SUMMARY
Hippo signaling and the activity of its transcriptional coactivator, Yorkie (Yki), are conserved and crucial regulators of tissue homeostasis. In the Drosophila midgut, after tissue damage, Yki activity increases to stimulate stem cell proliferation, but how Yki activity is turned off once the tissue is repaired is unknown. From an RNAi screen, we identified the septate junction (SJ) protein tetraspanin 2A (Tsp2A) as a tumor suppressor. Tsp2A undergoes internalization to facilitate the endocytic degradation of atypical protein kinase C (aPKC), a negative regulator of Hippo signaling. In the Drosophila midgut epithelium, adherens junctions (AJs) and SJs are prominent in intestinal stem cells or enteroblasts (ISCs or EBs) and enterocytes (ECs), respectively. We show that when ISCs differentiate toward ECs, Tsp2A is produced, participates in SJ assembly, and turns off aPKC and Yki-JAK-Stat activity. Altogether, our study uncovers a mechanism allowing the midgut to restore Hippo signaling and restrict proliferation once tissue repair is accomplished.
Graphical Abstract
In Brief
SJ assembly is a hallmark of EC differentiation in the Drosophila midgut. Xu et al. identify SJ proteins as potent tumor suppressors and uncover Hippo signaling as the surveillance mechanism for SJ deficiency. Specifically, they demonstrate that SJ protein Tsp2A facilitates the endocytic degradation of the Hippo-pathway-antagonizing molecule aPKC.
INTRODUCTION
Precise regulation of stem cell activity is crucial for tissue turnover and necessary to prevent hyperplasia. The digestive epithelium is an excellent system to study the activity of stem cells and how their proliferation and differentiation are regulated, especially in the context of damage. In the intestine, intestinal stem cells (ISCs) give rise to new ISCs and daughter cells known as enteroblasts (EBs), which are primed for differentiation toward either absorptive ECs (~90%) or secretory enteroendocrine cells (EEs; ~10%), depending on the activity of Notch signaling (He et al., 2018; Micchelli and Perrimon, 2006; Ohlstein and Spradling, 2006). Strikingly, ISCs/EBs dramatically accelerate proliferation and differentiation in order to replenish lost cells when tissue damage causes massive EC death (Amcheslavsky et al., 2009). A number of studies have demonstrated the conserved roles of core pathways, such as epidermal growth factor receptor (EGFR)-Ras-mitogen-activated protein kinase (MAPK) signaling (Jiang et al., 2011; Powell et al., 2012), Janus kinase signal transducer and activator of transcription (JAK-Stat) signaling (Grivennikov et al., 2009; Jiang et al., 2009), and Hippo signaling (Cai et al., 2010; Karpowicz et al., 2010; Ren et al., 2010; Shaw et al., 2010), in regulating ISCs. Although the role of these pathways in proliferation and differentiation of ISCs/ EBs has been characterized, less is known about the signals modulating their activities.
The Hippo signaling pathway is particularly essential for tissue homeostasis and cancer prevention. The core components of the Hippo pathway are conserved from Drosophila to mammals. In Drosophila, the kinase Hippo (Hpo) phosphorylates and activates the kinase Warts (Wts), with the aid of the scaffold proteins Mob as tumor suppressor (Mats) and Salvador (Sav). Activated Wts phosphorylates Yki, resulting in the cytoplasmic retention and inhibition of Yki. The mammalian Hippo network is similar but more complex, with two Hpo orthologs (MST1/2), two Wts orthologs (LATS1 or LATS2), two Mats orthologs (MOB1A or MOB1B), one Sav ortholog (SAV1), and two Yki orthologs (YAP or TAZ). When Hippo signaling is inhibited, Yki or YAP or TAZ is activated and induces transcriptional programs promoting tissue growth and inhibiting cell death (Hong and Guan, 2012; Huang et al., 2005; Zhao et al., 2008). In the digestive epithelium of both Drosophila (Karpowicz et al., 2010; Ren et al., 2010; Shaw et al., 2010) and mice (Cai et al., 2010; Gregorieff et al., 2015), the activity of Yki or YAP is induced to facilitate accelerated ISC proliferation following tissue damage. However, it is unclear how Yki or YAP activity is downregulated when tissue repair is complete.
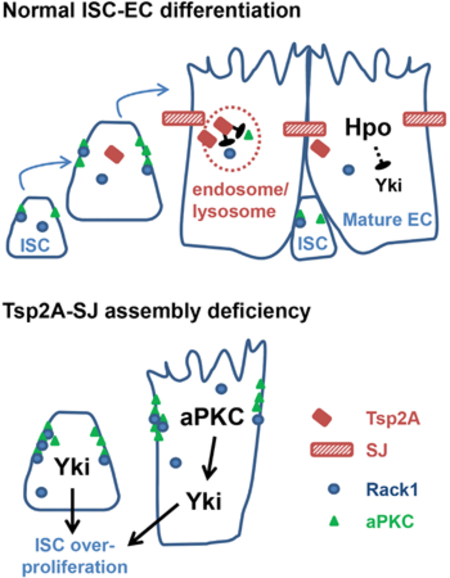
Many components of the Hippo pathway are localized near cell junctions, and their activities are regulated by subcellular localization (Sun and Irvine, 2016). Most notably, recruitment of Hpo to the subapical region close to adherens junctions (AJs) by Expanded (Ex), Merlin (Mer), and Kibra facilitates Hpo dimerization and activation (Deng et al., 2013). Moreover, the cell polarity determinant protein atypical protein kinase C (aPKC) antagonizes Hippo signaling by causing Hpo delocalization from the membrane and subsequent Hpo inactivation in both Drosophila and mammals (Archibald et al., 2015; Grzeschik et al., 2010). Interestingly, junction proteins often exhibit differential expression between stem cells and their differentiated progenies. For example, AJs are enriched among Drosophila ISCs/ EBs (Choi et al., 2011; Ohlstein and Spradling, 2006), Drosophila germline stem cells (Song et al., 2002), mouse hematopoietic stem cells (Zhang et al., 2003), and mammalian epidermal basal or stem cells (Green et al., 2010). In contrast, septate junctions (SJs) are mainly distributed between ECs in the Drosophila midgut (Resnik-Docampo et al., 2017), and tight junctions (TJs; analogous to Drosophila SJs) are localized between differentiated epithelial cell types in the mouse epidermis (Green et al., 2010) and trachea (Gao et al., 2015). Therefore, the AJ-SJ transition and de novo production of SJs might provide a pivotal link between Hippo signaling and stem cell differentiation.
In an RNAi screen to interrogate the function of transmembrane proteins in ISCs/EBs, we identified the SJ protein tetraspanin 2A (Tsp2A) as a tumor suppressor. Further characterization reveals that Tsp2A expression initiates in ISCs and Tsp2A protein assembles at the SJs in the progenitor cells that are differentiating toward ECs. Importantly, we found that Tsp2A undergoes active internalization from the SJs and mediates the degradation of the Hippo-antagonizing protein aPKC. Therefore, endocytic regulation by Tsp2A couples the process of EC maturation with the downregulation of Yki activity. Tsp2A belongs to the large family of four-pass transmembrane proteins that often function as scaffolding co-receptors. Similar to our observation with Tsp2A, previous studies have documented the endocytosis of a putative Tsp2A ortholog CD81 and claudins (TJ proteins) (Farquhar et al., 2012; Matsuda et al., 2004). While the internalization of Tsp2A ortholog and claudins has long been an intriguing observation in cultured cells, our finding about the Tsp2A-aPKC signaling uncovers a physiological function for the internalization of occluding junction (SJ or TJ) protein in vivo.
RESULTS
Tsp2A Acts as a Tumor Suppressor in the ISC-EC Lineage
Expression of Tsp2A RNAi (target region shown in Figure S1A; knockdown efficiency shown in Figure S1B) in ISCs and EBs using the esgGal4 UAS-GFP tubGal80ts (EGT) driver (Micchelli and Perrimon, 2006) causes severe hyperplasia; the midgut appears swollen, especially in the posterior region (Figures 1A and 1B), ISCs/EBs undergo massive expansion and overproliferation (Figures 1C and 1D), and the pseudostratified midgut epithelium becomes multilayered ( Figures 1Cʹ and 1Dʹ). A detailed time course analysis suggests that overproliferation starts when Tsp2A RNAi is expressed in ISCs/EBs for 3 days; Tsp2A RNAi expression in ISCs/EBs for 5 days or longer causes a significant induction of mitosis in the midgut (Figure 1E). Mosaic analysis with a repressible cell marker (MARCM) clones generated from ISCs expressing Tsp2A RNAi can grow much larger than control clones expressing Luc RNAi (Figures 3J and 3K), suggesting that Tsp2A is required within the lineage of an individual ISC to restrict its clonal expansion. In addition, RNAi lines targeting different regions of the Tsp2A transcript (Figure S1A) consistently cause a phenotype of midgut overproliferation (Figures 1F and S3F). Moreover, the RNAi line targeting the Tsp2A 3ʹ UTR can be rescued by the co-expression of Tsp2A cDNA in ISCs/EBs (Figures 1G–1I). Consistent with the RNAi phenotype, a pair of short guide RNAs (sgRNAs) targeting Tsp2A (region shown in Figure S1A) causes massive overproliferation and midgut hyperplasia when Cas9 is expressed in ISCs/EBs (Figures 1J–1L, 1Jʹ, and 1Kʹ). On the contrary to Tsp2A RNAi, expression of Tsp2A cDNA in ISCs/EBs (Figure S1C) causes moderate reduction of tissue damage-induced proliferation (Figure 1M).
Figure 1. Identification of Tsp2A as a Suppressor of Proliferation in ISCs/EBs.
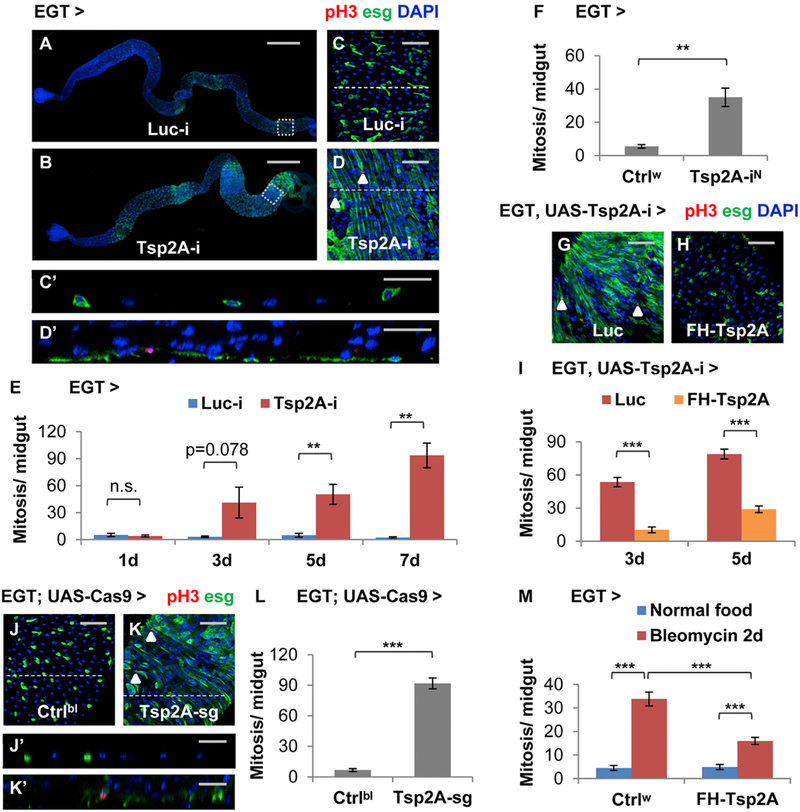
(A and B) Images showing the entire midgut with Luciferase (Luc) RNAi (the negative control) (A) or Tsp2A RNAi (B) expression in ISCs/EBs for 5 days. Scale bar, 500 mm. The GFP signal under the control of EGT labels ISCs/EBs and their recent progenies due to signal perdurance.
(C and D) The midgut expressing Luc RNAi or Tsp2A RNAi in ISCs/EBs for 5d were stained for the mitosis marker phospho-histone H3 (pH3). Scale bar, 50 μm. Examples of pH3+ cells were labeled with white arrowheads. (C) and (D) correspond to the posterior midgut regions encircled with white dashed squares in (A) and (B), respectively.
(Cʹ and Dʹ) The orthogonal projection images showing the cross-sections indicated by dashed lines in (C) and (D), respectively. Scale bar, 25 μm.
(E) Mitosis quantification of midguts expressing Luc RNAi or Tsp2A RNAi in ISCs/EBs for 1, 3, 5, or 7 days. For each genotype at each time point, at least 6 midguts were analyzed. Data are represented as mean ± SEM.
(F) Mitosis quantification of midguts expressing a different Tsp2A RNAi (from NIG, with superscript label ‘‘N’’) in ISCs/EBs for 7 days. Ctrlw (genotype: w1118) was used in genetic crosses as the control, because the genetic background is the same as NIG stocks. N = 7 or 6 midguts were analyzed for the genotype group of Ctrlw or Tsp2A-iN, respectively. Data are represented as mean ± SEM.
(G and H) pH3 staining of midguts expressing Tsp2A RNAi together with Luc cDNA (G) or FH-Tsp2A (H) in ISCs/EBs for 5 days. Scale bar, 50 μm. FH-Tsp2A is resistant to the knockdown of Tsp2A RNAi (the Bloomington stock), which targets the 3ʹ UTR region of Tsp2A. White arrowheads highlight examples of pH3+ cells.
(I) Mitosis quantification of midguts expressing Tsp2A RNAi together with Luc cDNA or FH-Tsp2A in ISCs/EBs for 3 or 5 days. N > 12 midguts were analyzed for each group. Data are represented as mean ± SEM.
(J–L) pH3 staining (K) and mitosis quantification (L) of midguts with ubiquitous expression of sgRNAs against Tsp2A and targeted expression of Cas9 in ISCs/EBs for 7 days. Scale bar, 50 μm. Flies with the same genetic background but only empty insertional landing sites (y v; attp2) were used as the control (Ctrlbl) (J) for sgRNA. White arrowheads highlight examples of pH3+ cells. N = 10 midguts were analyzed per genotype for quantification. Data are represented as mean ± SEM.
(Jʹ and Kʹ) The orthogonal projection images showing the cross sections indicated by dashed lines in (G) and (H), respectively. Scale bar, 25 μm.
(M) Mitosis quantification of midguts with or without FH-Tsp2A expression in ISCs/EBs for 5 days, with or without bleomycin feeding for the last 2 days before dissection. Ctrlw flies were used as the control in genetic crosses for FH-Tsp2A overexpression. N = 12 midguts were analyzed for each group. Data are represented as mean ± SEM.
Figure 3. Tsp2A Knockdown Results in the Accumulation of ISCs/EBs and Pre-ECs.
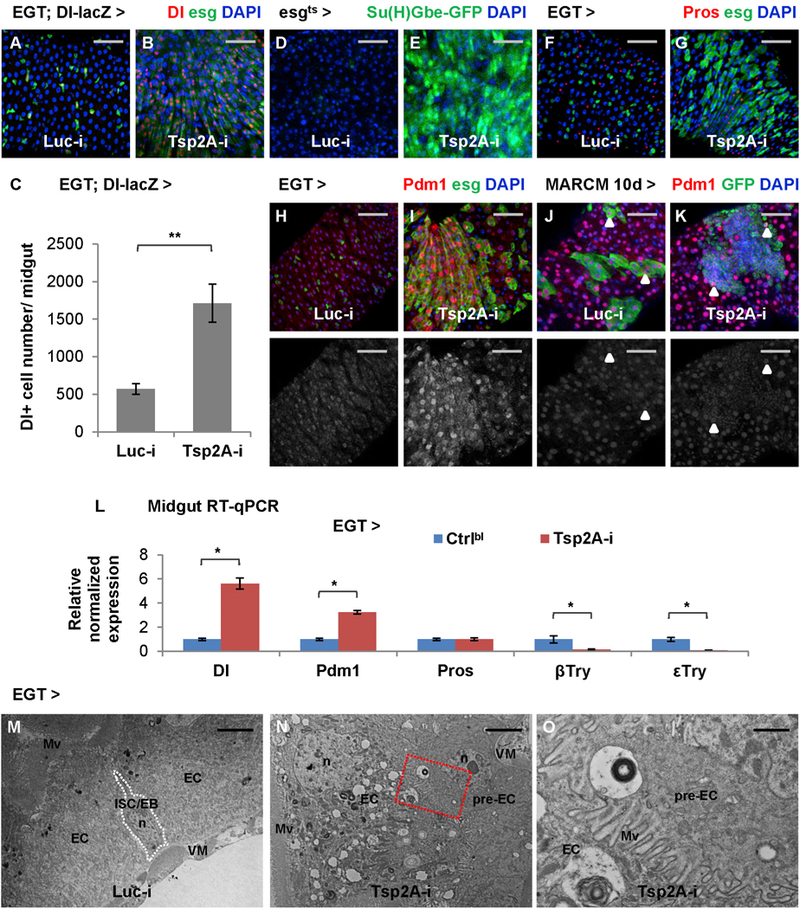
(A and B) Midguts expressing Luc RNAi (A) or Tsp2A RNAi (B) in ISCs/EBs for 3 days are stained for the ISC marker Dl-lacZ. Scale bar, 50 μm.
(C) Quantification of Dl+ cells in the midguts expressing Luc RNAi or Tsp2A RNAi in ISCs/EBs for 5d. N > 8 midguts were analyzed for each group. Data are represented as mean ± SEM.
(D–I) Midguts expressing Luc RNAi (D, F, and H) or Tsp2A RNAi (E, G, and I) in ISCs/EBs (driven by EGT or tubGal80ts; esgGal4 (esgGal4ts)) for 3 days are stained for the EB marker/Notch pathway reporter Su(H)Gbe-GFP (D and E), the EE marker Pros (F and G), or the EC marker Pdm1 (H and I). Scale bar, 50 μm. The red channel of Pdm1 signals is presented in grayscale below each of the merged images.
(J and K) Pdm1 staining of midgut with MARCM clones induced to express Luc RNAi (J) or Tsp2A RNAi (K) for 10 days. Scale bar, 50 μm. Arrowheads highlight examples of polyploid ECs in the lineage of randomly labeled ISCs that express Luc RNAi or Tsp2A RNAi. In the MARCM clone expressing Tsp2A RNAi, even the small nuclei are mostly stained positive for Pdm1.
(L) qRT-PCR quantification of midguts expressing Tsp2A RNAi in ISCs/EBs for 7 days for the different cell-type markers Dl, Pdm1, Pros, β-Trypsin, and ε-Trypsin. Data are represented as mean ± SEM.
(M and N) Electron micrographs of midguts expressing Luc RNAi (M) or Tsp2A RNAi (N) in ISCs/EBs for 5 days. Scale bar, 4 μm. Pre-ECs are the progenitor cells differentiating toward ECs, which exhibit mixed features of ISCs/EBs and ECs. Mv, microvilli; n, nucleus; VM, visceral muscle.
(O) A zoomed-in view of the region encircled with the red dashed box in (N) shows the formation of microvilli at the apical surface of a basally localized pre-EC. Scale bar, 1 μm.
Tsp2A is a SJ protein required for SJ assembly in the larval midgut (Izumi et al., 2016). A recent study found that depletion of the tricellular junction (TCJ) protein Gliotactin (Gli) impairs the intestinal barrier and causes infection-related c-Jun N-terminal kinase (JNK)-dependent induction of ISC proliferation in old flies (Resnik-Docampo et al., 2017). Because TCJ proteins might participate in SJ assembly, we asked whether Tsp2A RNAi-induced overproliferation is due to similar stress signaling. Strikingly, inhibition of JNK signaling by knocking down the kinase basket (bsk) or expressing a dominant-negative form of bsk (bskDN) cannot rescue the overproliferation phenotype caused by Tsp2A RNAi expression in ISCs/EBs (Figures S1D–S1H). In order to determine whether Tsp2A RNAi-induced overproliferation is a compensatory response to tissue damage, we stained the midguts for the apoptosis marker cleaved caspase-3 and detected minimal signs of cell death when there was apparent overproliferation following Tsp2A knockdown (Figures S1I–S1K). Therefore, unlike the case with Gli knockdown during aging, the Tsp2A knockdown phenotype is not caused by JNK stress signaling or tissue damage.
We further characterized the Tsp2A knockdown phenotype in different cell types of the midgut. Expression of Tsp2A RNAi in EBs with the Su(H)Gbe-Gal4, UAS-CD8-GFP; tubGal80ts (SGT) driver (Zeng et al., 2010) (Figures S1L–S1N) or in ECs with Myo1Ats (Figures S1O–S1Q) can induce overproliferation in the midgut, suggesting a non-autonomous effect. As it is the case with Tsp2A knockdown in ISCs/EBs, the Tsp2A knockdown phenotype in ECs can be rescued by an RNAi-resistant Tsp2A cDNA (Figure S1Q), but not by inhibition of JNK-mediated stress signaling with bsk RNAi (Figure S1R) or dominant-negative bsk (data not shown). Moreover, Tsp2A knockdown in ISCs with DlGal4ts also causes mild overproliferation (Figure S1S). In contrast, Tsp2A knockdown in EEs or visceral muscles does not affect proliferation (Figure S1S). Therefore, Tsp2A functions as a tumor suppressor in the ISC-EC lineage.
Characterization of Tsp2A Expression in the ISC-EC Lineage
For a comprehensive understanding of Tsp2A expression, we analyzed our adult midgut single-cell RNA sequencing (RNA-seq) data (Hung et al., 2018) and reconstructed the differentiation trajectories (Figure 2A) from the transcriptomes of 1,753 cells using the single-cell trajectories reconstruction, expression, and mapping (STREAM) method, which can disentangle and visualize complex branching trajectories from single-cell transcriptomic data without prior knowledge about the structure or the number of trajectories (Chen et al., 2018). As expected, the ISC-EC differentiation trajectory is characterized by the gradual loss of ISC markers such as headcase (hdc) (Resende et al., 2017) (Figure 2B) and the incremental expression of EC markers such as λ-Trypsin (Figure 2C). Moreover, as indicated in the 2D stream plots, the expression of shotgun, which encodes Drosophila AJ protein E-cadherin, is enriched in the ISCs/EBs and decreases when ISCs differentiates toward ECs (Figure 2D). In contrast, the expression levels of Tsp2A (Figure 2E) and other SJ protein-coding genes (Hung et al., 2018) gradually increase during ISC-EC differentiation. Consistent with the single-cell ISC-EC trajectory reconstruction, the ultrastructural analysis with transmission electron microscopy (TEM) captures the AJ-SJ transition in a differentiating progenitor cell that partially resembles an ISC/EB (small size, dense cytoplasm, and AJ connection to the neighboring ISC/EB) and partially resembles an EC (relatively more apical localization and microvilli formation at the apical surface) (Figures S2A–S2C). The mixed properties of ISCs and ECs indicate an intermediate status, which we refer to as premature EC (pre-EC). In summary, Tsp2A expression and SJ formation occur during ISC-EC differentiation and precede the complete maturation of ECs.
Figure 2. Tsp2A Expression Initiates in ISCs and Increases during ISC-EC Differentiation.
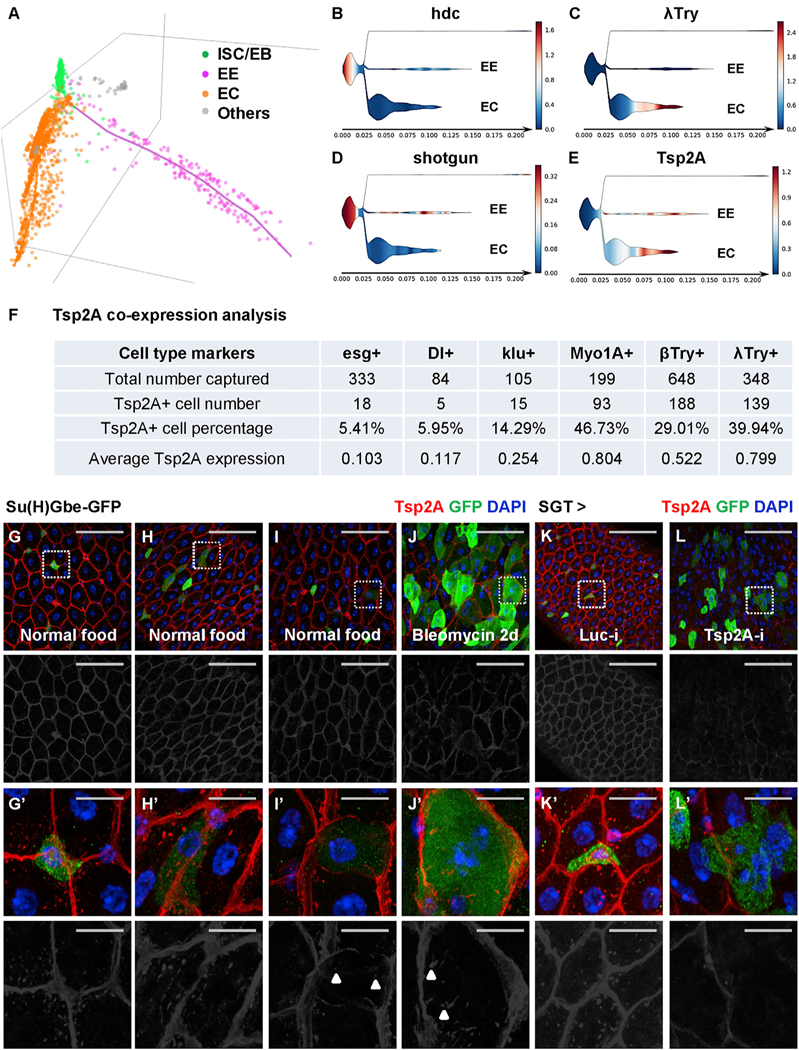
(A) STREAM analysis of single-cell RNA-seq reconstructs the trajectories of ISC differentiation toward the EE and EC lineages (Hung et al., 2018). Each dot in the 3D scattered plot represents a cell. The colored axes represent pseudotime for temporal trajectories. The different cell types (ISC/EB in green, EE in pink, EC in orange, and others in gray) were recognized by unsupervised clustering and the expression of known marker genes. The cell population at the distal end of EC differentiation is expected to be underrepresented in our analysis, because the large and presumably more mature ECs are likely to be filtered out when we used a 25 μm cell strainer to obtain dissociated single cells.
(B–E) STREAM plots in 2D projection map the expression of hdc (B), λ-Trypsin (C), shotgun (D), and Tsp2A (E) during ISC differentiation. The x axis indicates the hypothetical time in differentiation. The branch width in the y axis indicates the number of cells at a particular pseudotime in the branch. The ISCs/EBs are positioned on the left. The branches of EEs and ECs are positioned on the right. Other non-epithelial cells in the gut, shown in the top branch of each panel, are not derived from ISCs and thus barely connected to the rest of the plot. The color gradient scale bar indicating normalized expression levels (blue means low and red means high) is attached to the right of each panel.
(F) Tsp2A co-expression analysis based on single-cell RNA-seq (Table S1). We counted the number of cells expressing different cell markers and cells expressing Tsp2A together with each marker. The percentage of Tsp2A+ cells and the average normalized Tsp2A expression levels in each population sorted by these markers are also listed in the summary chart.
(G–J) Midguts from young adult flies fed with normal food (G–I) or bleomycin (J) for 2 days were co-stained for Tsp2A and the EB marker Su(H)Gbe-GFP. Scale bar, 50 μm. The red channel of Tsp2A staining signals is presented in grayscale below each of the merged images.
(Gʹ–Jʹ) High-magnification images, corresponding to the regions encircled with white dashed squares in G–J, exhibit different levels (Gʹ, no expression; Hʹ, weak; Iʹ, medium; Jʹ, strong) of Tsp2A expression in EBs. Scale bar, 10 μm.
(K and L) Tsp2A staining of midguts expressing Luc RNAi (K) or Tsp2A RNAi (L) in EBs with SGT driver for 7 days. SGT-driven GFP expression labels EBs. A zoom-in view of encircled regions is shown in (Kʹ and Lʹ).
The single-cell RNA-seq data allow us to perform co-expression analysis of Tsp2A with different cell-type markers and detect Tsp2A expression in ISCs/EBs (Figure 2F; Table S1). Whereas only a few Dl+ ISCs are expressing Tsp2A, a higher percentage of Tsp2A+ cells are detected among the cells expressing the EB marker klumpfuss (klu) (Hung et al., 2018). The Tsp2A+ cell percentage and average Tsp2A expression levels rise even higher among cells expressing the EC markers (Myo1A, β-Trypsin, and λ-Trypsin). Consistent with the single-cell analysis, our recent gene-expression profiling by targeted DamID detects a low level (0.74) of Tsp2A expression in esg+ cells and a high level (2.29) of Tsp2A expression in Myo1A+ cells (Doupe et al., 2018). In addition, a dramatic induction of Tsp2A mRNA levels is detected in ISCs/EBs by translating ribosome affinity purification (TRAP) (Thomas et al., 2012) and qRT-PCR quantification (Figures S2D–S2F) at very early stages following bleomycin feeding, which can stimulate ISC differentiation toward ECs (Amcheslavsky et al., 2009). Therefore, we conclude that Tsp2A expression initiates in ISCs and increases during ISC-EC differentiation.
To confirm Tsp2A protein expression and localization in EBs, we co-stained the midgut for Tsp2A and the Notch pathway activity reporter or EB marker Su(H)Gbe-GFP, which indicates the differentiation of midgut progenitor cells, especially toward ECs (Ohlstein and Spradling, 2007). Despite being expressed in a small number of cells under tissue homeostasis conditions, Su(H)Gbe-GFP labels EBs in a wide range of shapes with different levels of membrane-localized Tsp2A (Figures 2G–2I and 2Gʹ–2Iʹ). After bleomycin-induced tissue damage, ISC differentiation is accelerated, resulting in a greater number of Su(H) Gbe-GFP+ cells with Tsp2A assembly along the cell border (Figures 2J and 2Jʹ). Furthermore, Tsp2A RNAi expression in EBs reduces the overall staining of Tsp2A in the midgut (Figures 2K, 2L, 2Kʹ, and 2Lʹ). Therefore, Tsp2A protein is produced in EBs and assembled at SJs during ISC-EC differentiation.
Under tissue homeostasis conditions, Tsp2A staining is mostly observed in ECs and rarely in ISCs/EBs (Figures 2G–2I and S2G). However, when tissue damage induces ISC-EC differentiation, we could observe weak Tsp2A staining along cell borders of pre-ECs (as judged from their increased cell size) that retain EGT-driven GFP expression (Figure S2H). To investigate whether the overall staining of Tsp2A in the midgut depends on the regenerative activity of ISCs/EBs, we used EGT-driven reaper (rpr) expression to deplete ISCs/EBs (Figures S2I–S2M) and stained midguts for Tsp2A at different stages of tissue regeneration. The flies were fed bleomycin-containing food to cause midgut damage and returned to normal food for recovery. Strikingly, Tsp2A staining is disrupted by tissue damage but completely restored after tissue repair in the normal midgut with ISCs/EBs (Figures S2N–S2P). The post-damage recovery of Tsp2A staining is suppressed by ISC/EB depletion (Figures S2Q–S2S), highlighting the importance of ISC activity to regenerate mature ECs for the recovery of Tsp2A staining patterns following tissue damage.
Tsp2A Knockdown Causes the Accumulation of ISCs/EBs and Pre-ECs
To further investigate how Tsp2A knockdown affects ISC differentiation, we stained the midgut for different cell-type markers. Tsp2A RNAi expression in ISCs/EBs induces the ISC marker Dl-lacZ (Figures 3A–3C) and the EB marker Su(H)Gbe-GFP (Figures 3D and 3E). By contrast, Tsp2A knockdown does not affect the EE marker Pros (Figures 3F and 3G). Interestingly, Tsp2A RNAi expression in ISCs/EBs results in an increased number of pre-ECs that express the EC marker Pdm1 and retain some ISC/EB features (small nucleus or EGT > GFP expression) (Figures 3H and 3I). Similarly, Tsp2A RNAi expression induces the accumulation of Pdm1+ small nuclei pre-ECs in MARCM clones (Figures 3J and 3K). qRT-PCR quantification further demonstrates the induction of Dl and Pdm1, but not Pros, in the midgut following Tsp2A knockdown (Figure 3L). Therefore, Tsp2A knockdown induces ISC/EB expansion and ISC-EC differentiation.
Despite the induction of Su(H)Gbe-GFP and Pdm1, Tsp2A knockdown causes dramatically reduced expression of genes encoding trypsins (e.g., β-Trypsin and ε-Trypsin) in the midgut (Figure 3L), suggesting a lack of functional ECs. Furthermore, the ultrastructural analysis by TEM suggests that Tsp2A knockdown not only transforms the pseudostratified midgut epithelium into multilayers but also causes microvilli formation in many pre-ECs that maintain ISC/EB-like features (i.e., basal localization, small nucleus, and dense cytoplasm) (Figures 3M–3O). In conclusion, ISCs/EBs expressing Tsp2A RNAi cannot fully differentiate into mature ECs.
SJ Proteins Form an Interdependent Network Distinct from Other Junctions in the Midgut
Different types of junction proteins often depend on each other for their localization and function. For example, in Drosophila embryonic epithelia, the basolateral junction (BLJ) proteins are interdependent (Bilder et al., 2000) and required for the junctional localization of AJs (Bilder and Perrimon, 2000). We examined in the midgut whether the depletion of the SJ protein Tsp2A affects the localization of other junction proteins. Strikingly, whereas Tsp2A knockdown (Figures 4A and 4B) causes defects in junctional localization of other SJ proteins, such as Mesh, Snakeskin (Ssk), and Coracle (Cora) (Figures 4C–4H), it induces the expression of the AJ protein Armadillo (Arm) (Figures 4I and4J) or the BLJ protein Lethal giant larvae (Lgl) (Figures 4K and 4L). For the ultrastructural analysis of how Tsp2A knockdown affects cell junctions, we obtained electron micrographs and found that junctions are weakened but still present between ECs in midguts ubiquitously expressing Tsp2A RNAi (Figures 4M–4P). Moreover, as other non-SJ junction proteins are probably still present, blue dye fed to young adult flies (in the ‘‘Smurf assay’’; Rera et al., 2012) expressing Tsp2A RNAi in ISCs/EBs or ubiquitously does not leak out of the guts (Figures 4Q and4R), suggesting a functional intestinal barrier. Interestingly, a recent study found that the AJ protein E-cadherin is also dispensable for barrier function (Liang et al., 2017). Therefore, different junctions might function in a redundant manner to control the transepithelial permeability of the midgut in young adult flies.
Figure 4. SJ Proteins Are Interdependent and Distinct from Other Junctions for Their Membrane Localization in the Midgut Epithelium.
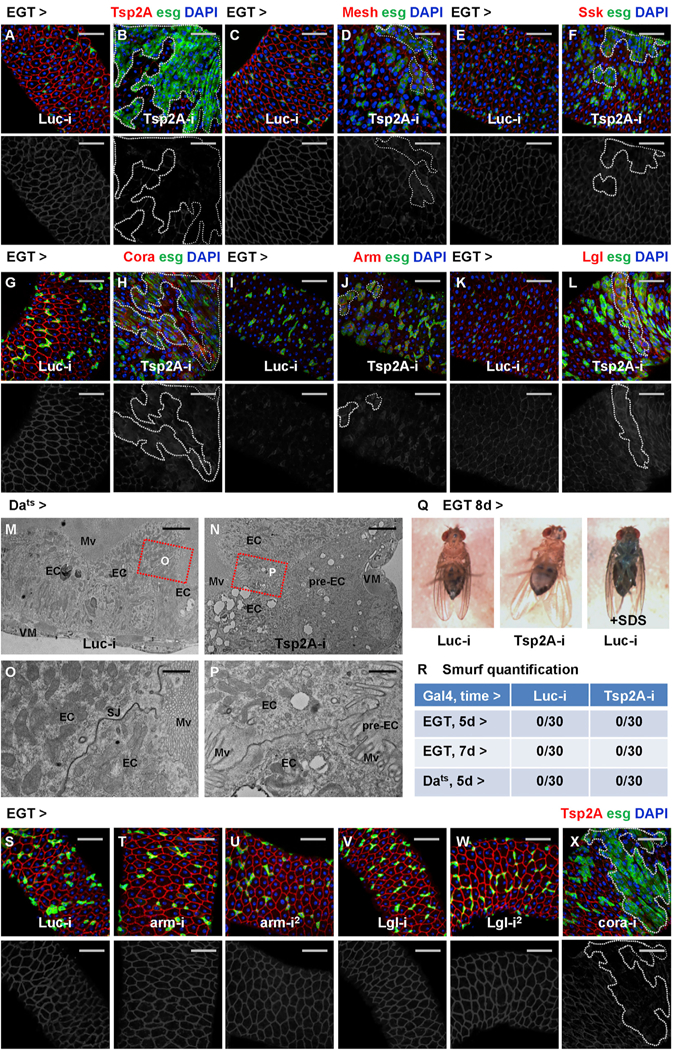
(A–L) Midguts expressing Luc RNAi (A, C, E, G, I, and K) or Tsp2A RNAi (B, D, F, H, J, and L) in ISCs/EBs for 3 days are stained for different junction proteins: Tsp2A (A and B), Mesh (C and D), Ssk (E and F), Cora (G and H), Arm (I and J), and Lgl (K and L). Scale bar, 50 μm. Dashed lines circle example regions (GFP+ ISCs/EBs and recent progenies due to perdurance) where Tsp2A RNAi is expressed. The red channel is presented in grayscale below each of the merged images (in A–L and S–X). Note that ISC tumors often initiate sporadically from a few ISCs/EBs expressing Tsp2A RNAi (Figures 4D, 4F, 4H, 4J, and 4L), which probably correspond to the relatively infrequent events of ISC-EC differentiation under normal feeding conditions.
(M and N) Electron micrographs of midguts ubiquitously expressing Luc RNAi (M) or Tsp2A RNAi (N) for 5 days. Scale bar, 4 μm.
(O and P) High-magnification images of midguts ubiquitously expressing Luc RNAi (O) or Tsp2A RNAi (P), corresponding to regions encircled with red dashed boxes in (M) or (N), show the cell junctions between ECs. Scale bar, 1 μm.
(Q) Smurf assay to evaluate the barrier function of midguts expressing Luc RNAi or Tsp2A RNAi in ISCs/EBs for 8 days. Flies fed with 1% SDS are used as a positive control. Note that although Tsp2A knockdown does not affect barrier function in young adult flies, prolonged RNAi expression (for more than 10 days; data not shown) could cause gut leakage in old flies.
(R) Quantification of Smurf-positive (leaky) flies among assayed flies expressing Luc RNAi or Tsp2A RNAi in ISCs/EBs or ubiquitously for 5–7 days. N = 30 flies are analyzed for each genotype at each time point.
(S–X) Tsp2A staining of midguts expressing Luc RNAi (S), arm RNAi (T and U, two different lines), Lgl RNAi (V and W, two different lines), or cora RNAi (X) in ISCs/ EBs. Scale bar, 50 mm. All RNAi lines are expressed for 7 days, except cora RNAi, for which expression for 3 days is sufficient to cause overproliferation and disrupt Tsp2A expression.
Next, we asked whether knockdown of other junction proteins affects Tsp2A localization. In addition to previous findings that Mesh and Ssk are required for Tsp2A assembly at SJs (Izumi et al., 2016), depletion of another SJ protein, Cora, but not Arm or Lgl, disrupts Tsp2A localization (Figures 4S–4X). Furthermore, we compared the localization of different junction proteins before and after tissue damage. After bleomycin feeding for 2 days, overall Mesh staining is disrupted and reduced in the midgut (Figures S3A and S3B), which is reminiscent of Tsp2A. In contrast, the expression of Arm increases among the expanding population of progenitor cells after tissue damage (Figures S3C and S3D). In summary, whereas SJ proteins (Tsp2A, Mesh, Ssk, Cora) depend on each other for junctional localization, Tsp2A does not appear to depend on or determine the membrane recruitment of other types of junctions in the midgut.
We further analyzed the knockdown of different junction proteins and compared their effects on ISC activity. Knockdown of different SJ genes (Tsp2A, Mesh, Ssk, and cora) causes a similar extent of ISC/EB expansion (Figures S3E–S3K). In contrast, knockdown of arm or Lgl in ISCs/EBs does not cause an easily discernible phenotype (Figures S3L–S3O), except that ISCs/EBs expressing Lgl RNAi exhibit enhanced regenerative proliferation after tissue damage (Figure S3O). Moreover, Gli knockdown does not affect midgut proliferation under either homeostatic or tissue damage conditions in young adult flies (Figures S3P–S3V). Therefore, the interdependent network of SJ proteins functions as a critical tumor suppressor in the midgut, which is different from other types of junction proteins such as Arm, Lgl, and Gli.
Tsp2A Participates in Endocytic Regulation
A detailed characterization of Tsp2A expression in the midgut reveals a striking pattern of subcellular localization in various intracellular punctae, in addition to the expected localization at SJs, in EBs and ECs (Figures 2Iʹ, 2Jʹ, 5A, and 5B). These Tsp2A punctae, when visualized by GFP-Tsp2A expression, appear to correspond to acidic compartments based on LysoTracker staining (Figure 5C). In addition, the endocytosis assay (Shravage et al., 2013) suggests that some Tsp2A punctae co-localize with internalized Texas-red-labeled avidin particles (Figure 5D). Moreover, these Tsp2A punctae also partially co-localize with the early endosome marker Hrs and the late endosome maker Rab7 (Riedel et al., 2016) (Figures 5E and 5F). Furthermore, TEM with immunogold labeling demonstrates the localization of GFP-Tsp2A at endosomes and lysosomes, in addition to the expected localization at SJs (Figures 5G–5I). Therefore, we propose that Tsp2A undergoes active internalization and endocytic degradation.
Figure 5. Tsp2A Participates in the Endocytic Cycle.
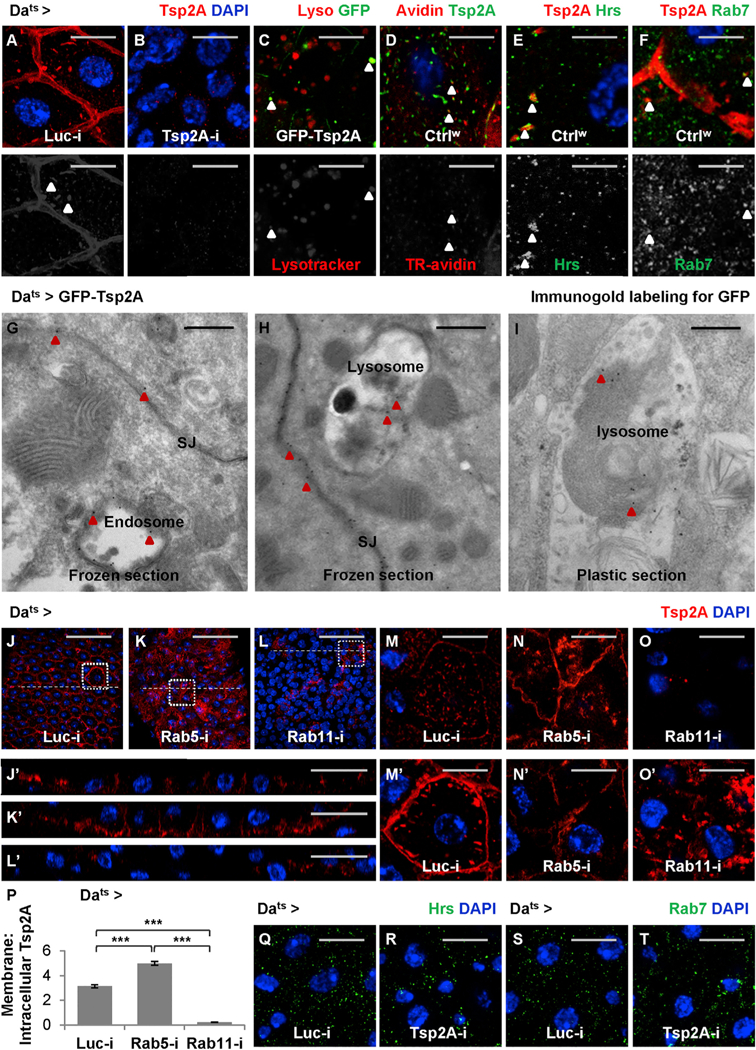
(A and B) Midguts expressing Luc RNAi (A) or Tsp2A RNAi (B) ubiquitously for 3 days are stained for Tsp2A. Scale bar, 10 μm. The Tsp2A staining found in intracellular punctae is not background noise, because it can be eliminated by Tsp2A knockdown. The red channel is presented in grayscale below the merged images (in A–D).
(C) Midguts ubiquitously expressing GFP-Tsp2A for 5 days are stained with LysoTracker red to label lysosomes or other acidic compartments. Scale bar, 10 μm. Arrowheads indicate the co-localization of GFP-labeled Tsp2A with LysoTracker. To avoid potential co-localization artifacts caused by overlaying different focal planes, a single z stack of confocal image is presented.
(D) Endocytosis assay. Wild-type midguts incubated with Texas-red-labeled avidin (TR-avidin) for 20 min are fixed and stained for Tsp2A. Scale bar, 5 μm. Arrowheads indicate Tsp2A punctae co-localization with internalized TR-avidin. A single z stack image (close to the cell surface) is presented. (E and F) Wild-type midguts are co-stained for Tsp2A and the early endosome marker Hrs (E) or the late endosome marker Rab7 (F). Scale bar, 5 μm. Arrowheads indicate Tsp2A punctae co-localization with Hrs or Rab7. Single z stack images are presented. The green channels are presented in grayscale below the merged images.
(G and H) Immunogold labeling and electron micrographs of frozen sections from midguts ubiquitously expressing GFP-Tsp2A for 5 days. Scale bar, 400 nm. Red arrowheads indicate examples of the 15 nm gold (black dots) labeling GFP-Tsp2A found in SJs (G and H), endosome (G), and lysosome (H).
(I) Immunoelectron microscopy (immuno-EM) of the plastic-embedded midgut section showing gold labeled GFP-Tsp2A (indicated by red arrowheads) in the lysosome. Scale bar, 400 nm. Compared to frozen sections, plastic sections perform better in maintaining the GFP antigen in lysosomes for immuno-EM.
(J–L) Tsp2A staining of midguts ubiquitously expressing Luc RNAi (J), Rab5 RNAi (K), or Rab11 RNAi (L) for 5 days. Scale bar, 50 μm.
(Jʹ–Lʹ) The orthogonal projection images showing the cross-sections indicated by dashed lines in J–L. Scale bar, 20 μm.
(M–O) High-magnification, single z stack images near the cell surface of midguts ubiquitously expressing Luc RNAi (M), Rab5 RNAi (N), or Rab11 RNAi (O), corresponding to regions encircled with white dashed squares in J–L. Scale bar, 10 μm.
(Mʹ–Oʹ) High-magnification, single z stack images near the cell center, corresponding to regions encircled with white dashed squares in J–L. Scale bar, 10 μm.
(P) Quantification of the ratios of membrane-localized to intracellular Tsp2A staining intensity in midguts ubiquitously expressing Luc RNAi, Rab5 RNAi, or Rab11 RNAi for 5 days. 24 cells from 3 midguts were analyzed for each genotype. Data are presented as mean ± SEM.
(Q–T) Hrs (Q and R) or Rab7 (S and T) staining of midguts expressing Luc RNAi (Q and S) or Tsp2A RNAi (R and T) ubiquitously for 3 days. Scale bar, 10 μm.
Next, we asked how Tsp2A expression is affected when different steps of the endocytic pathway are inhibited by manipulating different GTPases (Zhang et al., 2007). Inhibition of endocytosis (internalization from the plasma membrane) by Rab5 knockdown causes Tsp2A aggregation at the cell surface and a decrease in the number of intracellular Tsp2A punctae, whereas inhibition of endosome recycling by Rab11 knockdown causes Tsp2A accumulation in the intracellular punctae (Figures 5J–5O and 5Jʹ–5Oʹ, phenotype observed mostly in pre-ECs and ECs as judged from the size of their nuclei; quantification shown in Figure 5P). Consistent with the RNAi phenotype, the dominant-negative form of Rab5 (Rab5DN) or Rab11 (Rab11DN) causes the accumulation of Tsp2A staining at the cell surface or in the intracellular punctae, respectively (Figures S4A–S4F and S4Aʹ–S4Fʹ). Altogether, our data support the hypothesis that Tsp2A participates in the endocytic pathway.
Similar to Tsp2A knockdown, the expression of Rab5 RNAi or Rab5DN in ISCs/EBs causes dramatic midgut overproliferation, whereas the dominant-negative forms or RNAi lines of Rab7 (controlling endosome maturation) and Rab11 have much less or no influence on proliferation (Figures S4G–S4L). In contrast to the Rab5 loss-of-function phenotype, forcing endocytosis with the constitutively active form of Rab5 (Rab5ca) in ISCs/ EBs inhibits proliferation (Figures S4M–S4Q). Moreover, the overproliferation phenotype caused by Tsp2A RNAi expression in ISCs/EBs can be rescued by Rab5ca rather than the constitutively active form of Rab7 (Rab7ca) or Rab11 (Rab11ca) (Figures S4R–S4V). In addition, the co-expression of Rab5ca in ECs can suppress the overproliferation phenotype caused by Myo1Ats- driven Tsp2A RNAi expression (Figure S4W). Therefore, the role of Tsp2A as a tumor suppressor in the ISC-EC lineage might be related to the endocytosis process that implicates Tsp2A.
aPKC-Yki-JAK-Stat Mediates the Overproliferation Phenotype Caused by Tsp2A Knockdown
Although Tsp2A RNAi-induced overproliferation appears similar to the phenotype caused by Rab5 RNAi or Rab5DN, Tsp2A knockdown does not cause the overall elimination of early or late endosomes (Figures 5Q–5T). Therefore, the role of Tsp2A as a tumor suppressor could not simply be attributed to the broad inhibition of endocytosis. To identify the specific downstream signaling events regulated by Tsp2A and required for Tsp2A RNAi-induced overproliferation, we performed a suppressor screen among proteins that interact with Tsp2A or its putative orthologs (BioGRID; https://thebiogrid.org) (Table S2). Multiple different RNAi reagents targeting aPKC or its co-factor, Receptor of activated protein C kinase 1 (Rack1), inhibit Tsp2A RNAi- induced overproliferation (Figures 6A–6D and 6I; knockdown efficiency shown in Figures S5A and S5B). When expressed alone in ISCs/EBs, aPKC RNAi (using the reagent causing weak knockdown) or Rack1 RNAi cannot block tissue-damage-induced mitosis, whereas strong knockdown of aPKC completely inhibits mitosis (Figure S5C), suggesting that a minimal level of aPKC is required for proliferation. We further analyzed aPKC function in different cell types. Consistent with the observation in ISCs/ EBs, aPKC knockdown in EBs moderately suppresses tissue-damage-induced proliferation (Figure S5D). Moreover, aPKC RNAi also suppresses the overproliferation phenotype caused by Myo1Ats-driven Tsp2A RNAi expression in ECs (Figure S5E). Therefore, aPKC activity is required for the Tsp2A knockdown phenotype in the ISC-EC lineage.
Figure 6. aPKC Is Required for Tsp2A RNAi-Induced Overproliferation and Couples Tsp2A Knockdown with Yki and JAK-Stat Signaling.
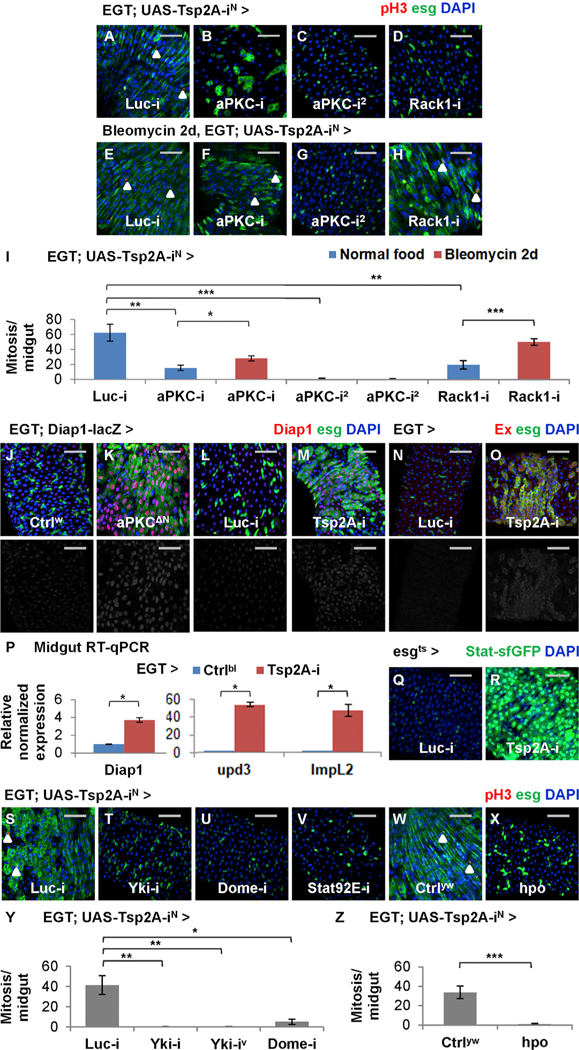
(A–H) pH3 staining of midguts expressing Tsp2A RNAi together with Luc RNAi (A and E), aPKC RNAi (B, C, F, and G, two different lines), or Rack1 RNAi (D and H) in ISCs/EBs for 5 days, with (E–H) or without (A–D) additional 2-day feeding with bleomycin. Scale bar, 50 μm. White arrowheads highlight examples of pH3+ cells.
(I) Mitosis quantification of midguts expressing Tsp2A RNAi together with Luc RNAi, aPKC RNAi, or Rack1 RNAi in ISCs/EBs for 7 days, with or without the last 2-day feeding with bleomycin. N > 7 midguts are analyzed for each group. Data are represented as mean ± SEM.
(J and K) Midguts with (K) or without (J) constitutively active aPKC (aPKCΔN) expression in ISCs/EBs for 3 days are stained for the Yki reporter Diap1-lacZ. Scale bar, 50 mm. The red channels are presented in grayscale, below the merged images.
(L and M) Midguts expressing Luc RNAi (L) or Tsp2A RNAi (M) in ISCs/EBs for 3 days are stained for Diap1-lacZ. Scale bar, 50 μm.
(N and O) Midguts expressing Luc RNAi (N) or Tsp2A RNAi (O) in ISCs/EBs for 3 days are stained for Ex, a transcriptional target of Yki. Scale bar, 50 μm.
(P) qRT-PCR measurement of Yki responsive genes (Diap1, upd3, and ImpL2) in midguts with or without Tsp2A RNAi expression in ISCs/EBs for 7 days. Data are represented as mean ± SEM.
(Q and R) Midguts expressing Luc RNAi (Q) or Tsp2A RNAi (R) in ISCs/EBs (driven by esgts) for 3 days are stained for the JAK-Stat pathway reporter Stat-sfGFP. Scale bar, 50 μm.
(S–V) pH3 staining of midguts expressing Tsp2A RNAi together with Luc RNAi (S), Yki RNAi (T), Dome RNAi (U), or Stat92E RNAi (V) in ISCs/EBs for 5 days. Scale bar, 50 μm.
(W and X) pH3 staining of midguts expressing Tsp2A RNAi alone (W) or Tsp2A RNAi together with hpo (X) in ISCs/EBs for 5 days. Scale bar, 50 mm. Ctrlyw (genotype: y w) flies have the same genetic background as the UAS-hpo line. White arrowheads highlight examples of pH3+ cells.
(Y) Mitosis quantification of midguts expressing Tsp2A RNAi together with Luc RNAi, Yki RNAi (two different lines), or Dome RNAi for 5 days. N > 5 midguts are analyzed for each group. Data are represented as mean ± SEM.
(Z) Mitosis quantification of midguts expressing Tsp2A RNAi alone or together with hpo in ISCs/EBs for 5 days. N > 7 midguts are analyzed for each group. Data are represented as mean ± SEM.
Knockdown of most candidates identified from the suppressor screen, such as myospheroid (mys, Drosophila ortholog of β integrin), Akt1, or EGFR, not only rescue Tsp2A RNAi-induced overproliferation, but also completely block ISC regenerative proliferation (Table S2). Although tetraspanins are often associated with integrins to modulate integrin-dependent cell adhesion and migration (Hemler, 2005), it is difficult to attribute the Tsp2A knockdown phenotype to integrin signaling, since mys is generally required for ISC proliferation (Lin et al., 2013), and we did not detect abnormal mys staining following Tsp2A knockdown in the midgut (data not shown). Similarly, Akt1 and EGFR are known as essential genes for ISC maintenance and proliferation (Jiang and Edgar, 2011). In contrast, ISCs/EBs expressing Tsp2A RNAi together with aPKC RNAi (the weak reagent) or Rack1 RNAi can still exhibit regenerative proliferation following tissue damage (Figures 6E–6I). Therefore, we focused on aPKC or Rack1 in pursuit of a mechanistic understanding about how Tsp2A knockdown affects proliferation.
Previous studies have suggested that aPKC induces Yki activity in the eye imaginal discs (Grzeschik et al., 2010). To examine whether aPKC also antagonizes Hippo signaling and activates Yki in the midgut, we expressed a constitutively active, N-terminally truncated form of aPKC (aPKCΔN) (Betschinger et al., 2003) in ISCs/EBs and stained for the Yki activity reporter Diap1-lacZ. The results suggest that aPKCΔN causes ISC/EB expansion and Yki activation (Figures 6J and6K). Consistent with the previous findings that Yki acts both autonomously in ISCs/EBs and non-autonomously in ECs to induce midgut proliferation (Karpowicz et al., 2010; Ren et al., 2010; Shaw et al., 2010), aPKCΔN expression in ECs also stimulates proliferation (Figure S5F). Furthermore, the overproliferation phenotype caused by the expression of constitutively active Yki (Yki3SA) in ISCs/EBs cannot be suppressed by aPKC RNAi or Rack1 RNAi (Figure S5G). In conclusion, aPKC or Rack1 regulates midgut proliferation via Yki activity.
To examine whether Tsp2A knockdown activates Yki, we stained midguts with two different Yki reporters (Diap1-lacZ and anti-Ex antibody; Maitra et al., 2006) and observed dramatically induced Yki activity in ISCs/EBs expressing Tsp2A RNAi (Figures 6L–6O). qRT-PCR confirmed that mRNA levels of Diap1, upd3, and ImpL2, all induced by Yki activation (Karpowicz et al., 2010; Kwon et al., 2015), increase in midguts expressing Tsp2A RNAi in ISCs/EBs (Figure 6P). Consistent with the induction of the JAK-Stat pathway ligand upd3, the reporter for JAK-Stat activity is induced following Tsp2A knockdown (Figures 6Q and 6R). Moreover, electron micrographs reveal the ultrastructural similarity between Yki activation and Tsp2A knockdown; the midgut epithelium expressing Yki3SA in ISCs/EBs becomes multilayered and forms microvilli in the pre-ECs that have not yet reached the lumen (Figures S5H–S5J). Furthermore, we compared the kinetics of Yki activity to Tsp2A-SJ assembly at different stages of tissue damage or repair by staining the midguts with the anti-Ex antibody. Consistent with the previous report (Karpowicz et al., 2010), Yki activity is induced in the expanding population of progenitor cells after tissue damage and downregulated to the normal levels after tissue repair (Figures S5K–S5M). Altogether, Yki activity is induced by Tsp2A knockdown and appears to inversely correlate with the kinetics of Tsp2A-SJ assembly following tissue damage.
To determine whether Yki activation or Hippo inhibition is required for Tsp2A RNAi-induced overproliferation, we expressed Yki RNAi or hpo cDNA together with Tsp2A RNAi in ISCs/EBs and found that both can rescue the overproliferation phenotype (Figures 6S, 6T, and 6W–6Z). Consistent with the previous findings that Yki depends on JAK-Stat activation to induce overproliferation (Karpowicz et al., 2010; Ren et al., 2010; Staley and Irvine, 2010), knockdown of JAK-Stat pathway components (dome, Stat92E, and hop) can rescue Tsp2A RNAi-induced overproliferation (Figures 6U, 6V, and 6Y; Table S2). Moreover, reduction of JAK-Stat activity by replacing one copy of wild-type hop with the amorphic allele (hop2 or hop27) not only rescues the Tsp2A knockdown phenotype but also allows ISCs expressing Tsp2A RNAi to respond to tissue damage by induced proliferation (Figure S5N). In summary, aPKC-Yki-JAK-Stat signaling mediates Tsp2A RNAi-induced overproliferation.
Tsp2A Interacts with aPKC or Rack1 to Mediate Their Endocytic Degradation
We further investigated the mechanism by which Tsp2A regulates aPKC and Rack1. The mammalian orthologs of Tsp2A are known to interact with protein kinase C and Rack1 (Perez-Hernandez et al., 2013; Zhang et al., 2001); whereas Rack1 is supposed to bind and stabilize activated protein kinase C (Vani et al., 1997). We confirmed the Tsp2A-aPKC and Tsp2A-Rack1 interactions (Figures 7A and7B), as well as an interaction between aPKC and Rack1 (Figures S6A and S6B), by co-immunoprecipitation (coIP) experiments in Drosophila S2R+ cells. When hemagglutinin (HA)-tagged aPKC-HA or Rack1-HA is ubiquitously expressed in the midgut, its signals can be found concentrated at intracellular Tsp2A punctae (Figures 7C and 7D). Although endogenous aPKC staining signals are rare, we also observed its co-localization with the Tsp2A punctae visualized by FLAG-HA-tagged FH-Tsp2A (Figure S6C). In conclusion, our data suggest a specific physical association between Tsp2A and aPKC or Rack1.
Figure 7. Tsp2A Functions as an Adaptor Facilitating aPKC/Rack Degradation.
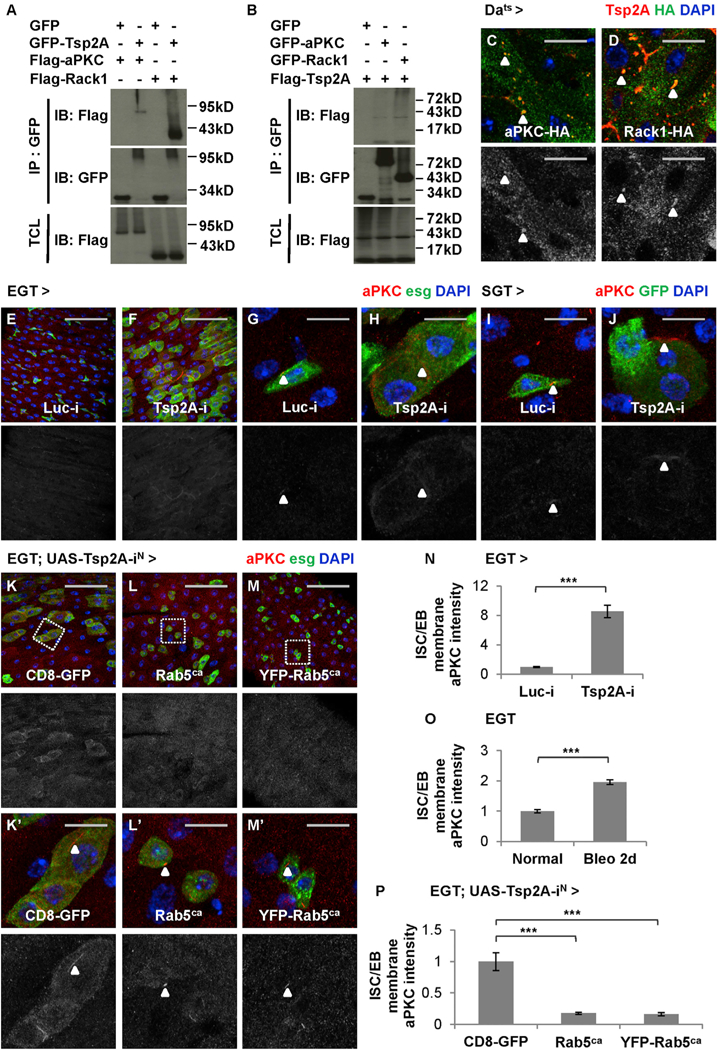
(A and B) S2R+ cell extracts expressing GFP or GFP fusion proteins are subjected to immunoprecipitation with GFP-Trap beads. IB, immunoblot; IP, immunoprecipitation; TCL, total cell lysate. GFP-Tsp2A co-precipitates with FLAG-aPKC and FLAG-Rack1 (A), whereas both GFP-aPKC and GFP-Rack1 co-precipitate with FLAG-Tsp2A (B). Note that the expected size of Tsp2A (without any tag) is 26.9 kDa and that the bands larger (appear as smears) or smaller than expected could represent modified or degraded forms of Tsp2A, according to a previous report (Izumi et al., 2016).
(C and D) Midguts expressing HA-tagged aPKC-HA (C) or Rack1-HA (D) are co-stained with anti-Tsp2A and anti-HA antibodies. Scale bar, 10 μm. Single z stack images are presented. The green channels of HA staining are presented in grayscale below the merged images. Arrowheads indicate examples of aPKC or Rack1 co-localization with Tsp2A punctae.
(E–H) Regular (E and F; scale bar, 50 μm) or high-magnification (G and H; scale bar, 10 μm) confocal images showing aPKC staining of midguts expressing Luc RNAi (E and G) or Tsp2A RNAi (F and H) in ISCs/EBs for 3 days. The red channels are presented in grayscale below the merged images (in E–M and Kʹ–Mʹ). Arrowheads highlight examples of membrane-localized aPKC staining.
(I and J) High-magnification images showing aPKC staining of midguts expressing Luc RNAi (I) or Tsp2A RNAi (J) in EBs for 7 days. Scale bar, 10 μm. Arrowheads indicate examples of concentrated aPKC staining.
(K–M) aPKC staining of midguts expressing Tsp2A RNAi together with CD8-GFP (control) (K) or Rab5ca (2 different lines, with, M, or without, L, YFP tag) in ISCs/ EBs for 5 days. Scale bar, 50 mm.
(Kʹ–Mʹ) High-magnification images of regions encircled with wash dashed squares in K–M. Scale bar, 10 mm. Arrowheads indicate aPKC staining.
(N) Quantification of relative aPKC signal intensity on the membrane of ISCs/EBs expressing Luc RNAi or Tsp2A RNAi for 3 days. N = 32 cells from 4 midguts were analyzed for each genotype. Data are represented as mean ± SEM.
(O) Quantification of relative aPKC staining intensity on the membrane of ISCs/EBs in midguts from young adult flies on normal food or on bleomycin food for 2 days before dissection. N = 30 cells from 3 midguts were analyzed for each group. Data are represented as mean ± SEM.
(P) Quantification of relative aPKC signal intensity on the membrane of ISCs/EBs expressing Tsp2A RNAi together with CD8-GFP, Rab5ca, or YFP-Rab5ca for 5 days. N = 18 cells from 3 midguts were analyzed for each genotype. Data are represented as mean ± SEM.
Next, we examined how Tsp2A knockdown affects the protein levels of Rack1 and aPKC by immunostaining with anti-aPKC (Dollar et al., 2005) and anti-Rack1 (Kadrmas et al., 2007) antibodies. Strikingly, the expression of Tsp2A RNAi in ISCs/EBs dramatically increases aPKC staining at the cell surface (Figures 7E–7H and S6D–S6G; two different RNAi lines used). With confocal microscopy at high magnification, it is quite apparent that aPKC staining is usually concentrated in tiny punctae on the membrane of wild-type ISCs/EBs (Figures 7G and S6F), whereas the signals increase and extend along the cell border when Tsp2A RNAi is expressed in ISCs/EBs (Figures 7H and S6G; quantification in Figures 7N and S6P). The induction of membrane-localized aPKC staining also occurs when Tsp2A RNAi is expressed in EBs or ECs under the control of SGT or Myo1Ats drivers (Figures 7I, 7J, S6H, and S6I), suggesting that Tsp2A regulates the protein levels of aPKC in different cell types in the ISC-EC lineage. Moreover, membrane-localized aPKC staining in ISCs/EBs increases significantly when the flies are fed with bleomycin food for 2 days (Figure 7O), which is consistent with the observation that most ISCs/EBs have not completed de novo Tsp2A-SJ assembly to become mature ECs at that time point following tissue damage (Figure S2O). The accumulation of aPKC on the cell membrane indicates a defect in endocytosis. Interestingly, a similar induction of membrane-localized aPKC is observed when endocytosis is nonspecifically inhibited with Rab5 RNAi expression in ISCs/EBs (Figures S6J, S6K, S6Jʹ, and S6Kʹ; quantification in Figure S6Q). In contrast, enhanced endocytosis by Rab5ca expression can prevent the induction of membrane-localized aPKC staining following Tsp2A knockdown in ISCs/EBs (Figures 7K–7M and 7ʹ–7Mʹ; quantification in Figure 7P). Finally, in addition to aPKC, Rack1 staining signals also increase following Tsp2A knockdown or tissue damage (Figures S6L–S6O). Altogether, Tsp2A appears to facilitate the endocytic degradation and inactivation of aPKC or Rack1 during ISC-EC differentiation.
DISCUSSION
Characterization of Tsp2A in the adult Drosophila midgut reveals a pivotal link between EC maturation and the restriction of ISC proliferation (Figure S7). Progenitor cells differentiating toward ECs undergo a series of changes (Xu et al., 2018), which include the increase of nucleus ploidy and cell size, the formation of SJs, as well as the loss of concentrated aPKC staining at the cell surface. aPKC activity in ISCs/EBs, which increases following tissue damage, can help promote and sustain proliferation via Yki-JAK-Stat signaling. Tsp2A assembly at SJs and its subsequent internalization facilitate aPKC or Rack1 degradation and downregulate aPKC activity. Therefore, in the regeneration process following tissue damage, Tsp2A-SJ assembly not only allows EC maturation but also signals ISCs to reduce proliferation activity when enough mature ECs have been produced for tissue repair. Defects in Tsp2A expression cause excessive aPKC-Yki-JAK-Stat activity and make the midgut epithelia highly proliferative, like a wound that cannot heal.
Endocytic Regulation and Tetraspanin Family Proteins
Endocytosis controls the abundance of transmembrane or membrane-associated molecules. Whereas most internalized proteins and lipids are recycled and return to the cell surface, some are delivered to late endosomes and eventually degraded in lysosomes (Maxfield and McGraw, 2004). Disruption of endocytic degradation often results in excessive accumulation of signaling molecules and overproliferation. For example, the leucine-rich repeat (LRR) protein Windpipe promotes endocytic degradation of Dome, the receptor of mitogenic JAK-Stat pathway, in both the eye discs and the midgut epithelium (Ren et al., 2015). Our study elucidated the functional relevance for the endocytosis of the SJ protein Tsp2A and identified specific components (i.e., aPKC and Rack1) that connect Tsp2A to Hippo signaling, the core pathway restricting ISC activity in the midgut.
There are 38 and 33 tetraspanin proteins encoded by the Drosophila and mammalian genomes, respectively, many of which are known to regulate endocytic processes (Han et al., 2012; Pols and Klumperman, 2009; Xu et al., 2004). In addition to Tsp2A, our RNAi screen included lines targeting 16 more tetraspanins and identified Tsp86D to be required in ISCs/EBs for midgut proliferation and Tsp42Ea to be a weak ISC/EB tumor suppressor (data not shown). Moreover, our suppressor screen revealed that overexpression of Tsp3A can enhance Tsp2A RNAi-induced overproliferation, whereas Tsp42Ef inhibits Tsp2A RNAi-induced overproliferation (Table S2). Both Tsp86D and Tsp3A are known to affect Notch activation by regulating membrane trafficking of the metalloprotease ADAM10 or Kuzbanian to the cell surface (Dornier et al., 2012). Tsp42Ea is an ortholog of human CD63, with the potential to promote endocytic degradation (Pols and Klumperman, 2009). Moreover, Tsp42Ef, localized at multivesicular bodies (MVBs), might also participate in endocytic regulation (Gross et al., 2012). Given that different tetraspanins often interact with each other (Charrin et al., 2014), future studies of these additional tetraspanins (i.e., Tsp86D, Tsp42Ea, Tsp3A, and Tsp42Ef), including the development of reagents to visualize their endogenous expression in the midgut, might lead to a comprehensive understanding of Tsp2A trafficking to the SJs, Tsp2A internalization, and/or Tsp2A downstream signaling.
Different Junction Proteins Implicated in Epithelial Tissue Organization and Signaling
Cell junctions between polarized epithelial cells mediate paracellular permeability and cell attachment. In both Drosophila and mammals, AJs are located at the apical side of the lateral membrane and consist of a cadherin-catenin complex connected to the cytoskeleton (Sun and Irvine, 2016). In mammals, TJs are positioned apically to AJs and form the paracellular diffusion barrier (Zihni et al., 2016). Although insect cells do not have TJs, many TJ protein orthologs are found at SJs (Wei and Ellis, 2001; Wu et al., 2004). In the Drosophila midgut, AJs are enriched in ISCs/EBs (Choi et al., 2011), whereas SJs are predominantly distributed among ECs (Resnik-Docampo et al., 2017). A previous study identified esg-binding sites in the promoter of Ssk and found that esg suppresses the expression of multiple SJ-encoding genes (Mesh, Ssk, and cora) (Korzelius et al., 2014). Our detailed characterization of Tsp2A expression and function during ISC-EC differentiation confirms earlier speculation that upregulation of SJ components might be an important early step in EC differentiation (Korzelius et al., 2014). Interestingly, during the revision of our manuscript, SJ proteins were reported to be required for epithelial polarity in the midgut (Chen et al., 2018). Our findings complement this study and provide further insight into the mechanisms of how the switch of cell polarity determinants from aPKC to SJs occurs during ISC-EC differentiation.
In addition to their structural roles in organizing the epithelium, junction proteins can participate in signal transduction. For example, shotgun knockdown in ISCs/EBs causes a mild increase in proliferation (Choi et al., 2011). shotgun knockdown in ECs induces rhomboid expression to facilitate secretion of mitogenic EGFs, causing feedback upregulation of EGFR-Ras-MAPK signaling and ISC proliferation (Liang et al., 2017). Moreover, a recent study found that disruption of SJs by Gli knockdown impairs intestinal barrier function in old flies and causes JNK-dependent induction of ISC proliferation (Resnik-Docampo et al., 2017). Unlike the work on Gli, we focused on the young adult stages and found that Tsp2A knockdown causes no defects in intestinal barrier function yet induces a strong overproliferation phenotype that cannot be rescued by JNK inhibition. In our study, we compared the knockdown effects of different junction proteins and demonstrated that the tumor suppressor role of SJ proteins is distinct from other types of junction proteins such as Arm, Lgl, and Gli.
In mammalian epithelial tissues such as the trachea, basal cells (stem cells) do not express the TJ proteins ZO1 and claudin-4 under normal conditions but can initiate TJ assembly when they are induced to differentiate after tissue damage (Gao et al., 2015). The kinetics of TJ protein expression in the process of tracheal progenitor cell differentiation is reminiscent of our observation with Tsp2A in the midgut. In the mammalian epithelium, the putative Tsp2A orthologs CD81, CD9, and CD151 localize to cell junctions (Lazo, 2007), whereas both CD81 and CD9 associate with claudins (Farquhar et al., 2012; Kovalenko et al., 2007). Moreover, CD81 and claudins can undergo internalization in mammalian epithelial cells (Farquhar et al., 2012; Matsuda et al., 2004). Given the similarity between Tsp2A and its mammalian orthologs, it will be interesting to examine whether the connection we have elucidated between SJ proteins and Hippo pathway could be generally applicable to TJ proteins in various mammalian epithelial tissues.
Tsp2A-aPKC-Hippo Signaling: Potential Implication in Cancer
Consistent with our discovery that Tsp2A suppresses epithelial cell proliferation, predicted Tsp2A orthologs such as CD81, CD9, and CD151 are often downregulated in carcinomas: CD81 is frequently deleted in cancers (Broad Institute Tumorscape Project; Beroukhim et al., 2010), with its expression reduced in bladder, liver, and gastric carcinomas (Inoue et al., 2001; Lee et al., 2015; Yoo et al., 2013); CD9 expression decreases in breast, ovarian, lung, esophageal, and colon carcinomas (Funakoshi et al., 2003; Houle et al., 2002; Miyake et al., 1996; Mori et al., 1998; Uchida et al., 1999); and CD151 is downregulated in colon cancers (Chien et al., 2008). Moreover, both aPKC and Rack1 are known to play an oncogenic role in various carcinomas (Li and Xie, 2015; Mosesson et al., 2008). Whereas previous studies mostly focused on the function of Tsp2A orthologs in cell migration and cancer metastasis (Hemler, 2014), our findings suggest that Tsp2A orthologs might also be implicated in endocytic regulation to affect tumorigenesis.
Many small molecules can be ligands or antagonists for tetraspanins. For example, human CD81 has a binding pocket for cholesterol (Zimmerman et al., 2016), raising the possibility that cholesterol could play a role in tumorigenesis by modulating the activity of CD81 or similar tetraspanins. Moreover, since CD81 is the receptor for hepatitis C virus (HCV) envelope protein and mediates HCV entry, there have been efforts to develop ligands or antibodies for CD81 or other putative Tsp2A orthologs (Rajesh et al., 2012). Based on our findings, products from these antiviral studies might be valuable for cancer research and treatment. Finally, our observation that tuning down aPKC or JAK-Stat activity can prevent ISCs expressing Tsp2A RNAi from overproliferating without blocking their normal regenerative response raises the possibility to treat cancer by converting hyperplastic ISCs into normal ISCs.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER | ||
|---|---|---|---|---|
| Antibodies | ||||
| rabbit anti-β-galactosidase | Cappel | Cat#0855976 | ||
| rabbit anti-pH3 | Millipore | Cat#06–570; RRID:AB_310177 | ||
| mouse anti-GFP | Invitrogen | Cat#A11120; RRID:AB_221568 | ||
| rabbit anti-GFP | Invitrogen | Cat#A6455; RRID:AB_221570 | ||
| mouse anti-Flag | Sigma | Cat#F3165; RRID:AB_259529 | ||
| rabbit anti-cleaved-caspase 3 | Cell Signaling | Cat#9661; RRID:AB_2341188 | ||
| rabbit anti-Tsp2A | Izumi et al., 2016 | N/A | ||
| rabbit anti-Mesh | Izumi et al., 2016 | N/A | ||
| rabbit anti-Ssk | Izumi et al., 2016 | N/A | ||
| rabbit anti-Cora | DSHB | Cat#C615.16; RRID:AB_1161644 | ||
| mouse anti-Arm | DSHB | Cat#N27A1; RRID:AB_528089 | ||
| rabbit anti-Lgl | Santa Cruz | Cat#sc-98260; RRID:AB_1564606 | ||
| rabbit anti-Pdm1 | Xiaohang Yang | N/A | ||
| mouse anti-Pros | DSHB | Cat#MR1A; RRID:AB_528440 | ||
| mouse anti-Hrs | DSHB | Cat#27–4; RRID:AB_2618261 | ||
| mouse anti-Rab7 | DSHB | Cat#Rab7; RRID:AB_2722471 | ||
| guinea pig anti-ex | Richard Fehon | Maitra et al., 2006 | ||
| rabbit anti-aPKC | Santa Cruz | Cat#sc-216 | ||
| rabbit anti-Rack1 | Julie Kadrmas | Kadrmas et al., 2007 | ||
| rat anti-HA (clone 3F10) | Sigma | Cat#11867423001; RRID:AB_390919 | ||
| donkey anti–rabbit IgG, Alexa Fluor 488 | Invitrogen | Cat#A21206; RRID:AB_141708 | ||
| donkey anti–rabbit IgG, Alexa Fluor 594 | Invitrogen | Cat#A21207; RRID:AB_141637 | ||
| donkey anti–mouse IgG, Alexa Fluor 488 | Invitrogen | Cat#A21202; RRID:AB_141607 | ||
| donkey anti–mouse IgG, Alexa Fluor 594 | Invitrogen | Cat#A21203; RRID:AB_141633 | ||
| donkey anti–guinea pig IgG, Alexa Fluor 594 | Jackson ImmunoResearch Labs | RRID:AB_2340474 | ||
| goat anti–rat IgG, Alexa Fluor 488 | Invitrogen | Cat#A11006; RRID:AB_141373 | ||
| rabbit anti-GFP | Abcam | Cat#6556; RRID:AB_305564 | ||
| rabbit anti-GFP | Invitrogen | Cat#A6455 | ||
| GFP-Trap agarose beads | Bulldog Biotechnology | Cat#GTA100 | ||
|
Chemicals, Peptides, and Recombinant Proteins | ||||
| Paraformaldehyde 16% solution | Electron Microscopy Sciences | Cat#15710 | ||
| PBS pH 7.4 | GIBCO | Cat#10010023 | ||
| Bleomycin | Calbiochem | Cat#203408 | ||
| blue dye no.1 | Spectrum Chemical | Cat#3844–45-9 | ||
| Texas Red-avidin | Invitrogen | Cat#A820 | ||
| Lysotracker Red DND-99 | Invitrogen | Cat#L-7528 | ||
| TRIzol Reagent | Invitrogen | Cat#15596018 | ||
| RNase-Free DNase I Set | QIAGEN | Cat#79254 | ||
| iScript Reverse Transcription Supermix | Bio-Rad | Cat#1708896 | ||
| iQ SYBR Green Supermix | Bio-Rad | Cat#1708880 | ||
| Cyclohexamide | Sigma | Cat#C7698 | ||
| RNaseOUT ribonuclease inhibitor | Invitrogen | Cat#10777019 | ||
| cOmplete Protease Inhibitor, EDTA-free | Roche | Cat#11836170001 | ||
| Gateway LR Clonase II Enzyme mix | Invitrogen | Cat#11791–020 | ||
| pENTR/D-TOPO Cloning Kit | Invitrogen | Cat#K2400–20 | ||
| Effectene Transfection Reagent | QIAGEN | Cat#301425 | ||
| GIBCO Schneider’s Drosophila Sterile Medium | Thermo Fisher | Cat#21720024 | ||
| Fetal Bovine Serum | Thermo Fisher | Cat#10437028 | ||
| Protease and phosphatase inhibitor cocktail | Pierce | Cat#78440 | ||
|
Critical Commercial Assays | ||||
| RNeasy Mini kit | QIAGEN | Cat#74104 | ||
| RNeasy MinElute Cleanup kit | QIAGEN | Cat#74204 | ||
| 4%–20% Mini-PROTEAN TGX Precast Protein Gels | Bio-Rad | Cat#4561096 | ||
|
Experimentl Models: Cell Lines | ||||
| D. melanogaster: Cell line S2R+ | Perrimon lab | N/A | ||
|
Experimental Models: Organisms/Strains | ||||
| D. melanogaster: UAS-Tsp2A sgRNA | This paper | N/A | ||
| D. melanogaster: UAS-FH-Tsp2A | This paper | N/A | ||
| D. melanogaster: UAS-GFP-Tsp2A | This paper | N/A | ||
| D. melanogaster: Dl-lacZ | BDSC | 11651 | ||
| D. melanogaster: y v; attP2 (Ctrlbl) | BDSC | 36303 | ||
| D. melanogaster: UAS-Luc RNAi | BDSC | 31603 | ||
| D. melanogaster: UAS-Tsp2A RNAi | BDSC | 40899 | ||
| D. melanogaster: UAS-Luc | BDSC | 35789 | ||
| D. melanogaster: FRT19A | BDSC | 1744 | ||
| D. melanogaster: UAS-bsk RNAi | BDSC | 31323, 32977 (#2) | ||
| D. melanogaster: UAS-bskDN | BDSC | 6409 | ||
| D. melanogaster: UAS-GFP-RpL10Ab | BDSC | 42683 | ||
| D. melanogaster: UAS-rpr | BDSC | 5823 | ||
| D. melanogaster: UAS-Hepca | BDSC | 6406 | ||
| D. melanogaster: UAS-CD8-GFP | BDSC | 32186 | ||
| D. melanogaster: UAS-Rab5ca | BDSC | 43335 | ||
| D. melanogaster: UAS-YFP-Rab5ca | BDSC | 9773 | ||
| D. melanogaster: UAS-Rab7ca | BDSC | 42707 | ||
| D. melanogaster: UAS-YFP-Rab11ca | BDSC | 50783 | ||
| D. melanogaster: UAS-Rab5DN | BDSC | 42704 | ||
| D. melanogaster: UAS-YFP-Rab7DN | BDSC | 9778 | ||
| D. melanogaster: UAS-YFP-Rab11DN | BDSC | 23261 | ||
| D. melanogaster: UAS-Rab5 RNAi | BDSC | 30518 | ||
| D. melanogaster: UAS-Rab7 RNAi | BDSC | 27051 | ||
| D. melanogaster: UAS-Rab11 RNAi | BDSC | 42709, 27730 (#2) | ||
| D. melanogaster: UAS-arm RNAi | BDSC | 31305, 31304 (#2) | ||
| D. melanogaster: UAS-Lgl RNAi | BDSC | 35773, 38989 (#2) | ||
| D. melanogaster: UAS-cora RNAi | BDSC | 28933, 51845 (#2) | ||
| D. melanogaster: UAS-Gli RNAi | BDSC | 38284, 58115 (#2) | ||
| D. melanogaster: UAS-aPKC RNAi | BDSC | 34332, 35001 (#2) | ||
| D. melanogaster: UAS-Rack1 RNAi | BDSC | 34694 | ||
| D. melanogaster: UAS-aPKCDN | BDSC | 51673 | ||
| D. melanogaster: UAS-Yki3SA | BDSC | 28817 | ||
| D. melanogaster: UAS-Yki RNAi | BDSC | 31965 | ||
| D. melanogaster: UAS-hpo | BDSC | 44254 | ||
| D. melanogaster: UAS-dome RNAi | BDSC | 32860 | ||
| D. melanogaster: UAS-Stat92E RNAi | BDSC | 33637 | ||
| D. melanogaster: hop2 | BDSC | 6032 | ||
| D. melanogaster: hop27 | BDSC | 8493 | ||
| D. melanogaster: y w; attP (Ctrlv) | VDRC | 60100 | ||
| D. melanogaster: UAS-Mesh RNAiv | VDRC | 108297 | ||
| D. melanogaster: UAS-Ssk RNAiv | VDRC | 11906, 105193 (#2) | ||
| D. melanogaster: UAS-Yki RNAiv | VDRC | 104523 | ||
| D. melanogaster: UAS-Tsp2A RNAiN | NIG | 11415R-2 | ||
| D. melanogaster: UAS-Rack1–3xHA | FlyORF | F001043 | ||
| D. melanogaster: UAS-aPKC-3xHA | FlyORF | F000876 | ||
| D. melanogaster: y w (Ctrlyw) | Perrimon lab | N/A | ||
| D. melanogaster: w1118 (Ctrlw) | Perrimon lab | N/A | ||
| D. melanogaster: Diap1-lacZ/TM6B | Karpowicz et al., 2010 | N/A | ||
| D. melanogaster: UAS-p35 (on 3rd chromosome) | Perrimon lab | N/A | ||
| D. melanogaster: esgGal4 UAS-GFP tubGal80ts (EGT) | Micchelli and Perrimon, 2006 | N/A | ||
| D. melanogaster: EGT; UAS-Cas9 (attP2) | Perrimon lab | N/A | ||
| D. melanogaster: esgGal4 tubGal80ts (esgts) | Perrimon lab | N/A | ||
| D. melanogaster: Myo1AGal4 tubGal80ts (Myo1Ats) | Perrimon lab | N/A | ||
| D. melanogaster: tubGal80ts; 24BGal4 (24Bts) | Perrimon lab | N/A | ||
| D. melanogaster: tubGal80ts; DaGal4 (Dats) | Perrimon lab | N/A | ||
| D. melanogaster: tubGal80ts; DlGal4 (Dlts) | Zeng et al., 2010 | N/A | ||
| D. melanogaster: tubGal80ts; Su(H)Gbe-Gal4 (Su(H)ts) | Zeng et al., 2010 | N/A | ||
| D. melanogaster: Su(H)Gbe-Gal4, UAS-CD8-GFP; tubGal80ts (SGT) | Zeng et al., 2010 | N/A | ||
| D. melanogaster: tubGal80ts; ProsGal4 (Prosts) | Perrimon lab | N/A | ||
| D. melanogaster: hsFlp tubGal80 FRT19A/FM7; tubGal4 UAS-mCD8::GFP/TM3 | Choi et al., 2011 | N/A | ||
|
Oligonucleotides | ||||
| Tsp2A sgRNAs | This paper | See Table S3 | ||
| RT-qPCR primers | This paper | See Table S3 | ||
|
Recombinant DNA | ||||
| cDNA RE51204 | Drosophila Genomics Resource Center | 9398 | ||
| cDNA FI03288 | Drosophila Genomics Resource Center | 1621950 | ||
| cDNA RE74715 | Drosophila Genomics Resource Center | 10113 | ||
| pENTR/D-TOPO-Tsp2A | This paper | N/A | ||
| pENTR/D-TOPO-aPKC | This paper | N/A | ||
| pENTR/D-TOPO-Rack1 | This paper | N/A | ||
| pAGW-Tsp2A | This paper | N/A | ||
| pAFW-Tsp2A | This paper | N/A | ||
| pAGW-aPKC | This paper | N/A | ||
| pAFW-aPKC | This paper | N/A | ||
| pAGW-Rack1 | This paper | N/A | ||
| pAFW-Rack1 | This paper | N/A | ||
|
Software and Algorithms |
||||
| ZEN 2 lite | Zeiss | N/A | ||
| ImageJ/Fiji | Schindelin et al., 2012 | N/A | ||
| CFX Manager | Bio-Rad | N/A | ||
| STREAM | Chen et al., 2018 | N/A | ||
| Prism | GraphPad Software | N/A | ||
|
Other | ||||
| Zeiss LSM780 microscope | Zeiss | N/A | ||
| CFX96 Real-Time System | Bio-Rad | N/A | ||
| BZ-9000 Fluorescence Microscope | Keyence | N/A | ||
| 1200-EX transmission electron microscope | JEOL | N/A | ||
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Norbert Perrimon (perrimon@receptor.med.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila culture
Flies were reared on standard cornmeal/agar medium. Fly food was changed every two days to keep fresh. Conditional expression in adult flies using tubGal80ts was achieved by maintaining flies at 18°C until 4–7 days after eclosion, and then shifting young adults to 29°C. For MARCM experiments, flies were reared at 18°C until 3–5 days after eclosion, heat-shocked at 37°C for 1 hr, and then maintained back at 18°C for 10d before dissection and analysis. For consistency, female adult flies, whose midguts are larger and easier to dissect, were analyzed in this paper. To induce tissue damage, fly cornmeal/agar food was melted and mixed well with a final concentration of 25 μg/ml bleomycin.
Transgenic flies
The cDNA of Tsp2A was cloned into pTFHW or pTGW expression vectors (the Drosophila Gateway Vector collection) to generate UAS-FH-Tsp2A and UAS-GFP-Tsp2A transgenic flies.
A pair of sgRNAs targeting the coding region of Tsp2A were designed and cloned into the double U6-sgRNA vector pCFD4 (Port et al., 2014) for site-specific insertion into attP2 on the 3rd chromosome. We obtained transgenic flies ubiquitously expressing two sgRNAs with the seed sequences listed in Table S3.
METHOD DETAILS
Staining and fluorescence imaging
For immunostainings, Drosophila midguts were dissected in PBS, fixed in 4% paraformaldehyde in PBS for 1 hr, incubated for 1–2 hr in Blocking Buffer [5% Normal Donkey Serum (with additional 5% Normal Goat Serum when goat-anti-rat secondary antibody was used), 0.3% Triton X-100, 0.1% BSA in PBS], and stained overnight at 4°C in PBST [0.3% Triton X-100, 0.1% BSA in PBS] with any of the following primary antibodies:
Rabbit anti-β-galactosidase (1:6000), rabbit anti-pH3 (1:3000), mouse anti-GFP (1:300), rabbit anti-GFP (Invitrogen; 1:500), mouse anti-Flag (1:1000), rabbit anti-cleaved-caspase 3 (1:500), rabbit anti-Tsp2A (1:2000), rabbit anti-Mesh (1:1000), rabbit anti-Ssk (1:1000), rabbit anti-Cora (1:50), mouse anti-Arm (1:20), rabbit anti-Lgl (1:300), rabbit anti-Pdm1 (1:500), mouse anti-Pros (1:50), mouse anti-Hrs (1:40), mouse anti-Rab7 (1:40), guinea pig anti-ex (1:2000), rabbit anti-aPKC (1:100), rabbit anti-Rack1 (1:500), rat anti-HA (1:1000).
After primary antibody staining, the midguts were washed 3 times with PBST, stained with DAPI (1:2000) and Alexa Fluor- conjugated donkey-anti-mouse, donkey-anti-rabbit, goat-anti-rat, or donkey-anti-guinea pig secondary antibodies (1:1000) in PBST at 22°C for 2 hr, washed 3 times with PBST, and mounted in Vectashield medium on microscopic slides.
For mitosis quantification, the number of pH3+ cells in the entire midgut was counted with a regular epi-fluorescence microscope. The images of the entire midgut were captured with a Keyence microscope.
The endocytosis assay was adapted from the existing protocol (Shravage et al., 2013). Dissected midguts were incubated ex vivo with Texas Red-avidin diluted in adult hemolymph-like buffer [2 mM CaCl2, 5 mM KCl, 5 mM HEPES, 8.2 mM MgCl2, 108 mM NaCl, 4 mM NaHCO3, 1 mM NaH2PO4, 10 mM Sucrose, 5 mM Trehalose, adjusted with NaOH to pH = 7.5] to a concentration of 80 μg/ml for 20 min, rinsed with PBS, fixed in 4% paraformaldehyde, and stained following a standard protocol.
Lysotracker staining was performed as previously described (Ren et al., 2009), midguts were dissected in PBS, incubated in 0.5 μM Lysotracker Red DND-99 for 3 min, rinsed, and then transferred to PBS on microscopic slides, and photographed immediately.
Images of the posterior midgut area were captured with a Zeiss LSM780 confocal microscope. A z stack of 10–20 images covering one layer of the epithelium from the apical to the basal side were acquired, adjusted, and assembled using NIH Fiji (ImageJ) (Schindelin et al., 2012), and shown as a maximum projection unless indicated otherwise.
For quantification of intracellular Tsp2A stainings, we encircled the intracellular region of an individual cell in the maximum projection image (excluding the surface stack) and measured with Fiji; For quantification of membrane-localized Tsp2A stainings in each cell, we took the sum of Tsp2A staining intensity at the cell surface Z stack and Tsp2A staining intensity at the cell border of the maximum projection image; For quantification of membrane-localized aPKC in each ISC/EB, we encircled the aPKC punctae or elongated aPKC staining along the cell border, and measured the total intensity using Fiji.
RT-qPCR
Total RNA was extracted from 15–20 midguts using TRIzol reagent, treated with DNase I, purified using the QIAGEN RNeasy Mini kit, and converted to cDNA using the iScript Reverse Transcription Supermix. The cDNA was analyzed by real-time quantitative PCR using the iQ SYBR Green Supermix. The sequences of RT-qPCR primers were listed in Table S3. GAPDH and rp49 were used as the internal controls. Each RT-qPCR was performed with three technical replicates, and at least two biological replicates. Representative data (analyzed by Bio-Rad CFX Manager) are presented as mean ± SEM. For qPCR experiments, single asterisk (*) indicates significant difference (p < 0.05) between different groups of technical replicates.
Translating Ribosome Affinity Purification (TRAP) for RT-qPCR analysis
For TRAP experiments, GFP-RpL10A expression was induced in ISCs/EBs with esgts or in ECs with Myo1Ats for 4d before dissection. N > 400 or N > 30 midguts were dissected in PBS for ISC/EB-specific or EC-specific TRAP, respectively. We used plastic Kimble Kontes pellet pestles to homogenize dissected midguts in 500 mL Extraction Buffer [20 mM HEPES (pH7.5), 150 mM KCl, 5 mM MgCl2, 1% Triton X-100, 0.5 mM DTT, 100 μg/ml Cyclohexamide, 100 U/ml RNaseOUT, 1× cOmplete Protease Inhibitor] on ice. To get rid of the debris, lysates were centrifuged at 4°C twice, first for 5 min at 2000 rpm, then for 10 min at maximum speed. Supernatants were incubated for 2 hr at 4°C on a rotator with 50 μL GFP-Trap agarose beads (Bulldog Bio) that have been pre-equilibrated in Extraction Buffer for at least 30 min. Next, the beads were centrifuged at 4°C at 2000 rpm for 2 min, and washed 3 times (for 10 min each time) in Wash Buffer [150 mM NaCl, 0.05% Triton X-100, 50 mM Tris, 5 mM MgCl2, 40 U/ml RNase OUT, 1× cOmplete Protease Inhibitor] at 4°C. After washing, the precipitated GFP-Trap beads were mixed and incubated with 100 μL TRIzol for 10 min, mixed thoroughly with 40 μL chloroform by repeated inverting, and centrifuged at 4○C at maximum speed for 10 min. The aqueous phase was used for RNA extraction using the QIAGEN RNeasy MinElute Cleanup kit (with on-column DNase I digestion). The RNA was converted to cDNA and used for qRT-PCR analysis. For TRAP RT-qPCR, α-tubulin was used as the internal control.
Smurf assay
Smurf assays were performed as previously described (Rera et al., 2012). Standard fly medium was mixed with blue dye no.1 at a concentration of 2.5% (wt/vol). Flies were maintained on dyed medium for 9 hr each time before analysis. A fly was counted as ‘‘Smurf positive’’ when blue dye could be observed outside of the digestive tract. Flies fed with food containing 1% SDS (Liang et al., 2017) for at least 3 days prior to the assay were used as positive controls.
Electron microscopy (EM) and EM with immunogold labeling
For regular EM, tissues of the posterior midgut were fixed in the fixative solution [2.5% Glutaraldehyde 1.25% Paraformaldehyde and 0.03% picric acid in 0.1 M sodium cacodylate buffer (pH 7.4)] for at least 2 hours at room temperature, washed in 0.1M cacodylate buffer and postfixed with 1% Osmiumtetroxide (OsO4)/1.5% Potassiumferrocyanide (KFeCN6) for 1 hour, washed 2× in water, 1× Maleate buffer (MB) 1x and incubated in 1% uranyl acetate in MB for 1hr followed by 2 washes in water and subsequent dehydration in grades of alcohol (10min each; 50%, 70%, 90%, 2×10min 100%). The samples were then put in propyleneoxide for 30 minutes and infiltrated ON in a 1:1 mixture of propyleneoxide and TAAB Epon (Marivac Canada Inc. St. Laurent, Canada). The following day the samples were embedded in TAAB Epon and polymerized at 60°C for 48 hr. Ultrathin plastic sections (~60nm) were cut on a Reichert Ultracut-S microtome, picked up on to copper grids, and contrasted with 0.3% lead citrate. Grids with the plastic sections were imaged using a JEOL 1200-EX transmission electron microscope operating at 80 kV with an AMT 2k CCD camera. Representative images from more than 3 different midgut sections are presented in figures.
For EM with immunogold labeling, tissues of the posterior midgut were fixed for 2 hr in 0.1 M sodium phosphate buffer (pH = 7.4) containing fresh 4% paraformaldehyde and 0.2% glutaraldehyde, rinsed in PBS, and then either processed with 2.3 M sucrose cryo-protection and frozen in liquid nitrogen, or processed with osmium fixation and plastic embedding. Frozen or plastic-embedded samples were cut into ~80 nm sections using a Reichert Ultracut S microtome and transferred to formvar-carbon coated copper grids. Unless specified otherwise, staining was performed at 22°C. Frozen sections were blocked in 1% BSA/PBS buffer for 10 min, incubated with anti-GFP antibody (1:30, Abcam 6556) in 1% BSA/PBS for 1 h, washed 4 times in PBS, incubated with protein A-conjugated 15 nm gold particles for 20 min, washed 2 times in PBS and 4 times in water, and incubated in a mixture of 0.3% uranyl acetate and 2% methyl cellulose for 5 min for contrasting/ embedding. Plastic sections were etched in saturated sodium m-periodate (SIGMA) for 3 min, washed 3 times in water, blocked in PBT buffer [0.1% Triton X-100, 1% BSA in PBS] for 30 min, incubated with anti-GFP antibody (Abcam; 1:50) in PBT at 4°C overnight, washed 4 times in PBS, incubated with protein A-conjugated 15 nm gold particles for 20 min, washed 2 times in PBS and 4 times in water, and stained with lead citrate. Grids with frozen or plastic sections were imaged the same way as regular EM.
Single-cell RNA-seq and data processing
Details on the adult midgut single-cell RNA-seq are described in (Hung et al., 2018). In brief, 1753 midgut cells with good sequencing quality were clustered the cells via Seurat analysis (Butler et al., 2018) into separate groups corresponding to 443 ISC/EBs, 196 EEs, 1053 ECs, and 61 other non-epithelial cells. Single-cell trajectory reconstruction was performed using STREAM (http://stream.pinellolab.org/), with the S2 state expressing high levels of ISC markers selected as the starting state. Trajectories were displayed as a 3D scattered plot and projected in 2D. The genes of interests are visualized as individual 2D STREAM plots.
Co-immunoprecipitation
cDNAs of Tsp2A (RE51204), aPKC (FI03288, isoform RA) and Rack1 (RE74715, isoform RA) were cloned into actin promoter-driven pAGW and pAFW expression vectors obtained from the Drosophila Gateway Vector collection. S2R+ cells were cultured in Schneider’s medium supplemented with 10% fetal bovine serum (FBS) at 25°C. DNA was transfected into S2R+ cells in a 10 cm plate using Effectene transfection reagent. After 3 d of incubation, cells were lysed with Lysis Buffer [30 mM pH = 7.5 Tris-HCl, 150 mM NaCl, 1% Brij97 (P6136, Sigma-Aldrich, St. Louis, MO, USA)] with halt protease and phosphatase inhibitor cocktail (Pierce). Lysates were incubated with GFP-Trap beads for 1–2 hr at 4°C to precipitate the protein complexes. Beads were washed 3 times with 1 mL lysis buffer. Protein complexes were detected by western blotting using rabbit anti-GFP (Invitrogen; 1:5000) and mouse anti-Flag (1:5000).
QUANTIFICATION AND STATISTICAL ANALYSIS
For statistical analyses in all but qPCR experiments, the average value and the standard error of the mean (SEM) of biological replicates were calculated with Microsoft Excel and shown in the figures. For comparison of any two data groups, p values were calculated by Student’s two-tail t test with unequal variances. Single asterisk (*) indicates a p value that is more than 0.01 but less than 0.05. Double asterisks (**) label a p value that is more than 0.001 but less than 0.01. Triple asterisks (***) label a p value that is less than 0.001. The p value of more than 0.1 is labeled as ‘‘not significant’’ (n.s.). For experiments with multiple data groups, one-way ANOVA with Bonferroni’s multiple comparisons test (using GraphPad Prism) was performed to confirm the significant differences (of less or equal p value) as labeled in Figures. No particular methods were used to determine whether the data met assumptions of the statistical approach. Sample sizes, determined empirically and by convenience, are documented in the figure legends. The raw data used for quantification in each Figure are documented in Table S4.
Supplementary Material
Highlights.
The assembly of septate junctions (SJs) occurs during ISC-EC differentiation
The SJ protein Tsp2A undergoes internalization and mediates aPKC degradation
Normal Tsp2A-SJ assembly ensures Hippo signaling to restrict ISC proliferation
Defective Tsp2A-SJ assembly causes aPKC accumulation and Yki hyperactivity
ACKNOWLEDGMENTS
We thank Allison Bardin, Steve Hou, and Ben Ohlstein for sharing fly stocks and Yasushi Izumi, Richard Fehon, Kim McCall, and Julie Kadrmas for antibodies. We thank Martin Hemler and Fernando Camargo for discussions, the Microscopy Resources on the North Quad (MicRoN) core at Harvard Medical School for confocal imaging support, and Maria Ericsson and the Harvard Medical School EM Facility for electron microscopy support. We also thank Perrimon lab members, especially Young Kwon, Phillip Karpowicz, Mary-Lee Deque´ ant, and Richelle Sopko, for discussion and technical instructions; Christians Villalta for transgenic fly injections; and Stephanie Mohr, Afroditi Petsakou, Richard Binari, David Doupé, and Li He for comments on the manuscript. Work in the Perrimon lab is supported by NIGMS (grant GM067761), the STARR consortium, and HHMI. N.P. is an Investigator of the Howard Hughes Medical Institute. H.-W.T. is supported by the Human Frontier Science Program. R.-J.H. is supported by the Jane Coffin Childs Foundation.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Amcheslavsky A, Jiang J, and Ip YT (2009). Tissue damage-induced intestinal stem cell division in Drosophila. Cell Stem Cell 4, 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald A, Al-Masri M, Liew-Spilger A, and McCaffrey L (2015). Atypical protein kinase C induces cell transformation by disrupting Hippo/Yap signaling. Mol. Biol. Cell 26, 3578–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. (2010). The land scape of somatic copy-number alteration across human cancers. Nature 463, 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, and Knoblich JA (2003). The Par complex directs asymmetric cell division by phosphorylating the cytoskeletal protein Lgl. Nature 422, 326–330. [DOI] [PubMed] [Google Scholar]
- Bilder D, and Perrimon N (2000). Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature 403, 676–680. [DOI] [PubMed] [Google Scholar]
- Bilder D, Li M, and Perrimon N (2000). Cooperative regulation of cell polarity and growth by Drosophila tumor suppressors. Science 289, 113–116. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Zhang N, Zheng Y, de Wilde RF, Maitra A, and Pan D (2010). The Hippo signaling pathway restricts the oncogenic potential of an intestinal regeneration program. Genes Dev 24, 2383–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charrin S, Jouannet S, Boucheix C, and Rubinstein E (2014). Tetraspanins at a glance. J. Cell Sci 127, 3641–3648. [DOI] [PubMed] [Google Scholar]
- Chen H, Albergante L, Hsu JY, Lareau CA, Bosco GL, Guan J, Zhou S, Gorban AN, Bauer DE, Aryee MJ, et al. (2018). STREAM: single-cell trajectories reconstruction, exploration and mapping of omics data. bioRxiv 10.1101/302554. [DOI] [PMC free article] [PubMed]
- Chen J, Sayadian AC, Lowe N, Lovegrove HE, and St Johnston D (2018). An alternative mode of epithelial polarity in the Drosophila midgut. PLoS Biol 16, e3000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CW, Lin SC, Lai YY, Lin BW, Lin SC, Lee JC, and Tsai SJ (2008). Regulation of CD151 by hypoxia controls cell adhesion and metastasis in colorectal cancer. Clin. Cancer Res 14, 8043–8051. [DOI] [PubMed] [Google Scholar]
- Choi NH, Lucchetta E, and Ohlstein B (2011). Nonautonomous regulation of Drosophila midgut stem cell proliferation by the insulin-signaling pathway. Proc. Natl. Acad. Sci. USA 108, 18702–18707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Matsui Y, Zhang Y, and Lai ZC (2013). Hippo activation through homodimerization and membrane association for growth inhibition and organ size control. Dev. Biol 375, 152–159. [DOI] [PubMed] [Google Scholar]
- Dollar GL, Weber U, Mlodzik M, and Sokol SY (2005). Regulation of Lethal giant larvae by Dishevelled. Nature 437, 1376–1380. [DOI] [PubMed] [Google Scholar]
- Dornier E, Coumailleau F, Ottavi JF, Moretti J, Boucheix C, Mauduit P, Schweisguth F, and Rubinstein E (2012). TspanC8 tetraspanins regulate ADAM10/Kuzbanian trafficking and promote Notch activation in flies and mammals. J. Cell Biol 199, 481–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doupe DP, Marshall OJ, Dayton H, Brand AH, and Perrimon N (2018). Drosophila intestinal stem and progenitor cells are major sources and regulators of homeostatic niche signals. Proc. Natl. Acad. Sci. USA 115, 12218–12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farquhar MJ, Hu K, Harris HJ, Davis C, Brimacombe CL, Fletcher SJ, Baumert TF, Rappoport JZ, Balfe P, and McKeating JA (2012). Hepatitis C virus induces CD81 and claudin-1 endocytosis. J. Virol 86, 4305–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funakoshi T, Tachibana I, Hoshida Y, Kimura H, Takeda Y, Kijima T, Nishino K, Goto H, Yoneda T, Kumagai T, et al. (2003). Expression of tetraspanins in human lung cancer cells: frequent downregulation of CD9 and its contribution to cell motility in small cell lung cancer. Oncogene 22, 674–687. [DOI] [PubMed] [Google Scholar]
- Gao X, Bali AS, Randell SH, and Hogan BL (2015). GRHL2 coordinates regeneration of a polarized mucociliary epithelium from basal stem cells. J. Cell Biol 211, 669–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KJ, Getsios S, Troyanovsky S, and Godsel LM (2010). Intercellular junction assembly, dynamics, and homeostasis. Cold Spring Harb. Perspect. Biol 2, a000125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregorieff A, Liu Y, Inanlou MR, Khomchuk Y, and Wrana JL (2015). Yap-dependent reprogramming of Lgr5(+) stem cells drives intestinal regeneration and cancer. Nature 526, 715–718. [DOI] [PubMed] [Google Scholar]
- Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, and Karin M (2009). IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell 15, 103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross JC, Chaudhary V, Bartscherer K, and Boutros M (2012). Active Wnt proteins are secreted on exosomes. Nat. Cell Biol 14, 1036–1045. [DOI] [PubMed] [Google Scholar]
- Grzeschik NA, Parsons LM, Allott ML, Harvey KF, and Richardson HE (2010). Lgl, aPKC, and Crumbs regulate the Salvador/Warts/Hippo pathway through two distinct mechanisms. Curr. Biol 20, 573–581. [DOI] [PubMed] [Google Scholar]
- Han SY, Lee M, Hong YK, Hwang S, Choi G, Suh YS, Park SH, Lee S, Lee SH, Chung J, et al. (2012). Tsp66E, the Drosophila KAI1 homologue, and Tsp74F function to regulate ovarian follicle cell and wing development by stabilizing integrin localization. FEBS Lett 586, 4031–4037. [DOI] [PubMed] [Google Scholar]
- He L, Si G, Huang J, Samuel ADT, and Perrimon N (2018). Mechanical regulation of stem-cell differentiation by the stretch-activated Piezo channel. Nature 555, 103–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemler ME (2005). Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol 6, 801–811. [DOI] [PubMed] [Google Scholar]
- Hemler ME (2014). Tetraspanin proteins promote multiple cancer stages. Nat. Rev. Cancer 14, 49–60. [DOI] [PubMed] [Google Scholar]
- Hong W, and Guan KL (2012). The YAP and TAZ transcription co-activators: key downstream effectors of the mammalian Hippo pathway. Semin. Cell Dev. Biol 23, 785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle CD, Ding XY, Foley JF, Afshari CA, Barrett JC, and Davis BJ (2002). Loss of expression and altered localization of KAI1 and CD9 protein are associated with epithelial ovarian cancer progression. Gynecol. Oncol 86, 69–78. [DOI] [PubMed] [Google Scholar]
- Huang J, Wu S, Barrera J, Matthews K, and Pan D (2005). The Hippo signaling pathway coordinately regulates cell proliferation and apoptosis by inactivating Yorkie, the Drosophila homolog of YAP. Cell 122, 421–434. [DOI] [PubMed] [Google Scholar]
- Hung R, Hu Y, Kirchner R, Li F, Xu C, Comjean A, Tattikota S, Song W, Sui S, and Perrimon N (2018). A cell atlas of the adult Drosophila midgut. bioRxiv 10.1101/410423. [DOI] [PMC free article] [PubMed]
- Inoue G, Horiike N, and Onji M (2001). The CD81 expression in liver in hepatocellular carcinoma. Int. J. Mol. Med 7, 67–71. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Motoishi M, Furuse K, and Furuse M (2016). A tetraspanin regulates septate junction formation in Drosophila midgut. J. Cell Sci 129, 1155–1164. [DOI] [PubMed] [Google Scholar]
- Jiang H, and Edgar BA (2011). Intestinal stem cells in the adult Drosophila midgut. Exp. Cell Res 317, 2780–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Patel PH, Kohlmaier A, Grenley MO, McEwen DG, and Edgar BA (2009). Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Grenley MO, Bravo M-J, Blumhagen RZ, and Edgar BA (2011). EGFR/Ras/MAPK signaling mediates adult midgut epithelial homeostasis and regeneration in Drosophila. Cell Stem Cell 8, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadrmas JL, Smith MA, Pronovost SM, and Beckerle MC (2007). Characterization of RACK1 function in Drosophila development. Dev. Dyn 236, 2207–2215. [DOI] [PubMed] [Google Scholar]
- Karpowicz P, Perez J, and Perrimon N (2010). The Hippo tumor suppressor pathway regulates intestinal stem cell regeneration. Development 137, 4135–4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzelius J, Naumann SK, Loza-Coll MA, Chan JS, Dutta D, Oberheim J, Gläßer C, Southall TD, Brand AH, Jones DL, and Edgar BA (2014). Escargot maintains stemness and suppresses differentiation in Drosophila intestinal stem cells. EMBO J 33, 2967–2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko OV, Yang XH, and Hemler ME (2007). A novel cysteine crosslinking method reveals a direct association between claudin-1 and tetraspanin CD9. Mol. Cell. Proteomics 6, 1855–1867. [DOI] [PubMed] [Google Scholar]
- Kwon Y, Song W, Droujinine IA, Hu Y, Asara JM, and Perrimon N (2015). Systemic organ wasting induced by localized expression of the secreted insulin/IGF antagonist ImpL2. Dev. Cell 33, 36–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo PA (2007). Functional implications of tetraspanin proteins in cancer biology. Cancer Sci 98, 1666–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MS, Kim JH, Lee JS, Yun SJ, Kim WJ, Ahn H, and Park J (2015). Prognostic significance of CREB-binding protein and CD81 expression in primary high grade non-muscle invasive bladder cancer: identification of novel biomarkers for bladder cancer using antibody microarray. PLoS ONE 10, e0125405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JJ, and Xie D (2015). RACK1, a versatile hub in cancer. Oncogene 34, 1890–1898. [DOI] [PubMed] [Google Scholar]
- Liang J, Balachandra S, Ngo S, and O’Brien LE (2017). Feedback regulation of steady-state epithelial turnover and organ size. Nature 548, 588–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, Zhang X, Ren J, Pang Z, Wang C, Xu N, and Xi R (2013). Integrin signaling is required for maintenance and proliferation of intestinal stem cells in Drosophila. Dev. Biol 377, 177–187. [DOI] [PubMed] [Google Scholar]
- Maitra S, Kulikauskas RM, Gavilan H, and Fehon RG (2006). The tumor suppressors Merlin and Expanded function cooperatively to modulate receptor endocytosis and signaling. Curr. Biol 16, 702–709. [DOI] [PubMed] [Google Scholar]
- Matsuda M, Kubo A, Furuse M, and Tsukita S (2004). A peculiar internalization of claudins, tight junction-specific adhesion molecules, during the intercellular movement of epithelial cells. J. Cell Sci 117, 1247–1257. [DOI] [PubMed] [Google Scholar]
- Maxfield FR, and McGraw TE (2004). Endocytic recycling. Nat. Rev. Mol. Cell Biol 5, 121–132. [DOI] [PubMed] [Google Scholar]
- Micchelli CA, and Perrimon N (2006). Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 439, 475–479. [DOI] [PubMed] [Google Scholar]
- Miyake M, Nakano K, Itoi SI, Koh T, and Taki T (1996). Motility-related protein-1 (MRP-1/CD9) reduction as a factor of poor prognosis in breast cancer. Cancer Res 56, 1244–1249. [PubMed] [Google Scholar]
- Mori M, Mimori K, Shiraishi T, Haraguchi M, Ueo H, Barnard GF, and Akiyoshi T (1998). Motility related protein 1 (MRP1/CD9) expression in colon cancer. Clin. Cancer Res 4, 1507–1510. [PubMed] [Google Scholar]
- Mosesson Y, Mills GB, and Yarden Y (2008). Derailed endocytosis: an emerging feature of cancer. Nat. Rev. Cancer 8, 835–850. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, and Spradling A (2006). The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 439, 470–474. [DOI] [PubMed] [Google Scholar]
- Ohlstein B, and Spradling A (2007). Multipotent Drosophila intestinal stem cells specify daughter cell fates by differential notch signaling. Science 315, 988–992. [DOI] [PubMed] [Google Scholar]
- Perez-Hernandez D, Gutiérrez-Vázquez C, Jorge I, López-Mart´ın S, Ursa A, Sánchez-Madrid F, Vázquez J, and Yáñez-Mo´ M (2013). The intracellular interactome of tetraspanin-enriched microdomains reveals their function as sorting machineries toward exosomes. J. Biol. Chem 288, 11649–11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pols MS, and Klumperman J (2009). Trafficking and function of the tetraspanin CD63. Exp. Cell Res 315, 1584–1592. [DOI] [PubMed] [Google Scholar]
- Port F, Chen HM, Lee T, and Bullock SL (2014). Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 111, E2967–E2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE, et al. (2012). The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 149, 146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajesh S, Sridhar P, Tews BA, Fénéant L, Cocquerel L, Ward DG, Berditchevski F, and Overduin M (2012). Structural basis of ligand interactions of the large extracellular domain of tetraspanin CD81. J. Virol 86, 9606–9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Finkel SE, and Tower J (2009). Conditional inhibition of autophagy genes in adult Drosophila impairs immunity without compromising longevity. Exp. Gerontol 44, 228–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren F, Wang B, Yue T, Yun E-Y, Ip YT, and Jiang J (2010). Hippo signaling regulates Drosophila intestine stem cell proliferation through multiple pathways. Proc. Natl. Acad. Sci. USA 107, 21064–21069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren W, Zhang Y, Li M, Wu L, Wang G, Baeg GH, You J, Li Z, and Lin X (2015). Windpipe controls Drosophila intestinal homeostasis by regulating JAK/STAT pathway via promoting receptor endocytosis and lysosomal degradation. PLoS Genet 11, e1005180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rera M, Clark RI, and Walker DW (2012). Intestinal barrier dysfunction links metabolic and inflammatory markers of aging to death in Drosophila. Proc. Natl. Acad. Sci. USA 109, 21528–21533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende LP, Truong ME, Gomez A, and Jones DL (2017). Intestinal stem cell ablation reveals differential requirements for survival in response to chemical challenge. Dev. Biol 424, 10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnik-Docampo M, Koehler CL, Clark RI, Schinaman JM, Sauer V, Wong DM, Lewis S, D’Alterio C, Walker DW, and Jones DL (2017). Tricellular junctions regulate intestinal stem cell behaviour to maintain homeostasis. Nat. Cell Biol 19, 52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedel F, Gillingham AK, Rosa-Ferreira C, Galindo A, and Munro S (2016). An antibody toolkit for the study of membrane traffic in Drosophila melanogaster. Biol. Open 5, 987–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RL, Kohlmaier A, Polesello C, Veelken C, Edgar BA, and Tapon N (2010). The Hippo pathway regulates intestinal stem cell proliferation during Drosophila adult midgut regeneration. Development 137, 4147–4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shravage BV, Hill JH, Powers CM, Wu L, and Baehrecke EH (2013). Atg6 is required for multiple vesicle trafficking pathways and hematopoiesis in Drosophila. Development 140, 1321–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Zhu CH, Doan C, and Xie T (2002). Germline stem cells anchored by adherens junctions in the Drosophila ovary niches. Science 296, 1855–1857. [DOI] [PubMed] [Google Scholar]
- Staley BK, and Irvine KD (2010). Warts and Yorkie mediate intestinal regeneration by influencing stem cell proliferation. Curr. Biol 20, 1580–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, and Irvine KD (2016). Cellular organization and cytoskeletal regulation of the Hippo signaling network. Trends Cell Biol 26, 694–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Lee PJ, Dalton JE, Nomie KJ, Stoica L, Costa-Mattioli M, Chang P, Nuzhdin S, Arbeitman MN, and Dierick HA (2012). A versatile method for cell-specific profiling of translated mRNAs in Drosophila. PLoS ONE 7, e40276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Shimada Y, Watanabe G, Li ZG, Hong T, Miyake M, and Imamura M (1999). Motility-related protein (MRP-1/CD9) and KAI1/CD82 expression inversely correlate with lymph node metastasis in oesophageal squamous cell carcinoma. Br. J. Cancer 79, 1168–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vani K, Yang G, and Mohler J (1997). Isolation and cloning of a Drosophila homolog to the mammalian RACK1 gene, implicated in PKC-mediated signalling. Biochim. Biophys. Acta 1358, 67–71. [DOI] [PubMed] [Google Scholar]
- Wei X, and Ellis HM (2001). Localization of the Drosophila MAGUK protein Polychaetoid is controlled by alternative splicing. Mech. Dev 100, 217–231. [DOI] [PubMed] [Google Scholar]
- Wu VM, Schulte J, Hirschi A, Tepass U, and Beitel GJ (2004). Sinuous is a Drosophila claudin required for septate junction organization and epithelial tube size control. J. Cell Biol 164, 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Lee SJ, Suzuki E, Dugan KD, Stoddard A, Li HS, Chodosh LA, and Montell C (2004). A lysosomal tetraspanin associated with retinal degeneration identified via a genome-wide screen. EMBO J 23, 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Ericsson M, and Perrimon N (2018). Understanding cellular signaling and systems biology with precision: a perspective from ultrastructural and organelle studies in the Drosophila midgut. Curr. Opin. Syst. Biol 11, 24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo TH, Ryu BK, Lee MG, and Chi SG (2013). CD81 is a candidate tumor suppressor gene in human gastric cancer. Cell Oncol. (Dordr.) 36, 141–153. [DOI] [PubMed] [Google Scholar]
- Zeng X, Chauhan C, and Hou SX (2010). Characterization of midgut stem cell- and enteroblast-specific Gal4 lines in Drosophila. Genesis 48, 607–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XA, Bontrager AL, and Hemler ME (2001). Transmembrane-4 superfamily proteins associate with activated protein kinase C (PKC) and link PKC to specific beta(1) integrins. J. Biol. Chem 276, 25005–25013. [DOI] [PubMed] [Google Scholar]
- Zhang J, Niu C, Ye L, Huang H, He X, Tong WG, Ross J, Haug J, Johnson T, Feng JQ, et al. (2003). Identification of the haematopoietic stem cell niche and control of the niche size. Nature 425, 836–841. [DOI] [PubMed] [Google Scholar]
- Zhang J, Schulze KL, Hiesinger PR, Suyama K, Wang S, Fish M, Acar M, Hoskins RA, Bellen HJ, and Scott MP (2007). Thirty-one flavors of Drosophila rab proteins. Genetics 176, 1307–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinaiyan AM, et al. (2008). TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22, 1962–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zihni C, Mills C, Matter K, and Balda MS (2016). Tight junctions: from simple barriers to multifunctional molecular gates. Nat. Rev. Mol. Cell Biol 17, 564–580. [DOI] [PubMed] [Google Scholar]
- Zimmerman B, Kelly B, McMillan BJ, Seegar TCM, Dror RO, Kruse AC, and Blacklow SC (2016). Crystal structure of a full-length human Tetraspanin reveals a cholesterol-binding pocket. Cell 167, 1041–1051.e1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


