Abstract
Summary
CharGer (Characterization of Germline variants) is a software tool for interpreting and predicting clinical pathogenicity of germline variants. CharGer gathers evidence from databases and annotations, provided by local tools and files or via ReST APIs, and classifies variants according to ACMG guidelines for assessing variant pathogenicity. User-designed pathogenicity criteria can be incorporated into CharGer’s flexible framework, thereby allowing users to create a customized classification protocol.
Availability and implementation
Source code is freely available at https://github.com/ding-lab/CharGer and is distributed under the GNU GPL-v3.0 license. Software is also distributed through the Python Package Index (PyPI) repository. CharGer is implemented in Python 2.7 and is supported on Unix-based operating systems.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
As genomic sequencing becomes increasingly important in research and translational medicine, obtaining clinical interpretation of variant predisposition for inherited disease phenotypes is critical. Currently, the prediction of variant pathogenicity is not standardized, and different clinical testing labs often produce conflicting results. For example, manual examination of 239 variants extracted from 1,000 exomes that were marked as pathogenic by the Human Gene Mutation Database (HGMD) revealed that a mere 7.5% were correctly classified as pathogenic according to literature sources (Dorschner et al., 2013). Recently, the American College of Medical Genetics and Genomics (ACMG) provided updated standards and guidelines by which to interpret germline variants (Richards et al., 2015).
There have been several efforts to develop automated variant interpretation tools. For example, one group created a browser-based ACMG variant classifier in which the ACMG criteria can be selectively applied (Kleinberger et al., 2016). Another group developed InterVar (Li and Wang, 2017), a tool that automates the initial variant interpretation but then relies on a manual review step to adjust the classification criteria based on prior information or domain knowledge before arriving at a final interpretation. Another group extended ACMG’s 33 rules with 108 refinements, including semi-qualitative aspects in classification, into a framework called Sherloc (Nykamp et al., 2017). However, its source code is not publicly available. The aim of this work is to provide an open-source framework for conducting a fully automated, systematic interpretation of germline variants based on ACMG guidelines and user-designed pathogenicity algorithms for customizing predictions for all inherited diseases.
CharGer (Characterization of Germline variants) provides a software implementation of the ACMG guidelines, with several unique modules not explicitly outlined by the ACMG, and a scoring system for predicting pathogenicity of germline variants in a fully automated way. It includes 16 scoring modules of evidence, consisting of 10 ACMG modules (8 pathogenic and 2 benign) and 6 custom modules (4 pathogenic and 2 benign). CharGer can generate variant pathogenicity classifications using two independent methods: one implements the original ACMG scoring system and the other extends the ACMG system to include custom modules and a user-adjustable scoring system as described below.
2 Materials and methods
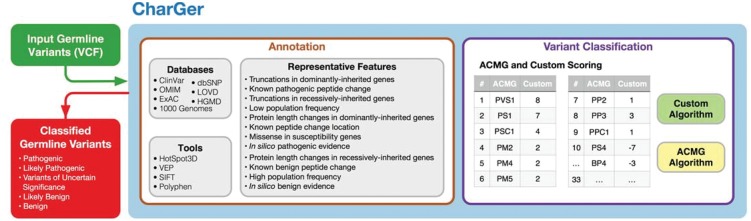
CharGer implements the ACMG guidelines for variant classification by sequentially performing annotation, scoring and classification (Fig. 1). A variant file in VCF or MAF format is the minimum input requirement. Examples of optional input files include lists of inherited genes, genes supporting pathogenicity/benignity, and de novo variants. Tailoring these additional inputs to the disease of interest allows users to increase the fidelity of the variant classifications.
Fig. 1.
CharGer workflow. Given an input variant file (and other optional inputs), CharGer performs variant annotation and then scores annotated variants according to matching ACMG and custom modules. Module scores are then processed through ACMG’s and CharGer’s classification algorithms to produce pathogenicity classification of variants
Variant annotations from Ensembl’s Variant Effect Predictor (VEP) (v81 or later) may be supplied within the input VCF file; otherwise they will be generated automatically by CharGer via a local installation of VEP and associated databases or via web accessions using the VEP ReST API (McLaren et al., 2016). ExAC population frequency annotations (Lek et al., 2016) can also be added by CharGer if not already provided by pre-annotation with VEP. To perform annotation using a snapshot of the ClinVar database, users can provide a single-allele version of TSV-formatted ClinVar, which provides consistent variants and annotations across the VCF, TXT, and XML versions of NCBI’s ClinVar downloads (Zhang et al., 2017). To check if variants are near mutation hotspots, CharGer can parse results from somatic mutation clusters produced by HotSpot3D (Niu et al., 2016).
CharGer operates by mapping ACMG modules to their corresponding ACMG tier (strong, moderate, etc.). In the ACMG algorithm, the modules that a variant satisfies are aggregated to produce a prediction of pathogenicity (Richards et al., 2015). At present (version 0.5.2), there are 16 total modules available for analyzing each variant (Supplementary Table S1). Depending on usage case, certain modules may be excluded from running (de novo vs. cohort).
CharGer’s classification system is flexible by allowing users to assign custom scores for modules and classification thresholds conveniently using command line options. The utility of the customizable scoring system can be demonstrated in cases where a disease presents with stronger indicators for particular modules compared to the default. Classification using the ACMG guideline, together with the customized system and ClinVar annotations, population frequency, VEP annotations, and module summaries, for each variant are recorded in an output file for downstream user analysis.
3 Results
We applied CharGer to classify 883 pediatric cancer germline variants (Supplementary Table S2) across 1120 pediatric cancer cases from the Pediatric Cancer Germline Project (PCGP) to obtain sensitivity and specificity estimates according to a ground truth set provided by a panel of clinical geneticists (Zhang et al., 2015). For the purpose of this analysis of cancer predisposition, we supplied additional input files to CharGer as follows: 152 cancer predisposition genes (Huang et al., 2018), 1819 cancer predisposition variants as curated by the TCGA PanCanAtlas Germline Analysis Working Group (Huang et al., 2018), and 17, 555 somatic mutation clusters computed from the TCGA MC3 MAF (Ellrott et al., 2018) using HotSpot3D (ver. 1.1.4, with the default settings except 10 Å cutoff and 20 Å maximum radius). We also used a tab-delimited ClinVar flat file (accessed 30 October 2017 from github.com/macarthur-lab/clinvar) and annotated using a local instance of VEP v76 with reference build GRCh37.
By grouping pathogenic and likely pathogenic variants (where the latter are called “probably pathogenic” in PCGP’s classification terms) vs. all other variants, the ACMG and CharGer custom algorithms identified, respectively, 80 and 97 out of 127 panel-determined pathogenic and likely pathogenic variants, and both algorithms classified 21 out of 756 PCGP panel-determined non-pathogenic variants as pathogenic or likely pathogenic (15 of which are labeled as having uncertain significance). These results translate to a sensitivity of 63% for ACMG guidelines and 76% for CharGer’s custom algorithm and a false-positive rate of 2.8% for both, demonstrating that CharGer’s automated classification can effectively prioritize variants for investigation (Supplementary Fig. S1).
Supplementary Material
Acknowledgements
The authors acknowledge Reyka Jayasinghe, Wen-wei Liang, Steven Foltz, Irena Lanc and Feng Chen for constructive comments.
Funding
This work was supported by the National Institutes of Health through National Cancer Institute [R01 CA178383, R01 CA18006, U24 CA211006] and National Human Genome Research Institute [R01 HG009711].
Conflict of Interest: none declared.
References
- Dorschner M.O., et al. (2013). Actionable, pathogenic incidental findings in 1, 000 participants’ exomes. Am. J. Hum. Genet ., 93(4), 631-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellrott K. et al. (2018) Scalable open science approach for mutation calling of tumor exomes using multiple genomic pipelines. Cell Sys., 6, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K-l. et al. (2018) Pathogenic germline variants in 10, 389 adult cancers. Cell, 173, 355–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinberger J. et al. (2016) An openly available online tool for implementing the ACMG/AMP standards and guidelines for the interpretation of sequence variants. Genet. Med., 18, 1165.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M. et al. (2016) Analysis of protein-coding genetic variation in 60, 706 humans. Nature, 536, 285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wang K. (2017) InterVar: clinical interpretation of genetic variants by the 2015 ACMG-AMP guidelines. Am. J. Hum. Genet., 100, 267–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W. et al. (2016) The Ensembl variant effect predictor. Genome. Biol., 17, 122.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B. et al. (2016) Protein-structure-guided discovery of functional mutations across 19 cancer types. Nat. Genet., 48, 827–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykamp K. et al. (2017) Sherloc: a comprehensive refinement of the ACMG-AMP variant classification criteria. Genet. Med., 19, 1105–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S. et al. (2015) Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med., 17, 405–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. et al. (2015) Germline mutations in predisposition genes in pediatric cancer. N. Engl. J. Med., 373, 2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. et al. (2017) ClinVar data parsing. Wellcome Open Res, 2, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.