Abstract
Background
The E-cadherin gene (CDH1) is frequently mutated in diffuse gastric cancer and lobular breast cancer, and germline mutations predispose to the cancer syndrome Hereditary Diffuse Gastric Cancer. We are taking a synthetic lethal approach to identify druggable vulnerabilities in CDH1-mutant cancers.
Methods
Density distributions of cell viability data from a genome-wide RNAi screen of isogenic MCF10A and MCF10A-CDH1−/− cells were used to identify protein classes affected by CDH1 mutation. The synthetic lethal relationship between selected protein classes and E-cadherin was characterised by drug sensitivity assays in both the isogenic breast MCF10A cells and CDH1-isogenic gastric NCI-N87. Endocytosis efficiency was quantified using cholera toxin B uptake. Pathway metagene expression of 415 TCGA gastric tumours was statistically correlated with CDH1 expression.
Results
MCF10A-CDH1−/− cells showed significantly altered sensitivity to RNAi inhibition of groups of genes including the PI3K/AKT pathway, GPCRs, ion channels, proteosomal subunit proteins and ubiquitinylation enzymes. Both MCF10A-CDH1−/− and NCI-N87-CDH1−/− cells were more sensitive than wild-type cells to compounds that disrupted plasma membrane composition and trafficking, but showed contrasting sensitivities to inhibitors of actin polymerisation and the chloride channel inhibitor NS3728. The MCF10A-CDH1−/− cell lines showed reduced capacity to endocytose cholera toxin B. Pathway metagene analysis identified 20 Reactome pathways that were potentially synthetic lethal in tumours. Genes involved in GPCR signalling, vesicle transport and the metabolism of PI3K and membrane lipids were strongly represented amongst the candidate synthetic lethal genes.
Conclusions
E-cadherin loss leads to disturbances in receptor signalling and plasma membrane trafficking and organisation, creating druggable vulnerabilities.
Electronic supplementary material
The online version of this article (10.1007/s10120-018-0859-1) contains supplementary material, which is available to authorised users.
Keywords: Diffuse gastric cancer, Synthetic lethality, E-cadherin, Plasma membrane, CDH1
Introduction
CDH1 is a tumour suppressor gene that encodes E-cadherin, a homophilic cell-to-cell adhesion protein that is localised to the adherens junction on the basolateral surface of epithelial cells. In addition to cell adhesion, E-cadherin plays central roles in differentiation, cell polarity, migration, the detection of cell–cell tension and cell survival signalling [1]. Somatic mutations in CDH1 occur frequently in diffuse gastric cancer (DGC) and lobular breast cancer (LBC) and its downregulation is a hallmark of the epithelial–mesenchymal transition [2, 3]. In addition, germline CDH1 mutations genetically characterise the inherited cancer syndrome Hereditary Diffuse Gastric Cancer (HDGC) [4]. Carriers of pathogenic CDH1 mutations from HDGC families have a ~ 70% lifetime risk of developing advanced DGC and female carriers have an additional ~ 40% risk of LBC [5, 6]. To develop novel drugs for the chemoprevention and treatment of E-cadherin-deficient tumours, we are taking a synthetic lethal (SL) approach to identify vulnerabilities in these cancers. We have previously conducted a genome-wide siRNA screen and a 4000 compound ‘known drug’ screen in isogenic breast MCF10A cells with and without E-cadherin expression to identify vulnerabilities in the MCF10A-CDH1−/− cells that could be exploited in an SL manner [7]. MCF10A is a non-malignant breast cell line with relatively few cytogenetic abnormalities and mutations [8, 9]. It is therefore a suitable reference line for studies that explore normal protein interactions and also an informative model for HDGC chemoprevention studies, particularly since equivalent non-malignant stomach cell lines are not available. Compared to MCF10A cells, MCF10A-CDH1−/− cells have modest changes in cell morphology but show striking differences in the organisation of both the actin and microtubule cytoskeletons [10]. Our genome-wide siRNA screen on this isogenic line identified large numbers of potential SL interactions with E-cadherin, particularly in GPCR signalling and cytoskeletal functions. MCF10A-CDH1−/− cells were also more sensitive to multiple drugs including antagonists of PI3K, c-SRC and histone deacetylases. However, a unifying SL mechanism was not established. To elucidate the mechanisms underpinning E-cadherin’s diverse SL interactions and the subsequent vulnerabilities in CDH1-null cancers, we have now extended our analysis of isogenic cell line pairs and applied a novel bioinformatic approach that statistically queries tumour genome-wide expression data for potential SL pathways. Our data supports a model in which rearrangement of the actin cytoskeleton following abrogation of the adherens junction [10] leads to disruption of normal plasma membrane organisation and dynamics. We provide evidence which suggests that this disruption leads to wide-ranging disturbances in receptor signalling and plasma membrane trafficking, which in turn create druggable synthetic lethal vulnerabilities in E-cadherin-null cells.
Materials and methods
Cell viability ratios
Data on cell viability after gene knockdown were previously generated via a high-throughput siRNA screen in the breast MCF10A and MCF10A-CDH1−/− isogenic lines [7]. For each gene, a viability ratio (MCF10A/MCF10A-CDH1−/−) was calculated to indicate the relative impact of gene knockdown on cell viability in the two cell lines.
Visualisation of viability ratios
The statistical software R was used to generate density plots summarising the distribution of viability ratios for specific lists of genes [11]. Lists were obtained from a variety of sources and are noted in the figure legends. Extracellular ligands were not included in the signalling pathway lists because of the artificial cell culture environment. To identify statistical differences between the viability ratio distributions for the selected gene sets and the distribution for all 18,120 genes included in the siRNA screen, the Kolmogorov-Smirnov test was used.
Cancer genome atlas gene expression data
RNA-seq data from the Cancer Genome Atlas (TCGA) Stomach Adenocarcinoma (STAD) project was downloaded from the International Cancer Genome Consortium data portal (version 25, September 2017). Per-gene read count data were processed in R [11], using the RNA-seq workflow described in the limma package users guide. Normal tissue samples were removed from the analysis, leaving expression data for 20,502 genes across 415 gastric tumours.
Metagene creation
The reactome.db R package was used to obtain information about gene membership of Reactome pathways [12]. For each pathway comprising at least five genes, singular value decomposition (SVD) was used to generate a metagene (with per-gene expression data standardised to have mean 0 and standard deviation 1 prior to performing SVD) using the first eigenvector for that pathway.
Computational identification of SL relationships
To identify pathway metagenes that exhibited a potential synthetic lethal association with CDH1, samples were binned into tertile groups (bottom 1/3, middle 1/3, top 1/3) for each quantity of interest (metagene value or CDH1 expression), and a chi-squared statistic was calculated as a measure of association between metagene tertiles and CDH1 tertiles. To account for inter-gene correlation that was not pathway-specific, resampling was used to generate a null distribution of the chi-squared statistics. For each pathway size, k (i.e., number of genes in the pathway), a random sample of k genes was taken, and their expression values were used to generate a metagene as described above. A chi-squared statistic was then generated for the resampled data, and this process was repeated 500,000 times per pathway to generate a null distribution for each pathway metagene. Empirical p values were then calculated for each pathway metagene (by counting the number of times that the resampled chi-squared statistics exceeded the observed value), and these p values were then adjusted for multiple testing using the Benjamini and Hochberg False Discovery Rate (FDR) controlling procedure. Pathway metagenes with a FDR-adjusted p value below 0.2 were considered to exhibit a significant association with CDH1 levels. Hierarchical clustering was carried out using the heatmap.2 function inside the ggplot2 package.
Drug titrations
Latrunculin B, cytochalasin D, methyl-β-cyclodextrin and amphotericin B were obtained from Sigma-Aldrich and bafilomycin A1, atorvastatin, AZD5363, otenabant and PI103 from SelleckChem. NS3728 was synthesised at the Ferrier Institute. The NCI-N87 gastric cancer cell line was obtained from ATCC. NCI-N87-CDH1−/− was generated by homozygous frameshift deletion of base c.1553 in exon 10 using CRISPR-Cas9 (A Chen, manuscript in preparation). Drug treatments were largely carried out as previously described [7]. Briefly, MCF10A and MCF10A-CDH1−/− cells were seeded in black-walled, clear-bottom 96 well plates (Corning, Corning, NY) at a density of 4000 cells/well in a volume of 100 µl. NCI-N87 and NCI-N87-CDH1−/− (passage ≤ 10) were seeded at 10,000 cells/ml. After a 24 h incubation, 10 µl drug (dissolved in vehicle to a 20 mM stock and diluted in complete media) was added to each well. After a further 48 h, cells were fixed and stained with 1 µg/mL Hoechst 33,342, 0.25% paraformaldehyde and 0.075% saponin in PBS [13]. Six fields/well at 4× magnification were captured using the Cytation 5 imager (Biotek). Nuclei were counted using Gen5 (Biotek) and normalised to the vehicle control for each cell line.
Cholera toxin endocytosis assay
MCF10A and MCF10A-CDH1−/− cells were seeded at 4000 cells per well in 96-well, black-walled, clear-bottom tissue culture plates in 100 µL cholera toxin-deficient complete growth medium. Plates were incubated at RT for 30 min then at 37 °C (5% CO2) for a further 48 h. The medium was then replaced with 50 µL cholera toxin-deficient growth medium supplemented with 10 µg/mL Alexa Fluor 488-Cholera Toxin Subunit B (Thermo Fisher Scientific, Waltham, MA) and plates incubated for 30 min at 37 °C (5% CO2). Cells were washed twice with PBS and fixed with 0.25% paraformaldehyde in PBS with 1 µg/mL Hoechst 33,342 (Thermo Fisher Scientific) overnight at RT in the dark. Fluorescent microscopy was performed at 10× magnification using a Nikon Eclipse Ti inverted microscope, and at least 15 images per well were acquired with a DS-QiMc camera. Hoechst-stained nuclei were quantified for a total cell count using CellProfiler™ (Broad Institute). ImageJ was used to measure the fluorescent intensity of cholera toxin and subtract background fluorescence [14].
Results
Different protein classes show contrasting SL distributions
We previously carried out a genome-wide siRNA screen in isogenic breast MCF10A cells with and without E-cadherin expression and observed that GPCRs and cytoskeletal proteins frequently displayed an SL relationship with E-cadherin [7]. To elucidate the mechanisms underpinning synthetic lethality in E-cadherin-deficient cells, we have now extended our analysis of the siRNA screen data to include genes with the reverse effect to synthetic lethality, that is, genes whose siRNAs reduced the viability of MCF10A cells more than MCF10A-CDH1−/− cells. We have termed these genes ‘reverse synthetic lethal’ (RSL). We hypothesise that RSL proteins have direct or indirect interactions with E-cadherin, but CDH1−/− cells have compensated for the loss of this interaction, making them less sensitive to knockdown of RSL proteins (relative to wild-type cells). This adaption is most likely to have occurred through the activation of one or more proteins with overlapping function (i.e., functional redundancy). In contrast to these RSL proteins, we hypothesise that MCF10A-CDH1−/− cells have been unable to fully compensate for the loss of the interaction between E-cadherin and any given SL protein, exposing a vulnerability (summarised in Fig. 1a). Although RSL proteins may not represent useful drug targets on their own, compounds which also inhibit the functional homologue may prove to be SL. Regardless, the identity of RSL proteins provides a fuller understanding of the impact of E-cadherin loss on epithelial cells. 1648 genes met our RSL threshold of a MCF10A-CDH1−/−/MCF10A viability ratio of ≥ 1.3. Using the DAVID Functional Annotation Clustering tool v6.8 [15], the most enriched functional cluster identified amongst RSL genes was a group of terms associated with phosphatases (enrichment score = 14.88; Table 1). Of 239 phosphatases in the Human Dephosphorylation Database, 224 were included in our siRNA screen. A density distribution of the MCF10A-CDH1−/−/MCF10A viability ratios for these 224 phosphatases produced a single peak with the highest density occurring at a viability ratio of 1.34 (Fig. 1b). This distribution was shifted significantly (p < 2.2 × 10−16) in the RSL direction compared to the distribution of all 18,120 genes in the siRNA screen which displayed a single peak centered around a MCF10A-CDH1−/−/MCF10A ratio of 1.00. In contrast, the overall density distribution of 496 protein kinases in the siRNA screen showed a small but highly significant shift (p < 2.2 × 10−16) towards synthetic lethality with the peak of the density distribution at 0.95 (Fig. 1c). The contrasting cell viability distribution of kinase and phosphatase siRNAs is likely to be related to the greater functional promiscuity of phosphatases [16]. Ion channels were also highly enriched in the gene set analysis of RSL genes (enrichment score = 10.93; Table 1). Consistent with this observation, the MCF10A-CDH1−/−/MCF10A density distribution of 161 voltage-gated ion channels in the siRNA screen was significantly right-shifted from the overall distribution of all 18,120 genes with a peak at 1.14 (p < 2.2 × 10−16; Fig. 1d). In contrast, the distribution of 370 solute carrier genes was not significantly different to the distribution of all 18,120 genes (p = 0.45; Fig. 1e). The RSL phenotype of the ion channels was observed in most voltage-gated ion channel subgroups with the exception of small families of gap junction (n = 20), bestrophin (n = 4) and leucine-rich repeat channel (n = 5) proteins which showed a contrasting trend towards a SL phenotype (Fig. 1f). Terms associated with ribosomes, splicing and proteasomes were also enriched in the RSL gene set analysis (Table 1). Cytoplasmic ribosomal proteins (n = 78) and spliceosome proteins (n = 132) showed a corresponding significant RSL shift in the MCF10A-CDH1−/−/MCF10A density distributions (Fig. 1g, h), as did ubiquitin-specific peptidases (n = 55), E2 ubiquitin conjugating enzymes (n = 36), HECT E3 ubiquitin ligases (n = 25) and proteasome complex proteins (n = 43) (Fig. 1i, j, k, m). In contrast, the RING E3 ubiquitin ligases (n = 299) were not RSL, but instead featured a large SL shoulder in the MCF10A-CDH1−/−/MCF10A ratios centered at approximately 0.8 (p < 2.2 × 10−16) (Fig. 1l). Surprisingly, GPCR signalling terms were highly enriched amongst the RSL gene functions (enrichment score = 14.33; Table 1) just as they were in the SL gene functions [7]. Of the 1409 GPCRs identified by the Human Gene Nomenclature Committee, 721 were represented in the siRNA screen, including 245 non-sensory, 86 orphan, 363 olfactory GPCRs and 27 taste GPCRs. Consistent with the gene ontology analyses, the MCF10A-CDH1−/−/MCF10A density distributions for the 245 non-sensory GPCRs had a bimodal distribution, with peaks at viability ratios of 0.82 and 1.37 (p = 1.0 × 10−12; Fig. 2a). The left peak contained the SL GPCRs identified previously and the right peak comprised the set of RSL GPCRs. A similar bimodal distribution was also observed for the 86 orphan GPCRs (peaks at 0.82 and 1.39; Fig. 2b). In contrast, the viability ratio distribution of the taste and olfactory GPCRs (which have negligible expression in breast cells) predictably presented as single peaks centered at viability ratios of 0.97 (Fig. 2c) and 1.07, respectively. The density distributions for a set of 35 guanine nucleotide binding protein (G-protein) genes showed a single major peak that was shifted in the SL direction (max = 0.92, p = 0.05; Fig. 2d). Together, the above results demonstrate that whole classes of membrane receptors, channels and protein-modifying enzymes are influenced by E-cadherin loss.
Fig. 1.
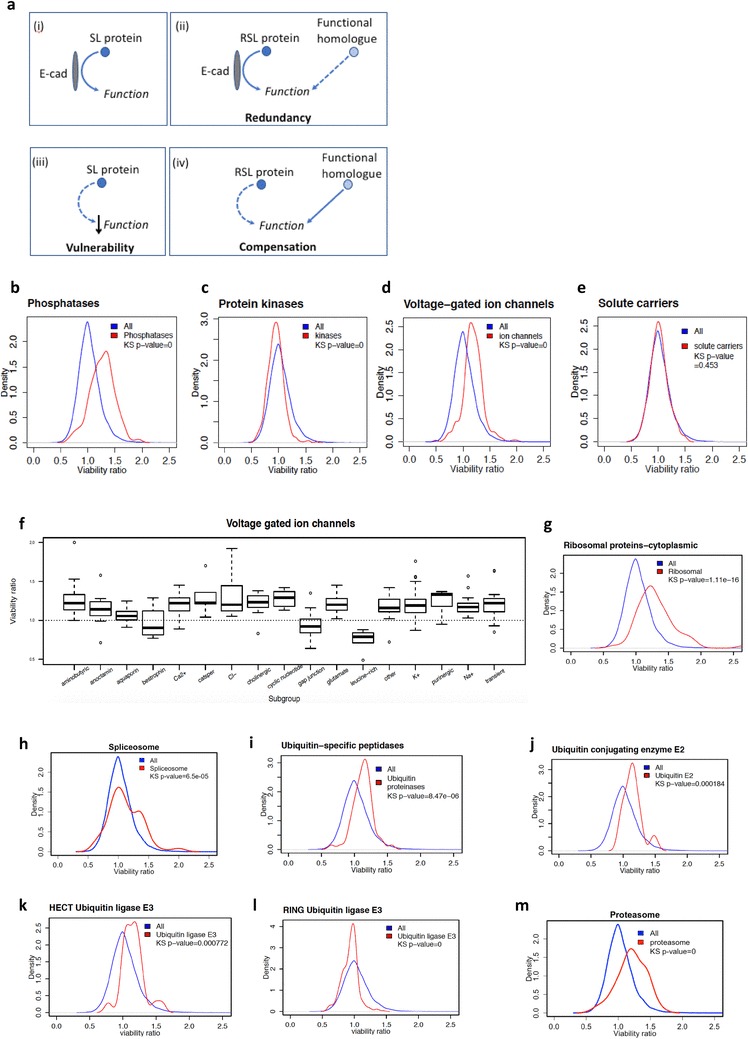
a Model of SL and RSL interactions. SL and RSL proteins both have functions affected by E-cadherin activity; however, RSL proteins have the potential to be compensated for by functional homologues (i–ii). When E-cadherin is lost, inhibition of an SL protein leads to reduced cell fitness (iii), whereas the inhibition of an RSL protein has less effect because its function is compensated for by the increased activity of the functional homologue (iv). b–e Density distributions of MCF10A-CDH1−/−/MCF10A viability ratios following siRNA knockdown of families of phosphatases, protein kinases, voltage-gated ion channels and solute carriers. Lists were obtained from a variety of sources: phosphatases from the human dephosphorylation database (http://www.Koehn.embl.de/depod), protein kinases from UniProt (http://www.uniprot.org) and voltage-gated ion channels and solute carriers from the Human Gene Nomenclature Committee (http://www.genenames.org). Distributions were generated in R using knockdown data from the MCF10A and MCF10A-CDH1−/− cell line pair. The distribution of all 18,120 genes in the siRNA screen is represented by the blue lines (‘All’). The statistical significance of any differences between the distributions of all data (blue) and the functional group (red) was determined using the Kolmogorov-Smirnov test. KS p value = 0 equates to a p value of < 2.2 × 10−16. f Boxplot expanding the viability ratios of voltage-gated ion channels into functional subgroups. With the exception of the bestrophin, gap junction and leucine-rich subgroups, all groups were RSL, with a median viability ratio > 1. g–m Density distributions of cytoplasmic ribosomal proteins, spliceosome proteins, ubiquitin-specific peptidases, E2 ubiquitin-conjugating enzymes, HECT family ubiquitin E3 ligases, RING family ubiquitin E3 ligases and proteasome subunits. Lists were obtained from sources including ribosomal proteins—the ribosomal protein database (ribosome.med.miyazaki-u.ac.jp), ubiquitin-associated peptidases and E2 ubiquitin-conjugating enzymes—the Human Gene Nomenclature Committee (http://www.genenames.org), and E3 ubiquitin ligases from the National Heart Lung and Blood Institute (https://hpcwebapps.cit.nih.gov/ESBL/Database/E3-ligases/)
Table 1.
Pathway enrichment analysis of reverse synthetic lethal gene families
| Enrichment score | UniProt keywords or sequence features | Adj. p value |
|---|---|---|
| 14.88 | Protein phosphatase | 4.2E−38 |
| 14.33 | G-protein coupled receptor | 1.2E−17 |
| Receptor | 2.5E−17 | |
| 10.93 | Ion channel | 3E−18 |
| Ion transport | 1.4E−10 | |
| 10.55 | Receptor | 2.5E−17 |
| Glycoprotein | 1E−11 | |
| 9.34 | Ribonucleoprotein | 3E−11 |
| Ribosomal protein | 9.4E−10 | |
| 9.04 | Glycoprotein | 1E−11 |
| Disulfide bond | 7.3E−11 | |
| 5.2 | Spliceosome | 1.3E−07 |
| mRNA splicing | 3.5E−06 | |
| 4.63 | Metal ion-binding site: manganese 1; via carbonyl | 6.3E−03 |
| Manganese | 2.2E−01 | |
| 4.46 | Protease | 3.5E−06 |
| Zymogen | 1.5E−05 | |
| 4.06 | Metal ion-binding site: iron | 8.3E−01 |
| Metal ion-binding site: manganese | 9.8E−01 | |
| 4.01 | Palmitate | 1.8E−03 |
| Lipoprotein | 4.7E−02 | |
| 4.00 | Threonine protease | 1.0E−06 |
| Proteasome | 1.1E−04 |
The most significant UniProt Keywords for each enrichment cluster from a DAVID ontology analysis of genes with a CDH1−/−/MCF10A viability ratio ≥ 1.3 are shown. Where no UniProt Keyword was listed, the UniProt Sequence Feature was used
Fig. 2.
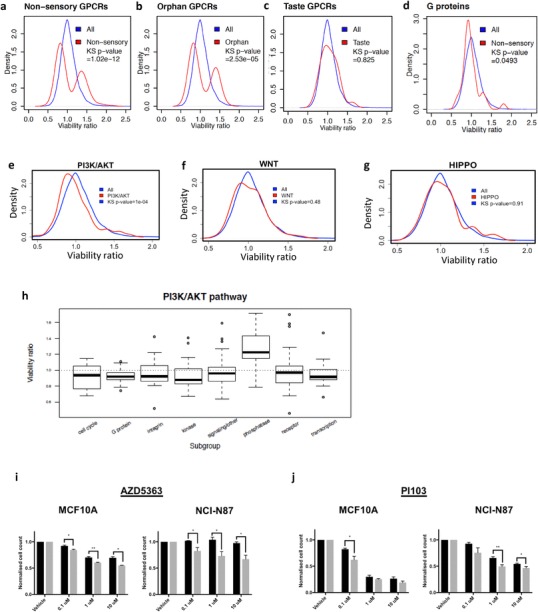
a–d Density distributions of the CDH1−/−/MCF10A viability ratios for non-sensory GPCRs, orphan GPCRs, taste GPCRs and G proteins. e–g Density distributions of the CDH1−/−/MCF10A viability ratios for signalling pathways associated with intact adherens junctions: PI3K/AKT, WNT pathway and HIPPO pathway. Pathway genes were obtained from KEGG. The statistical significance was determined using the Kolmogorov–Smirnov test. h Boxplot of the functional subgroups in the PI3K/AKT pathway. Note that all subgroups with the exception of the phosphatases are shifted in the synthetic lethal direction (viability ratio < 1). i–j Normalised cell counts 48 h after treatment with serial dilutions of AZD5363 and PI103. For each drug, both the MCF10A and NCI-N87 isogenic pair are shown (wild-type black bars; CDH1−/− grey bars). Six fields/well at 4× magnification were captured using the Cytation 5 imager (Biotek). Nuclei were counted using Gen5 (Biotek) and normalised to the vehicle control for each cell line
The PI3K/AKT pathway is an important mediator of E-cadherin synthetic lethality
To determine whether we could also detect SL effects in major signalling pathways, we first examined the MCF10A-CDH1−/−/MCF10A viability ratio density distributions for three cell survival pathways that are associated with E-cadherin-mediated cell-to-cell adhesion: PI3K/AKT, WNT and HIPPO [17–22]. Using KEGG pathway genes, the PI3K/AKT pathway (n = 224) showed a significant SL shift (peak = 0.90, p = 1.3 × 10− 4; Fig. 2e) while no significant shift in distribution was observed for either the WNT or HIPPO pathways (p = 0.48 and 0.91, respectively; Fig. 2f, g). Separation of the PI3K/AKT pathway genes into functional subgroups showed that the SL effect occurred across most functional groups in the pathway including plasma membrane receptors, integrins and G proteins (Fig. 2h). The only exception to these SL effects was phosphatases which were strongly RSL. The NFKB and MAPK pathways, which are indirectly associated with adherens junction signalling, showed distributions that were not significantly different to the overall 18,120 gene distribution (p = 0.18 and 0.86, respectively; results not shown). Of the other major KEGG cancer pathways, MTOR, RAS, TNF, JAK-STAT and focal adhesion signalling pathways also showed significant SL distributions, but this synthetic lethality could be largely attributed to genes which overlapped with the canonical PI3K/AKT pathway (Suppl. Fig. 1). The observed synthetic lethality was further validated by testing the PI3K antagonist PI103 and the pan-AKT antagonist NS3728 on both the MCF10A isogenic cell line pair and a recently derived CDH1 isogenic gastric NCI-N87 pair (A. Chen, manuscript in preparation). Both drugs caused a significant SL effect in the two cell line pairs (Fig. 2i, j), consistent with the siRNA data.
CDH1−/− cells show changes in membrane trafficking
The highly significant SL and RSL effects of diverse GPCR’s, voltage-gated ion channels, and ubiquitinylation enzymes with roles in vesicle trafficking and receptor recycling, prompted us to investigate the possibility that disruption of plasma membrane organisation or dynamics in E-cadherin-null cells might unify these distinct effects. To do this, we first examined the density distributions of groups of proteins involved in membrane trafficking, using the lists provided in the KEGG Membrane Trafficking hierarchies [23]. 36 functional groups (with a minimum of 20 proteins/group) from the exocytosis, SNARE, endocytosis, endocytosis-lysozyme transport, protein recycling and endosome-golgi transport hierarchies were examined. The three most SL distributions, (i.e., the distributions with the lowest MCF10A-CDH1−/−/MCF10A density distribution peaks) comprised the family of ‘endocytosis/ARF GTPases and associated proteins’ (n = 23; peak = 0.85; p = 0.007; Fig. 3a) and two membrane curvature protein families: F-BAR (n = 20; peak = 0.84; p = 0.09; Fig. 3b) and ESCRT (n = 24, peak = 0.87; p = 0.02; Fig. 3c). These associations prompted us to test directly for the presence of an SL interaction between E-cadherin and endocytosis, by measuring the uptake of fluorescently labelled cholera toxin B in the MCF10A isogenic cell line pair. The B subunit of cholera toxin, which is non-toxic, is frequently used to measure internalisation by various endocytic mechanisms [24]. After 30 min, uptake of Alexa Fluor 488-labelled cholera toxin B was significantly lower (p = 0.04) in the CDH1−/− cells compared to the wild-type MCF10A cells (Fig. 3d). Together, these data demonstrate that membrane trafficking is perturbed in MCF10A-CDH1−/− cells, perhaps due to an underlying disruption of plasma membrane organisation and dynamics. Comparable disruption of exocytosis may explain the accumulation of mucins in signet ring cells, a common feature of DGC and a hallmark of early stage HDGC (Fig. 3e).
Fig. 3.
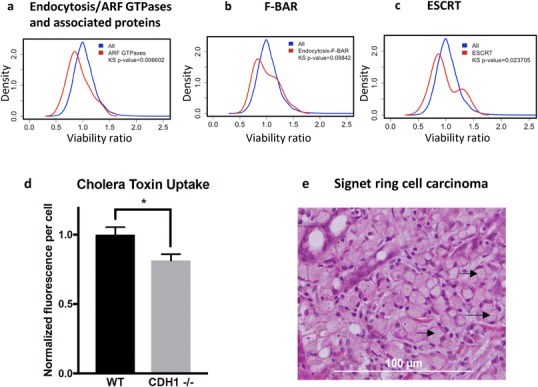
Density distributions of CDH1−/−/MCF10A viability ratios for a endocytosis/ARF GTPases and associated proteins, b F-BAR curvature proteins and c ESCRT curvature proteins. d Differential uptake of fluorescently labelled cholera toxin subunit B in MCF10A and CDH1−/− cells. Cells were grown in medium deprived of cholera toxin (a standard component of MCF10A growth media) for 48 h before addition of Alexa Fluor 488-labelled cholera toxin B. After 30 min, cells were washed and fixed before cell counting and measurement of fluorescence intensity. Fluorescence was normalised to both the total cell count and the MCF10A result. e Haematoxylin and eosin stain of gastric stage T1a carcinoma from a germline CDH1 mutation carrier showing mucin-filled signet ring cells. Three examples are indicated with black arrows
Drug inhibition of membrane/cytoskeletal functions in breast and gastric isogenic cell lines
To determine whether the disrupted cytoskeletal and membrane function of CDH1-deficient cells was reflected in altered drug sensitivity, we treated the isogenic MCF10A and gastric NCI-N87 cell lines with selected compounds that impact on cytoskeletal function, plasma membrane dynamics and lipid raft composition. The inhibitors of actin polymerisation, cytochalasin D and latrunculin B caused a significant SL differential (i.e., greater inhibition of the CDH1-deficient line) in the isogenic MCF10A cells but an RSL differential (i.e., greater inhibition of the CDH1-expressing line) was observed in the isogenic NCI-N87 cells (Fig. 4a, b). The endocytosis inhibitor bafilomycin A1, an antagonist of vacuolar ATPase [25], caused an SL differential in both the isogenic MCF10A and NCI-N87 lines (Fig. 4c). Lipid rafts are dynamic, relatively ordered membrane nanodomains that are enriched in cholesterol and sphingolipids and have a probable role in signal transduction [26]. Disruption of these domains with the membrane cholesterol-depleting drug methyl-β-cyclodextrin (MβCD) and the inhibitor of cholesterol synthesis, atorvastatin, caused an SL differential in both the MCF10A and NCI-N87 isogenic lines (Fig. 4d, e). Amphotericin B, an antifungal agent that binds to cholesterol and leads to perforation of the plasma membrane [27] showed contrasting effects, with an RSL effect in the MCF10A pair and an SL effect on the NCI-N87 isogenic lines (Fig. 4f). Regardless of the direction of the effect (SL or RSL), the differential activity of membrane-disrupting compounds strongly suggest that E-cadherin loss is leading to changes in plasma membrane dynamics. A fundamental shift in plasma membrane organisation or dynamics is consistent with our observation that RNAi inhibition has either an SL or RSL effect on the majority of GPCR and ion channel proteins. To explore the possible utility of pharmacological inhibition of these classes of membrane proteins, we tested the sensitivity of our isogenic cell line pairs to the chloride channel inhibitor NS3728 and the CB1 cannabinoid receptor antagonist Otenabant. NS3728 was SL in the MCF10A lines but RSL in the NCI-N87 lines, whereas Otenabant showed a significant SL effect on both isogenic pairs (Fig. 4g, h). These results point to the potential utility of selected GPCR or ion channel inhibitors for the treatment of CDH1-deficient cancers but highlight the importance of cellular context.
Fig. 4.
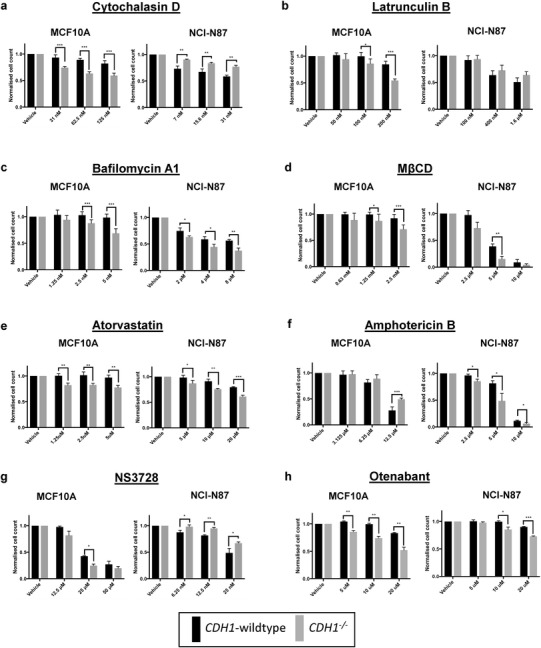
Normalised cell counts 48 h after treatment with serial dilutions of a cytochalasin D, b latrunculin B, c bafilomycin A1, d methyl β cyclodextrin, e atorvastatin, f amphotericin B, g NS3728 and h otenabant. For each drug, both the MCF10A and NCI-N87 isogenic pairs are shown (wild-type black bars; CDH1−/− grey bars). Six fields/well at 4× magnification were captured using the Cytation 5 imager (Biotek). Nuclei were counted using Gen5 (Biotek) and normalised to the vehicle control for each cell line
Candidate synthetic lethal pathways identified using gastric tumour pathway metagenes
The isogenic gastric cancer and non-malignant breast cell line data provide valuable insights into possible targets for the chemoprevention and treatment of CDH1-deficient tumours. To enable us to prioritise functions and pathways that are most likely to be robust clinical targets, we statistically interrogated tumour gene expression data for patterns that were consistent with E-cadherin-associated synthetic lethality. In this approach, we hypothesised that if a genuine SL interaction exists between E-cadherin and any given pathway or protein, there would be fewer tumours than expected by chance with low expression (relative to all tumours) of both CDH1 and the pathway or protein. RNA-seq data from 415 gastric cancers was obtained from TCGA [28] and expression metagenes were generated for each of the 2069 pathways in Reactome [29] (version 58) by taking the first eigenvector of the singular value decomposition [30]. Pathway metagene values were separated into tertiles, and a chi-squared test statistic was used to measure association with CDH1 expression tertiles. Empirical p values were calculated for each pathway via resampling (500,000 iterations per pathway), and statistical significance was assessed after FDR correction (Fig. 5a). Using an adjusted p value threshold of 0.2, 20 pathways were identified as being associated with CDH1 expression (Table 2), although several of these pathways overlapped in terms of gene composition (Fig. 5b). The majority of these candidate SL pathways were involved in cell–cell adhesion, membrane trafficking, membrane lipid composition, apoptosis of cell adhesion proteins and GPCR signalling. Consistent with the metagene selection method, hierarchical clustering of gene expression data from each of these pathways showed variably sized clusters of genes with either strikingly high or low expression in CDH1-deficient tumours (Fig. 5c). The expression changes were most pronounced in the 10% of tumours with the lowest CDH1 expression by rank. Notably, the 10% rank level corresponded to a distinct inflexion point in the absolute CDH1 expression of all samples (Fig. 5d). The 10% rank position corresponds to 36% of the overall median expression of CDH1 and may mark a functional threshold for E-cadherin expression [31]. The low expression of CDH1 in this bottom 10% of tumours cannot simply be attributed to high stromal cell contamination of DGCs, since tumours included in the TCGA analysis were restricted to those with a minimum tumour nuclei content of 60% [28]. The genes in the hierarchical clustering regions with the highest relative expression in CDH1-deficient tumours contained 92 known receptors or enzymes (excluding orphan GPCRs). The MCF10A/MCF10A CDH1−/− RNAi cell viability ratios for these 92 genes showed a bimodal distribution, with the major SL peak occurring at a viability ratio of 0.87 (p = 0.002; Fig. 5e). This distribution demonstrates that our two approaches to identify candidate SL proteins produce an overlapping set of SL candidates which can now be prioritised for further validation (Suppl. Table 1).
Fig. 5.
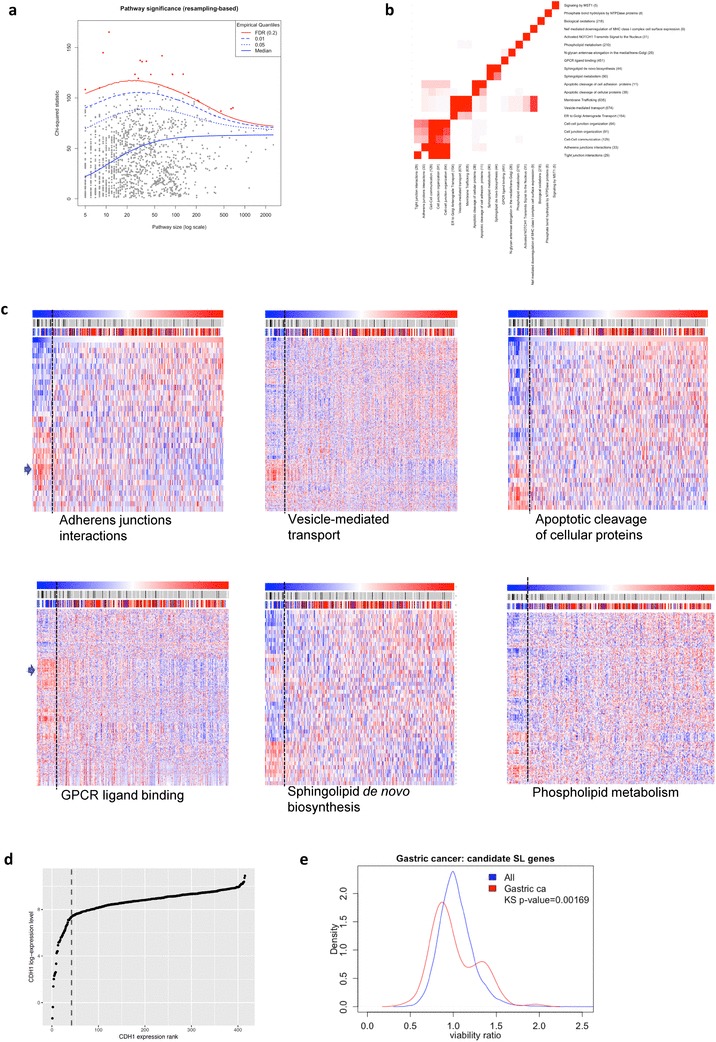
a Per-pathway chi-squared statistics for association between metagene and CDH1 expression tertiles. Blue lines indicate quantiles of the test statistic distribution under the null hypothesis, derived via resampling (500,000 iterations). Solid red line indicates significance threshold relating to control of the False Discovery Rate (FDR) at 0.2. Red dots represent pathways with FDR-adjust p values below this threshold. b Overlap in gene content between each candidate SL pathway. c Hierarchical clustering of gene expression patterns from each of six candidate SL pathways. Samples (columns) are ordered from left to right based on CDH1 expression (low to high) and the individual genes are in the rows. The Lauren classification of each tumour is marked in the bar immediately above the heatmap: blue-diffuse; red-intestinal; black-mixed; clear-undetermined. The middle bar shows CDH1 mutation status for each sample: black, mutation (of any type); grey, no mutation; clear, missing data. The vertical dashed line shows the 10% rank position for tumour sample CDH1 expression. The blue arrows mark two examples of clusters of genes with high relative expression in CDH1-deficient tumours. d Relative CDH1 expression (log2) across the TCGA stomach cancer dataset. The 10% rank position is shown by the dashed line. Note, this line marks an inflexion point in the absolute E-cadherin expression level. e Density distributions of CDH1−/−/MCF10A viability ratios for 92 receptors and enzymes (excluding orphan GPCRs) that were present in the gene clusters with the highest relative expression in CDH1-deficient tumours
Table 2.
Gastric cancer pathways significantly associated with E-cadherin synthetic lethality
| Reactome ID | Reactome pathway | Gene numbera | FDR-adjusted P value |
|---|---|---|---|
| R-HSA-421270 | Cell–cell junction organisation | 64 | 0.0168 |
| R-HSA-351906 | Apoptotic cleavage of cell adhesion proteins | 11 | 0.0168 |
| R-HSA-1500931 | Cell–cell communication | 129 | 0.0228 |
| R-HSA-164940 | Nef-mediated downregulation of MHC class I complex cell surface expression | 9 | 0.0228 |
| R-HSA-5653656 | Vesicle-mediated transport | 674 | 0.0228 |
| R-HSA-199991 | Membrane trafficking | 635 | 0.0228 |
| R-HSA-428157 | Sphingolipid metabolism | 90 | 0.0315 |
| R-HSA-446728 | Cell junction organisation | 91 | 0.0315 |
| R-HSA-418990 | Adherens junctions interactions | 33 | 0.0315 |
| R-HSA-2122948 | Activated NOTCH1 Transmits Signal to the Nucleus | 31 | 0.0315 |
| R-HSA-111465 | Apoptotic cleavage of cellular proteins | 38 | 0.0315 |
| R-HSA-1660661 | Sphingolipid de novo biosynthesis | 44 | 0.1146 |
| R-HSA-199977 | ER to Golgi Anterograde Transport | 154 | 0.1442 |
| R-HSA-500792 | GPCR ligand binding | 451 | 0.1453 |
| R-HSA-975576 | N-glycan antennae elongation in the medial/trans-Golgi | 26 | 0.1453 |
| R-HSA-8852405 | Signalling by MST1 | 5 | 0.1669 |
| R-HSA-420029 | Tight junction interactions | 29 | 0.1842 |
| R-HSA-8850843 | Phosphate bond hydrolysis by NTPDase proteins | 8 | 0.197 |
| R-HSA-211859 | Biological oxidations | 218 | 0.197 |
| R-HSA-1483257 | Phospholipid metabolism | 210 | 0.197 |
aThe number of genes in the pathway that were represented in the TCGA data
Discussion
E-cadherin loss leads to distinct changes in plasma membrane organisation and cytoskeletal architecture. These changes are likely to be caused by disturbance of the close interaction between the membrane and the cortical actin network subsequent to E-cadherin loss [10, 32]. Actin interactions are required for membrane deformation processes such as endocytosis, exocytosis, autophagy and receptor/channel recycling [33–35]. They are also important for the maintenance of membrane characteristics such as membrane tension, ion channel activity and the correct partitioning of the membrane into lipid rafts or other nanostructures [32, 36–38]. We hypothesise that disorganisation of the cortical cytoskeleton in E-cadherin-deficient cells undermines the efficiency of these different processes, establishing numerous vulnerabilities that can be exposed with specific RNAi and chemical antagonists. We propose a model of E-cadherin synthetic lethality in which the function of both SL and RSL proteins is influenced by the presence of E-cadherin. For SL proteins, the loss of E-cadherin reduces functionality leading to an increased sensitivity to further inhibition of the SL protein (with either drug or RNAi). RSL proteins also experience reduced functionality in E-cadherin-deficient cells, but this loss in activity is compensated for by related proteins. As a result, further inhibition of an RSL protein has less effect on the viability of E-cadherin-deficient cells compared to E-cadherin-expressing cells. Predictions from this model include (i) non-specific antagonists of an RSL protein may inhibit related proteins and cause an SL effect; (ii) different cell types will differ in their repertoires of proteins with overlapping functions, resulting in the potential for contrasting SL or RSL effects with the same inhibitor; (iii) protein classes that are relatively promiscuous are more likely to be RSL (e.g. phosphatases vs kinases [16] and ubiquitin-specific peptidases vs RING E3 ubiquitin ligases), and (iv) protein complexes with functionally redundant subunits are likely to be RSL, consistent with our observations on spliceosomes, ribosomes and proteasomes. This model may not be exclusive. For example, the striking bimodality in the CDH1−/−/MCF10A viability ratios for the non-sensory GPCRs could be caused by biased agonism, a process whereby the same receptor can activate different downstream pathways depending on the ligand or allosteric transducers or modifiers proximal to the receptor [39, 40]. For GPCRs, the bias is typically between canonical G-protein signalling and signalling through a β-arrestin-mediated pathway [41]. Several allosteric modifiers have been linked to GPCR bias, including plasma membrane lipid composition [42, 43], membrane compartmentalisation [44], mechanical stretch [45] and the stoichiometry of downstream signalling components [46]. In conclusion, our insights into the mechanisms underpinning E-cadherin synthetic lethality obtained in both in vitro models and whole gastric tumours provide a framework for the rational selection of drugs that could be used to selectively target E-cadherin-deficient cancers. In particular, the disruption of PI3K/AKT signalling, membrane organisation and vesicle trafficking, points to important areas for future research.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by the New Zealand Health Research Council (project grant contract 15/247 and programme contract 17/610; Principal Investigator PJ Guilford) and University of Otago PhD scholarships awarded to ST Kelly, TP Brew and AB Single.
Conflict of interest
The authors declare no conflict of interest.
Human or animal rights
This article does not contain any studies with human or animal subjects performed by any of the authors.
References
- 1.Lecuit T, Yap AS. E-cadherin junctions as active mechanical integrators in tissue dynamics. Nat Cell Biol. 2015;17:533–539. doi: 10.1038/ncb3136. [DOI] [PubMed] [Google Scholar]
- 2.Jeanes A, Gottardi CJ, Yap AS. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene. 2008;27:6920–6929. doi: 10.1038/onc.2008.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCart Reed AE, Kutasovic JR, Lakhani SR, Simpson PT. Invasive lobular carcinoma of the breast: morphology, biomarkers and ‘omics. Breast Cancer Res. 2015;17:12. doi: 10.1186/s13058-015-0519-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guilford P, Hopkins J, Harraway J, McLeod M, McLeod N, Harawira P, et al. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392:402–405. doi: 10.1038/32918. [DOI] [PubMed] [Google Scholar]
- 5.Hansford S, Kaurah P, Li-Chang H, Woo M, Senz J, Pinheiro H, et al. Hereditary diffuse gastric cancer syndrome: CDH1 mutations and beyond. JAMA Oncol. 2015;1:23–32. doi: 10.1001/jamaoncol.2014.168. [DOI] [PubMed] [Google Scholar]
- 6.van der Post RS, Vogelaar IP, Carneiro F, Guilford P, Huntsman D, Hoogerbrugge N, et al. Hereditary diffuse gastric cancer: updated clinical guidelines with an emphasis on germline CDH1 mutation carriers. J Med Genet. 2015;52:361–374. doi: 10.1136/jmedgenet-2015-103094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Telford B, Chen A, Beetham H, Single A, Frick J, Brew T, et al. Synthetic lethal screens identify vulnerabilities in GPCR signalling and cytoskeletal organisation in E-cadherin-deficient cells. Mol Cancer Ther. 2015;14:1213–23. doi: 10.1158/1535-7163.MCT-14-1092. [DOI] [PubMed] [Google Scholar]
- 8.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/S1046-2023(03)00032-X. [DOI] [PubMed] [Google Scholar]
- 9.Soule HD, Maloney TM, Wolman SR, Peterson WD, Jr, Brenz R, McGrath CM, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- 10.Chen A, Beetham H, Black MA, Priya R, Telford BJ, Guest J, et al. E-cadherin loss alters cytoskeletal organization and adhesion in non-malignant breast cells but is insufficient to induce an epithelial-mesenchymal transition. BMC Cancer. 2014;14:552. doi: 10.1186/1471-2407-14-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.R Development Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing. Computing RFfS, editor. Vienna 2011.
- 12.Ligtenberg W. Reactome.db: A set of annotation maps for Reactome. 1.58.0 ed2016. p. R package.
- 13.Single A, Beetham H, Telford BJ, Guilford P, Chen A. A comparison of real-time and endpoint cell viability assays for improved synthetic lethal drug validation. J Biomol Screen. 2015;20:1286–1293. doi: 10.1177/1087057115605765. [DOI] [PubMed] [Google Scholar]
- 14.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 16.Smoly I, Shemesh N, Ziv-Ukelson M, Ben-Zvi A, Yeger-Lotem E. An asymmetrically balanced organization of kinases versus phosphatases across eukaryotes determines their distinct impacts. PLoS Comput Biol. 2017;13:e1005221. doi: 10.1371/journal.pcbi.1005221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Santis G, Miotti S, Mazzi M, Canevari S, Tomassetti A. E-cadherin directly contributes to PI3K/AKT activation by engaging the PI3K-p85 regulatory subunit to adherens junctions of ovarian carcinoma cells. Oncogene. 2009;28:1206–1217. doi: 10.1038/onc.2008.470. [DOI] [PubMed] [Google Scholar]
- 18.Pece S, Chiariello M, Murga C, Gutkind JS. Activation of the protein kinase Akt/PKB by the formation of E-cadherin-mediated cell-cell junctions. Evidence for the association of phosphatidylinositol 3-kinase with the E-cadherin adhesion complex. J Biol Chem. 1999;274:19347–19351. doi: 10.1074/jbc.274.27.19347. [DOI] [PubMed] [Google Scholar]
- 19.Kovacs EM, Ali RG, McCormack AJ, Yap AS. E-cadherin homophilic ligation directly signals through Rac and phosphatidylinositol 3-kinase to regulate adhesive contacts. J Biol Chem. 2002;277:6708–6718. doi: 10.1074/jbc.M109640200. [DOI] [PubMed] [Google Scholar]
- 20.Benham-Pyle BW, Pruitt BL, Nelson WJ. Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and beta-catenin activation to drive cell cycle entry. Science. 2015;348:1024–1027. doi: 10.1126/science.aaa4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim NG, Koh E, Chen X, Gumbiner BM. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc Natl Acad Sci U S A. 2011;108:11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCrea PD, Maher MT, Gottardi CJ. Nuclear signaling from cadherin adhesion complexes. Curr Top Dev Biol. 2015;112:129–196. doi: 10.1016/bs.ctdb.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353-D61. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torgersen ML, Skretting G, van Deurs B, Sandvig K. Internalization of cholera toxin by different endocytic mechanisms. J Cell Sci. 2001;114:3737–3747. doi: 10.1242/jcs.114.20.3737. [DOI] [PubMed] [Google Scholar]
- 25.Huss M, Wieczorek H. Inhibitors of V-ATPases: old and new players. J Exp Biol. 2009;212:341–346. doi: 10.1242/jeb.024067. [DOI] [PubMed] [Google Scholar]
- 26.Sezgin E, Levental I, Mayor S, Eggeling C. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat Rev Mol Cell Biol. 2017;18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaminski DM. Recent progress in the study of the interactions of amphotericin B with cholesterol and ergosterol in lipid environments. Eur Biophys J. 2014;43:453–467. doi: 10.1007/s00249-014-0983-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cancer Genome Atlas Research N Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Croft D, Mundo AF, Haw R, Milacic M, Weiser J, Wu G, et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2014;42:D472-7. doi: 10.1093/nar/gkt1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.West M, Blanchette C, Dressman H, Huang E, Ishida S, Spang R, et al. Predicting the clinical status of human breast cancer by using gene expression profiles. Proc Natl Acad Sci U S A. 2001;98:11462–11467. doi: 10.1073/pnas.201162998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vleminckx K, Vakaet L, Mareel M, Fiers W, van Roy F. Genetic manipulation of E-cadherin expression by epithelial tumor cells reveals an invasion suppressor role. Cell. 1991;66:107–119. doi: 10.1016/0092-8674(91)90143-M. [DOI] [PubMed] [Google Scholar]
- 32.Yap AS, Gomez GA, Parton RG. Adherens junctions revisualized: organizing cadherins as nanoassemblies. Dev Cell. 2015;35:12–20. doi: 10.1016/j.devcel.2015.09.012. [DOI] [PubMed] [Google Scholar]
- 33.Papadopulos A. Membrane shaping by actin and myosin during regulated exocytosis. Mol Cell Neurosci. 2017;84:93–99. doi: 10.1016/j.mcn.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 34.Diz-Munoz A, Fletcher DA, Weiner OD. Use the force: membrane tension as an organizer of cell shape and motility. Trends Cell Biol. 2013;23:47–53. doi: 10.1016/j.tcb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kast DJ, Dominguez R. The cytoskeleton-autophagy connection. Curr Biol. 2017;27:R318-R26. doi: 10.1016/j.cub.2017.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pontes B, Monzo P, Gauthier NC. Membrane tension: a challenging but universal physical parameter in cell biology. Semin Cell Dev Biol. 2017;71:30–41. doi: 10.1016/j.semcdb.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 37.Sokabe M, Sawada Y, Kobayashi T. Ion channels activated by mechanical forces in bacterial and eukaryotic cells. Subcell Biochem. 2015;72:613–626. doi: 10.1007/978-94-017-9918-8_28. [DOI] [PubMed] [Google Scholar]
- 38.Head BP, Patel HH, Insel PA. Interaction of membrane/lipid rafts with the cytoskeleton: impact on signaling and function: membrane/lipid rafts, mediators of cytoskeletal arrangement and cell signaling. Biochim Biophys Acta. 2014;1838:532–545. doi: 10.1016/j.bbamem.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith JS, Lefkowitz RJ, Rajagopal S. Biased signalling: from simple switches to allosteric microprocessors. Nat Rev Drug Discov. 2018;17:243–260. doi: 10.1038/nrd.2017.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freed DM, Bessman NJ, Kiyatkin A, Salazar-Cavazos E, Byrne PO, Moore JO, et al. EGFR ligands differentially stabilize receptor dimers to specify signaling kinetics. Cell. 2017;171:683–695. doi: 10.1016/j.cell.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith JS, Rajagopal S. The beta-arrestins: multifunctional regulators of g protein-coupled receptors. J Biol Chem. 2016;291:8969–8977. doi: 10.1074/jbc.R115.713313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zocher M, Zhang C, Rasmussen SG, Kobilka BK, Muller DJ. Cholesterol increases kinetic, energetic, and mechanical stability of the human beta2-adrenergic receptor. Proc Natl Acad Sci U S A. 2012;109:E3463-72. doi: 10.1073/pnas.1210373109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dawaliby R, Trubbia C, Delporte C, Masureel M, Van Antwerpen P, Kobilka BK, et al. Allosteric regulation of G protein-coupled receptor activity by phospholipids. Nat Chem Biol. 2016;12:35–39. doi: 10.1038/nchembio.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu CH, Gong Z, Liang ZL, Liu ZX, Yang F, Sun YJ, et al. Arrestin-biased AT1R agonism induces acute catecholamine secretion through TRPC3 coupling. Nat Commun. 2017;8:14335. doi: 10.1038/ncomms14335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang W, Strachan RT, Lefkowitz RJ, Rockman HA. Allosteric modulation of beta-arrestin-biased angiotensin II type 1 receptor signaling by membrane stretch. J Biol Chem. 2014;289:28271–28283. doi: 10.1074/jbc.M114.585067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Onfroy L, Galandrin S, Pontier SM, Seguelas MH, N’Guyen D, Senard JM, et al. G protein stoichiometry dictates biased agonism through distinct receptor-G protein partitioning. Sci Rep. 2017;7:7885. doi: 10.1038/s41598-017-07392-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


