Abstract
Denitrifying bacteria carry out nitrate and nitrite respiration inside and outside the cell, respectively. In Thermus thermophilus, nitrate and nitrite transport processes are carried out by major facilitator superfamily (MFS) transporters. The sequence of the nar operon of nitrate-only respiring strains of T. thermophilus includes two tandemly organized MFS transporter genes (narK and narT) of the NarK1 and NarK2 families. Both can function as nitrate/nitrite antiporters, but NarK has been proposed as more specific for nitrate whereas NarT more specific for nitrite. In some nitrate- and nitrite-respiring strains of the same species, a single MFS transporter (NarO) belonging to a different MFS subfamily appears. To analyze the role of this single MFS in the same genetic context, we transferred the two types of nar operon to the aerobic strain HB27, and further included in both of them the ability to respire nitrite. The new denitrifying strains HB27dn, with two MFS, and HB27dp, with a single one, were used to isolate mutants devoid of transporters. Through in trans complementation experiments, we demonstrate that the NarO single MFS works efficiently in the transport of both nitrate and nitrite.
Electronic supplementary material
The online version of this article (10.1007/s10123-018-0023-0) contains supplementary material, which is available to authorized users.
Keywords: Thermus, Denitrification, Nitrate, Nitrite, Transport, MFS
Introduction
The use of nitrate either as substrate for nitrogen assimilation or as electron acceptor under anaerobic conditions requires its active transport through the cytoplasmic membrane to reach the concentrations required for the assimilative or respiratory nitrate reductases to function. These reductases produce nitrite which cannot accumulate due to its toxicity for the cells. Nitrite elimination can be achieved through the activity of the assimilatory nitrite reductase, which produces ammonium, or through its extrusion by specific transporters in the case of nitrate respiration, as further steps in the denitrification pathway are catalyzed in the periplasm.
In most denitrifying bacteria, nitrate and nitrite transport processes are carried out by transmembrane transporters belonging to the major facilitator superfamily (MFS) of proteins, forming a distinct subfamily known as NarK (Clegg et al., 2002; Moir & Wood, 2001; Pao et al., 1998). Like all MSF members, NarK transporters generally include 12 transmembrane-spanning helices, where helices 1, 2, 4, 5, 7, 8, 10, and 11 form the central transport pore. Sequence comparisons have revealed the existence of two major NarK types of proteins, where NarK1 subtype clusters with proteins linked to nitrate assimilation operons, whereas the NarK2 subtype includes putative nitrate/nitrite antiporters associated to nitrate reductase operons (Goddard et al., 2017; Moir & Wood, 2001; Wood et al., 2002). To this latter type belong the NarU and NarK proteins of E. coli for which structural X-ray models are available (Yan et al., 2013; Zheng et al., 2013). However, whereas NarK has been suggested to function as a nitrate/nitrite antiporter, the NarU homolog is proposed to function as a nitrate/cation symporter, showing that there is not a clear-cut way to define the actual role of these transporters even knowing their 3D structure.
In some denitrifying bacteria, like Pseudomonas aeruginosa PA01 (Sharma et al., 2006) or in nitrate respiring strains of Thermus thermophilus (Ramirez et al., 2000), two NarK proteins, one of the NarK1 and the other of the NarK2 subtypes, appear encoded in tandem near to or as part of the operon for the respiratory nitrate reductase. In other bacteria like Pseudomonas denitrificans or Paracoccus denitrificans, these two proteins are fused in a single polypeptide that contains 24 alpha-helices divided in two domains that have been shown to be functional when expressed separately, despite some apparent interdependence has been detected (Goddard et al., 2008). Either as separate proteins or as a single fusion, the first protein or protein domain belongs to the NarK1 subtype of MFS transporters, whereas the second belongs to the NarK2 subtype. Recently, it has been proven by complementation assays in Paracoccus denitrificans that the NarK1-like domain of a fusion NarK1-NarK2 protein functions basically as nitrate transporter but that it is still able to function as nitrate/nitrite symporter, whereas the NarK2-like domain is more specialized in nitrate/nitrite antiport (Goddard et al., 2017).
Thermophilic organisms have to deal with low oxygen availability due to its low solubility at the high temperatures of their natural habitats. Actually, even in thermophilic genera described as strictly aerobes like Thermus thermophilus, horizontally transferable genetic islands exist that encode for the ability to denitrify (Alvarez et al., 2014). In fact, many natural isolates of this species are able to grow anaerobically by nitrate respiration with the accumulation of nitrite as the final respiration product, whereas other strains of the same species can further reduce nitrite to produce water insoluble N2O (Alvarez et al., 2014; Cava et al., 2008).
The last two genes of the nitrate respiration operon (narCGHJIKT) of the T. thermophilus NAR1 strain encode NarK1- and NarK2-like transporters, re-named as NarK and NarT, respectively. Mutants of this strain lacking both proteins cannot grow anaerobically with nitrate (Ramirez et al., 2000), thus supporting the absence of alternative transporters for nitrate under these conditions. Although with different efficiencies, single narT or narK deletion mutants can grow anaerobically with nitrate and also accumulate nitrite upon addition of nitrate to the growth medium, supporting that both of them can function as nitrate/nitrite antiporters, despite a major role for NarT (NarK2) in this antiporter activity was deduced from its higher phenotypic affection (Ramirez et al., 2000).
Unexpectedly, the genomes of some recently sequenced denitrifying strains of Thermus spp. encode a single member of the NarK family, named NarO, which shows a low identity towards NarK1 or NarK2 subfamilies and actually clusters in a separate group of MSF transporters. In this work, we analyze the role of the NarO protein in comparison to the in tandem NarK1–2 genes in the same genetic context. Our data show that the NarO protein can replace the NarK1 and NarK2 types of transporters, resulting in restoration of the wild-type phenotype in a nitrate/nitrite transporter defective mutant. These data support that the NarO type of nitrate transporters are actually flexible bifunctional nitrate/proton symporters and nitrate/nitrite antiporters, and suggest that the presence of two transporters may constitute a selective trait to avoid nitrite toxicity in those strains in which this nitrogen oxide constitutes the final product of anaerobic respiration.
Materials and methods
Bacterial strains and growth conditions
The strains of Thermus thermophilus used in this work are indicated in Table 1. T. thermophilus HB27 grows only under areobic conditions, whereas strains T. thermophilus NAR1 and PRQ25 can grow anaerobically by nitrate respiration and denitrification respectively (Cava et al., 2008). The strains T. thermophilus HB27dn and HB27dp are denitrifying derivatives of HB27 that encode the nitrate respiration cluster from the NAR1 strain (HB27dn) or from the PRQ25 strain (HB27dp) and the nitrite respiration cluster (nic) from the PRQ25 strain (Alvarez et al., 2011). For aerobic growth, the strains were incubated on TB medium (Ramírez-Arcos et al., 1998) at 60 °C with mild shaking (150 r.p.m.). Anaerobic growth was achieved in Eppendorf tubes containing 2 ml of TB medium supplemented with potassium nitrate (20 mM) overlaid by mineral oil and incubated statically at 60 °C. Agar (1.5% w/v) was added to the TB medium for growth on plates, which were incubated at 60 °C in a water-saturated atmosphere. Escherichia coli DH5α [supE44 Δ(lacZYA-argF)U169 Φ80lacZΔM15 hsdR17 recA1 endA1 gyrA96 thi-1 relA1] was used for plasmid construction. E. coli was routinely grown at 37 °C in liquid or solid LB medium. Ampicillin (100 μg/ml), kanamycin (30 μg/ml), or hygromycin B (100 μg/ml) were added when needed.
Table 1.
Thermus thermophilus strains used in this work
| Strain name | Phenotype and genotype | Reference or source |
|---|---|---|
| HB27 | Wild type aerobic | Dr. Koyama |
| NAR1 | Nitrate respiring | (Cava et al., 2008; Vieira & Messing, 1982) |
| PRQ25 | Denitrifying | (Manaia et al., 1994) |
| HB27dn | Denitrifying with NCE from NAR1, NarK, and NarT transporters | (Alvarez et al., 2011) |
| HB27dp | Denitrifying with NCE from PRQ25, single NarO transporter | (Alvarez et al., 2011) |
| HB27dn ∆narKT::kat | HB27dn devoid of transporters, KanR | This work |
| HB27dn ∆narKT | HB27dn devoid of transporters | This work |
| HB27dp ∆narO::kat | HB27dp devoid of transporters, KanR | This work |
| HB27dp ∆narO | HB27dp devoid of transporters | This work |
Sequence analysis and DNA methods
A draft sequence from T. thermophilus PRQ25 was obtained through pyrosequencing in a Roche-454 system (Lifesequencing, Valencia, Spain). The genes encoding enzymes implicated in the denitrification process were identified in the contigs by BLAST sequence comparisons (Alvarez et al., 2011). The sequence of the nar operon of this strain has been deposited in the GenBank database with the accession number MH158735.
Phylogenetic comparisons
Homolog search was performed with BLAST (Altschul et al., 1990) against a non-redundant protein database. A protein data set of 34 sequences was aligned using CLUSTAL OMEGA version 1.2.4 (Sievers et al., 2011). The data set includes reference sequences from E. coli, P. aeruginosa, or P. denitrificans. For visualization of the generated tree, the Interactive Tree of Life (iTOL) v3 tool was used (Letunic & Bork, 2016). Protein sequences used in this analysis are listed in supplemental Table S1.
DNA techniques
Plasmid purification, restriction analysis, plasmid construction, polymerase chain reaction (PCR), and routine DNA sequencing were carried out by standard methods (Sambrook et al., 1989). Plasmids used are listed in Table 2.
Table 2.
Plasmids used in this work
| Plasmid | Description or use | Reference or source |
|---|---|---|
| pKT1 | Source of kat cassette | (Lasa et al., 1992) |
| pUC119 | Suicide plasmid in Thermus | (Vieira & Messing, 1982) |
| pUCΔnarKT | Mutation of narKT | This work |
| pUCΔnarKT::kat | Mutation of narKT | This work |
| pUCΔnarO | Mutation of narO | This work |
| pUCΔnarO::kat | Mutation of narO | This work |
| pMK184 | Expression plasmid in Thermus | (Cava et al., 2007) |
| pMK184narK | Expression of NarK | This work |
| pMK184narT | Expression of NarT | This work |
| pMK184narO | Expression of NarO | This work |
| pMH184 | Expression plasmid in Thermus | (Cava et al., 2007) |
| pMH184narK | Expression of NarK | This work |
| pMH184narT | Expression of NarT | This work |
| pMH184narO | Expression of NarO | This work |
Construction of HB27-denitrifying derivatives 27dn and 27dp
Genomic DNA from T. thermophilus PRQ25 strain was isolated by standard procedures. For transformation, genomic DNA (200 ng) from the PRQ25 strain was added to 0.5 ml of exponential cultures (optical density at 550 nm = 0.2), allowing incubation at 60 °C to continue for 4 h with standard shaking. For the selection of the HB27dn strain, a derivative of the nitrate-respiring HB27c strain was used as recipient strain and selection was carried out under anaerobic conditions at 60 °C for 48 h with sodium nitrite (5 mM). Individual colonies were isolated on TB plates and subjected to nitrate reduction/nitrite consumption assays (Alvarez et al., 2011). Nitrite-respiring transformants were selected. For the selection of the HB27dp strain, the HB27 strain was transformed as above, and anaerobic growth selection was carried out with potassium nitrate (20 mM) as above. Upon the isolation of nitrate-respiring colonies from plates, a further transformation step was carried out with selection by nitrite respiration as described above.
The presence of the nar cluster from NAR1 in 27dn or from PRQ25 in 27dp was confirmed by amplification of the narO and narKT genes respectively. A scheme of the denitrification clusters of each of the new strains is shown in supplemental Fig. S1.
Isolation and complementation of mutants
Mutation of narO or narKT was performed by homologous recombination with DNA constructions containing approximately 1000-bp-long flanking regions of each targeted gene. The upstream regions were amplified by PCR using the primers mutnarOKTupdir, mutnarOuprev, and mutnarKTuprev (Table 3), while primers drpArbsXbaIdir and drpBstopEcoRIrev were used for the amplification of the downstream regions (Table 3). The PCR products were sequentially cloned between the SalI-XbaI-EcoRI restriction sites of plasmid pUC119 (Vieira & Messing, 1982), an Escherichia coli plasmid that does not replicate in T. thermophilus. The kanamycin resistance kat cassette (Lasa et al., 1992) was inserted into the XbaI site, oriented downstream to avoid polar effects on the expression of the nitrate reductase complex. This way, the HB27dn ΔnarKT::kat and the HB27dp ΔnarO::kat mutants were obtained. Further derivatives were obtained in which the kat cassette was deleted through additional recombination with linearized DNA encoding the recombination arms without the kat cassette. Kanamycin-sensitive mutant strains HB27dn ΔnarKT and HB27dp ΔnarO were obtained (Supplemental Fig. 1). Transformation and mutant selection were carried out as described (Alvarez et al., 2011). All the mutants isolated were checked by PCR.
Table 3.
Oligonucleotides used in this work
| Oligonucleotide | Sequence (5′-3′) | Purpose |
|---|---|---|
| katXbaIdir | AAAATCTAGACCCGGGAGTATAACAGA | Kat cassette |
| katXbaIrev | AAAATCTAGACGTTCAAAATGGTATGCGTTTTGA | Kat cassette |
| mutnarOKT up dir | AAAAGTCGACAGCTCTACACTCGCACCCT | pUCΔnarO::kat, pUCΔnarKT::kat |
| mutnarO up rev | AAATTCTAGATCAAAGGGTCTCCCGCC | pUCΔnarO::kat |
| mutnarKT up rev | AAATTCTAGATCACCTCGCCAGCTTGC | pUCΔnarKT::kat |
| drpArbsXbaIdir | AAAATCTAGAGGGCCTAGGGA | pUCΔnarO::kat, pUCΔnarKT::kat |
| drpBstopEcoRIrev | AAAAGAATTCCTAGCCCCCCT | pUCΔnarO::kat, pUCΔnarKT::kat |
| narOdir | CCAACCTTTCGGGCTCCCTC | Check narO |
| narOrev | CCGAGGGCGTAGAAGACGAG | Check narO |
| narTdir | CTTGGCGCACCCTCTGGATC | Check narKT |
| narTrev | GTACCCCGAGAAGTGCCCTC | Check narKT |
| narKrbsXbaIdir | AAAATCTAGACGAGGTGAGCTATGATCCAC | Overexpression NarK, pMK, and pMHnarK |
| narKstopEcoRIrev | AAATGAATTCTCAGCATGGACGGTCTCCTT | Overexpression NarK, pMK, and pMHnarK |
| narTrbsXbaIdir | AAAATCTAGAGGAGACCGTCCATGCTGAAG | Overexpression NarT, pMK, and pMHnarT |
| narTstopEcoRIrev | AATTGAATTCTTAGCAGGGCTTCTCCGCC | Overexpression NarT, pMK, and pMHnarT |
| narOrbsXbaIdir | AAAATCTAGAGGAGGTGCGCCATGTCCA | Overexpression NarO, pMK, and pMHnarO |
| narOstopEcoRIrev | AATTGAATTCTTAGGCCTTCCTGTCCCTCA | Overexpression NarO, pMK, and pMHnarO |
Derivatives of the bifunctional plasmids pMK184 (KmR) and pMH184 (HygR) were constructed for the expression in trans of the narO, narK, and narT genes. For this purpose, the corresponding genes and their Shine Dalgarno sequence were amplified by PCR with the primers indicated in Table 3 and cloned into the XbaI and EcoRI sites of these plasmids. In these constructs, transcription of the constitutive promoter PslpA that drives the expression of the kat or hyg cassettes transcribes also the cloned genes, allowing moderate levels of constitutive expression. A scheme of the complementation plasmids is shown in supplemental Fig. S2.
Nitrite production
To determine the amount of excreted nitrite, cells were grown at 60 °C in a shaker bath to an OD550 of 0.3. After the addition of potassium nitrate (20 mM), the cultures were incubated at 60 °C without stirring for an additional 2 h period. After washing thrice with 50 mM sodium phosphate buffer (pH 7.5), cells were resuspended to an OD550 of 0.5 in nitrate-free preheated medium and separated into 2 ml aliquots. These were incubated at 70 °C with increasing concentrations of nitrate (0, 0.01, 0.1, 1, 10, and 20 mM) and the nitrite excreted was determined at different times using previously described methods (Snell & Snell, 1949).
Nitrate and nitrite sensitivity assays
Sensitivity to nitrite and nitrate was assayed on TB plates and liquid medium in the presence of hygromycin B. For plate assays, a bunch of Thermus HB27 colonies harboring, either an empty plasmid (pMH184) or a plasmid expressing a MFS transporter (pMH184NarK, pMH184NarT, and pMH184NarO) were picked on liquid TB medium supplemented with hygromicyn (100 μg/ml) and grown at 65 °C until reaching OD600 of 0.3. Serial dilutions (1/10) were carried out on the same medium and 10 μL samples of each dilution where dot plotted on TB plates with hygromycin and either KNO3 (200 mM) or NaNO2 (150 mM). Colony growth was checked after 48 h of incubation at 60 °C.
For liquid assays, three individual colonies transformed with each plasmid were grown overnight on TB with hygromycin (100 μg/ml). Tubes containing 3 ml of the same medium and increasing concentrations of r KNO3 (0–500 mM) or NaNO2 (0–140 mM) were inoculated to reach an OD600nm of 0.05. After 24 h of incubation at 65 °C under shaking (170 rpm), the final OD 600nm was measured.
Results and discussion
NarO belongs to a new family of NarK transporters
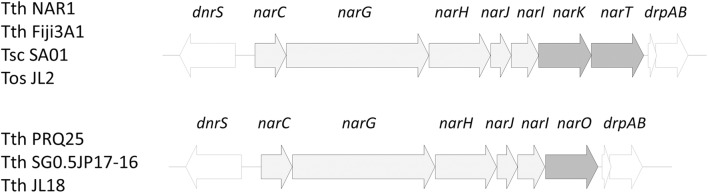
The nar operon from T. thermophilus PRQ25, SG0.5JP17-16 and JL18 encodes a single transporter of the NarK family, NarO, instead of the two in tandem transporters of the NarK1 (NarK) and NarK2 (NarT) subfamilies encoded within the T. thermophilus strains NAR1, Fiji3A1, T. oshimae JL2, and T. scotoductus SA01 (Alvarez et al., 2014) (Fig. 1).
Fig. 1.
One or two MSF transporters are encoded within the nitrate reductase operon. Organization of the nar operon in the indicated strains of Thermus spp.: T. thermophilus NAR1, T. thermophilus Fiji3A1, T. oshimai JL2, T. scotoductus SA01, T. thermophilus PRQ25, T. thermophilus SG0.5JP17-16, and T. thermophilus JL18
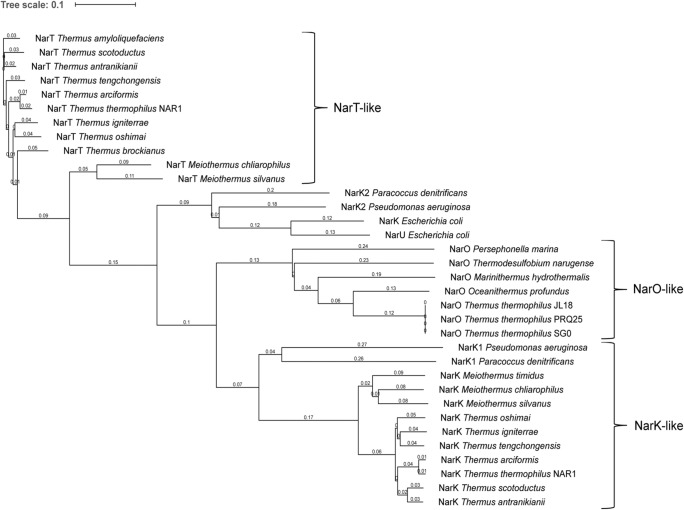
NarO sequence identities of the PRQ25 strain ranged between 31% with NarK and 27% with NarT of the NAR1 strain, whereas NarK and NarT were 30% identical. Clustal comparisons with nar-associated transporters of different bacteria, including Thermales and Proteobacteria indicate that NarO-like transporters constitute a separate subfamily within the NarK superfamily in addition to those represented by NarK- and NarT-like proteins (Fig. 2).
Fig. 2.
Cladogram of the nitrate/nitrite transporters subfamily. The identification of homologs to the nitrate/nitrite putative transporters was performed by searching with BLAST (Altschul et al., 1990) against a non-redundant protein database. The protein data set was aligned using CLUSTAL OMEGA (Sievers et al., 2011). iTOL (Letunic & Bork, 2016) was used for the visualization of the generated tree. Branch-distance is indicated. The proteins used for the comparison are listed in Supplemental Table S1
The presence of in tandem or fused transporters of the MFS family in nitrate respiration clusters of model denitrifying bacteria has been proposed to be related with preferential roles as nitrate/H+ symporters for the first protein or protein domain encoded, and as nitrate/nitrite antiporters for the second component (Goddard et al., 2017). In this context, it is interesting to note that the NAR1 and the Fiji3A1 strains are partial denitrifiers that accumulate high concentrations of nitrite extracellularly, whereas the PRQ25 and SG0.5JP17-16 strains are complete denitrifiers that eliminate nitrite from the medium. Therefore, it is tempting to speculate that the presence of two transporters could be more efficient in keeping out nitrite at high concentrations, whereas a single transporter coupled to nitrite respiration could be enough to detoxify for a full denitrifying bacterium.
The presence of at least one MSF transporter is required for anaerobic growth
To check the hypothesis above in a similar background in the absence of alternative MFS transporters, and also to escape from the low transformation efficiency of T. thermophilus PRQ25 (Alvarez et al., 2011; Cava et al., 2008), we constructed two genetically amenable denitrifying derivatives of the aerobic strain HB27 (“Materials and Methods”). One of them contained the nar operon from the NAR1 strain (HB27dn), which includes the genes for the NarK and NarT transporters, and the other the nar operon from the PRQ25 strain (HB27dp), which contains the gene for the NarO transporter. In addition, both of these strains encode the nitrite respiration cluster (nic, from the PRQ25 strain), allowing further detoxification of nitrite. Under aerobic conditions, these two strains grew at similar rates and with similar yield as the aerobic HB27 strain (data not shown), demonstrating that the presence of the denitrification apparatus does not imply a significant burden for aerobic growth.
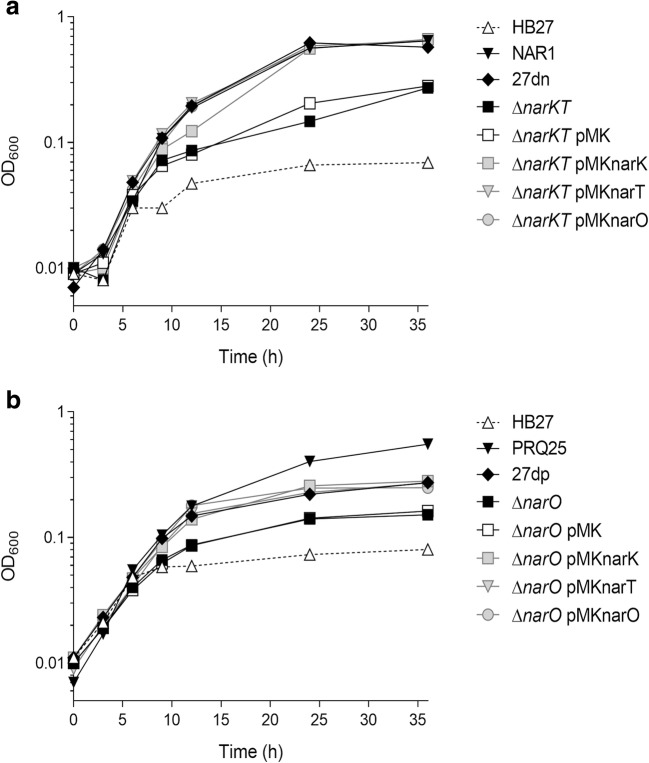
When compared with the native denitrifying strains under nitrate respiration conditions, the HB27dn strain grew also similarly to the NAR1 strain (Fig. 3a), whereas the HB27dp strain grew initially at similar rates as the natural PRQ25 but reaching lower cell yields (Fig. 3b). These data suggest that nitrate respiration shows similar efficiency in the HB27 derivatives as in the parental strain, but that further steps of the denitrification (reduction of nitrite to N2O) do not provide the same energy in these lab-generated HB27-derived strains as in the natural PRQ25 strain. As expected, the parental HB27 strain stopped growing after the residual oxygen present in the medium was consumed.
Fig. 3.
Anaerobic growth with nitrate of the mutants in the nitrate/nitrite transporters. a Anaerobic growth curves of strains HB27, NAR1, HB27dn (27dn), HB27dn ∆narKT (∆narKT), and the HB27dn ∆narKT mutant harboring empty plasmid pMK184 (pMK) or overexpressing NarK (pMKnarK), NarT (pMKnarT), or NarO (pMKnarO). b Anaerobic growth curves of strains HB27, PRQ25, HB27dp (27dp), HB27dp ∆narO (∆narO), and the HB27dp ∆narO mutant harboring empty plasmid pMK184 (pMK) or overexpressing NarK (pMKnarK), NarT (pMKnarT), or NarO (pMKnarO)
The analysis of the relevance of the transporters was studied by comparison of the growth of these two strains with that of deletion mutants lacking them. For this, a HB27dp ΔnarO single mutant and a HB27dn ΔnarKT double mutant were isolated by homologous recombination (Materials and Methods). In both cases, growth under anaerobic conditions with nitrate was greatly impaired with growth yields of 23 and 63% of the ΔnarKT and ΔnarO mutants respect to their parental HB27dn and HB27dp strains after 24 h of incubation (Fig. 3a, b ). Interestingly, when either NarO, NarK, or NarT were expressed in trans from a plasmid in the single or in the double mutant, the complemented strains were able to grow anaerobically with nitrate in a similar way as their parental strain. As anaerobic growth with nitrate requires both the entrance of nitrate and the extrusion of nitrite, these data suggest that the three proteins can function as nitrate/nitrite transporters under these experimental conditions.
NarO can transport nitrate and nitrite
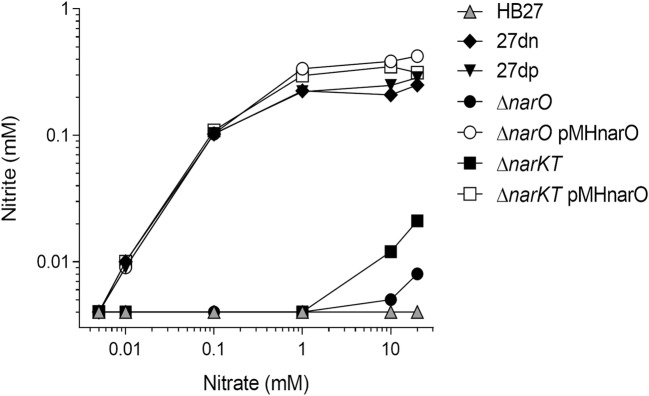
To determine whether the defective anaerobic growth in the mutants was due to a defect in nitrate and/or nitrite transport, cells treated for 2 h under anoxic conditions with nitrate 20 mM were washed and resuspended in nitrate-free medium. Aliquots were incubated at 70 °C in preheated medium with different concentrations of nitrate, and the nitrite excreted to the supernatant was measured after 20 min (Fig. 4). As it can be observed, HB27dp ∆narO and HB27dp ∆narKT mutants did not excrete nitrite when low nitrate concentrations (< 1 mM) were present in the medium. Only when the external concentration of nitrate reached 10 mM was a little amount of nitrite (< 0.05 mM) detected in the medium, likely due to leakage. By contrast, their parental HB27dn and HB27dp counterparts efficiently secreted nitrite, supporting the full conversion of nitrate to nitrite and the efficient transport of both nitrate and nitrite. Above 1 mM nitrate, the amount of nitrite secreted remained constant suggesting that either the transporters or the enzyme or both were saturated. Interestingly, the expression of NarO from a plasmid was able to recover the nitrite extrusion rate not only in the HB27dp ∆narO background as it could be expected, but also in the HB27dn ∆narKT double mutant strain.
Fig. 4.
NarO transports both nitrate and nitrite. Nitrite production (mM) in the different strains after 20 min of incubation in the presence of different concentrations of nitrate (0–20 mM). Symbols: ▲ HB27; ♦ 27dn; ▼ 27dp; ● 27dp ∆narO; ○ 27dp ∆narO pMHnarO; ■ 27dn ∆narKT; □ 27dn ∆narKT pMHnarO
These data clearly show that at least under the conditions assayed, the NarO protein is as efficient as nitrate/nitrite transporter (antiporter) as the combination of NarK and NarT.
Protection provided by the MFS transporters against nitrate and nitrite toxicity
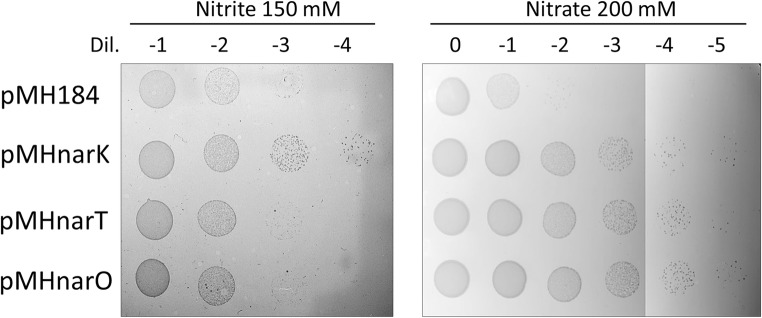
All the above results support for the three subtypes of transporters a role as nitrate and nitrite antiporters in denitrifying strains under the conditions assayed (Figs. 3 and 4). In order to check the putative role in warding off against toxicity of the resulting nitrogen oxides in a context in which biological conversion by reduction is minimal, we expressed the three genes in the aerobic strain T. thermophilus HB27 and compared the resistance to nitrate (200 mM) and nitrite (150 mM) of the transformants with that provided by an empty plasmid. As can be observed in the qualitative resistance assay on plates shown in Fig. 5, expression of NarK produced resistance to nitrite, allowing the transformant to grow much better (around 100-fold) than the strain with the empty plasmid. By contrast, expression of NarT or NarO did not result in a significant increased resistance to the assayed concentration of nitrite in these plate-based growth assays. When the same strains were plated onto plates containing 200 mM of nitrate, all the three proteins produced a 1000-fold increase in resistance compared with the strain carrying the empty plasmid.
Fig. 5.
Resistance to nitrite and nitrate provided by MFS transporters on plate assays. Dot plate assays with serial 1:10 dilutions of cultures of T. thermophilus HB27 transformed with the indicated plasmids. TB plates with hygromycin B containing sodium nitrite or potassium nitrate 150 and 200 mM, respectively, were used
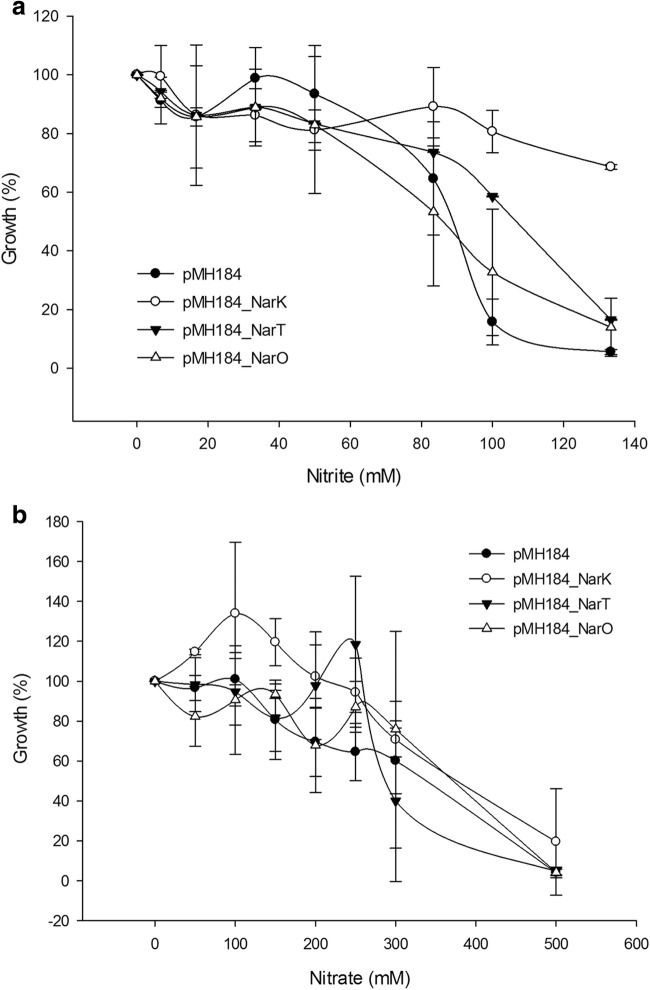
To evaluate these resistances in liquid medium in a more quantitative manner, identical amounts of cells of the HB27 strain transformed with the plasmids expressing each of the transporters and with the empty plasmid were inoculated in TB medium with increasing concentrations of nitrite, allowing them to grow for 24 h at 65 °C. The results of Fig. 6a show that expression of NarK from a plasmid allows the cell to grow above 120 mM of nitrite, a concentration at which the wild type with the empty plasmid cannot grow at all. It is also interesting to note that NarT and NarO provide some resistance to lower nitrite concentrations. Thus, we concluded that in addition to the role as nitrate/nitrite antiporters, these three proteins, and specially NarK, can work as efficient nitrate extrusion pumps under the conditions assayed.
Fig. 6.
Liquid growth inhibition by nitrite and nitrate. Cultures of the aerobic T. thermophilus HB27 strain transformed with the indicated plasmid were grown for 24 h at 65 °C on TB medium with hygromycin (100 μg/ml) in the presence of the indicated concentrations of nitrite (a) or nitrate (b). The mean percentage of growth (OD600) of three different colonies respect to the controls without nitrogen oxide is represented
Results with nitrate in similar experiments supported that the three proteins also provide resistance to nitrate at a critical concentration between 200 and 300 mM, suggesting that the proteins could also pump out nitrate (Fig. 6b).
Conclusions
Previous work in T. thermophilus showed that absence of NarT (NarK2 subfamily) had more severe effects at the levels of anaerobic growth rate, nitrate transport, and extracellular nitrite accumulation than the absence of NarK in the nitrate-respiring NAR1 strain (Ramirez et al., 2000). Through a series of analyses, we concluded that despite both proteins being able to function as nitrate/nitrite antiporters, NarK showed preference for nitrate transport and NarT for nitrite extrusion. A similar conclusion was further reached by other groups working on the NarK1 and NarK2 transporters of Pseudomonas aeruginosa PA01 (Sharma et al., 2006) and the N- and C-terminal domains of the double-NarK fusion protein of Paracoccus denitrificans (Goddard et al., 2008). In this work, we wondered why in several denitrifying strains of T. thermophilus a single transporter of a different MFS family was present. For this, we isolated mutants lacking any nitrate/nitrite transporters in denitrifying derivatives of the HB27 strain that were genetically amenable proxies of the natural nitrate-respiring (NAR1) and nitrate-denitrifying (PRQ25) strains. We showed that these mutants lacking nitrate/nitrite transporters were defective in anaerobic growth with nitrate and that expression of any of the three classes of transporters, NarK, NarT, or NarO, was capable of complementing the anaerobic growth. Therefore, NarO, the new class of transporter, functions efficiently as nitrate/nitrite antiporter being able to replace the NarK-NarT two transporter system found in many nar operon of T. thermophilus.
A putative advantage of having two transporters over a single one could be related to an increased efficiency in nitrite detoxification in partial denitrifying isolates. In this sense, it is interesting to note that the expression of these transporters in a strict aerobic genetic context, in which reduction of nitrite or nitrate was not possible, allowed us to identify NarK as a good provider of resistance to high levels of nitrite. In contrast to previous hypothesis that suggested a major role as nitrate transporter, these data support that NarK is a very efficient nitrite extrusion protein, a role that cannot be performed efficiently by NarT or NarO which could be better as nitrate/nitrite antiporters.
A yet difficult to understand question is the resistance to nitrate provided by the overexpression of the three transporters to the aerobic strain. As nitrite cannot be produced in significant amounts in this aerobic context, and high resistance to nitrite is only provided by NarK, the most likely explanation for these results is that the three enzymes can also function as nitrate extrusion proteins in the absence of nitrite. Therefore, our data suggest that these MFS subfamilies are actually proton: nitrate/nitrite antiporters that have further been adapted to transport nitrate in denitrifying strains.
Electronic supplementary material
(PNG 71 kb)
(PNG 120 kb)
(DOCX 19 kb)
Acknowledgements
This work was supported by grant BIO2016-77031-R from the Spanish Ministry of Economy and Competitiveness. An institutional grant from Fundación Ramón Areces to the CBMSO is also acknowledged.
Compliance with ethical standards
Competing interests
The authors declare that they have no conflict of interest.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Alvarez L, Bricio C, Blesa A, Hidalgo A, Berenguer J. Transferable denitrification capability of Thermus thermophilus. Appl Environ Microbiol. 2014;80:19–28. doi: 10.1128/AEM.02594-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez L, Bricio C, Gomez MJ, Berenguer J. Lateral transfer of the denitrification pathway genes among Thermus thermophilus strains. Appl Environ Microbiol. 2011;77:1352–1358. doi: 10.1128/AEM.02048-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cava F, Laptenko O, Borukhov S, Chahlafi Z, Blas-Galindo E, Gomez-Puertas P, Berenguer J. Control of the respiratory metabolism of Thermus thermophilus by the nitrate respiration conjugative element NCE. Mol Microbiol. 2007;64:630–646. doi: 10.1111/j.1365-2958.2007.05687.x. [DOI] [PubMed] [Google Scholar]
- Cava F, Zafra O, da Costa MS, Berenguer J. The role of the nitrate respiration element of Thermus thermophilus in the control and activity of the denitrification apparatus. Environ Microbiol. 2008;10:522–533. doi: 10.1111/j.1462-2920.2007.01472.x. [DOI] [PubMed] [Google Scholar]
- Clegg S, Yu F, Griffiths L, Cole JA. The roles of the polytopic membrane proteins NarK, NarU and NirC in Escherichia coli K-12: two nitrate and three nitrite transporters. Mol Microbiol. 2002;44:143–155. doi: 10.1046/j.1365-2958.2002.02858.x. [DOI] [PubMed] [Google Scholar]
- Goddard AD, Bali S, Mavridou DA, Luque-Almagro VM, Gates AJ, Dolores Roldan M, Newstead S, Richardson DJ, Ferguson SJ. The Paracoccus denitrificans NarK-like nitrate and nitrite transporters-probing nitrate uptake and nitrate/nitrite exchange mechanisms. Mol Microbiol. 2017;103:117–133. doi: 10.1111/mmi.13546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard AD, Moir JW, Richardson DJ, Ferguson SJ. Interdependence of two NarK domains in a fused nitrate/nitrite transporter. Mol Microbiol. 2008;70:667–681. doi: 10.1111/j.1365-2958.2008.06436.x. [DOI] [PubMed] [Google Scholar]
- Lasa I, Caston JR, Fernandez-Herrero LA, de Pedro MA, Berenguer J. Insertional mutagenesis in the extreme thermophilic eubacteria Thermus thermophilus HB8. Mol Microbiol. 1992;6:1555–1564. doi: 10.1111/j.1365-2958.1992.tb00877.x. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manaia CM, Hoste B, Gutierrez MC, Gillis M, Ventosa A, Kersters K, da Costa MS. Halotolerant Thermus strains from marine and terrestrial hot springs belong to Thermus thermophilus (ex Oshima and Imahori, 1974) nom. rev. emend. Syst Appl Microbiol. 1994;17:526–532. doi: 10.1016/S0723-2020(11)80072-X. [DOI] [Google Scholar]
- Moir JW, Wood NJ. Nitrate and nitrite transport in bacteria. Cell Mol Life Sci : CMLS. 2001;58:215–224. doi: 10.1007/PL00000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pao SS, Paulsen IT, Saier MH., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Arcos S, Fernandez-Herrero LA, Berenguer J. A thermophilic nitrate reductase is responsible for the strain specific anaerobic growth of Thermus thermophilus HB8. Biochim Biophys Acta. 1998;1396:215–227. doi: 10.1016/S0167-4781(97)00183-8. [DOI] [PubMed] [Google Scholar]
- Ramirez S, Moreno R, Zafra O, Castan P, Valles C, Berenguer J. Two nitrate/nitrite transporters are encoded within the mobilizable plasmid for nitrate respiration of Thermus thermophilus HB8. J Bacteriol. 2000;182:2179–2183. doi: 10.1128/JB.182.8.2179-2183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sharma V, Noriega CE, Rowe JJ. Involvement of NarK1 and NarK2 proteins in transport of nitrate and nitrite in the denitrifying bacterium Pseudomonas aeruginosa PAO1. Appl Environ Microbiol. 2006;72:695–701. doi: 10.1128/AEM.72.1.695-701.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell FD, Snell CT. Colorimetric methods of analysis. New York: Van Nostrand; 1949. [Google Scholar]
- Vieira J, Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982;19:259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wood NJ, Alizadeh T, Richardson DJ, Ferguson SJ, Moir JW. Two domains of a dual-function NarK protein are required for nitrate uptake, the first step of denitrification in Paracoccus pantotrophus. Mol Microbiol. 2002;44:157–170. doi: 10.1046/j.1365-2958.2002.02859.x. [DOI] [PubMed] [Google Scholar]
- Yan H, Huang W, Yan C, Gong X, Jiang S, Zhao Y, Wang J, Shi Y. Structure and mechanism of a nitrate transporter. Cell Rep. 2013;3:716–723. doi: 10.1016/j.celrep.2013.03.007. [DOI] [PubMed] [Google Scholar]
- Zheng H, Wisedchaisri G, Gonen T. Crystal structure of a nitrate/nitrite exchanger. Nature. 2013;497:647–651. doi: 10.1038/nature12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PNG 71 kb)
(PNG 120 kb)
(DOCX 19 kb)