at cytosine–guanine dinucleotides, known as “CpG” sites. It was noted in the early 1980s that many animal genomes (mostly vertebrates) are depleted of CpG dinucleotides. Bird (1980) proposed that this lack of CpG sites was due to DNA methylation. The underlying mechanism is that methylated cytosines rapidly undergo spontaneous deamination reactions, which then result in CpG to TpG mutations on methylated strands (Coulondre et al. 1978) (Fig. 1A). Bird (1980) demonstrated that the degree of DNA methylation and the degree of depletion of CpG sites across diverse animal species were highly correlated. The levels of CpG depletion also exhibit regional variability: the most famous examples are the so-called “CpG islands.” These regions were first identified from mammalian genomes as being largely devoid of DNA methylation in contrast to a high genome-wide level of methylation. They also maintain a high content of CpG dinucleotides compared with the rest of the genome (Cooper et al. 1983; Bird 1986; Zeng et al. 2014).
Fig. 1.
(A) Methyl-cytosines generated by DNA methylation can undergo a spontaneous deamination to become thymines. Since DNA methylation in animals typically occurs within a CpG context, this will cause mutations from CpG to TpG. Methylated genomic regions will thus gradually lose CpG dinucleotides. (B) Normalized CpG content (CpG O/E) measures the depletion of CpG dinucleotides. In the honey bee, genes can be separated into two groups based upon the degree of depletion of CpG. Unmethylated genes of fruit flies, in contrast, do not show such bimodality. Figure modified from Elango et al. (2009). (C) CpG O/E and experimentally measured methylation in the honeybee are negatively correlated. Data on methylation are from Zemach et al. (2010).
Nonetheless, it is now known that depletion of CpG alone is not a sufficient indicator of DNA methylation at the level of the whole genome in all species. For example, genomes of hymenopteran species generally do not exhibit any depletion of CpG dinucleotides. In fact, CpGs are over-abundant in these genomes (The Honeybee Genome Sequencing Consortium 2006; The International Aphid Genome Consortium 2010; The Nasonia Genome Working Group 2010). On the other hand, these genomes encode functional DNA-methylation systems and exhibit variable levels of experimentally determined DNA methylation. It is likely that other factors than DNA methylation affect CpG composition of these genomes (Zeng and Yi 2010; Kent et al. 2012; Hunt et al. 2013b). Nevertheless, using the relative depletion of CpGs compared to the genomic background appears to be an efficient indicator for inferring regional differences of DNA methylation (Yi and Goodisman 2009) (Fig. 1B and C). For example, analyses of honey bee genomes have led to the finding that honey bees’ genes consist of methylated and non-methylated genes that also differ in their broad functional roles (Elango et al. 2009; Foret et al. 2009; Wang and Leung 2009).
An influx of experimentally generated data on DNA methylation from diverse organisms in recent years provides an opportunity to examine just how well this evolutionary measure of DNA methylation (CpG depletion) predicts the actual DNA methylation of specific genomic regions. Remarkably, experimentally measured levels of DNA methylation from multiple laboratories and studies are highly correlated with the degree of CpG depletion (Suzuki et al. 2007; Weber et al. 2007; Sarda et al. 2012; Wang et al. 2013) (Fig. 1C). However, it should be noted that DNA methylation is not the only force that affect the depletion of CpG dinucleotides: for example, biased gene conversion and stochastic effects can also significantly impact the degree of CpG depletion (Cohen et al. 2011). Nevertheless, the fact that the rather crude evolutionary signature of genomic DNA methylation corresponds well to the current patterns of DNA methylation strongly suggests that, at least for DNA methylation, the current epigenetic signals can be used as an indicator of past epigenetic patterns.
Conversely, we may be able to use evolutionary signatures of DNA methylation in genomic sequences to infer past evolutionary changes of DNA methylation. However, to do this, it is necessary to understand the rate of CpG to TpG mutation caused by DNA methylation, as well as the amount of time germ cells are subject to DNA methylation. Currently, such information is unavailable for most species. However, for mammals it is thought that germ cells are methylated for most of the organism’s lifetime, except for the short amount of time after fertilization during which DNA methylation signals are erased and then re-established (Monk et al. 1987; Reik et al. 2001). Consequently, mutations that originate from DNA methylation may accumulate throughout the lifetime of the organisms. In accord with this idea, mutations at CpG sites in genomes of primates show clock-like accumulation, in contrast to other mutations whose rates are more in line with the numbers of germ cell divisions (Kim et al. 2006).
Indeed, due to the recent developments in ancient DNA methods the reconstruction of past epigenetic patterns is now feasible. In a recent study (Pedersen et al. 2014), the nucleosome map of a 4,000 year old Paleo-Eskimo human was reconstructed based on sequence read coverage, as nucleosome DNA is preferentially conserved. Patterns of DNA methylation were also inferred from the post-mortem occurrence CpG to TpG rates (Pedersen et al. 2014; Gokhman et al. 2014). These studies open the exciting possibility to study the epigenomes of extinct populations or species, providing valuable information about evolutionary dynamics of epigenetic modifications and their impacts on regulation of gene expression.
Deep phylogenetic conservation of DNA methylation
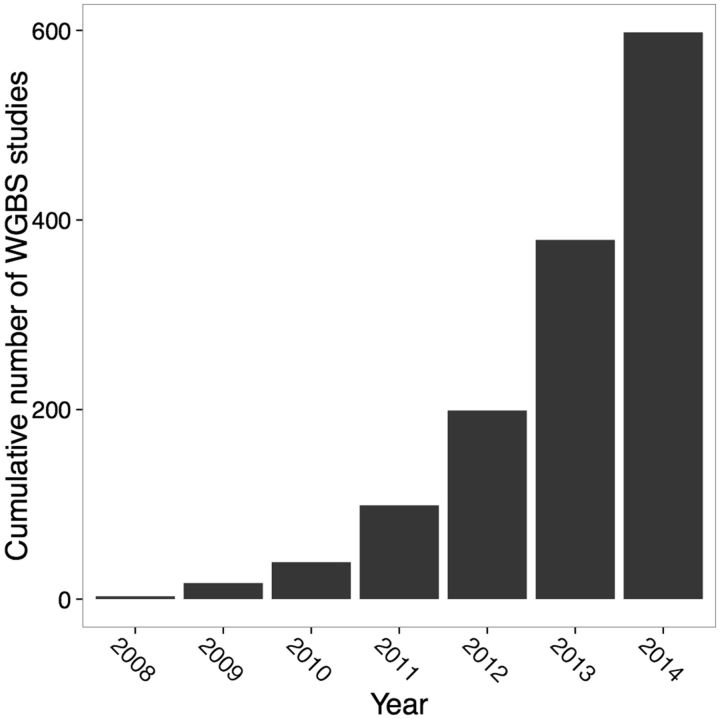
Given the high correlation between evolutionary signatures of DNA methylation and current methylation within genomes, a critical question is then “how do epigenetic patterns change over evolutionary time?” The first step in answering this question is to identify how epigenetic divergence between species occurs. The development of genome-wide methods of analyzing DNA methylation that combine the conversion of genomic DNA using sodium bisulfite with next-generation sequencing has opened tremendous new opportunities in this regard (Bock et al. 2010; Harris et al. 2010). In principle, this method can be applied to generate and examine maps of whole-genome methylation from any species with a reference genome. The number of studies employing such high-throughput sequencing methods of genomic DNA methylation is rapidly increasing, and is expected to reach 600 by the end of this year (Fig. 2 and Supplementary Table S1).
Fig. 2.
Rapid increase in the number of whole-genome methylation studies via bisulfite sequencing (WGBS) deposited in the Gene Expression Omnibus (GEO). Results were collected from the search “methylation profiling by high-throughput sequencing [Dataset Type]”. The 2014 estimate was extrapolated from the number of submissions submitted by January 30, 2014 (18 studies).
With these new genomic resources at hand, several recent studies have investigated how genomic DNA methylation changes over evolutionary timescales (Table 1). For example, Sarda et al. (2012) examined four highly divergent animal species that collectively span over a billion years of evolution (honey bee, silkworm, sea anemone, and sea squirt). DNA methylation in these species is restricted to gene bodies (Suzuki et al. 2007; Zemach et al. 2010), reflecting the ancestral pattern of DNA methylation in genomes of animals (Elango and Yi 2008; Suzuki and Bird 2008). Sarda et al. (2012) first classified genes into methylated and non-methylated genes, based on the patterns of experimentally determined DNA methylation. Then, using this binary classification of gene-methylation status, they examined how often methylation of genes changes between the two statuses (“methylated” versus “non-methylated”) across the phylogeny. Over 75% (429/563) of orthologs identified in this study were classified as “methylated” in all lineages, indicating a strong conservation of DNA methylation across an extremely long evolutionary timescale. These genes were highly enriched in basic “housekeeping” cellular functions (Sarda et al. 2012).
Table 1.
Comparative studies of DNA methylation suggest conservation of DNA methylation across different evolutionary timescales
| Species | Divergence time | Conservation patterns | |
|---|---|---|---|
| Zemach et al. (2010) | Several invertebrates, vertebrates, and plants | >Billions of years | Gene-body methylation is conserved in invertebrates and plants |
| Feng et al. (2010) | Several invertebrates, vertebrates, and plants | >Billions of years | This study, together with the above-mentioned study of Zemach et al. (2010), are the first studies to generate maps of whole-genome methylation from multiple, diverged species |
| Bonasio et al. (2012) | Two ant species, Camponotus floridanus and Harpegnathos saltator | >100 millions of years | The number of genes exhibiting conserved patterns of methylation across species are significantly higher than expected by chance |
| Sarda et al. (2012) | Honey bee, silkworm, sea squirt, sea anemone | >1 billion of years | 429/593 genes are consistently methylated in all species |
| Hunt et al. (2013a, b) | Honey bee, fire ant | ∼60 millions of years | 93% of orthologous genes methylated in both species |
| Wang et al. (2013) | Jewel wasp, honey bee | ∼180 millions of years | 1.23-fold (42.2/34.3%) enrichment of co-methylated genes between the two species |
Notes: For convenience, we discuss studies with focus on genomes of animals. These studies all used whole-genome sequencing of bisulfite converted genomic DNA to generate DNA-methylation maps.
A secondary conclusion is that methylated genes tend to be more conserved than non-methylated genes at the sequence level because orthologs are identified using sequence-divergence. Indeed, methylated genes are generally more conserved at the sequence-level than are non-methylated genes (Takuno and Gaut 2011; Sarda et al. 2012; Wang et al. 2013). Similarly, between the honeybee and the fire ant, 93% of orthologous genes are methylated in both species (Hunt et al. 2013a). Wang et al. (2013) found that a significantly higher number of genes than expected are co-methylated between honey bees and jewel wasps. In addition, differential methylation between castes of the same species is also somewhat conserved across species (Bonasio et al. 2012). When examined in detail, regional patterns of DNA methylation within a gene (heavy methylation of 5′ regions compared with 3′ regions) are also conserved between hymenopteran insects (Hunt et al. 2013c). Similar findings were also reported for plants (e.g., Takuno and Gaut 2013).
Methods for identifying methylated versus non-methylated genes and DNA-methylation levels vary between studies, rendering direct comparisons difficult. Nevertheless, these studies demonstrate that, on the whole, patterns of DNA methylation exhibit a surprising degree of evolutionary conservation across divergent species. One caveat of these studies is that methylation data typically are generated from whole bodies, rather than from specific cell types. Thus, the patterns of methylation in these data may reflect those of predominant cell types in each organism. Although it is unlikely that the genome-wide patterns across long evolutionary timescales would be systematically biased due to this reason, tissue-type, and cell-type-specific patterns of DNA methylation should be taken into account if we want to study the evolution of DNA methylation in detail.
Functional specificity of divergence of DNA methylation between species
Specifically, comparisons of DNA methylation of homologous tissues between closely related species could provide insights into the nature of genes or genomic regions that change methylation among species, as well as the underlying evolutionary mechanisms. Recent studies are beginning to address these questions using humans and closely related primates, thereby reflecting the growing interest of epigenetic divergence as a potential molecular mechanism underlying human-specific evolution (Table 2). These studies generally have found that levels of DNA methylation of specific regions are highly similar between humans and chimpanzees, supporting the strong conversation of regional patterns of DNA methylation across evolutionary time. At the same time, they have revealed foci of DNA-methylation divergence that often coincide with developmental specializations.
Table 2.
Analyses of the divergence of whole-genome methylation between humans and closely related primates
| Species | Tissue/cell type | Method | Patterns of conservation and divergence | |
|---|---|---|---|---|
| Pai et al. (2011) | Human, chimpanzee | Heart, liver, kidney | Illumina 27K Chip | Substantial inter-tissue conservation across species. 12–18% of interspecies differences in expression are associated with promoter methylation differences |
| Molaro et al. (2011) | Human, chimpanzee | Sperm | WGBS | Average correlation between the two species is 0.87 |
| Reported that human-specific hypo-methylated regions occur near genes encoding neurological functions | ||||
| Martin et al. (2011) | Human, chimpanzee, orangutan | Neutrophils | HpaII digestion and MethylSeq | High conservation methylome-wide except for ∼10% CpG island-like regions |
| Hodges et al. (2011) | Human, chimpanzee | HSPC, B cells, neutrophils | WGBS | Hypo-methylated regions show significant overlap between species. Inter-tissue variability of region-length is similar in humans and chimpanzees |
| Zeng et al. (2012) | Human, chimpanzee | Prefrontal cortex | WGBS | 3.5% (474/13,454) promoters are differentially methylated |
| Wang et al. (2012) | Human, macaque | Prefrontal cortex | MeDIP-Chip and SEQUENOM MassARRAY | Patterns of methylation of the brain are similar, with only few differentially methylated regions identified. |
| Identified two differentially methylated regions with protein-conservation involved in neural functions | ||||
| Hernando-Herraez et al. (2013) | Human, chimpanzee, bonobo, gorilla, orangutan | Peripheral blood | Illumina 450K Chip | Correspondence between protein sequence and gene regulation except for ∼800 genes.184 genes perfectly conserved at protein level show epigenetic differences between humans and chimpanzees |
For example, Pai et al. (2011) investigated the patterns of DNA methylation of multiple humans and chimpanzees from three distinctive tissues (heart, kidney, and liver) using arrays of DNA methylation designed for humans. A unique advantage of their experimental design is that the degree of methylation-divergence among tissues and among species could be investigated simultaneously. Using this method, they demonstrated a greater tissue-specific methylation-divergence compared with the species-specific methylation-divergence. In other words, according to Pai et al. (2011), DNA methylation appears to play a highly conserved tissue-specific role in humans and chimpanzees. In fact, in another study, it was shown that tissue-specific patterns of DNA methylation appear to be conserved between even more distantly related taxa: humans and mice (diverged ∼80 Ma) (Irizarry et al. 2009). Nevertheless, Pai et al. (2011) also identified many differentially methylated regions between humans and chimpanzees, and these regions were enriched in the category of “development” gene ontology. In particular, they demonstrated that differentially methylated regions between humans and chimpanzees exhibit particularly strong correlations with gene expression, indicating the significance of methylation-divergence for gene-expression divergence.
One potential caveat of using pre-developed arrays to analyze DNA methylation is that it may fail to capture the dynamic nature of DNA methylation of sites that are not pre-selected by specific arrays. In addition, underlying sequence differences between species can affect the efficiency of hybridization to arrays using probes designed for humans. More recent studies thus begin to employ sequencing of whole-genome bisulfite converted genomic DNA. These studies, focusing on specific tissues in humans and closely related primates, further indicate that the divergence of DNA methylation may be related to the divergence of functional specificity. For example, Zeng et al. (2012) analyzed whole-genome methylation maps of the prefrontal cortex from several humans and chimpanzees. They found that levels of DNA methylation of gene bodies (transcription units) were mostly conserved between the two species. However, hundreds of promoters exhibited highly divergent methylation levels between humans and chimpanzees (Fig. 3). The within-species variation in DNA methylation was generally much lower compared with between-species variation (Fig. 3). This pattern is indicative of potentially species-level divergence in DNA methylation for these regions. Interestingly, these differentially methylated promoters included many of those for genes that are associated with neuropsychiatric disorders and neurological development (Zeng et al. 2012). Another recent study compared DNA methylation in the peripheral blood of humans, chimpanzees, bonobos, gorillas, and orangutans using a methylation array designed for humans (Hernando-Herraez et al. 2013). Genes identified as having undergone human-specific changes in DNA methylation were enriched in circulatory-system functions. Together, these studies indicate that we can identify genomic regions that are differentially methylated between closely related primate species, and that these regions appear to be related to regulation of tissue-specific functions.
Fig. 3.
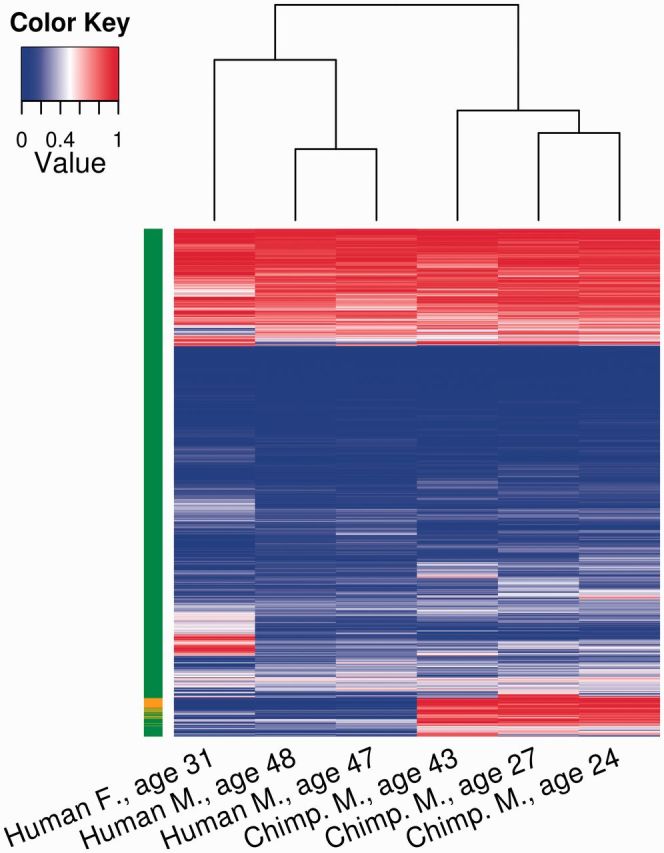
Levels of DNA methylation of 12,063 orthologous promoters in three humans and three chimpanzees analyzed by Zeng et al. (2012). Hierarchical clustering both within and between species was performed using the average methylation of promoters to generate a heatmap. The column to the left of the heatmap designates promoters as similarly methylated (green) or as highly diverged between species (orange).
An interesting, yet presently unexplained, observation is that, as reported by several studies, genes associated with neurological functions exhibit human-specific patterns of DNA methylation, even in non-neurological tissues. Molaro et al. (2011) examined whole-genome methylation maps of human and chimpanzee sperm. Interestingly, genes near human-specific hypomethylated regions were enriched for neurological functions (Molaro et al. 2011). Similarly, regions of human-specific DNA methylation in the peripheral blood also included many genes involved in neurological functions (Hernando-Herraez et al. 2013).
Speciation and epigenetic divergence: evidence from flies and mice
Epigenetic divergence has been shown to directly affect the divergence of species by conferring hybrid incompatibility. Genetic determinants of hybrid incompatibilities are often referred to as “speciation genes” (Orr et al. 2004). For example, one of most famous genetic determinants of hybrids’ male sterility in Drosophila is a gene named Odysseus Homeobox (OdsH), which causes male sterility in closely related Drosophila simulans–Drosophila mauritiana hybrids (Ting et al. 1998). Even though this gene is a major locus underlying hybrid sterility, its normal function in conspecific context shows at most a modest effect (Sun et al. 2004), and the mechanism by which it causes hybrid sterility has remained unclear until recently. It was shown that OdsH binds to repetitive DNA in heterochromatic regions, and subsequently affects chromosome condensation (Bayes and Malik 2009). The binding targets of this gene have sufficiently differentiated between D. simulans and D. mauritiana to cause divergent patterns of heterochromatin condensation between these species (Bayes and Malik 2009). Consequently, divergence at the level of chromatin condensation causes premeiotic defects in hybrids (Bayes and Malik 2009). In other words, incompatibility at the level of chromatin packaging explains male sterility of hybrids in at least some Drosophila species. This example showcases that genetic changes causing an epigenetic alteration with little phenotypic effect in one species may still have a large effect for inter-specific hybrids.
Another well-known example of a speciation gene is found in the crosses of the mouse subspecies Mus musculus musculus and Mus musculus domesticus. These interspecific hybrids are sterile due to spermatogenic failure. A major determinant of this hybrid sterility is the Prdm9 locus (Mihola et al. 2009). This gene encodes a histone H3 lysine 4 (H3K4) trimethyltransferase (Mihola et al. 2009), thus capable of modulating a key epigenetic modification of histone-tails (H3K4me3, often associated with “active” transcriptional domains). The detailed mechanism of how this gene causes sterility of hybrids is not completely resolved yet, but it is known to involve chromosome disjunctions and malformation in inter-specific hybrids and few phenotypic effects in the parental species (Flachs et al. 2012; Bhattacharyya et al. 2013).
Evolutionary implications of epigenetic patterns
The epigenome constitutes the interface between an organisms’ genome and its environment. Environmental factors such as chemicals, nutritional factors, or pathogens can alter the epigenetic landscape. A well-known example is how nutrition can determine the caste system in honey bees (Kucharski et al. 2008). In addition, epigenetic drift (errors accumulated during mitotic divisions), epimutation (spontaneous loss or gain of epigenetic marks), and genetic polymorphism can also generate epigenetic variation (see Richards 2008 for a review). How then are these epigenetic variations inherited to the next generation?
One mechanism is a direct inheritance of epigenetic modifications themselves to the next generation. Indeed, identifying “trans-generational inheritance”, loosely defined as inheritance of new epigenetic patterns across generations, constitutes a currently active area of research (Schmitz et al. 2011; Daxinger and Whitelaw 2012; Flores et al. 2013; Crews 2014). However, in mammals, epigenetic marks are erased in early embryos before being re-established, a process referred to as “reprogramming” (Reik et al. 2001; Hochedlinger and Jaenisch 2006; Feng et al. 2010). The epigenetic reprogramming also occurs in flowering plants, where it may be linked to silencing of transposable elements (Feng et al. 2010). Because of this reprogramming, a possible evolutionary role of epigenetic marks has been traditionally neglected, especially in vertebrate evolution. Recently however, studies from diverse plant and animal taxa have begun to show that some epigenetic marks escape epigenetic reprogramming. Molecular mechanisms of potential trans-generational epigenetic inheritance may involve transmission of non-coding RNA molecules and chromatin states in the germ-lines, such as transposon-silencing Piwi-interacting RNAs in fruit fly egg and histone modifications in human sperm (Brennecke et al. 2008; Hammoud et al. 2009, respectively).
However, determining the actual mechanisms of trans-generational inheritance is not without difficulties. Among other confounders, studies of trans-generational inheritance need to rule out the contribution of DNA sequence mutations, which can be challenging (e.g., mutations in genomic regions containing structural variation can be difficult to detect by current methods). Consequently, examples of trans-generational inheritance are still scarce and the evolutionary implications of heritable epigenetic alleles in organisms’ phenotypes, and ultimately, in their survival are yet to be determined. Most probably, instances of inherited epigenetic variation differ largely among taxa, these being more frequent in plants than in vertebrates and especially mammals, where the reestablishment of epigenetic state is almost complete in the new generation.
Genetic determinants of epigenetic patterns
On the other hand, evidence is accumulating that DNA sequence may control inter-individual epigenetic variation and its functional consequences in natural populations, with the caveat that these conclusions are largely from datasets from humans. First, several studies have measured the genome-wide heritability of CpG methylation using twins. These studies demonstrate modest, yet substantial, levels of genome-wide heritabilities, ranging from 5% to 19% (Boks et al. 2009; Kaminsky et al. 2009; Bell et al. 2012; Gordon et al. 2012; Grundberg et al. 2013). Further evidence for the genetic control of methylation comes from associations between common single-nucleotide polymorphisms (SNPs) (>5% frequency) and DNA methylation at individual CpG sites. Thousands of SNPs have been identified to act as quantitative trait loci affecting DNA methylation (referred to as “mQTLs”) in a variety of human tissues (Table 3). Most identified mQTLs were found within 1 Mb of the CpG sites, but some mQTLs could be located much farther away or even reside in different chromosomes (Gibbs et al. 2010; Zhang et al. 2010; Bell et al. 2011). A major limitation of these studies is that they are based on arrays and use small sample sizes. Therefore, the figures for both heritability and the number of mQTLs may be underestimates of the true genetic control of variation in methylation.
Table 3.
Studies on variation in inter-individual methylation in human populations, describing association with genetic variants
| Ancestrya | Tissue/cell type (N) | Method (methylation/genotype chips) | # CpG | |
|---|---|---|---|---|
| Zhang et al. (2010) | European | Cerebellum (153) | Illumina 27K/Affymetrix 5.0K | 748 (out of 8.6K) |
| Gibbs et al. (2010) | European | Cerebellum (108) | Illumina 27K/Illumina 550K | >1K |
| Frontal cortex (133) | ||||
| Temporal cortex (127) | ||||
| Caudal pons regions (125) | ||||
| Bell et al. (2011) | African (Hapmap YRI) | Lymphoblastoid cell lines (referred to as “LCL”) (77) | Illumina 27K/HapMap (3.8 M) | 217 |
| Gutierrez-Arcelus et al. (2013) | European | Fibroblasts (107) | Illumina 450K/Illumina 2.5M | >14K |
| LCL (111) | ||||
| T-cells (66) | ||||
| Fraser et al. (2012) | African, European (Hapmap YRI,CEU) | LCL (90 per population) | Illumina 27K/HapMap (3.8 M) | >49 |
| Moen et al. (2013) | African, European (Hapmap YRI,CEU) | LCL (>60 per population) | Illumina 450K/HapMap (3.8 M) | 1.8K (out of 37K)b |
| Heyn et al. (2013) | African, East Asian, European | LCL (>72 per population) | Illumina 450K/Illumina 550K and 650K | 298 (out of 439)b |
Notes: Associations are given as approximate number of CpGs (“#CpG” column) that show statistical significance for association with one or more SNPs. Please note that the methods for detecting significant associations differ among studies and thus, numbers are not directly comparable. If more than one tissue or population is tested, numbers indicate the smallest value per tissue or population.
aAncestry following the designation of the International HapMap Project (Thorisson et al. 2005). YRI: Yoruba in Ibadan, Nigeria. CEU: Utah residents with ancestry from northern and western Europe.
bThese studies only search associations on a subset of CpGs that show significant differences of DNA methylation between populations.
Recent studies also demonstrate that levels of CpG methylation vary significantly across human populations, and that this variation is, at least partially, genetically determined. For example, Heyn et al. (2013) analyzed the patterns of whole-blood methylation from African-American, Asian-American, and European-American individuals and concluded that genetic variation could account for two-thirds of the population-specific CpG methylations (Heyn et al. 2013). Interestingly, these population-specific CpGs are enriched for other epigenetic marks such as histone modifications and the binding of transcription factors. Moreover, some of these CpGs show signatures of accelerated evolution in some human populations since the divergence of humans and chimpanzees (Heyn et al. 2013). Another study by Moen et al. (2013) detected mQTLs in the HapMap lymphoblastoid cell-lines of European and African ancestries. Interestingly, many of the detected mQTLs were involved in diseases that are present at different prevalence in the two ancestries, such as cardiovascular and autoimmune disorders. More recently, a study of duplicate genes in the human genome illustrated that differential DNA methylation between duplicate genes are surprisingly consistent across 10 distinct human tissues, suggesting that underlying determinants of methylation states exist at the genomic locations examined (Keller and Yi 2014). Overall, these studies provide evidence that variation in DNA methylation is at least partially heritable, functionally relevant for transcriptional regulation and for susceptibilities to disease, and potentially subject to natural selection. Consequently, they support the view that epigenetic patterns are to a substantial degree subject to the well-defined evolutionary rules of mutation and natural selection because they are ultimately determined by genomic variants.
Conclusions and future directions
The strong correlation between evolutionary signatures and current patterns of DNA methylation, as well as the robust conservation of gene-specific DNA methylation in deep phylogenies, indicate that patterns of DNA methylation are evolutionarily stable for many genomic regions. What are the underlying molecular bases for such conservation of DNA methylation, and what are the functional and evolutionary consequences? Functional analyses of genes and genomic regions with respect to their evolutionary epigenetic conservation may provide clues to these pressing questions. Several studies conclude that genes that are consistently methylated in deep phylogenies are enriched in essential, “housekeeping” cellular functions (Hunt et al. 2010; Sarda et al. 2012; Takuno and Gaut 2013).
One caveat of these comparative studies of DNA methylation is that they often use whole bodies. Consequently, cellular heterogeneity among different organisms can bias the observations. Recent studies of humans and closely related primates begin to address this issue by focusing on epigenetic variation in specific tissues and/or cell types. These studies indicate that differentially methylated regions between closely species may be implicated in developmental specializations, and potentially, adaptive evolution. Also, two of the best-studied examples of the so-called speciation genes involve epigenetic incompatibilities (Mihola et al. 2009; Bayes and Malik 2009). Synthesizing these diverse observations, the emerging picture is that epigenetic conservation in evolutionary timescale is linked to functional conservation, whereas epigenetic divergence between closely related species may be related to functional specificity.
An outstanding question is how to reconcile the timescale of epigenetic phenomena (typically confined within an organism’s lifespan) with the multiple-generation timescale of evolution. One possibility is that trans-generational inheritance of epigenetic marks could operate with no influence of genomic determinants (Morgan et al. 1999; Dias and Ressler 2014). Further efforts must be made to study cases of epigenetic inheritance and to characterize the underlying mechanisms. Non-coding RNA and histone modification transmission through the germ-line may constitute rare but clear examples in which inherited epigenetics changes could have a role in evolution. On the other hand, genetic variation between individuals may also affect epigenetic patterns, which in turn cause functional consequences that are subject to natural selection. Indeed, many different types of studies indicate the presence of genomic variants that affect epigenetic patterns. For example, studies of human populations provide evidence that a large number of SNPs are associated with variation in DNA methylation (Table 3). Future studies with increased sample sizes and systematic control for confounders, such as heterogeneous cell populations and age, will help elucidate the details of the association between genomic and epigenomic factors. Furthermore, environmental effects could modulate epigenetic patterns (such as DNA methylation) and indirectly impact genomic features, by means of increasing local transposition, mutation rates, or recombination rates (Richards 2008). Elucidating the detailed mechanisms of genome and epigenome interactions is a promising lead to understanding the true significance of epigenetics on evolution.
Supplementary data
Supplementary Data available at ICB online.
References
- Bayes JJ, Malik HS. Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science. 2009;326:1538–41. doi: 10.1126/science.1181756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, Gilad Y, Pritchard JK. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12: R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell JT, Tsai PC, Yang TP, Pidsley R, Nisbet J, Glass D, Mangino M, Zhai G, Zhang F, Valdes A, et al. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLoS Genet. 2012;8:e1002629. doi: 10.1371/journal.pgen.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya T, Gregorova S, Mihola O, Anger M, Sebestova J, Denny P, Simecek P, Forejt J. Mechanistic basis of infertility of mouse intersubspecific hybrids. Proc Natl Acad Sci USA. 2013;110:E468–E477. doi: 10.1073/pnas.1219126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res. 1980;8:1499–1504. doi: 10.1093/nar/8.7.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–13. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- Bock C, Tomazou EM, Brinkman AB, Muller F, Simmer F, Gu H, Jager N, Gnirke A, Stunnenberg HG, Meissner A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 2010;28:1106–14. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boks MP, Derks EM, Weisenberger DJ, Strengman E, Janson E, Sommer IE, Kahn RS, Ophoff RA. The relationship of DNA methylation with age, gender and genotype in twins and healthy controls. PLoS One. 2009;4:e6767. doi: 10.1371/journal.pone.0006767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonasio R, Li Q, Lian J, Mutti NS, Jin L, Zhao H, Zhang P, Wen P, Xiang H, Ding Y, et al. Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr Biol. 2012;22:1755–1764. doi: 10.1016/j.cub.2012.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–92. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burggren WW, Crews D. Epigenetics in comparative biology: why we should pay attention. Integr Comp Biol. 2014;54:7–20. doi: 10.1093/icb/icu013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Taggart MH, Bird AP. Unmethylated domains in vertebrate DNA. Nucleic Acids Res. 1983;11:647–58. doi: 10.1093/nar/11.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen NM, Kenigsberg E, Tanay A. Primate CpG Islands are maintained by heterogeneous evolutionary regimes involving minimal selection. Cell. 2011;145:773–86. doi: 10.1016/j.cell.2011.04.024. [DOI] [PubMed] [Google Scholar]
- Coulondre C, Miller JH, Farabaugh PJ, Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978;274:775–80. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Crews D. Epigenetic synthesis, or how environmental contamination has changed the course of evolution. 2014 [Google Scholar]
- Daxinger L, Whitelaw E. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet. 2012;13:153–62. doi: 10.1038/nrg3188. [DOI] [PubMed] [Google Scholar]
- Dias BG, Ressler KJ. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat Neurosci. 2014;17:89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elango N, Hunt BH, Goodisman MAD, Yi SV. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proc Natl Acad Sci USA. 2009;106:11206–11211. doi: 10.1073/pnas.0900301106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elango N, Yi SV. DNA methylation and structural and functional bimodality of vertebrate promoters. Mol Biol Evol. 2008;25:1602–8. doi: 10.1093/molbev/msn110. [DOI] [PubMed] [Google Scholar]
- Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci USA. 2010;107:8689–94. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flachs P, Mihola O, Simecek P, Gregorová S, Schimenti JC, Matsui Y, Baudat F, de Massy B, Pialek J, Forejt J, et al. Interallelic and intergenic incompatibilities of the Prdm9 (Hst1) gene in mouse hybrid sterility. PLoS Genet. 2012;8:e1003044. doi: 10.1371/journal.pgen.1003044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores KB, Wolschin F, Amdam GV. The role of methylation of DNA in environmental adaptation. Integr Comp Biol. 2013;53:359–72. doi: 10.1093/icb/ict019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foret S, Kucharski R, Pittelkow Y, Lockett G, Maleszka R. Epigenetic regulation of the honey bee transcriptome: unravelling the nature of methylated genes. BMC Genomics. 2009;10:472. doi: 10.1186/1471-2164-10-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB, Lam LL, Neumann SM, Kobor MS. Population-specificity of human DNA methylation. Genome Biol. 2012;13:R8. doi: 10.1186/gb-2012-13-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JR, van der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, Arepalli S, Dillman A, Rafferty IP, Troncoso J, et al. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhman D, Lavi E, Prüfer K, Fraga MF, Riancho JA, Kelso J, Pääbo S, Meshorer E, Carmel L. Reconstructing the DNA methylation maps of the neandertal and the denisovan. Science. 2014;344:523–7. doi: 10.1126/science.1250368. [DOI] [PubMed] [Google Scholar]
- Gordon L, Joo JE, Powell JE, Ollikainen M, Novakovic B, Li X, Andronikos R, Cruickshank MN, Conneely KN, Smith AK, et al. Neonatal DNA methylation profile in human twins is specified by a complex interplay between intrauterine environmental and genetic factors, subject to tissue-specific influence. Genome Res. 2012;22:1395–406. doi: 10.1101/gr.136598.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundberg E, Meduri E, Sandling JK, Hedman AK, Keildson S, Buil A, Busche S, Yuan W, Nisbet J, Sekowska M, et al. Global analysis of DNA methylation variation in adipose tissue from twins reveals links to disease-associated variants in distal regulatory elements. Am J Hum Genet. 2013;93:876–90. doi: 10.1016/j.ajhg.2013.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, Buil A, Ongen H, Yurovsky A, Bryois J, Giger T, Romano L, Planchon A, et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife. 2013;2:e00523. doi: 10.7554/eLife.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrel DT, Cairns BR. Distinctive chromatin in human sperm packages genes to embryo development. Nature. 2009;460:473–8. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RA, Wang T, Coarfa C, Nagarajan RP, Hong C, Downey SL, Johnson BE, Fouse SD, Delaney A, Zhao Y, et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotech. 2010;28:1097–105. doi: 10.1038/nbt.1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–24. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- Hernando-Herraez I, Prado-Martinez J, Garg P, Fernandez-Callejo M, Heyn H, Hvilsom C, Navarro A, Esteller M, Sharp AJ, Marques-Bonet T. Dynamics of DNA methylation in recent human and great ape evolution. PLoS Genet. 2013;9:e1003763. doi: 10.1371/journal.pgen.1003763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn H, Moran S, Hernando-Herrae I, et al. DNA methylation contributes to natural human variation. Genome Research. 2013;23:1363–72. doi: 10.1101/gr.154187.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochedlinger K, Jaenisch R. Nuclear reprogramming and pluripotency. Nature. 2006;441:1061–7. doi: 10.1038/nature04955. [DOI] [PubMed] [Google Scholar]
- Hodges E, Molaro A, Dos Santos CO, Thekkat P, Song Q, Uren PJ, Park J, Butler J, Rafii S, McCombie WR, et al. Directional DNA methylation changes and complex intermediate states accompany lineage specificity in the adult hematopoietic compartment. Mol Cell. 2011;44:17–28. doi: 10.1016/j.molcel.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt BG, Brisson JA, Yi SV, Goodisman MAD. Functional conservation of DNA methylation in the pea aphid and the honeybee. Genome Biol Evol. 2010;2:719–28. doi: 10.1093/gbe/evq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt BG, Glastad K, Yi SV, Goodisman MAD. Patterning and regulatory associations of DNA methylation are mirrored by histone modifications in insects. Genome Biol Evol. 2013a;5:591–8. doi: 10.1093/gbe/evt030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt BG, Glastad KM, Goodisman MAD. Genome composition, caste, and molecular evolution in eusocial insects. Proc Natl Acad Sci USA. 2013b;110:E445–6. doi: 10.1073/pnas.1220586110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt BG, Glastad KM, Yi SV, Goodisman MAD. The function of intragenic DNA methylation: insights from insepct epigenomes. Integr Comp Biol. 2013c;53:319–28. doi: 10.1093/icb/ict003. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293:1068–70. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- Kaminsky ZA, Tang T, Wang SC, Ptak C, Oh GH, Wong AH, Feldcamp LA, Virtanen C, Halfvarson J, Tysk C, et al. DNA methylation profiles in monozygotic and dizygotic twins. Nature Genet. 2009;41:240–5. doi: 10.1038/ng.286. [DOI] [PubMed] [Google Scholar]
- Keller TE, Yi SV. DNA methylation and evolution of duplicate genes. Proc Natl Acad Sci USA. 2014;111:5932–7. doi: 10.1073/pnas.1321420111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent CF, Minaei S, Harpur BA, Zayed A. Recombination is associated with the evolution of genome structure and worker behavior in honey bees. Proc Natl Acad Sci USA. 2012;109:18012–7. doi: 10.1073/pnas.1208094109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Elango N, Warden CW, Vigoda E, Yi SV. Heterogeneous genomic molecular clocks in primates. PLoS Genet. 2006;2:e163. doi: 10.1371/journal.pgen.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–30. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- Martin DIK, Singer M, Dhahbi J, Mao G, Zhang L, Schroth GP, Pachter L, Boffelli D. Phyloepigenomic comparison of great apes reveals a correlation between somatic and germline methylation states. Genome Res. 2011;21:2049–57. doi: 10.1101/gr.122721.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihola O, Trachtulec Z, Vlcek C, Schimenti JC, Forejt J. A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science. 2009;323:373–5. doi: 10.1126/science.1163601. [DOI] [PubMed] [Google Scholar]
- Moen EL, Zhang X, Mu W, Delaney SM, Wing C, McQuade J, Myers J, Godley LA, Dolan ME, Zhang W. Genome-wide variation of cytosine modifications between European and African populations and the implications for complex traits. Genetics. 2013;194:987–96. doi: 10.1534/genetics.113.151381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaro A, Hodges E, Fang F, Song Q, McCombie WR, Hannon GJ, Smith AD. Sperm methylation profiles reveal features of epigenetic inheritance and evolution in primates. Cell. 2011;146:1029–41. doi: 10.1016/j.cell.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk M, Boubelik M, Lehnert S. Temporal and regional changes in DNA methylation in the embryonic, extraembryonic and germ cell lineages during mouse embryo development. Development. 1987;99:371–82. doi: 10.1242/dev.99.3.371. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HGE, Martin DIK, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–8. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- Nabel CS, Manning SA, Kohli RM. The curious chemical biology of cytosine: deamination, methylation,and oxidation as modulators of genomic potential. ACS Chem Biol. 2011;7:20–30. doi: 10.1021/cb2002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA, Masly JP, Presgraves DC. Speciation genes. Curr Opin Genet Dev. 2004;14:675–9. doi: 10.1016/j.gde.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Pai AA, Bell JT, Marioni JC, Pritchard JK, Gilad Y. A genome-wide study of DNA methylation patterns and gene expression levels in multiple human and chimpanzee tissues. PLoS Genet. 2011;7:e1001316. doi: 10.1371/journal.pgen.1001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen JS, Valen E, Velazquez AMV, Parker BJ, Rasmussen M, Lindgreen S, Lilje B, Tobin DJ, Kelly TK, Vang S, et al. Genome-wide nucleosome map and cytosine methylation levels of an ancient human genome. Genome Res. 2014;24:454–66. doi: 10.1101/gr.163592.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–93. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Richards EJ. Population epigenetics. Curr Opin Genet Dev. 2008;18:221–6. doi: 10.1016/j.gde.2008.01.014. [DOI] [PubMed] [Google Scholar]
- Sarda S, Zeng J, Hunt BG, Yi SV. The evolution of invertebrate gene body methylation. Mol Biol Evol. 2012;29:1907–16. doi: 10.1093/molbev/mss062. [DOI] [PubMed] [Google Scholar]
- Schmitz RJ, Schultz MD, Lewsey MG, O’Malley RC, Urich MA, Libiger OJ, Schork NJ, Ecker JR. Transgenerational epigenetic instability is a source of novel methylation variants. Science. 2011;334:369–73. doi: 10.1126/science.1212959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Ting C-T, Wu C-I. The normal function of a speciation gene, odysseus, and its hybrid sterility effect. Science. 2004;305:81–3. doi: 10.1126/science.1093904. [DOI] [PubMed] [Google Scholar]
- Suzuki MM, Kerr ARW, De Sousa D, Bird A. CpG methylation is targeted to transcription units in an invertebrate genome. Genome Res. 2007;17:625–31. doi: 10.1101/gr.6163007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–76. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- Takuno S, Gaut BS. Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol Biol Evol. 2011;29:219–27. doi: 10.1093/molbev/msr188. [DOI] [PubMed] [Google Scholar]
- Takuno S, Gaut BS. Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. Proc Natl Acad Sci USA. 2013;110:1797–802. doi: 10.1073/pnas.1215380110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–49. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International Aphid Genome Consortium. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Nasonia Genome Working Group. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327:343–8. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorisson GA, Smith AV, Krishnan L, Stein LD. The international HapMap project web site. Genome Res. 2005;15:1592–3. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting C-T, Tsaur S-C, Wu M-L, Wu C-I. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science. 1998;282:1501–4. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- Wang X, Wheeler D, Avery A, Rago A, Choi J-H, Colbourne JK, Clark AG, Werren JH. Function and evolution of DNA methylation in Nasonia vitripennis. PLoS Genet. 2013;9:e1003872. doi: 10.1371/journal.pgen.1003872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Leung FCC. In silico prediction of two classes of honeybee genes with CpG deficiency or CpG enrichment and sorting according to gene ontology classes. J Mol Evol. 2009;68:700–5. doi: 10.1007/s00239-009-9244-3. [DOI] [PubMed] [Google Scholar]
- Wang J, Cao X, Zhang Y, Su B. Genome-wide DNA methylation analyses in the brain reveal four differentially methylated regions between humans and non-human primates. BMC Evol Biol. 2012;12:144. doi: 10.1186/1471-2148-12-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, Schübeler D. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–66. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- Yi SV, Goodisman MAD. Computational approaches for understanding the evolution of DNA methylation in animals. Epigenetics. 2009;4:551–6. doi: 10.4161/epi.4.8.10345. [DOI] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–40. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- Yu M, Hon GC, Szulwach KE, Song C-X, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368–80. doi: 10.1016/j.cell.2012.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328:916–9. doi: 10.1126/science.1186366. [DOI] [PubMed] [Google Scholar]
- Zeng J, Konopka G, Hunt BG, Preuss TM, Geschwind D, Yi SV. Divergent whole-genome methylation maps of human and chimpanzee brains reveal epigenetic basis of human regulatory evolution. Am J Hum Genet. 2012;91:455–65. doi: 10.1016/j.ajhg.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Nagrajan HK, Yi SV. Fundamental diversity of human CpG Islands at multiple biological levels. Epigenetics. 2014;9 doi: 10.4161/epi.27654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng J, Yi S. DNA methylation and genome evolution in honeybee: gene length, expression, functional enrichment co-vary with the evolutionary signature of DNA methylation. Genome Biol Evol. 2010;2:770–80. doi: 10.1093/gbe/evq060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Cheng L, Badner JA, Chen C, Chen Q, Luo W, Craig DW, Redman M, Gershon ES, Liu C. Genetic control of individual differences in gene-specific methylation in human brain. Am J Hum Genet. 2010;86:411–9. doi: 10.1016/j.ajhg.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.