Abstract
Summary
PathwayConnector is a web-tool that facilitates the construction of complementary pathway-to-pathway networks and subnetworks of them, based on a reference pathway network derived from the rich information available either in KEGG or Reactome database for pathway mapping. Specifically, for a given set of pathways, PathwayConnector (i) finds all the direct connections between them, (ii) adds a minimum set of complementary pathways required to achieve connectivity between the pathways, leading to informative fully connected networks and (ii) provides a series of clustering methods for the further grouping of pathways in to sub-clusters. The proposed web-tool is a simple yet informative tool towards identifying connected groups of pathways that are significantly related to specific diseases.
Availability and implementation
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Pathway-based analysis allows for a comprehensive understanding of the molecular mechanisms related to complex diseases. Although there are several available pathway analysis methods (Jin et al., 2014), the way in which human pathways are functionally linked within an overall network of existing human pathways is largely unexplored. In classical pathway analysis gene lists, usually obtained from any experimental-computational method, can be further analyzed by relevant software tools, that allow enrichment analysis to be performed based on prior knowledge gene-set libraries and pathways connected to them (Kuleshov et al., 2016). Such tools may provide significant score-based information on how genes are involved into pathways and pathways to a disease, but also have several limitations that may fall in to the following criticisms. Firstly, it is not clear whether these pathways are functionally linked. Second, when filtering the pathways with a specific threshold on their score, important yet not statistically significant pathways might be excluded whereas non-relevant yet statistically important pathways might be included. Third, when dealing with large lists of pathways it is not always straightforward to relate them to a specific biological status building fully functional stories based on them. To this extend, the proposed PathwayConnector is a web-tool that provides an easy way for rapidly relating pathways together, by creating complementary networks of pathways related to a specific biological status. PathwayConnector is able to provide with respect to a specific reference network: (i) direct connections between pathways of interest (first-neighbors approach), (ii) complementary networks that show the shortest paths between pathways of interest and the intermediate pathways involved between them and (iii) additional clustering approaches to highlight communities (clusters) of pathways.
2 Implementation
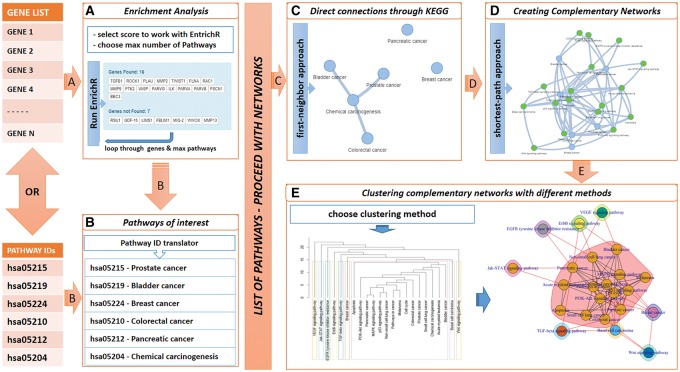
For a number of species where data are available, an overall reference pathway-to-pathway network has been developed, that covers all the possible connections that exist between all the available pathways, as referenced in two major pathway databases: KEGG (Kanehisa, 2002) and Reactome (Fabregat et al., 2018). The derived pathway-pathway network acts as a reference network map for retrieving information about the functional interconnections between pathways of interest. For this we focused on the connectivity information included in the selected database (KEGG: biochemical relationships, Reactome: relations of signaling and metabolic organized into biological pathways and processes). The output of a data mining process over the relationships that each database offers, provided us with all the links between pathways, which were used as edges to construct the undirected, unweighted pathway network. This network is regularly updated and works as the main pathway repository for the services/methods PathwayConnector draws from. Users can provide two types of inputs, as shown in Figure 1A and B: (i) lists of genes as notated in either KEGG’s or Reactome’s repository, and (ii) lists of pathway IDs, that can be found in either KEGG’s or Reactome’s website. When the web-tool detects a gene list it automatically applies enrichment analysis, by means of (i) the EnrichR package required for enrichment analysis of human model organism (Kuleshov et al., 2016), (ii) the “clusterProfiler” package (Yu et al., 2012), required for enrichment analysis of non-human species employed in KEGG’s repository and (iii) the “ReactomePA” package (Yu and He, 2016), required for the enrichment of non-human species included in Reactome’s repository. In this way, users are able to obtain the top-scored pathway list required for the network analysis. Users can either examine whether the input genes have been correctly enriched, or re-run the enrichment process with different gene synonyms and scoring methods (see Supplementary Fig. S1). Both types of input lead to an initial network of pathways, as shown Figure 1C, showing the way in which six pathways are directly connected (or not) to each other within the reference pathway network. Herein the methodology that follows, draws from the graph theory as well as the “igraph” R package (Csardi and Nepusz, 2006) that has been employed for the implementation of this web-tool.
Fig. 1.
Schematic workflow of the PathwayConnector web-tool
2.1 The missing pathway approach
Following the initial network construction based on the given (or the derived) pathway list, our proposed methodology identifies and adds the key nodes which ensure the minimal connectivity of the network, i.e. each node in the original list is connected to at least another original node. For this, a specific algorithm finds and calculates all the shortest paths within the reference network that interconnect these nodes together. Then, the algorithm chooses those nodes which belong to the shortest path-length to be included in the final network. For more than one shortest paths of the same path-length, these paths are also included in to the final network. Figure 1D depicts the complementary added 17 nodes to the initial 6-node network which contained two standalone nodes.
2.2 Clustering the complementary network
For small and well connected pathway networks, the latter methodology is often sufficient for further investigation. However, for large and disconnected/sparse pathway networks, the missing pathway approach often leads to even larger complementary networks whose analysis and interpretation with respect to a certain biological status becomes more challenging. To address this, as part of this web-tool we provide the option of clustering the pathways in sub-clusters through a series of community structure detection algorithms (clustering algorithms, further detailed in the Supplementary Material and Supplementary Fig. S2) as shown in Figure 1E. Clustering can be used to separate the complementary networks into sub-networks, and in effect to provide more comprehensive information related to a biological status.
3 Novelty and applications
The described methodology has been applied, with noteworthy results to pathways related to Alzheimer’s Disease (AD) (Zachariou et al., 2018), in a framework which: (i) integrates knowledge, (ii) prioritises genes and (iii) utilizes the PathwayConnector to optimize pathway selection for further interpretation with respect to pathway mechanisms underlying AD. Additional success stories include its application to Huntington’s disease (HD) and Spastic Ataxia (SA) (Kakouri et al., in press). PathwayConnector, when applied in post-pathway analysis on results concerning HD, SA and AD provided us with pathways that were not included in the classical pathway analysis. Specifically, in the case of HD, three pathways identified that were not included in the enrichment analysis results. In the case of SA, the shortest path algorithm provided 14 additional pathways, which were not initially included in the pathway list of the classical pathway enrichment analysis. In the case of Alzheimer’s disease seven additional pathways were provided. By considering the topological information provided by the reference pathway–pathway network through PathwayConnector, we can ensure that key pathways functionally connected to the top selected pathways are not lost due to an ad-hoc cutoff or over-representation statistics in the classical pathway enrichment analysis. We expect that the proposed PathwayConnector will be a valuable tool to complement and enhance classical pathway analysis and offer further insights on the functional inter-relation of pathways of interest.
Funding
This work was supported by the European Commission Research Executive Agency Grant BIORISE (No. 669026), under the Spreading Excellence, Widening Participation, Science with and for Society Framework. This work was partly supported by H2020- WIDESPREAD-04-2017-Teaming Phase 1, Grant Agreement 763781, Integrated Precision Medicine Technologies.
Conflict of Interest: none declared.
Supplementary Material
References
- Csardi G., Nepusz T. (2006) The igraph software package for complex network research. InterJournal, 1695, 1–9. [Google Scholar]
- Fabregat A. et al. (2018) Reactome diagram viewer: data structures and strategies to boost performance. Bioinformatics, 34, 1208–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L. et al. (2014) Pathway-based analysis tools for complex diseases: a review. Genomics Proteomics Bioinformatics, 12, 210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakouri A. et al. (2018) Revealing Clusters of Connected Pathways through Multisource Data Integration in Huntington’s Disease and Spastic Ataxia. IEEE Journal of Biomedical and Health Informatics, in press. [DOI] [PubMed]
- Kanehisa M. (2002) The KEGG database. Novartis Found. Symp., 247, 91–101. [PubMed] [Google Scholar]
- Kuleshov M.V. et al. (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res., 44, W90–9W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., He Q.Y. (2016) ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol. BioSyst., 12, 477–479. [DOI] [PubMed] [Google Scholar]
- Yu G. et al. (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS, 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariou M. et al. (2018) Integrating multi-source information on a single network to detect disease-related clusters of molecular mechanisms. J. Proteomics. doi: 10.1016/j.jprot.2018.03.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.