Abstract
The digital world is generating data at a staggering and still increasing rate. While these ‘Big Data’ have unlocked novel opportunities to understand public health, they hold still greater potential for research and practice. This review explores several key issues arising around big data. First, we propose a taxonomy of sources of big data in order to clarify terminology and identify threads common across some subtypes of big data. Next, we consider common public health research and practice uses for big data, including surveillance, hypothesis-generating research, and causal inference, while exploring the role that machine learning may play in each use. We then consider the ethical implications of the big data revolution with particular emphasis on maintaining appropriate care for privacy in a world in which technology is rapidly changing social norms regarding the need for (and even the meaning of) privacy. Finally, we make suggestions regarding structuring teams and training to succeed in working with big data in research and practice.
Keywords: Big Data, Machine Learning, Privacy, Training
Introduction
As measurement techniques, data storage equipment, and the technical capacity to link disparate datasets develop, increasingly large volumes of information are available for public health research and decision-making.(11) Numerous authors have described and made predictions about the role of this ‘big data’ in health care, (13; 93) epidemiology,(59; 92) surveillance,(62; 113) and other aspects of population health management.(88; 95) This review first describes types of big data, then describes methods appropriate for core functions of public health: surveillance, hypothesis-generating discovery, and causal inference, and finally addresses maintaining care for privacy and structuring teams and training to succeed in working with big data.
Taxonomy of Big Data in Public Health
Most big data used by public health researchers and practitioners fits one of five descriptions. Big public health datasets usually include one or more of a) measures of participant biology, as in genomic or metabolomic datasets, b) measures of participant context, as in geospatial analyses (84; 91), c) administratively collected medical record data that incorporates more participants than would be feasible in a study limited to primary data collection,(93; 104) d) participant measurements taken automatically at extremely frequent intervals as by a GPS device or FitBit,(39) or e) measures compiled from the ‘data effluent’ created by life in an electronic world, such as search term records,(67) social media postings,(7) or cell phone records.(1; 137)
While data collection from each of these sources leverages emerging technologies to collect larger volumes of data than was available prior to the technological development, each form of data has fundamentally different implications for public health research and practice, as noted in Table 1. ‘Wider’ datasets (i.e. datasets in category (a) or (b), measuring many potential relevant aspects of each subject at each measurement time) typically require reducing the number of dimensions in the dataset to a more interpretable number, either selecting specific variables of greater interest for further analysis (as in selecting candidate biomarkers from a metabolomics dataset or identifying ‘eigengenes’(3)) or by identifying variance patterns within these variables (as by a principal component analysis identifying patterns of gut bacteria). (125) By contrast, ‘taller’ datasets (i.e. categories (c) and (d)) may require more work to filter out irrelevant or low quality observations (e.g. health records of clinical visits unrelated to the hypothesis of interest) or to condense observations into a more tractable, yet information-rich summary.(37) Effluent data offers access to constructs that have heretofore been extremely difficult to measure directly, such as social network structure (1; 49) or racial animus.(89)
Table 1:
Types of Big Data for Public Health
| Source | Examples | Aspect of bigness1 | Key technical issues | Typical uses |
|---|---|---|---|---|
| -omic/biological | Whole exome profiling, metabolomics | Wide | Lab effects, informatics pipeline | Etiologic research, screening |
| Geospatial | Neighborhood characteristics | Wide | Spatial autocorrelation | Etiologic research, surveillance |
| Electronic health records | Records of all patients with hypertension | Tall, often also Wide | Data cleaning, natural language | Clinical research, surveillance |
| Personal monitoring | Daily GPS records, Fitbit readings | Tall | Redundancy, inferring intentions | Etiologic research, potentially clinical decision-making |
| Effluent data | Google Search Results, Reddit | Tall | Selection biases, natural language | Surveillance, screening, identifying hidden social networks. |
‘Wide’ datasets have many columns; ‘tall’ datasets have many rows.
Each subtype of data poses unique challenges. Biological data is subject to lab effects (where one or more observations may be strongly affected by lab procedures hidden from the analyst) and geospatial data is subject to auto-correlation (wherein spatial units near each other tend to be more correlated), electronic health record data is subject to potentially large standardization and quality-related challenges. ‘Effluent data’, wherein a hypothesis test focuses on analyzing data not originally collected for research purposes, may require substantial attention to the way the data were initially collected (e.g. using 311 records for noise or graffiti complaints as a marker of neighborhood characteristics requires careful understanding of the factors leading residents to call 311, and whether these factors are demographically patterned).(139) Broadly, data collected automatically, as in personal monitoring and effluent data, are often of interest to behavioral researchers, but typically obscure intention, frustrating attempts at truly understanding behavior.
While this taxonomy is intended to categorize sources of big data, a given dataset may of course include more than one, as when a hospital’s data warehouse includes not only electronic medical records of a given patient’s visits but also the results from sequencing her whole genome. Indeed, such merged datasets may be the key to identifying etiologic links that have heretofore perplexed researchers, such as gene-environment interactions.
Big Data Surveillance using Machine Learning
Public health surveillance systems monitor trends in disease incidence, health behaviors, and environmental conditions in order to allocate resources to maintain healthy populations.(121) While some of the highest profile uses of big data for surveillance relate to effluent data (e.g. Google Flu Trends), all five categories of big data may contribute to informing authorities about the state of public health. However, the scale of these novel sources of data poses analytic challenges as well. Within the data science field, the “curse of dimensionality”(14) associated with wide datasets has been somewhat alleviated through the adoption of machine learning models, particularly in contexts where prediction or hypothesis generation rather than hypothesis-testing is the analytic goal. We review here some inroads machine learning has made in public health, with particular emphasis on surveillance, and provide a glossary of terminology as used in machine learning for public health researchers (Table 2).
Table 2.
A glossary of terms used in data science and machine learning for public health researchers and practitioners
| Data Science Term | Related Public Health Research Term or Concept |
|---|---|
| Accuracy | Proportion of results correctly classified (i.e. (true positives plus true negatives) divided by total number of results predicted) |
| Data mining | Exploratory analysis |
| Ensemble learning | A machine learning approach involving training multiple models on data subsets and combining results from these models when predicting for unobserved inputs |
| Features | Measurements recorded for each observation (e.g. participant age, sex, and BMI are each features) |
| Label | Observed or computed value of an outcome or other variable of interest |
| Labeling | The process of setting a label for a variable, as opposed to leaving the variable’s value unknown |
| Learning algorithm | The set of steps used to train a model automatically from a data set (not to be confused with the model itself, e.g. there are many algorithms to train a neural network, each with different bounds on time, memory and accuracy). |
| Natural language | Working with words as data, as in qualitative or mixed-methods research (generally, human-readable but not readily machine-readable) |
| Noisy labels | Measurement error |
| Out-of-sample | Applying a model fitted to one dataset to make predictions in another |
| Overfitting | Fitting a model to random noise or error instead of the actual relationship (either due to having a small number of observations or a large number of parameters relative to the number of observations) |
| Pipeline | (From bioinformatics) The ordered set of tools applied to a dataset to move it from its raw state to a final interpretable analytic result |
| Precision | Positive predictive value |
| Recall | Sensitivity |
| Semi-supervised learning | An analytic technique used to fit predictive models to data where many observations are missing outcome data. |
| Small-n, large-p | A wide but short dataset: n = number of observations, p= number of variables for each observation |
| Supervised learning | An analytic technique in which patterns in covariates that are correlated with observed outcomes are exploited to predict outcomes in a data set or sets in which the correlates were observed but the outcome was unobserved. For example, linear regression and logistic regression are both supervised learning techniques. |
| Test dataset | A subset of a more complete dataset used to test empirical performance of an algorithm trained on a training dataset |
| Training | Fitting a model |
| Training dataset | A subset of a more complete dataset used to train a model whose empirical performance can be tested on a test dataset |
| Unsupervised learning | An analytic technique in which data is automatically explored to identify patterns, without reference to outcome information. Latent class analysis (when used without covariates) and k-means clustering are unsupervised learning techniques. |
Broadly, machine learning is an umbrella term for techniques that fit models algorithmically by adapting to patterns in data. These techniques can be classified as one of: a) supervised learning, b) unsupervised learning, and c) semi-supervised learning. Supervised learning is defined by identifying patterns that relate variables to measured outcomes and maximize accuracy when predicting those outcomes. For example, an automatically fitted regression model (including any form of generalized linear model) is a supervised learning technique. By contrast, unsupervised learning exploits innate properties of the input data set to detect trends and patterns without explicit designation of one column as the outcome of interest. For example, principal component analysis, which identifies underlying covariance structures in observed data, is unsupervised. Semi-supervised learning, a sort of hybrid, is used in contexts where prediction is a goal but the majority of data points are missing outcome information.(148) Semi-supervised and unsupervised methods are often used in the “data mining” phase as precursors to supervised approaches intended for prediction or more rigorous statistical analyses in a follow-up.
While machine learning has been more broadly adopted within data science, some public health researchers and practitioners have embraced machine learning as well. For example, unsupervised learning has been used for spatial and spatio-temporal profiling,(4; 134) outbreak detection and surveillance,(38; 146) identifying patient features associated with clinical outcomes (47; 142) and environmental monitoring.(26; 65) Semi-supervised variants of existing learning algorithms (Table 3) have been utilized to build an early warning system for adverse drug reactions from social media data, (145) detect falls from smartphone data (33) and identify outlier air pollutants, (18) among other applications. Supervised learning has been used to predict hospital readmission, (32; 44) tuberculosis transmission, (87) serious injuries in motor vehicle crashes (61) and Reddit users shifting towards suicidal ideation, (28) among many other applications. Table 3 reviews some specific applications of machine learning techniques to address public health problems.
Table 3:
Selected machine learning approaches that have been applied to big data in public health
| Approach | Learning type | Usage Examples |
|---|---|---|
| K-means clustering | Unsupervised | Hot spot detection(5) |
| Retrospective Event Detection | Unsupervised | Case ascertainment(34) |
| Content Analysis | Unsupervised | Public health surveillance(38) |
| K-nearest neighbors clustering | Supervised | Spatio-temporal hot spot detection;(133) Clinical outcomes from genetic data; falls from wearable sensors |
| Naïve Bayes | Supervised | Acute gastrointestional syndrome surveillance; (51) |
| Neural Networks | Supervised | Identifying microcalcification clusters in digital mammograms;(100) predicting mortality in head trauma patients;(31) predicting influenza vaccination outcome(127) |
| Support Vector Machines | Supervised | Diagnosis of diabetes mellitus; (12) detection of depression through Twitter posts(27) |
| Decision trees | Supervised | Identifying infants at high risk for serious bacterial infections;(9) comparing cost-effectiveness of different influenza treatments;(116) and physical activity from wearable sensors(101) |
Using Machine Learning for Hypothesis Generation from Big Data
Machine learning has also been used in big data settings for hypothesis generation. Algorithmic identification of the measures associated with an outcome of interest allows researchers to focus on independent validation and interpretation of these associations in subsequent studies. Techniques to identify subsets of more strongly associated covariates, referred to within machine learning as ‘feature selection’, can broadly be divided into three groups: wrapper methods, filter methods, and embedded methods. Wrapper methods involve fitting machine learning models (such as those used for prediction) on different subsets of variables. Based on differences in how well models fit when variables are included, a final set of variables can be selected as the most predictive. For example, the familiar stepwise regression technique is one such wrapper method.(30; 128) By contrast, filter methods leverage conventional measures such as correlation, mutual information, or P-values from statistical tests to filter out features of lower relevance. Filter methods are often favored over wrapper methods for their simplicity and lower computational costs.(24) Finally, embedded methods embed the variable selection step into the learning algorithm. Embedded methods such as least absolute shrinkage and selection operator (LASSO), (123) elastic nets (150) and regularized trees (29) have been used to select features for the prediction of “successful aging”, (54) flu trends (112) and lung cancer mortality, (63) among others. Scalable approaches to feature selection in extremely large feature spaces (“ultra-wide” data sets) constitute an active area of research.(119; 144)
Analysis of Big Data for Causation
Causal inference from observational data is notoriously challenging,(45) and yet remains a cornerstone of public health research, particularly epidemiology. Within the public health community, it is well known that the conditions under which an observed statistical association in observational data can be explained only as the effect of manipulating the exposure of interest cannot typically be ensured, regardless of the scale of data.(107) Moreover, confounding, selection bias, and measurement error, all common threats to valid causal inference, are independent of sample size. However, there are four key ways big data and the machine learning techniques developed in part to work with big data may improve causally-focused research.
First, novel sources of exposure data increase the availability of potential instrumental variables. In instrumental variable analyses, an upstream exposure that causes an outcome only by manipulating a downstream exposure of interest can be used to estimate the causal effect of the downstream exposure.(46) For example, it is plausible that changes to compulsory schooling laws change all-cause mortality only by affecting years of schooling completed.(79) Under this ‘instrumental variable assumption’, compulsory schooling laws can be used as an instrument to estimate the effect of education on all-cause mortality. Instrumental variables have been used extensively for Mendelian randomization studies (in which a genetic variant acts as the instrumental variable).(115; 131) Recent developments in analytic techniques combining estimates from using multiple genetic variants, which may be considered a form of meta-analysis, are a particularly intriguing use of big data.(19; 55) However, we caution that the instrumental variable assumption for any given instrument variable must be considered carefully and the assumption requires specific background knowledge.(36) As such, proliferation of potential instruments is not in itself beneficial; it is only proliferation of valid instruments that can improve causal research.
Second, wider datasets with more measured covariates offer opportunities to use negative controls (76) more extensively to estimate the potential magnitude of residual confounding, measurement error, or selection bias.(8) For example, an analyst using electronic medical records to estimate the impact of BMI in early adulthood in relation to risk of adult onset diabetes might be concerned about confounding by socio-economic status (acting as a fundamental cause through health-orientation, health literacy, etc.(75)) and might control for the best available proxy measure of socio-economic status (e.g. median income in reported ZIP code). While this measure is likely imperfect and thus may leave residual confounding, she might take advantage of the breadth of outcomes available in electronic medical records that might act as negative controls by, for example, assessing whether BMI is associated with mammography screening after controlling for the socio-economic proxy. If an association exists before controlling for ZIP code median income but drops close to zero after controlling, the analyst may conclude that residual confounding due to error in her socio-economic status measure is unlikely to result in strong bias in her primary analysis because such error would need to be uncorrelated with screening status (though residual confounding can never be ruled out). The use of negative controls has been described extensively in the epidemiologic methods literature (76; 77) but remains relatively uncommon.
Third, the availability of more covariates may allow for more precise causal mediation estimates (130) allowing stronger “causal explanation” tests of hypotheses regarding health production.(42) For example, studies exploring residential proximity to fast food as a cause of obesity (e.g.(25)) typically hypothesize that the exposure (proximity) affects the outcome (obesity) as mediated by consuming fast food. Such a study could benefit from linked GPS-based personal monitoring data that allow researchers to consider whether study subjects actually visited the fast food restaurants proximal to their residential location.
Finally, machine learning is increasingly being integrated into causal inference techniques, particularly in contexts where prediction or discovery is a component of an inferential process. For example, analysts using target maximum likelihood estimation (TMLE) to estimate causal treatment effects frequently use SuperLearner, an ‘ensemble’ supervised learning technique (i.e. one that combines estimates from multiple machine learning algorithms), as a portion of the targeting phase. In TMLE, the targeting step requires a predictive model incorporating information from covariates but imposes no functional form on that model; thus, tunable predictive models such as SuperLearner are ideal.(129) Similarly, methodologists have recently proposed techniques using machine learning to identify the strata in which a randomized intervention has the strongest effect. In this case, machine learning is being used for discovery, as an efficient search over set of potential groupings too large to test each one independently.(2; 132)
Big Data and Privacy
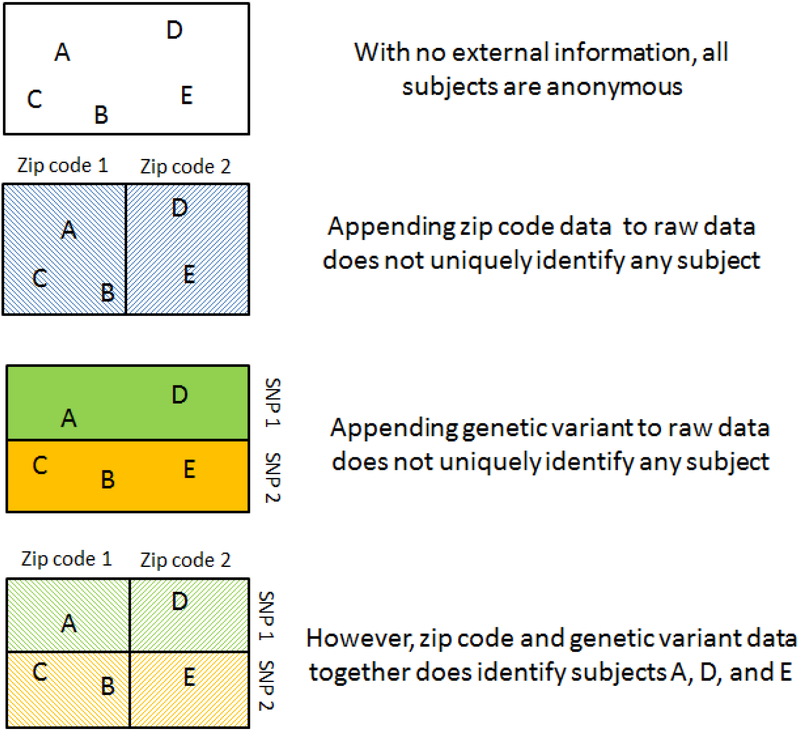
The proliferation and availability of big data, especially effluent data, has already fostered privacy concerns among the general public, and these concerns are expected to grow and diversify.(86) With respect to public health research and practice, big data raises three key issues: 1) the risk of inadvertent disclosure of personally identifying information (e.g. by the use of online tools(10)), 2) the potential for increasing dimensionality of data to make it difficult to determine if a dataset is sufficiently de-identified to prevent ‘deductive disclosure’ of personally identifying information (Figure 1), 3) the challenge of identifying and maintaining standards of ethical research in the face of emerging technologies that may shift the generally accepted norms regarding privacy (e.g. GPS, drones, social media, etc.).
Figure 1.
A schematic illustration of deductive disclosure: merging two datasets that are each successfully anonymized may result in a dataset in which subjects can be personally identified.
Although avoiding disclosure of study participants’ private information is a key principle of research ethics mandated in the United States by the Health Insurance Portability and Accountability Act (HIPAA),(69; 97) inadvertent disclosure of publically identifying information by health researchers has occurred repeatedly.(94) Indeed, inadvertent disclosure has become increasingly commonplace as increasing volumes of personally identifying data are stored in massive data warehouses. (81) While such disclosure can occur owing to malicious acts by malefactors, it may occur more frequently due to misunderstandings of well-meaning individuals.(94) For example, researchers may be unaware that using online geographic tools such as Google Maps to identify contextual features of subjects’ neighborhood constitutes a violation of typical terms of Institutional Review Board conditions.(10) Similarly, researchers who report pooled counts or allele frequencies in genome-wide association studies may inadvertently reveal the presence of an individual in that study sample to anyone who knows that person’s genotype.(20; 48)
Secondly, increasing columns of data may create a form of fingerprint such that subjects in de-identified datasets containing could be re-identified, a process known as deductive disclosure.(110; 126) Whereas institutional review board terms have conventionally treated the 18 columns of data specified by the HIPAA privacy rule to be the personally identifying ones (e.g. name, phone number), they often consider data derived from these identifying measures to connote anonymity (e.g. mean household income among census respondents living within a 1 km radius of the subject, or a specific variant of a given SNP taken from the whole exome dataset), formally, HIPAA specifies that data is considered identifiable if there is a way to identify an individual regardless of the columns included. Merged datasets containing many columns of big data from different domains that are themselves de-identified may still combine to make subjects re-identifiable (e.g. neighborhood median income plus ARDB2 Gln27Glu variant may be sufficient to identify a subject who would not be identifiable through neighborhood median income or Gln27Glu variant alone). Figure 1 is a schematic representation of this deductive disclosure that may occur as a result of merging. Techniques to protect confidentiality in the face of data merges (see sidebar on Data Perturbation for one such example), may become a key component of future data sharing agreements, though such techniques induce precision costs.
Finally, in part because of changing technologies including social media, drone surveillance, and open data in general, some ethicists suggest accepted norms around privacy may change.(143; 149) Changing privacy norms have a long history: formal definitions of privacy have been inconsistent, from “the right to be left alone”(135) in 1890 to the late 1960’s idea that privacy amounted to control over the information one produces (138) to more recent notions defining non-intrusion, seclusion, limitation, and control as separate categories of privacy.(120) A recurring theme in discussions of privacy, even prior to the big data era, is that the notion of ownership of information is problematic because nearly all data-producing actions, from clinical visits to social media postings to lab-based gene expression measurement, involve the work of more than one person, each of whom have created and therefore have some rights to the data.(85; 117) If anything, one constant theme regarding privacy is that no single clear definition suffices,(122) and we may expect the waters to get muddier as more people are involved in the data creation and collation process. For public health, there are no proscriptive answers; rather, we must follow and contribute to the societal discussion of privacy norms while remaining true to principles of using fair procedures to determine acceptable burdens imposed by our decisions.(58)
Big Data, Public Health Training, and Future Directions
The use of big data in public health research and practice calls for new skills to manage and analyze these data, though it does not remove the need for the skills traditionally considered part of public health training, such as statistical principles, communication, domain knowledge, and leadership.(124) However, the training and effort required to gain and maintain current knowledge of recent advances in algorithmic and statistical frameworks is non-trivial.
Two specific skills may become important to foster for all big data users. First, it may be important to develop the capacity to ‘think like a computer’ when working with data. For example, while it is comparatively easy for a person to guess that records showing a “Bob Smith” and “Robert Smirh” living at the same address probably represent the same person, it is a much more complex leap for a simple name-matching algorithm that naively compares one letter at a time, to recognize not only that Bob is a common nickname for Robert, but also that t and r look similar in some fonts and are next to each other on a keyboard. Such ‘computational thinking’, wherein an analyst can recognize which problems pose greater algorithmic challenges, runs deeper than simply knowing how to program, run software, or build hardware, and has been suggested as a supplement to reading, writing, and arithmetic early in a child’s life.(141) But even public health trainees without childhood computational education may benefit from being able to “think like a computer” when faced with data sets that are time- and resource-intensive. We refer the reader to important reviews (41; 83) that have concretized the two core principles in computational thinking: abstraction and automation.
Second, quantitative bias analysis and related techniques will likely become a more important part of public health training, especially within epidemiology and biostatistics. As complex public health data sets become more integrated, more studies are expected to use secondary data. However, because systematic biases are harder to rule out in contexts where the investigator was not part of the data collection process, techniques that can explore the probability of incorrect inference under different assumptions of bias will be important to retain confidence in substantive conclusions.(60) Similarly, decisions about choice and evaluation of methods often involve tradeoffs between correctness on specific data points and probabilistic notions of correctness on the whole data set, e.g. gene-specific vs. genome-wide predictive models (106) and will require deep understanding of probability and statistics.
These two core skills are only a subset of the overall data science skills needed to work with public health big data, including an understanding of health informatics, data engineering, computational complexity, and adaptive learning. However, because these skills require substantial investment to master, we submit that training in more advanced data science techniques should be available but not required of public health students, analogous to other optional but important skills such as community-based health assessment.(74) This cultivation of specialized skills will necessitate diverse teams, a model already familiar to public health practitioners but less incorporated in training to date. Sidebar 5 summarizes how specialization in training has shaped bioinformatics education, which may provide a template for public health education. Numerous other perspectives on data science education may also be helpful.(40; 99)
As both specialized and generalized big data skills become more common in the public health workforce, these skills should be used to optimize data collection procedures. A biostatistician comfortable with real-time data processing may be more likely to push for data-adaptive trial protocols,(6) for example, or an informatics specialist with experience using natural language processing techniques to extract data from clinician notes might help a clinician understand how to frame her notes to be most efficient for clinical and research use. Epidemiologists comfortable with stepped wedge designs(118) may be more likely to suggest them to policy makers rolling out public health initiatives. Broadly, learning new ways to work with data effectively will and should shape not only which data we will choose to collect but also how we choose to collect it.
Limitations and open issues in the use of Machine Learning for Big Data Public Health
Appropriate use of both big data and machine learning rely on understanding several key limitations of each. First, we observe that machine learning’s capacity to overcome the curse of dimensionality requires tall data sets.(43) Small and/or biased training sets can lead to overfitting (Table 2) which limits the problems that current machine learning methods can address. Second, machine learning models are often described as “black boxes” whose opacity precludes interpretability or sanity-checking of key assumptions by non-experts.(109) While recent work has partially addressed this limitation (Sidebar 4),the problem persists. Third, in some instances, observers assume that models that learn automatically from data are more objective therefore more accurate than human-constructed models. Although data-driven models frequently can predict outcomes better than theory-driven models, data-driven model building also involves subjective decisions, such as choice of training and evaluation data sets, choice of pre-processing criteria, and choice of learning algorithms and initial parameters. These decisions cumulatively result in biases and prejudices that may be obscured from casual users.(17; 21) Fourth, data quantity often comes at the expense of quality. This is an issue for any big data analysis, but may be especially pernicious in the context of machine learning methods that use a test set to estimate prediction accuracy in the broader world. If data collection artifacts render training and test sets overly similar to each other but different from those of the data sets that the model would typically be applied to, overfitting may lead to unanticipatedly poor prediction accuracy in the real world.(15; 23) Finally, because big data studies often requires linking secondary-use data from heterogeneous sources, discrepancies between these data sources can induce biases, including demographically patterned bias (e.g. linking by name more frequently misses women who change surname after marriage.(16))
Conclusions
As the big data revolution continues, public health research and practice must continue to incorporate novel data sources and emerging analytical techniques, while contributing to knowledge, infrastructure, methodologies, and retaining a commitment to the ethical use of data. We feel this is a time to be optimistic: all five sources of big data identified in this review hold considerable potential to answer previously unanswerable questions, perhaps especially with the use of modern machine learning techniques. Such successes may arrive more quickly and more rigorously to the extent that the public health community can embrace a specialized, team science model in training and practice.
Sidebar 1. Measurement Error and Big Data
Although larger sample sizes afforded by big data reduce the probability of bias due to random error, bias due to measurement error is independent of sample size. (50; 56; 92) While some have argued the decrease in random error allows researchers to tolerate more measurement error,(88) this perspective implicitly assumes that hypothesis testing rather than estimation is an analyst’s goal, a perspective which has repeatedly been rejected within the public health literature.(35; 103) Indeed, measurement error may be more problematic in big data analyses,(64) because analysts working with secondary or administrative data may not have access to knowledge about potential data artifacts. For example, metabolomic datasets are vulnerable to measurement error related to timing of sample collection,(108; 111) but if the timing of sample collection was not included the dataset, an analyst will be unable to assess the potential impact of this error. Emerging machine learning techniques accounting for measurement error (known within that literature as ‘noisy labels’) may also be informative.(52; 53; 90; 96)
Sidebar 2. Data Perturbation
Data perturbation is a technique in which random noise is added to potentially identifying observed variables in order to prevent study participants from being identified while attempting to minimize information loss.(57) For example, a data perturbation algorithm might replace identifying information (e.g. birthdate) with values sampled from observed distribution of that variable. This idea has been developed extensively within the computer science data mining literature,(72; 78; 114) but relatively less explored within public health research to date (with some notable exceptions, including the National Health Interview Survey (80)).
Sidebar 3. Specialization in Bioinformatics Training
Bioinformatics curricula are typically framed to support three roles: (a) scientists, who use existing tools and domain expertise to develop and test hypotheses, (b) users, who consume information generated through bioinformatics research but typically do not apply the tools directly (e.g. genetic counselors, clinicians, etc.) (c) engineers, who develop novel bioinformatics tools to address problems that may or may not be specific to a domain.(136) Although many individuals act more than one of these roles at some point in an informatics career, identifying the core competencies of each role helps to frame the training need to specialize in each. For example, whereas engineers require strong algorithmic and programming skills, users need only a conceptual understanding of algorithms (but require much stronger interpretive and translational skills).
Sidebar 4. Interpretability of Machine Learning Models
While interpretability is not the primary goal of machine learning, some algorithms (e.g. decision trees) are inherently more interpretable than others. Broadly, interpretation of models is an area of active research, wherein one key idea involves the separation of the predictive model and the interpretation methodology itself. For instance, a naive approach involves the post hoc ranking of features based on empirical P-values calculated against a null distribution for each feature.(71) A modification of this involves ranking features in terms of their actual values in situations where they can be interpreted as probabilities.(102) More sophisticated approaches such as LIME (105) and Shap (82) provide general yet simple linear explanations of how features are weighted when a prediction is made, irrespective of the underlying model.
Sidebar 5. Future Directions in Machine Learning for Big Data in Public Health
There are three developments in machine learning that may be of interest to public health researchers and practitioners. First, machine learning has recently begun to formally confront outcome measurement error, (52; 53; 90; 96) particularly for datasets with a low-sensitivity outcome measure.(22; 98; 147) Second, several machine learning approaches designed for real-time prediction learn through a penalty-reward system based on feedback on its predictions rather than by fitting a model to a previously collected dataset.(140) This class of approaches, known as reinforcement learning, could be used in online data collection tools and surveillance. Finally, ‘deep learning’ approaches, which use large volumes of data and computational power to identify common but abstract components for automated classification (without the need for human guidance), have been used extensively in image classification and natural language processing.(68) It is expected that they will gain increased application to health data in the future as computational costs decrease.(66)
Acknowledgments:
We thank Stephanie Shiau for her thoughtful comments on an earlier version of this manuscript. S.J.M. was supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development grant 5T32HD057833-07. V.P. was supported by the Washington Research Foundation Fund for Innovation in Data-Intensive Discovery and the Moore/Sloan Data Science Environments Project at the University of Washington.
Contributor Information
Stephen J Mooney, Harborview Injury Prevention & Research Center, University of Washington, Seattle, sjm2186@uw.edu.
Vikas Pejaver, Department of Biomedical Informatics and Medical Education, Research Associate, The eScience Institute, University of Washington, Seattle, vpejaver@uw.edu.
References
- 1.Aiello AE, Simanek AM, Eisenberg MC, Walsh AR, Davis B, et al. 2016. Design and methods of a social network isolation study for reducing respiratory infection transmission: The eX-FLU cluster randomized trial. Epidemics 15:38–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alaa AM, van der Schaar M. 2017. Bayesian Inference of Individualized Treatment Effects using Multi-task Gaussian Processes. arXiv preprint arXiv:1704.02801 [Google Scholar]
- 3.Alter O, Brown PO, Botstein D. 2000. Singular value decomposition for genome-wide expression data processing and modeling. Proceedings of the National Academy of Sciences 97:10101–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson TK. 2009. Kernel density estimation and K-means clustering to profile road accident hotspots. Accident; analysis and prevention 41:359–64 [DOI] [PubMed] [Google Scholar]
- 5.Anderson TK. 2009. Kernel density estimation and K-means clustering to profile road accident hotspots. Accident Analysis & Prevention 41:359–64 [DOI] [PubMed] [Google Scholar]
- 6.Angus DC. 2015. Fusing randomized trials with big data: the key to self-learning health care systems? Jama 314:767–8 [DOI] [PubMed] [Google Scholar]
- 7.Aramaki E, Maskawa S, Morita M. Twitter catches the flu: detecting influenza epidemics using Twitter Proc. Proceedings of the conference on empirical methods in natural language processing, 2011:1568–76: Association for Computational Linguistics [Google Scholar]
- 8.Arnold BF, Ercumen A, Benjamin-Chung J, Colford JM Jr. 2016. Brief Report: Negative Controls to Detect Selection Bias and Measurement Bias in Epidemiologic Studies Epidemiology (Cambridge, Mass: ) 27:637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bachur RG, Harper MB. 2001. Predictive model for serious bacterial infections among infants younger than 3 months of age. Pediatrics 108:311–6 [DOI] [PubMed] [Google Scholar]
- 10.Bader MD, Mooney SJ, Rundle AG. 2016. Protecting personally identifiable information when using online geographic tools for public health research. American Public Health Association; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bansal S, Chowell G, Simonsen L, Vespignani A, Viboud C. 2016. Big Data for Infectious Disease Surveillance and Modeling. Journal of Infectious Diseases 214:S375–S9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barakat NH, Bradley AP, Barakat MN. 2010. Intelligible support vector machines for diagnosis of diabetes mellitus. IEEE transactions on information technology in biomedicine : a publication of the IEEE Engineering in Medicine and Biology Society 14:1114–20 [DOI] [PubMed] [Google Scholar]
- 13.Bates DW, Saria S, Ohno-Machado L, Shah A, Escobar G. 2014. Big data in health care: using analytics to identify and manage high-risk and high-cost patients. Health Affairs 33:1123–31 [DOI] [PubMed] [Google Scholar]
- 14.Bellman RE. 2015. Adaptive control processes: a guided tour. Princeton university press [Google Scholar]
- 15.Bernau C, Riester M, Boulesteix AL, Parmigiani G, Huttenhower C, et al. 2014. Cross-study validation for the assessment of prediction algorithms Bioinformatics (Oxford, England: ) 30:i105–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bohensky MA, Jolley D, Sundararajan V, Evans S, Pilcher DV, et al. 2010. Data linkage: a powerful research tool with potential problems. BMC health services research 10:346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolukbasi T, Chang K-W, Zou J, Saligrama V, Kalai A. 2016. Quantifying and reducing stereotypes in word embeddings. arXiv preprint arXiv:1606.06121 [Google Scholar]
- 18.Bougoudis I, Demertzis K, Iliadis L, Anezakis V-D, Papaleonidas A. Semi-supervised Hybrid Modeling of Atmospheric Pollution in Urban Centers Proc. International Conference on Engineering Applications of Neural Networks, 2016:51–63: Springer [Google Scholar]
- 19.Bowden J, Smith GD, Burgess S. 2015. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. International journal of epidemiology 44:512–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braun R, Rowe W, Schaefer C, Zhang J, Buetow K. 2009. Needles in the haystack: identifying individuals present in pooled genomic data. PLoS Genet 5:e1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caliskan A, Bryson JJ, Narayanan A. 2017. Semantics derived automatically from language corpora contain human-like biases. Science 356:183–6 [DOI] [PubMed] [Google Scholar]
- 22.Calvo B, Larrañaga P, Lozano JA. 2007. Learning Bayesian classifiers from positive and unlabeled examples. Pattern Recognition Letters 28:2375–84 [Google Scholar]
- 23.Castaldi PJ, Dahabreh IJ, Ioannidis JP. 2011. An empirical assessment of validation practices for molecular classifiers. Briefings in bioinformatics 12:189–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandrashekar G, Sahin F. 2014. A survey on feature selection methods. Computers & Electrical Engineering 40:16–28 [Google Scholar]
- 25.Davis B, Carpenter C. 2009. Proximity of fast-food restaurants to schools and adolescent obesity. American Journal of Public Health 99:505–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Davis HT, Aelion CM, McDermott S, Lawson AB. 2009. Identifying natural and anthropogenic sources of metals in urban and rural soils using GIS-based data, PCA, and spatial interpolation. Environmental pollution (Barking, Essex : 1987) 157:2378–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Choudhury M, Gamon M, Counts S, Horvitz E. Predicting depression via social media. Proc. ICWSM, 2013:2: [Google Scholar]
- 28.De Choudhury M, Kiciman E, Dredze M, Coppersmith G, Kumar M. Discovering shifts to suicidal ideation from mental health content in social media Proc. Proceedings of the 2016 CHI Conference on Human Factors in Computing Systems, 2016:2098–110: ACM; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng H, Runger G. Feature selection via regularized trees Proc. Neural Networks (IJCNN), The 2012 International Joint Conference on, 2012:1–8: IEEE [Google Scholar]
- 30.Efroymson M 1960. Multiple regression analysis. Mathematical methods for digital computers 1:191–203 [Google Scholar]
- 31.Eftekhar B, Mohammad K, Ardebili HE, Ghodsi M, Ketabchi E. 2005. Comparison of artificial neural network and logistic regression models for prediction of mortality in head trauma based on initial clinical data. BMC medical informatics and decision making 5:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Egger ME, Squires MH 3rd, Kooby DA, Maithel SK, Cho CS, et al. 2015. Risk stratification for readmission after major hepatectomy: development of a readmission risk score. Journal of the American College of Surgeons 220:640–8 [DOI] [PubMed] [Google Scholar]
- 33.Fahmi P, Viet V, Deok-Jai C. Semi-supervised fall detection algorithm using fall indicators in smartphone Proc. Proceedings of the 6th International Conference on Ubiquitous Information Management and Communication, 2012:122: ACM [Google Scholar]
- 34.Fisichella M, Stewart A, Denecke K, Nejdl W. Unsupervised public health event detection for epidemic intelligence Proc. Proceedings of the 19th ACM international conference on Information and knowledge management, 2010:1881–4: ACM [Google Scholar]
- 35.Gardner MJ, Altman DG. 1986. Confidence intervals rather than P values: estimation rather than hypothesis testing. Br Med J (Clin Res Ed) 292:746–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Glymour MM, Tchetgen Tchetgen EJ, Robins JM. 2012. Credible Mendelian randomization studies: approaches for evaluating the instrumental variable assumptions. American journal of epidemiology 175:332–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldsmith J, Liu X, Jacobson J, Rundle A. 2016. New Insights into Activity Patterns in Children, Found Using Functional Data Analyses. Medicine and science in sports and exercise 48:1723–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gomide J, Veloso A, Meira W Jr, Almeida V, Benevenuto F, et al. Dengue surveillance based on a computational model of spatio-temporal locality of Twitter Proc. Proceedings of the 3rd international web science conference, 2011:3: ACM [Google Scholar]
- 39.Graham DJ, Hipp JA. 2014. Emerging technologies to promote and evaluate physical activity: cutting-edge research and future directions. Frontiers in public health 2:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greene AC, Giffin KA, Greene CS, Moore JH. 2016. Adapting bioinformatics curricula for big data. Brief. Bioinform 17:43–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grover S, Pea R. 2013. Computational thinking in K–12: A review of the state of the field. Educ. Res 42:38–43 [Google Scholar]
- 42.Hafeman DM, Schwartz S. 2009. Opening the Black Box: a motivation for the assessment of mediation. International Journal of Epidemiology:dyn372 [DOI] [PubMed] [Google Scholar]
- 43.Halevy A, Norvig P, Pereira F. 2009. The unreasonable effectiveness of data. IEEE Intelligent Systems 24:8–12 [Google Scholar]
- 44.Hasan O, Meltzer DO, Shaykevich SA, Bell CM, Kaboli PJ, et al. 2010. Hospital readmission in general medicine patients: a prediction model. Journal of general internal medicine 25:211–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hernan MA, Robins JM. 2010. Causal inference. CRC Boca Raton, FL: [Google Scholar]
- 46.Hernán MA, Robins JM. 2006. Instruments for causal inference: an epidemiologist’s dream? Epidemiology 17:360–72 [DOI] [PubMed] [Google Scholar]
- 47.Holmes E, Loo RL, Stamler J, Bictash M, Yap IK, et al. 2008. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature 453:396–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Homer N, Szelinger S, Redman M, Duggan D, Tembe W, et al. 2008. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet 4:e1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hunter RF, McAneney H, Davis M, Tully MA, Valente TW, Kee F. 2015. “Hidden” Social Networks in Behavior Change Interventions. American Journal of Public Health 105:513–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ioannidis JP. 2013. Informed consent, big data, and the oxymoron of research that is not research. The American Journal of Bioethics 13:40–2 [DOI] [PubMed] [Google Scholar]
- 51.Ivanov O, Wagner MM, Chapman WW, Olszewski RT. Accuracy of three classifiers of acute gastrointestinal syndrome for syndromic surveillance Proc. Proceedings of the AMIA Symposium, 2002:345: American Medical Informatics Association; [PMC free article] [PubMed] [Google Scholar]
- 52.Jain S, White M, Radivojac P. Estimating the class prior and posterior from noisy positives and unlabeled data Proc. Advances in Neural Information Processing Systems, NIPS 2016, Barcelona, Spain, December 2016:2693–701: [Google Scholar]
- 53.Jain S, White M, Radivojac P. February 2017. Recovering true classifier performance in positive-unlabeled learning In AAAI Conference on Artificial Intelligence, AAAI 2017, pp. 2066–72. San Francisco, California, U.S.A [Google Scholar]
- 54.Jeste DV, Savla GN, Thompson WK, Vahia IV, Glorioso DK, et al. 2013. Association between older age and more successful aging: critical role of resilience and depression. The American journal of psychiatry 170:188–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kang H, Zhang A, Cai TT, Small DS. 2016. Instrumental variables estimation with some invalid instruments and its application to Mendelian randomization. Journal of the American Statistical Association 111:132–44 [Google Scholar]
- 56.Kaplan RM, Chambers DA, Glasgow RE. 2014. Big data and large sample size: a cautionary note on the potential for bias. Clinical and translational science 7:342–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kargupta H, Datta S, Wang Q, Sivakumar K. On the privacy preserving properties of random data perturbation techniques Proc. Data Mining, 2003. ICDM 2003. Third IEEE International Conference on, 2003:99–106: IEEE [Google Scholar]
- 58.Kass NE. 2001. An ethics framework for public health. American journal of public health 91:1776–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khoury MJ, Ioannidis JP. 2014. Big data meets public health. Science 346:1054–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kochenderfer MJ, Reynolds HJD. 2015. Decision making under uncertainty: theory and application. MIT press [Google Scholar]
- 61.Kononen DW, Flannagan CA, Wang SC. 2011. Identification and validation of a logistic regression model for predicting serious injuries associated with motor vehicle crashes. Accident; analysis and prevention 43:112–22 [DOI] [PubMed] [Google Scholar]
- 62.Kostkova P A roadmap to integrated digital public health surveillance: the vision and the challenges Proc. Proceedings of the 22nd International Conference on World Wide Web, 2013:687–94: ACM [Google Scholar]
- 63.Kovalchik SA, Tammemagi M, Berg CD, Caporaso NE, Riley TL, et al. 2013. Targeting of low-dose CT screening according to the risk of lung-cancer death. The New England journal of medicine 369:245–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kwan M-P. 2016. Algorithmic geographies: Big data, algorithmic uncertainty, and the production of geographic knowledge. Annals of the American Association of Geographers 106:274–82 [Google Scholar]
- 65.Larson T, Gould T, Simpson C, Liu LJ, Claiborn C, Lewtas J. 2004. Source apportionment of indoor, outdoor, and personal PM2.5 in Seattle, Washington, using positive matrix factorization. Journal of the Air & Waste Management Association (1995) 54:1175–87 [DOI] [PubMed] [Google Scholar]
- 66.Lasko TA, Denny JC, Levy MA. 2013. Computational phenotype discovery using unsupervised feature learning over noisy, sparse, and irregular clinical data. PloS one 8:e66341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lazer D, Kennedy R, King G, Vespignani A. 2014. The parable of Google Flu: traps in big data analysis. Science 343:1203–5 [DOI] [PubMed] [Google Scholar]
- 68.LeCun Y, Bengio Y, Hinton G. 2015. Deep learning. Nature 521:436–44 [DOI] [PubMed] [Google Scholar]
- 69.Lee LM, Gostin LO. 2009. Ethical collection, storage, and use of public health data: a proposal for a national privacy protection. Jama 302:82–4 [DOI] [PubMed] [Google Scholar]
- 70.Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, et al. 2010. Tackling the widespread and critical impact of batch effects in high-throughput data. Nature reviews. Genetics 11:733–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, et al. 2009. Automated inference of molecular mechanisms of disease from amino acid substitutions Bioinformatics (Oxford, England: ) 25:2744–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li H, Muralidhar K, Sarathy R, Luo XR. 2014. Evaluating Re-Identification Risks of Data Protected by Additive Data Perturbation. Journal of Database Management (JDM) 25:52–74 [Google Scholar]
- 73.Li Y, Ngom A. Data integration in machine learning Proc. Bioinformatics and Biomedicine (BIBM), 2015 IEEE International Conference on, 2015:1665–71: IEEE [Google Scholar]
- 74.Lichtveld MY. 2016. A Timely Reflection on the Public Health Workforce. LWW; [DOI] [PubMed] [Google Scholar]
- 75.Link BG, Phelan J. 1995. Social conditions as fundamental causes of disease. Journal of health and social behavior:80–94 [PubMed] [Google Scholar]
- 76.Lipsitch M, Tchetgen ET, Cohen T. 2010. Negative controls: a tool for detecting confounding and bias in observational studies Epidemiology (Cambridge, Mass: ) 21:383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lipsitch M, Tchetgen ET, Cohen T. 2012. Negative control exposures in epidemiologic studies. Epidemiology 23:351–2 [DOI] [PubMed] [Google Scholar]
- 78.Liu K, Giannella C, Kargupta H. 2008. A survey of attack techniques on privacy-preserving data perturbation methods. Privacy-Preserving Data Mining:359–81 [Google Scholar]
- 79.Lleras-Muney A 2005. The relationship between education and adult mortality in the United States. The Review of Economic Studies 72:189–221 [Google Scholar]
- 80.Lochner K, Hummer RA, Bartee S, Wheatcroft G, Cox C. 2008. The public-use National Health Interview Survey linked mortality files: methods of reidentification risk avoidance and comparative analysis. American Journal of Epidemiology 168:336–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lord N 2017. The History of Data Breaches. https://digitalguardian.com/blog/history-data-breaches
- 82.Lundberg S, Lee S-I. 2016. An unexpected unity among methods for interpreting model predictions. arXiv preprint arXiv:1611.07478 [Google Scholar]
- 83.Lye SY, Koh JHL. 2014. Review on teaching and learning of computational thinking through programming: What is next for K-12? Comput. Human Behav. 41:51–61 [Google Scholar]
- 84.Lynch SM, Mitra N, Ross M, Newcomb C, Dailey K, et al. 2017. A Neighborhood-Wide Association Study (NWAS): Example of prostate cancer aggressiveness. PloS one 12:e0174548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mai J-E. 2016. Big data privacy: The datafication of personal information. The Information Society 32:192–9 [Google Scholar]
- 86.Mai J-E. 2016. Three Models of Privacy. NORDICOM:171 [Google Scholar]
- 87.Mamiya H, Schwartzman K, Verma A, Jauvin C, Behr M, Buckeridge D. 2015. Towards probabilistic decision support in public health practice: predicting recent transmission of tuberculosis from patient attributes. Journal of biomedical informatics 53:237–42 [DOI] [PubMed] [Google Scholar]
- 88.Mayer-Schönberger V, Cukier K. 2013. Big data: A revolution that will transform how we live, work, and think. Houghton Mifflin Harcourt [Google Scholar]
- 89.McKetta S, Hatzenbuehler ML, Pratt C, Bates L, Link BG, Keyes KM. 2017. Does Social Selection Explain the Association between State-Level Racial Animus and Racial Disparities in Self-Rated Health in the United States? Annals of Epidemiology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Menon A, Rooyen BV, Ong CS, Williamson B. Learning from corrupted binary labels via class-probability estimation. Proc. Proceedings of the 32nd International Conference on Machine Learning (ICML-15), 2015:125–34: [Google Scholar]
- 91.Mooney SJ, Joshi S, Cerdá M, Kennedy GJ, Beard JR, Rundle AG. 2017. Contextual Correlates of Physical Activity among Older Adults: A Neighborhood-Environment Wide Association Study (NE-WAS). Cancer Epidemiology and Prevention Biomarkers:cebp 0827.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mooney SJ, Westreich DJ, El-Sayed AM. 2015. Epidemiology in the era of big data Epidemiology (Cambridge, Mass: ) 26:390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Murdoch TB, Detsky AS. 2013. The inevitable application of big data to health care. Jama 309:1351–2 [DOI] [PubMed] [Google Scholar]
- 94.Myers J, Frieden TR, Bherwani KM, Henning KJ. 2008. Ethics in public health research: privacy and public health at risk: public health confidentiality in the digital age. American Journal of public health 98:793–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Naimi AI, Westreich DJ. 2014. Big Data: A revolution that will transform how we live, work, and think. Oxford University Press [Google Scholar]
- 96.Natarajan N, Dhillon IS, Ravikumar PK, Tewari A. Learning with noisy labels. Proc. Advances in neural information processing systems, 2013:1196–204: [Google Scholar]
- 97.Ness RB, Committee JP. 2007. Influence of the HIPAA privacy rule on health research. Jama 298:2164–70 [DOI] [PubMed] [Google Scholar]
- 98.Nguyen MN, Li X-L, Ng S-K. Positive unlabeled leaning for time series classification. Proc. IJCAI, 2011, 11:1421–6: [Google Scholar]
- 99.Otero P, Hersh W, Jai Ganesh AU. 2014. Big data: are biomedical and health informatics training programs ready? Contribution of the IMIA working group for health and medical informatics education. Yearb. Med. Inform 9:177–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Papadopoulos A, Fotiadis DI, Likas A. 2002. An automatic microcalcification detection system based on a hybrid neural network classifier. Artificial intelligence in medicine 25:149–67 [DOI] [PubMed] [Google Scholar]
- 101.Parkka J, Ermes M, Korpipaa P, Mantyjarvi J, Peltola J, Korhonen I. 2006. Activity classification using realistic data from wearable sensors. IEEE transactions on information technology in biomedicine : a publication of the IEEE Engineering in Medicine and Biology Society 10:119–28 [DOI] [PubMed] [Google Scholar]
- 102.Pejaver V, Urresti J, Lugo-Martinez J, Pagel KA, Lin GN, et al. 2017. MutPred2: inferring the molecular and phenotypic impact of amino acid variants. bioRxiv:134981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Poole C 2001. Low P-values or narrow confidence intervals: which are more durable? Epidemiology 12:291–4 [DOI] [PubMed] [Google Scholar]
- 104.Psaty BM, Breckenridge AM. 2014. Mini-Sentinel and regulatory science--big data rendered fit and functional. The New England journal of medicine 370:2165. [DOI] [PubMed] [Google Scholar]
- 105.Ribeiro MT, Singh S, Guestrin C. Why Should I Trust You?: Explaining the Predictions of Any Classifier Proc. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2016:1135–44: ACM [Google Scholar]
- 106.Riera C, Padilla N, de la Cruz X. 2016. The complementarity between protein-specific and general pathogenicity predictors for amino acid substitutions. Hum. Mutat 37:1013–24 [DOI] [PubMed] [Google Scholar]
- 107.Robins JM. 2001. Data, design, and background knowledge in etiologic inference. Epidemiology 12:313–20 [DOI] [PubMed] [Google Scholar]
- 108.Rocke DM, Durbin B. 2001. A model for measurement error for gene expression arrays. Journal of computational biology 8:557–69 [DOI] [PubMed] [Google Scholar]
- 109.Rost B, Radivojac P, Bromberg Y. 2016. Protein function in precision medicine: deep understanding with machine learning. FEBS letters 590:2327–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rothstein MA. 2010. Is deidentification sufficient to protect health privacy in research? The American Journal of Bioethics 10:3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sampson JN, Boca SM, Shu XO, Stolzenberg-Solomon RZ, Matthews CE, et al. 2013. Metabolomics in epidemiology: sources of variability in metabolite measurements and implications. Cancer Epidemiology and Prevention Biomarkers 22:631–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Santillana M, Zhang DW, Althouse BM, Ayers JW. 2014. What can digital disease detection learn from (an external revision to) Google Flu Trends? American journal of preventive medicine 47:341–7 [DOI] [PubMed] [Google Scholar]
- 113.Sedig K, Ola O. 2014. The challenge of big data in public health: an opportunity for visual analytics. Online journal of public health informatics 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shah A, Gulati R. Evaluating applicability of perturbation techniques for privacy preserving data mining by descriptive statistics Proc. Advances in Computing, Communications and Informatics (ICACCI), 2016 International Conference on, 2016:607–13: IEEE [Google Scholar]
- 115.Smith GD, Ebrahim S. 2004. Mendelian randomization: prospects, potentials, and limitations. International journal of epidemiology 33:30–42 [DOI] [PubMed] [Google Scholar]
- 116.Smith KJ, Roberts MS. 2002. Cost-effectiveness of newer treatment strategies for influenza. The American journal of medicine 113:300–7 [DOI] [PubMed] [Google Scholar]
- 117.Solove DJ. 2008. Understanding privacy. [Google Scholar]
- 118.Spiegelman D 2016. Evaluating public health interventions: 2. Stepping up to routine public health evaluation with the stepped wedge design. American journal of public health 106:453–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tan M, Tsang IW, Wang L. 2014. Towards ultrahigh dimensional feature selection for big data. Journal of Machine Learning Research 15:1371–429 [Google Scholar]
- 120.Tavani HT. 2007. Philosophical theories of privacy: Implications for an adequate online privacy policy. Metaphilosophy 38:1–22 [Google Scholar]
- 121.Teutsch SM, Churchill RE. 2000. Principles and practice of public health surveillance. Oxford University Press, USA [Google Scholar]
- 122.Thomson JJ. 1975. The right to privacy. Philosophy & Public Affairs:295–314 [Google Scholar]
- 123.Tibshirani R 1996. Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society. Series B (Methodological):267–88 [Google Scholar]
- 124.Tilson H, Gebbie KM. 2004. The public health workforce. Annu. Rev. Public Health 25:341–56 [DOI] [PubMed] [Google Scholar]
- 125.Titiunik R 2015. Can Big Data solve the fundamental problem of causal inference? PS: Political Science & Politics 48:75–9 [Google Scholar]
- 126.Tolich M 2004. Internal confidentiality: When confidentiality assurances fail relational informants. Qualitative Sociology 27:101–6 [Google Scholar]
- 127.Trtica-Majnaric L, Zekic-Susac M, Sarlija N, Vitale B. 2010. Prediction of influenza vaccination outcome by neural networks and logistic regression. Journal of biomedical informatics 43:774–81 [DOI] [PubMed] [Google Scholar]
- 128.Vacek JL, Vanga SR, Good M, Lai SM, Lakkireddy D, Howard PA. 2012. Vitamin D deficiency and supplementation and relation to cardiovascular health. The American journal of cardiology 109:359–63 [DOI] [PubMed] [Google Scholar]
- 129.Van der Laan MJ, Polley EC, Hubbard AE. 2007. Super learner. Statistical applications in genetics and molecular biology 6 [DOI] [PubMed] [Google Scholar]
- 130.VanderWeele T 2015. Explanation in causal inference: methods for mediation and interaction. Oxford University Press [Google Scholar]
- 131.VanderWeele TJ, Tchetgen EJT, Cornelis M, Kraft P. 2014. Methodological challenges in Mendelian randomization Epidemiology (Cambridge, Mass: ) 25:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wager S, Athey S. 2017. Estimation and inference of heterogeneous treatment effects using random forests. Journal of the American Statistical Association [Google Scholar]
- 133.Wang J, McMichael AJ, Meng B, Becker NG, Han W, et al. 2006. Spatial dynamics of an epidemic of severe acute respiratory syndrome in an urban area. Bulletin of the World Health Organization 84:965–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang J, McMichael AJ, Meng B, Becker NG, Han W, et al. 2006. Spatial dynamics of an epidemic of severe acute respiratory syndrome in an urban area. Bulletin of the World Health Organization 84:965–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Warren SD, Brandeis LD. 1890. The right to privacy. Harvard law review:193–220 [Google Scholar]
- 136.Welch L, Lewitter F, Schwartz R, Brooksbank C, Radivojac P, et al. 2014. Bioinformatics curriculum guidelines: toward a definition of core competencies. PLoS Comput. Biol 10:e1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wesolowski A, Metcalf C, Eagle N, Kombich J, Grenfell BT, et al. 2015. Quantifying seasonal population fluxes driving rubella transmission dynamics using mobile phone data. Proceedings of the National Academy of Sciences 112:11114–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Westin AF. 1967. Special report: legal safeguards to insure privacy in a computer society. Communications of the ACM 10:533–7 [Google Scholar]
- 139.White A, Trump K-S. 2016. The Promises and Pitfalls of 311 Data. Urban Affairs Review:1078087416673202 [Google Scholar]
- 140.Wiering M, Van Otterlo M. 2012. Reinforcement learning. [Google Scholar]
- 141.Wing JM. 2006. Computational thinking. Commun. ACM 49:33–5 [Google Scholar]
- 142.Wright A, Chen ES, Maloney FL. 2010. An automated technique for identifying associations between medications, laboratory results and problems. Journal of biomedical informatics 43:891–901 [DOI] [PubMed] [Google Scholar]
- 143.Xafis V 2015. The acceptability of conducting data linkage research without obtaining consent: lay people’s views and justifications. BMC medical ethics 16:79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yamada M, Tang J, Lugo-Martinez J, Hodzic E, Shrestha R, et al. 2016. Ultra High-Dimensional Nonlinear Feature Selection for Big Biological Data. arXiv preprint arXiv:1608.04048 [Google Scholar]
- 145.Yang M, Kiang M, Shang W. 2015. Filtering big data from social media--Building an early warning system for adverse drug reactions. Journal of biomedical informatics 54:230–40 [DOI] [PubMed] [Google Scholar]
- 146.Yang W, Mu L. 2015. GIS analysis of depression among Twitter users. Applied Geography 60:217–23 [Google Scholar]
- 147.Zhao Y, Kong X, Philip SY. Positive and unlabeled learning for graph classification Proc. Data Mining (ICDM), 2011 IEEE 11th International Conference on, 2011:962–71: IEEE [Google Scholar]
- 148.Zhu X 2005. Semi-supervised learning literature survey. [Google Scholar]
- 149.Zimmer M 2010. “But the data is already public”: on the ethics of research in Facebook. Ethics and information technology 12:313–25 [Google Scholar]
- 150.Zou H, Hastie T. 2005. Regularization and variable selection via the elastic net. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 67:301–20 [Google Scholar]