Abstract
The Erk mitogen-activated protein kinase plays diverse roles in animal development. Its widespread reuse raises a conundrum: when a single kinase like Erk is activated, how does a developing cell know which fate to adopt? We combine optogenetic control with genetic perturbations to dissect Erk-dependent fates in the early Drosophila embryo. We find that Erk activity is sufficient to ‘posterior-ize’ 88% of the embryo, profoundly altering gene expression and morphogenetic movements in all cells within this region. Posterior fate adoption requires at least 1 h of signaling, whereas a 30 min Erk pulse specifies a distinct cell type, intermediate neuroblasts. In contrast to the persistence detector model of Erk interpretation, cell fates are controlled by the cumulative load of signaling, not the duration of a single pulse. The fly embryo thus harbors a classic example of dynamic control, where the temporal profile of Erk signaling selects between distinct physiological outcomes.
ETOC Blurb
Johnson and Toettcher use optogenetics to dissect Erk-dependent responses in the early Drosophila embryo, discovering an endoderm/ectoderm cell fate switch that depends on the dynamics of Erk activity.
Graphical Abstract
One of the great mysteries of animal development is how a small number of intracellular signals can be reused at different positions and times to coordinate a wide range of cell fate decisions. A classic paradigm for this one-to-many mapping is the idea of a morphogen, a substance whose concentration varies with embryonic position and where different concentrations are sufficient to induce different cell fates (Gurdon, et al., 1998). Alternatively, cell fates may be specified by combinatorial control: the spatial overlap between particular combinations of patterning cues (Rahimi, et al., 2016). A third model, dynamic control, holds that a single signal could select among cellular responses based on features such as the amplitude, duration, or frequency of pathway activation (Imayoshi, et al., 2013; Purvis and Lahav, 2013). Although all three paradigms have been proposed to explain cell fate decisions, directly demonstrating which underlies particular developmental decisions has been extremely challenging. Researchers typically lack the ability to vary a single feature, such as the concentration, duration, or spatial range of a signal, while holding others constant.
Here, we set out to dissect cell fate control in a model developmental context: the Erk-dependent control of cellular responses in the early Drosophila embryo. Two factors make the early embryo ideal for such a study. First, Erk activity is required for cells to adopt distinct fates at three different positions (Figure 1A). Erk activation by the Torso receptor patterns head structures at the anterior pole and gut endoderm at the posterior pole, whereas Erk activation by EGFR on the embryo’s lateral surface is required to form intermediate neuroblasts, a subpopulation of neuronal progenitor cells. Second, we have previously shown that Erk signaling can be precisely controlled using optogenetics, enabling one to directly test how specific signal features map to gene expression and cell fate (Bugaj, et al., 2018; Johnson, et al., 2017; Wilson, et al., 2017; Toettcher, et al., 2013).
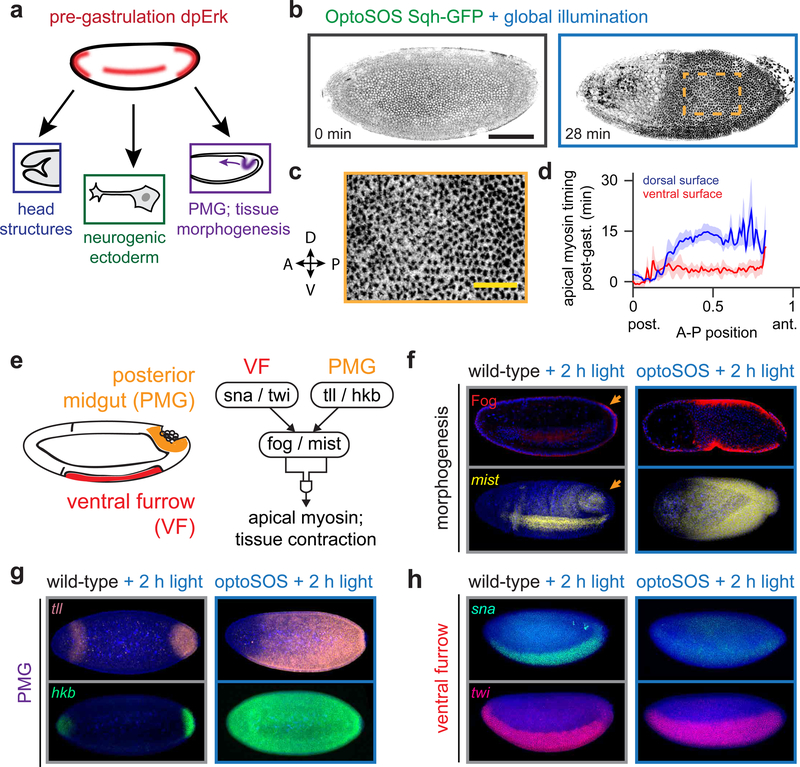
Figure 1. Optogenetic Erk signaling induces gut endoderm gene expression and tissue morphogenesis.
(a) Erk activity is present at three locations within the blastula and coordinates distinct fates. Of these, only the posterior normally undergoes apical constriction and invagination at the start of gastrulation. (b) Maximum-projected images from an OptoSOS-SqhGFP embryo under continuous blue light during nuclear cycle 14 (left) and during gastrulation (right). Scale bar: 100 μm. Dark regions indicate apical myosin localization (See also Movie S2). (c) Detail of boxed region from b, showing a uniform distribution of myosin puncta across the embryo surface. Scale bar: 30 μm. (d) Time of initial apical myosin appearance as a function of anteroposterior (A-P) and dorsoventral (D-V) position from 5 individual OptoSOS embryos. (e) Schematic of the genetic network controlling tissue contractility in the ventral furrow and posterior midgut. (f-h) RNA FISH for mist (f), tll/hkb (g), sna/twi (h), and immunostaining for Fog (in f) in gastrulating Histone-GFP (“wild-type”) and OptoSOS embryos. All embryos were illuminated for 2 h prior to fixation. Scale bar: 100 μm.
By combining optogenetic stimuli with classic genetic perturbations, we find that different thresholds of Erk signaling trigger cells to adopt posterior or lateral fates. High, sustained Erk activity induces gut endoderm differentiation, defined by expression of posterior genes and morphogenetic movements during gastrulation. This decision can be triggered at all but the most anterior positions in the embryo, where it is inhibited by Bicoid. In contrast, a 30 min pulse of Erk activity expands the population of ind-expressing neurogenic ectoderm cells. Systematically varying the blue light stimulus reveals that differentiation into gut endoderm and neurogenic ectoderm is triggered by the total integrated dose of Erk activity, not other signal parameters such as the amplitude, duration or time of light delivery. We propose that Erk dose may be sensed by a two-step accumulator-and-thresholder mechanism, and find evidence that the induction of distinct Erk target genes reflects these two operations.
Results
Light-activated Erk ‘posteriorizes’ the embryo, profoundly disrupting tissue morphogenesis
We first sought to characterize the molecular and phenotypic consequences of high levels of Erk signaling throughout the early embryo. To do so, we took advantage of the OptoSOS system, in which exposure to modest intensities of blue light (0.5–2 mW/cm2) induces Erk phosphorylation to 150–200% of the maximum level reached in wild-type embryos within minutes at all positions in the embryo (Johnson, et al., 2017). This result is different from the modest increase in Erk phosphorylation induced by gain-of-function mutations in the pathway (Figure S1A-C), possibly as a result of feedback inhibition that is triggered by long-term pathway activation in these mutants (Goyal, et al., 2017). Brightfield imaging of gastrulating OptoSOS embryos after light stimulation revealed that these embryos exhibited profound morphogenesis defects (Johnson, et al., 2017), but the precise nature of these defects has not yet been determined.
To better characterize the morphogenesis phenotype of global Erk activation, we set out to image tissue movements in gastrulating OptoSOS embryos at single-cell spatial resolution. We generated OptoSOS embryos that expressed a fluorescent myosin light chain, Sqh-GFP, which redistributes to the apical surface of invaginating cells during gastrulation (Martin, et al., 2009; Royou, et al., 2002). Dark-incubated OptoSOS-SqhGFP embryos gastrulated normally, exhibiting apical myosin redistribution in two tissues that normally invaginate, the ventral furrow and at the posterior pole (Movie S1; Figure S2A). In contrast, OptoSOS-SqhGFP embryos exposed to 2 h of saturating blue light (1 mW/cm2 at 450 nm) massively expanded the domain of apical myosin localization across the majority of the embryo (Figures 1B and S2B; Movie S2–3). We observed apical myosin appearing in puncta over the entire contractile domain without any cell-sized gaps (Figure 1B; inset shown in Figure 1C). To assess whether the decision to contract was cell-autonomous or could be propagated between cells, we stimulated OptoSOS-SqhGFP embryos with a narrow, 8-cell wide stripe of light at the mid-embryo (Figure S2C, Movie S3). We found that light stimulation only induced apical myosin and constriction within the stimulated region, without propagating outwards from regions of Erk stimulation, unlike the recent observation of EGFR-driven contractility waves in the tracheal placode (Nishimura, et al., 2007).
To assess the spatiotemporal dynamics of Erk-induced tissue morphogenesis, we quantified apical myosin accumulation as a function of position and time in five stimulated OptoSOS embryos (see STAR Methods and Figures S2D-F for details). Apical myosin appeared at all but the anterior-most positions, covering 88% of the total embryo. The timing of contraction varied with position: we observed a posterior-to-anterior wave of myosin recruitment and cell movement that was slower and more pronounced on the dorsal than ventral surface (Figure 1D). These differences in timing persisted despite the fact that blue light was simultaneously delivered to the entire embryo. Our observations begin to paint a picture for Erk-induced cell fate determination in the early embryo. Contraction is triggered only in illuminated cells (Figure S2C) and all illuminated cells exhibit apical myosin localization without gaps (Figure 1C). This data suggests that within the posterior 88% of the embryo, Erk is both necessary and sufficient to trigger contractile cell fates. However, while the decision to contract is Erk-dependent, its timing is position-specific and set independently of Erk (Figure 1D).
We reasoned that Erk-induced contractility may reflect the ectopic formation of a tissue that normally contracts during gastrulation: the ventral furrow (VF) or posterior midgut (PMG) (Figure 1E). In both cases, tissue contractility is thought to be driven by the localized expression of folded gastrulation (fog) and mist, a secreted ligand and its cognate G protein coupled receptor (GPCR) (Manning, et al., 2013; Dawes-Hoang, et al., 2005). Although fog and mist are involved in both VF and PMG movements, their expression in the VF is regulated by snail (sna) and twist (twi) and in the PMG by tailless (tll) and huckebein (hkb).
To test whether Erk triggers the expansion of VF- or PMG-like tissue, we stained light-stimulated OptoSOS embryos for the molecular hallmarks of contractility and tissue identity. (For these and other immunofluorescence experiments, Histone-GFP embryos were illuminated and stained alongside OptoSOS embryos as controls for non-specific effects of blue light illumination; see Figures 1F-H.) We found that the domains of Fog protein and mist mRNA expression exactly overlapped the light-stimulated contractile domain (Figure 1F). They also matched a concomitant expansion in the domain of PMG gene expression: tll expression was expanded to match the contractile domain, with hkb expression extending even further to the anterior pole (Figure 1G). In contrast, VF markers were either unaffected (in the case of twi) or eliminated (sna), data which are consistent with the reported repression of sna by hkb (Figure 1H) (Reuter and Leptin, 1994). We also do not observe ventral furrow invagination in regions of OptoSOS embryos that were exposed to high light doses; this may be partially due to the loss of Snail expression and partially due to the global expression of Fog and Mist preventing local invagination just along the furrow.
These results can be readily interpreted in the current model of terminal signaling, which already holds that Erk signaling is necessary for posterior specification (Schupbach and Wieschaus, 1986). We additionally find that Erk activity is sufficient to drive cells to adopt a contractile, posterior fate at all but the anterior-most positions in the embryo. This sufficiency is masked in gain-of-function mutants in Erk signaling (e.g., MEK F53S and Tor D4021) which only partially expand the posterior contractile domain and posterior gap gene expression, likely as a result of Erk signaling that is not fully activated in these mutants (Figure S1) (Goyal, et al., 2017; de las Heras and Casanova, 2006).
Light-triggered posterior fates are blocked by Bicoid at the anterior pole
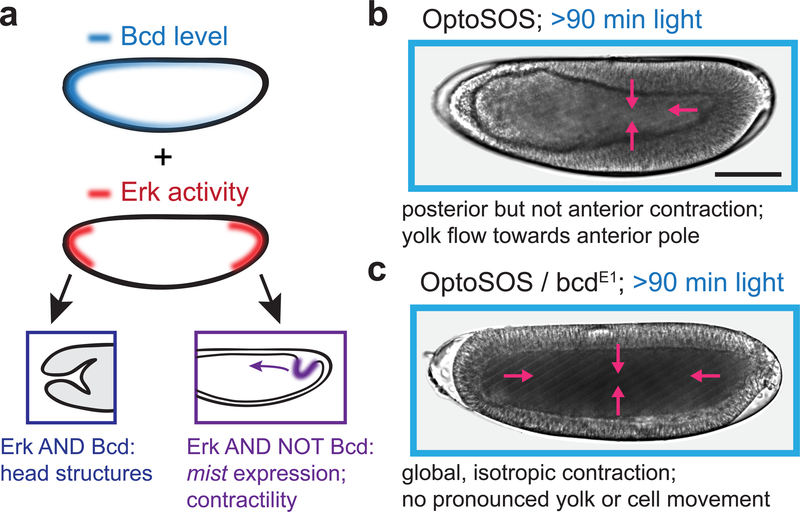
Even in globally-illuminated OptoSOS embryos, posteriorization fails to occur at the anterior pole. This result mirrors normal development, where high levels of Bicoid (Bcd) at the anterior pole are required for the formation of anterior structures; indeed, bcd mutant embryos exhibit mirror-image posterior invaginations during gastrulation (Driever, et al., 1990). These observations suggest that in the absence of Bicoid, global OptoSOS activation may lead to posterior fates and contractility at all positions across the embryo (Figure 2A). To test this prediction, we generated embryos from OptoSOS mothers that are also homozygous for the bcdE1 loss-of-function allele (termed OptoSOS-bcd; for strain details see STAR Methods) and compared their gastrulation to OptoSOS embryos using differential interference contrast (DIC) microscopy.
Figure 2. Erk-induced contractility is global and isotropic in the absence of Bicoid activity.
(a) Schematic illustrating the conceptual model that anterior Bicoid activity may be combinatorially interpreted with Erk to repress posterior fates, including light-induced tissue contractility. (b-c) Differential interference contrast (DIC) images of gastrulating OptoSOS (b) and OptoSOS-bcd (c) embryos that were stimulated with at least 90 min of continuous light prior to gastrulation (see also Movie S4). In b, contraction extends everywhere except the anterior pole (purple arrows), leading to large-scale cell rearrangements towards the posterior pole and yolk movement towards the anterior pole. In c, contraction is isotropic (purple arrows) and blocks virtually all tissue reorganization. Scale bar: 100 μm.
OptoSOS embryos with functional Bcd contracted everywhere but the anterior pole, leading to the flow of yolk toward the anterior pole and a thinning of the epithelial monolayer there (Figure 2B; Movie S4, top). In contrast, blue-light-illuminated OptoSOS-bcd embryos exhibited uniform, isotropic contraction during gastrulation (Figure 2C; Movie S4, bottom). Normally, the asymmetric contractility of OptoSOS embryos leads to a massive anterior-to-posterior movement of cells and posterior-to-anterior flow of yolk. In contrast, the synchronized, isotropic contraction observed in OptoSOS-bcd embryos suppressed virtually all of these flows and movements. These embryos were still subjected to strong compressive forces, as many OptoSOS-bcd embryos popped, ejecting yolk and cells out of one or both poles (Movie S5). We thus conclude that in the absence of Bicoid, Erk-induced endoderm specification is complete, with all cells adopting a posterior morphogenesis program during gastrulation. Due to their ability to drive coordinated tissue movements at any embryonic position in response to light, OptoSOS-bcd embryos may prove useful in future studies that aim to quantitatively relate mechanical forces to tissue/embryo morphogenesis (Izquierdo, et al., 2018; Guglielmi, et al., 2015).
Distinct Erk dynamics trigger cells to adopt either lateral or posterior cell fates
Although our experiments so far focused on adoption of posterior fates, they also reveal a puzzling dual role for Erk along the embryo’s lateral surface. In wild-type embryos, lateral Erk activity is required to form intermediate neuroblasts, a population of ectoderm-derived neuronal progenitors (Schweitzer, et al., 1995). These cells exhibit high levels of endogenous Erk signaling but do not normally contract at the start of gastrulation. Yet light-stimulated Erk is still able to induce their contraction and posterior gene expression (Figure 1). How can Erk signaling specify two distinct responses from the same cell population?
We hypothesized that the distinct responses are the result of dynamic control. According to such a model, differences in some feature of Erk activity over time would select between lateral and posterior fates. Intriguingly, prior studies revealed that Erk dynamics differ between the embryo’s termini and lateral regions in two ways. First, the developmental timing of Erk activation differs: at the posterior, Erk signaling is initiated during the earliest nuclear cycles, whereas lateral Erk is activated shortly before gastrulation (Lim, et al., 2015). Second, the duration of Erk signaling also differs: lateral Erk is activated in a transient 30 min pulse, whereas terminal signaling is sustained for over 1 h (Lim, et al., 2015; Coppey, et al., 2008).
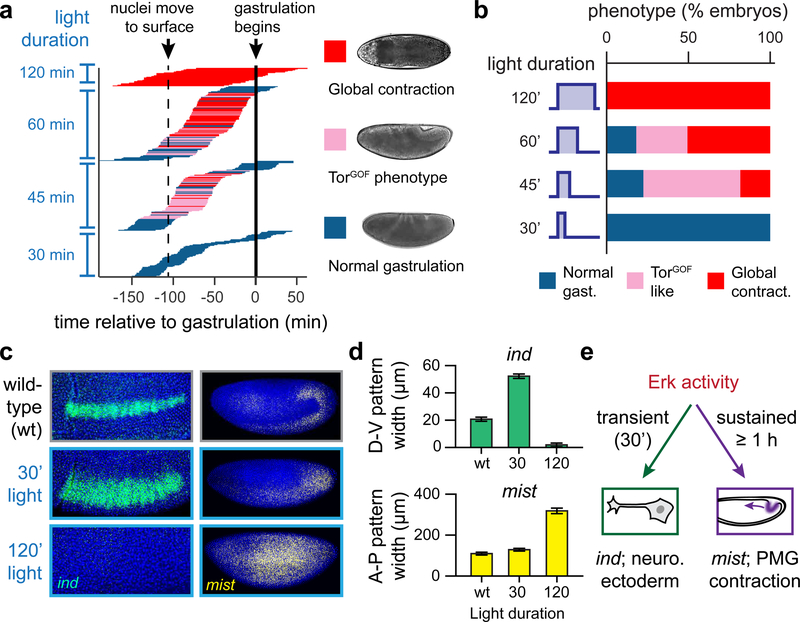
To determine whether the timing or duration of Erk activity influences cell fates, we systematically varied the light inputs delivered to OptoSOS embryos. We stimulated 288 individual OptoSOS embryos of varying ages with either 30, 45, 60, or 120 min of blue light and imaged their progression through gastrulation (Figure S3A-B). Far-red (740 nm) light was used for imaging to avoid additional optogenetic stimulation, and embryos were scored based on their gastrulation phenotypes, which indicated the size of the domain of contractile posterior endoderm. The results of this experiment are shown in Figure 3A.
Figure 3. Erk dynamics control a cell fate switch in the early Drosophila embryo.
(a) Experimental data showing the phenotypes of 288 individual OptoSOS embryos that were imaged after stimulating with light at various developmental times and with different durations of light (See Figure S3A,B for experimental workflow). Each horizontal line represents one DIC-imaged embryo, and the length and position of the line represents the time of light application. The color of each line represents its gastrulation phenotype according to the legend shown. The time at which stimulation was applied was deduced after aligning all embryos at the experimentally-measured start of gastrulation (solid line); the time of nuclei moving to the surface (dashed line) is approximate and shown for reference. (b) The fraction of embryos exhibiting each gastrulation phenotype is plotted as a function of stimulus duration for the embryos in a. (c) Representative images of RNA FISH for ind (at the start of gastrulation) and mist (just prior to gastrulation) for Histone-GFP (“wild-type”) embryos and OptoSOS embryos that were illuminated with 450 nm light for the indicated durations. Scale bars: 100 μm. (d) Quantification of the width of each expression pattern (D-V for ind; A-P for mist) for embryos stimulated as in c. (e) Conceptual model of how Erk-induced neurogenic and contractile fates are distinguished. Transient Erk activity gives rise to ind expression and neurogenic fates, while sustained activity programs mist expression and tissue contractility.
All embryos that were subjected to a 30 min pulse of light gastrulated normally (Figure 3A, bottom). However, these embryos were not completely unaffected by Erk: cuticle preparations revealed abdominal segment fusions similar to those induced by gain-of-function MEK mutants, likely due to disruption of the segment-patterning gene network through increased expression of the terminal gap genes tll and hkb (Figure S3C). At the other extreme, 60 min of illumination triggered a full contractile response in the majority of illuminated embryos, with 100% of embryos contracting after 120 min of light (Figure 3A, top). Lastly, a 45 min pulse of light caused a majority of embryos to adopt an intermediate phenotype similar to what is observed in embryos expressing a constitutively active Torso receptor (TorGOF embryos): a partial expansion of the posterior domain and incomplete germ band extension, leading to lethality shortly after gastrulation (Figure 3A, middle; for details of this phenotype see also STAR Methods, Figure S3D-E, and Movie S6). Although the mechanism underlying partial expansion of the posterior domain is not completely clear, we conjecture that it may arise from the combined dose of Erk activity summed from our light input and the endogenous gradient, which is initially broad and subsequently narrows to the poles (Coppey, et al., 2008).
These observations were highly informative about how Erk activity is interpreted into a cell fate response. The same light intensity was used for all experiments, yet led to outcomes varying from normal gastrulation to global tissue contraction. Thus, cell fate is not simply encoded in the amplitude of Erk activation, as would be expected for a classical morphogen. The developmental time at which light was delivered also had no clear effect provided it was delivered before gastrulation and after nuclear cycle 10, when nuclei are at the embryo surface and are presumably first able to respond to Erk signaling. This insensitivity to timing rules out the possibility that posterior fates are only established during a specific temporal window. In contrast, the duration of Erk activation was quite predictive of gastrulation and cuticle phenotypes (Figure 3B; Figure S4). It thus appears that some feature of Erk activation that correlates with its duration – such as its pulse length, total time on, or area under the curve – is sufficient to program gut endoderm at non-terminal positions.
For Erk dynamics to truly control a cell fate switch, cells must be capable of mounting distinct responses in response to different temporal signals. Thus far we only probed a single fate: the ability for sustained Erk signaling to induce posterior fates throughout the embryo. If dynamic control governs the switch between posterior and lateral fates, then a short pulse of light-activated Erk would be expected to expand the population of intermediate neuroblasts, with a long pulse switching the same cells to endodermal fates. To test this prediction, we stimulated OptoSOS cells with 0, 30 min or 120 min of light and stained for intermediate neuroblasts defective (ind), a gene that marks the Erk-dependent neuroblast population at lateral embryonic positions, and mist, which marks posterior contractile cells. Stimulation with a transient, 30 minute light pulse expanded ind expression dorsally but did not induce mist expression in those cells (Figure 3C). In contrast, 120 min of light completely abolished ind expression, instead inducing mist expression in the same lateral cells (Figure 3C). We quantified the expansion of ind and mist patterns, finding that transient illumination could expand the ind stripe by almost three-fold along the dorsoventral axis without substantially altering the extent of the mist pattern; conversely, ind was abolished and mist was expanded along the anteroposterior axis in response to sustained illumination (Figure 3D).
It is noteworthy that in these experiments we only observed a widening of the lateral stripe of intermediate neuroblasts, not global induction of ind expression similar to that which we observed for endoderm expansion. This spatial restriction is to be expected, due to the well-established co-requirement of Erk and intermediate levels of Dorsal for ind expression (Lim, et al., 2013). After transient light stimulation, the expanded domain of Erk activity overlaps the region of Dorsal expression in a thicker lateral stripe. In sum, optogenetic stimulation and gene expression analyses reveal that lateral cells can be switched between three outcomes – no Erk-triggered response, neurogenic endoderm, and gut ectoderm – simply by increasing the duration of activity of a single signaling pathway (Figure 3E).
Cell responses are triggered by the cumulative dose of Erk signaling
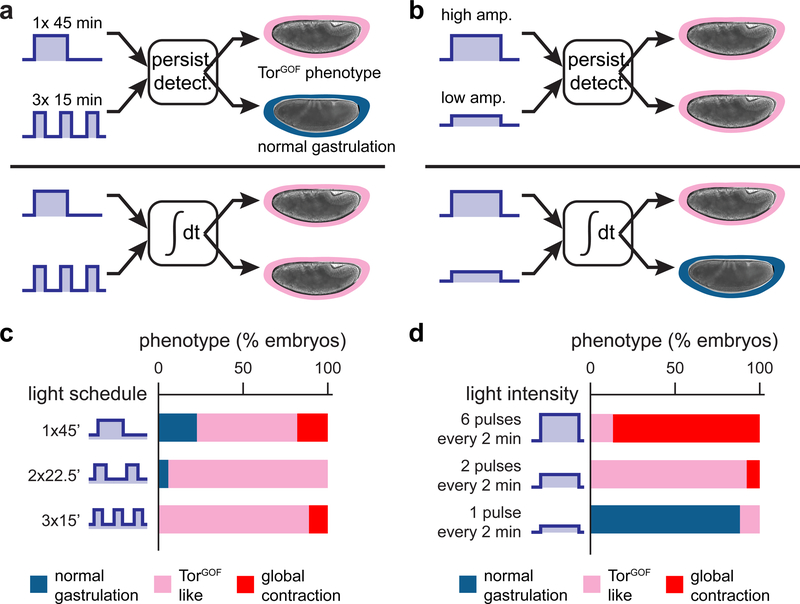
What feature of Erk activity over time is sensed by cells to determine their fate? Differences in signaling dynamics have long been hypothesized to select among Erk-dependent cell fates (Marshall, 1995; Bishop, et al., 1994), and at least two distinct mechanisms for decoding Erk dynamics have been proposed. The classic model is that of a persistence detector, where downstream genes sense the duration of a single Erk pulse (as is thought to be the case for gene products such as c-fos) (Nakakuki, et al., 2010; Murphy, et al., 2004). An alternative model is that of cumulative load detection, where distinct fates are triggered when the total integrated signal crosses a threshold (Gillies, et al., 2017; Hannanta-Anan and Chow, 2016). Reasoning that these different types of dynamic decoding could be distinguished by their responses to different time-varying stimuli, we next set out to assess cell fates after different light schedules (Figure 4A,B).
Figure 4. Embryo phenotypes are determined by the cumulative load of Erk signaling.
Gastrulation phenotypes may be triggered by two classes of dynamic decoders, a persistence detector or a cumulative load sensor. These two decoders can be discriminated by their responses to certain light patterns. (a) A persistence detector could be triggered by a single light bolus but not when that signal is partitioned into multiple short light bouts; a cumulative load sensor would fire in response to both equal-dose inputs. (b) A cumulative load sensor could be triggered by high-intensity illumination but not a lower-intensity input of the same duration; a persistence detector would respond similarly to both equal-duration inputs. (c) Gastrulation phenotypes for the light schedules in a. OptoSOS embryos were exposed to a single 45 min pulse of blue light or the same 45 min dose split into two or three equal pulses and delivered over a 90 min period. Each bar represents the percentage of embryos exhibiting different gastrulation phenotypes according to the legend shown. (d) Gastrulation phenotypes for the light schedules shown in b. OptoSOS embryos were exposed to different numbers of 1 sec light pulses every 2 min for a total of 60 min. Bars represent phenotypes as in c.
We reasoned that a cumulative load sensor and a persistence detector would behave differently if the same light dose was delivered as a single bolus or multiple short pulses (Figure 4A), inputs which share the same cumulative load but have different pulse durations. We chose a total illumination time of 45 min because it induces the TorGOF-like phenotype, an intermediate outcome that could be altered either by an increase or decrease in the strength of signaling. Over a 90 min time period prior to gastrulation we stimulated embryos with either a single 45 min pulse, two 22.5 min pulses separated by a 45 min gap, or three 15 min pulses separated by 15 min gaps. Regardless of the pulse schedule used, a majority of embryos in each stimulus condition adopted an identical TorGOF phenotype, consistent with cumulative load but not persistence detection determining the gut endoderm fate switch (Figure 4C). These data also show that the accumulated Erk dose is accurately ‘remembered’ even across multiple nuclear division cycles: two 22.5 min pulses separated by a 45 min gap induce the same phenotype as a single 45 min light dose. We obtained similar results using a second phenotypic assay: cuticle preparations to assess segmentation of the body plan (Figure S4A). In this assay, a similar proportion of embryos exhibited segment fusions in response to 15 min of continuous illumination or three 5 min pulses delivered over 90 min, suggesting that the segmentation gene network is also sensitive to the total Erk dose. Additionally, a majority of embryos stimulated with a single 45 min pulse or three 15 min pulses lacked cuticle structures altogether (Figure S4A), consistent with these embryos’ failure to properly gastrulate (Figure 4C).
To further probe whether the total dose of Erk controls embryonic phenotypes, we reasoned that different ways of varying the dose should elicit similar phenotypes, such as by varying signaling duration at a fixed amplitude or varying amplitude over a fixed duration (Figure 4B). To attain intermediate Erk amplitudes, we adopted a strategy of varying the number of brief, bright light pulses delivered every 2 min. We previously showed that the ERK pathway gradually turns on and off over ~4 min, so that these fast, frequent stimuli are averaged to an intermediate activity level (Toettcher, et al., 2013). This strategy, termed pulse width modulation, is easily transferable between assays with different light sources or physical configurations where intensity is difficult to control and has been applied in a growing number of optogenetic contexts (Chen, et al., 2017; Davidson, et al., 2013).
Indeed, we found that varying the effective intensity of a 60 min light stimulus resulted in the same sequence of gastrulation phenotypes as when we varied the duration of a single pulse (compare Figure 4D to Figure 3A,B). These data are also consistent with the behavior of MEKGOF and TorGOF mutant embryos, which activate Erk to a low, constant level away from the termini (Figure S1A-C) and induce segmentation and gastrulation phenotypes that are similar to those obtained by 30 or 45 min light pulses (Figure S3D-E; Figure S4B). We thus conclude that the decision to adopt posterior, contractile cell fates is primarily controlled by the overall dose (i.e. duration × time) of Erk signaling.
Distinct Erk target genes accumulate gradually or are triggered above a stimulus threshold
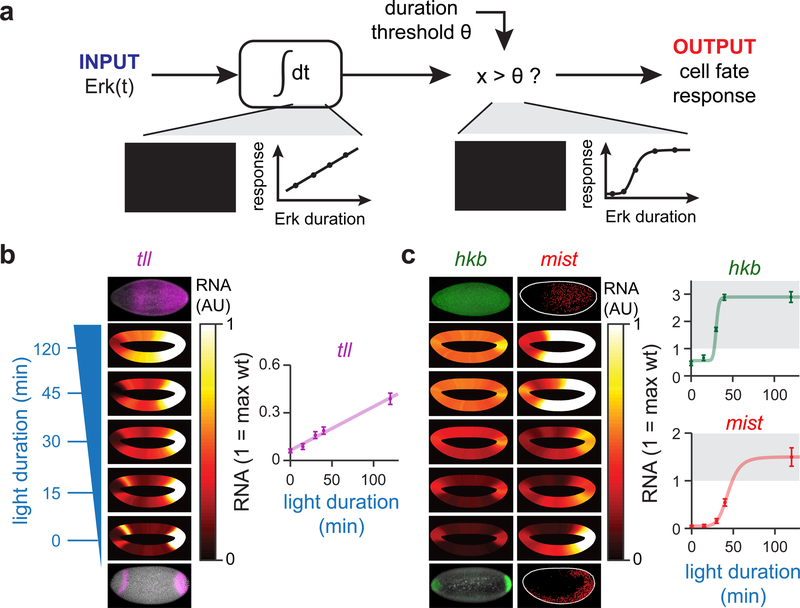
How might the cumulative load of a signal trigger an all-or-none cell fate switch? The simplest model requires two signal processing components, an ‘accumulator’ and ‘thresholder’ (Figure 5A). The accumulator component passively integrates Erk signal over time, providing memory of the total amount of Erk signaling that has been delivered. The thresholder would act downstream of the accumulator, comparing the accumulated signal to a fixed threshold and triggering a response only after the threshold is crossed. Such a model would predict two distinct behaviors among Erk-dependent genes: some might act as accumulators whose levels rise in linear proportion to the total light input, whereas others would act as thresholders that respond abruptly above a stimulus threshold.
Figure 5. Distinct target genes act as accumulators and thresholders of Erk activity.
(a) Model of cumulative load sensing by an accumulator/ thresholder circuit. An accumulator node would increase linearly in response to Erk activity until a critical threshold θ is reached, at which point a thresholder node would turn on in a switch-like fashion. (b,c) Analysis of RNA FISH data from OptoSOS embryos exposed to varying durations of blue light and stained for (b) tll and (c) hkb/mist. The mean expression levels around the surface the embryo is shown in each embryo-shaped heatmap; at least 50 embryos were analyzed for each light duration in b and c. The heatmaps are saturated so that the white color is equivalent to the maximum level seen in a wild-type embryo (full plots are shown in Figure S5). For the inset plots in c-d, the gene expression of cells at the dorsal-most cap of the embryo is plotted as a function of light duration, where a value of 1 is set to the maximum RNA signal seen in wild-type embryos.
To test if distinct target genes act as accumulators and thresholders of Erk signaling, we stained for tll, hkb and mist in response to different durations of light stimulation in at least 50 embryos per condition. We then quantified RNA levels around the circumference of each embryo in order to obtain a quantitative map of target gene induction at all anterior/posterior and dorsal/ventral coordinates (expression heat maps shown in Figure 5B,C, with line graphs plotted in Figure S5). To ascertain how gene expression varies with light dose, we analyzed a region on the dorsal surface at 50% embryo length (Figure 5B,C, right panels). This region was chosen because wild-type embryos exhibit no detectable Erk activity or target gene expression there, so all activity is solely a function of our light stimulus. We found that tll responded as would be predicted for an accumulator gene: its levels increased linearly as a function of the duration of Erk signaling (Figure 5B). In contrast, hkb and mist acted as threshold genes that turned on after 30 min and 45 min of signaling, respectively (Figure 5C; for comparison of linear and ultrasensitive model fits see Figure S5E,F). The difference we observe between tll and hkb correlates well with prior mutant data showing that in embryos with weak terminal signaling, hkb expression is lost before tll expression (Furriols, et al., 1996). The threshold duration at which mist is switched on (between 45 and 60 min) coincides exactly with the threshold for large-scale tissue contractility, consistent with mist’s essential role in this process.
Although our data reveals some candidate accumulator and thresholder nodes, a fully-defined transcriptional network for Erk dynamic interpretation remains to be elucidated. Other attractive candidates for these functions also exist, including the Erk-regulated transcriptional repressor Capicua (Cic). In the now-classic model of terminal signaling, Cic is phosphorylated by Erk, leading to Cic nuclear export and degradation followed by de-repression of tll and hkb (Jimenez, et al., 2000). It is possible that the gradual loss of Cic thus integrates the total Erk dose, triggering gene expression only after its removal. However, a few lines of evidence suggest that this is not the whole story. The gastrulation phenotype of cic-null embryos is less severe than that of illuminated OptoSOS embryos, suggesting that Erk drives further posteriorization independently of Cic. Also, staining for Cic protein in globally-illuminated OptoSOS embryos or GOF mutant embryos reveals that Cic is not fully degraded even in regions where tll and hkb expression are expressed at high, uniform levels (Johnson, et al., 2017; de las Heras and Casanova, 2006).
Discussion
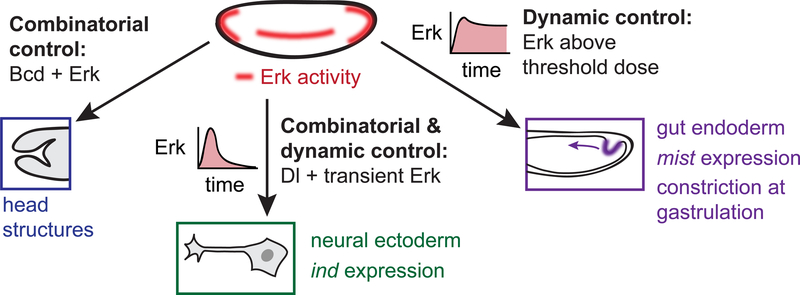
Through a combination of genetic perturbations and time-varying optogenetic stimuli, the current study begins to define a model for how Erk is capable of programming three distinct cell fates the early embryo (Figure 6). At the posterior, sustained Erk signaling induces gut endoderm, tissue that is characterized by expression of the Fog/Mist receptor-ligand pair which leads to apical constriction and tissue invagination. The boundary of this tissue is determined by the total Erk dose, and sustained Erk activity at almost any embryonic position is sufficient to trigger gut endoderm gene expression and contractility. A notable exception is the anterior pole, where the combination of Bcd and Erk switches cells to anterior fates. In the middle of the embryo, the combination of transient Erk and Dorsal normally induces the formation of ectoderm-derived neuroblasts, a fate that can be overridden by additional, ectopic Erk activity. By isolating the Erk pathway and titrating the inputs we deliver, we further show that some cells in the embryo can adopt three distinct responses at different signaling thresholds. Lateral cells shift from no response to form intermediate neuroblasts (characterized by ind expression) or gut endoderm (marked by mist expression and contractility) as a single input parameter – the duration of Erk signaling – is increased. These three transitions are reminiscent of the requirements that define a morphogen, a substance whose concentration determines multiple distinct fates (Gurdon, et al., 1998). Yet in the case of Erk it is signaling dynamics, not instantaneous concentration, which is interpreted into a cellular response.
Figure 6. A conceptual model of Erk-dependent cell fate control in the early embryo.
At the anterior pole, the presence of Bicoid prevents light-induced endoderm specification and tissue contraction, regardless of Erk dose. Along the ventral midline, transient Erk activity induces cells to adopt an intermediate neuroblast fate, provided that they are also exposed to intermediate Dorsal signaling. In the posterior, sustained Erk activity induces posterior midgut differentiation, contraction at gastrulation, and suppression of neurogenic fates. This model reveals that lateral cells may be switched between two fates, posterior midgut and neurogenic ectoderm – depending on their cumulative dose of Erk activity.
In contrast to the now-classic models for how Erk dynamics are decoded in cultured cells (Nakakuki, et al., 2010; Murphy, et al., 2004), we find that the early Drosophila embryo does not read the duration of a single persistent stimulus, but rather senses the cumulative load of Erk signaling. Two lines of evidence support this conclusion. First, the same overall Erk dose can be delivered in a single bolus or divided into discrete pulses spread out over a 2 h window, leading to the same effect. Second, long low-amplitude light stimuli (as well as gain-of-function mutants that activate Erk to low levels) achieve the same phenotypes as short high-amplitude light pulses. But does this distinction between duration and cumulative load sensing matter? We would argue that it is quite important, as the putative network architectures that perform these two signal processing functions can be quite different. Persistence detection is thought to rely on network motifs like the coherent feedforward loop (Mangan and Alon, 2003), whereas cumulative load detection can be implemented by combining long-term integration with an ultrasensitive downstream step. Obtaining the dynamic input-output response of a biological network can thus be a crucial first step towards a complete understanding of its network architecture and subsequent identification of molecular components (Mettetal, et al., 2008). Such insights are sorely needed, as we still broadly lack a mechanistic understanding of how signaling pathway activity is decoded into precise, reproducible patterns of gene expression.
There are still many unresolved questions regarding Erk-dependent cell fate choices in the early embryo. We have shown Bicoid is sufficient to prevent posterior fates at the anterior pole, but future work must be done to dissect how Erk dynamics interact with the Bicoid gradient to pattern the formation of different anterior structures. A complete picture of anterior fate choices may benefit from precise control over both Bicoid and Erk using multi-color optogenetics (Huang, et al., 2017). Moreover, the Erk-dependent terminal gap genes tll and hkb do much more than specify terminal fates, and participate in complex interactions with other gap and segmentation genes. Indeed, we find that brief 15–30 min light stimuli do not affect gastrulation but lead to abdominal segment fusion, suggesting that segmentation gene network is quite sensitive to perturbations in the spatiotemporal profile of Erk signaling. The use of light to deliver quantitative spatial and temporal perturbations to gap gene expression could prove instrumental for a deeper understanding of the segmentation circuit (Schroeder, et al., 2004).
The Ras/Erk pathway is only one of many signaling outputs from receptor tyrosine kinases (RTKs), and in general it remains an open question whether light-induced Erk fully reproduces all the nuances of receptor-level stimulation. To further probe this question in the early embryo, we tested whether OptoSOS stimulation recapitulates a classic genetic epistasis result: in embryos expressing a gain-of-function Torso RTK, loss of tll can suppress the TorGOF phenotype (Klingler, et al., 1988). Indeed, we found that the expected proportion of OptoSOS-tll treated with 45 min of light exhibit the tailless phenotype, not the TorGOF-like phenotype normally observed in OptoSOS embryos (Figure S4C; STAR Methods). Interestingly, suppression is lost at higher light doses, suggesting that sufficiently strong Erk activity can posteriorize embryos even in the absence of tll (Figure S4C). Looking forward, the recent development of light-controlled RTKs (Dine, et al., 2018; Grusch, et al., 2014; Kim, et al., 2014) opens the door to a full, systematic comparison between stimulation at the levels of the receptor versus Ras, and we eagerly await a quantitative comparison between these approaches.
The relationship between Erk dynamics and cellular responses has long been studied in cultured mammalian cells (Marshall, 1995; Bishop, et al., 1994). Here we find that principles of dynamic control also operate to control Erk-dependent cell fates in a developing organism. We would argue that the early Drosophila embryo is an ideal model system for dissecting dynamic control: cell fates specification occurs within 3 hours and is highly reproducible between embryos, gastrulation movements can be observed by brightfield microscopy and provide a spatially-localized readout of cell fate, the list of ‘downstream’ candidates for decoding dynamics is limited to a few dozen active zygotic genes in the early embryo (De Renzis, et al., 2007), and the combination of optogenetic and classical genetic tools enable complex perturbations of network components. Moreover, Erk signaling in the early embryo is likely to be only one of many examples of dynamic cell fate control in vivo. The approaches outlined here could prove useful in many additional contexts for dissecting how developmental cell fates are specified.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Jared Toettcher (toettcher@princeton.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila melanogaster stocks
Transgenic UAS-optoSOS flies were produced as described previously (Johnson et al., 2017) by ϕC31-based integration at either CH III 68A4 or CH II 25C6 and are driven maternally using P(mata-GAL-VP16)mat15 for all experiments (Hunter and Wieschaus, 2000). Other stocks used include BcdE1 (FBal0001080; a gift from Eric Wieschaus), TorD4021 (FBal0016921), Sqh-GFP (FBti0073027), and UAS MEK F53S (Goyal et al., 2017). A Histone-GFP stock (FBtp0012478) was used for all wild-type comparisons in co-immunostaining experiments (Figures 1F-H and 3C-D). The Sqh-GFP;optoSOS fly was generated by double balancing and crossing Sqh-GFP with optoSOS CHIII. These flies were then crossed to 67;15 to drive optoSOS maternally. To generate OptoSOS embryos lacking maternal Bicoid we crossed bcdE1 double-balanced flies with optoSOS CH II double-balanced flies. These flies were then crossed to 67;15 bcdE1 tsl to drive optoSOS in the presence of the bcdE1 mutant maternally. To generate optoSOS-tll flies we recombined optoSOS with tllL10 (FBal0016889), screening for the tll cuticle phenotype, these were then crossed with 67;15 to drive optoSOS, with 25% of the progeny being homozygous for the mutation.
METHOD DETAILS
Light illumination and light stimulation experiments
For all experiments carried out on bulk embryos (e.g. cuticle preparations), we used a custom-built panel of 30 individual 450 nm blue LEDs placed ~5 cm from the embryos and wrapped in foil. The light intensity at the sample location was measured with a MQ-510 Quantum light meter with separate sensor (Apogee Instruments) and determined to be approximately 1 mW / cm2 of 450 nm light. Note that for all staining experiments comparing wild-type and light-induced OptoSOS stimulation (e.g. embryos in Figures 1 and 3), we co-incubated Histone-GFP (for wild-type) and OptoSOS embryos under the same light source, stained them together, and analyzed them separately post facto based on whether or not they expressed GFP.
Microscopy
Cuticles were prepared for imaging (Johnson, et al., 2017) and imaged on Nikon Eclipse Ni at 10X objective using dark-field microscopy. In live fluorescence imaging experiments, embryos were dechorionated in 50% bleach for 2 min and rinsed in water before mounting and imaging. Microscopy of live and fixed fluorescent samples was performed using a Nikon A1 RS point-scanning confocal microscope (Princeton Confocal Microscopy Core).
DIC imaging was performed on our Nikon Eclipse Ti spinning-disk confocal microscope. A 740–760nm band-pass filter (Chroma) was placed in the brightfield illumination light path to prevent unwanted optogenetic stimulation during imaging. During times at which global stimulation was desired, the 750 nm band-pass filter was replaced with a 450 nm band-pass filter, and light intensity was adjusted to deliver ~1 mW / cm2 of 450 nm light. Light intensity was measured with a MQ-510 Quantum light meter with separate sensor (Apogee Instruments) placed at the sample location. Patterned optogenetic illumination was performed using a Mightex Polygon digital micromirror device using an X-Cite XLED 450-nm blue-light.
Immunostaining and Fluorescence In Situ Hybridization
Fluorescence in situ hybridization (FISH) and immunostaining was performed as described previously (Kosman et al., 2004). Primary antibodies used were rat anti-Fog (a gift from Eric Wieschaus), sheep anti-GFP (1:1,000, Bio-Rad), sheep anti-digoxigenin, (DIG) (1:125; Roche), and mouse anti-biotin (1:125; Jackson Immunoresearch). DAPI (1:10,000; Vector Laboratories) was used to stain for nuclei, and Alexa Fluor conjugates (1:500; Invitrogen) were used as secondary antibodies. For pairwise comparisons of wild-type and optogenetic stimulation, embryos were collected, stained, and imaged together under the same experimental conditions.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical details (e.g. whether standard deviation or standard error is shown, the number of samples and replicates) are included in the figure legend for the corresponding experiment.
Analysis of myosin timing
All analyses of myosin activity began from maximum intensity-projected images of the gastrulating embryo (representative images are shown in Figures 1B and S2D). We separately validated that the myosin increases occurred solely on the apical surface by examining the original, un-projected 3D Z-stacks. Additionally, axial projections of our 3D image stacks enabled us to clearly separate the basal myosin accumulation during cellularization from apical myosin during gastrulation (Figure S2E). All analyses were performed in MATLAB® (The MathWorks, Natick, Massachusetts) and code is available on GitHub.
For analysis of the spatial domain and timing of apical myosin, images of Sqh-GFP embryos were analyzed in a method similar to (Asokan, et al., 2015), adapted to the much larger size “puncta” that we observe on the embryo’s apical surface compared to migrating mammalian cells. We took advantage of the observation that myosin accumulation occurs in small, intense spots on the apical surface of cells, with regions of low myosin accumulation between these puncta (Figure 1C). This enabled us to filter regions of myosin accumulation from the surrounding ‘background’ by subtracting a 36 μm2 2-dimensional median filtered copy of the image, resulting in a background-subtracted image IMsub (Figure S2F; top panel). A mask of the embryo was also created by thresholding the original image, followed by morphological closing to fill ‘holes’ in the embryo and erosion to remove small non-embryo structures. Together, these resulted in a binary mask of the embryo IMmask.
We next sought to process IMmask to obtain a ‘ring’ of apical pixels around the embryo in which to analyze myosin intensity. We first eroded the posterior-most portion of IMmask to exclude the non-contractile germ cells while preserving the contractile posterior epithelium. We then performed morphological erosion on IMmask using a 30 μm-radius disk, and subtracted it from whole embryo to create a 30 μm-radius ‘ring’ around the embryo; Figure S2F; bottom left panel. The average myosin intensity in IMsub was measured in angular bins around this ring at each time-point, thus creating a “map” of myosin intensity as a function of time and angle around the embryo (e.g. 0° = anterior; 90° = dorsal; 180° = posterior; Figure S2F; top right panel).
We then logically compared the intensity of this myosin map to its mean intensity, so as to obtain a binary map which revealed the times and positions at which myosin accumulation exceeded the mean across the experiment (Figure S2F; bottom right panel). We set the time of the first appearance of apical myosin (at any position) as t = 0, and the time delay of the first appearance of myosin in the other angular bins was calculated relative to this first-appearance time. The result for five light-stimulated OptoSOS embryos is shown in Figure 1D.
We also used the binary map to determine the length of embryo which apical myosin was induced. To do so, we queried the binary map for the maximum embryo-length at which myosin was observed at both the dorsal and ventral surface. This analysis resulted in an estimated extent of constriction of 88.4 ± 0.3% embryo length.
Analysis of gene expression profiles
For analysis of the width of patterns in Figure 3D was measured in ImageJ (National Institutes of Health). For ind, a line was drawn in the across the pattern in the middle of the embryo and its width was calculated. An ind pattern width of zero was recorded if no pattern was observed. The distance of basal membrane from the edge of the embryo was used to estimate the age of the embryos; embryos were only analyzed if they shared widths with control embryos for which endogenous patterns were observed. To assess expansion of the mist pattern from the posterior pole, the length from posterior to anterior that mist was found on both sides the embryo in NC12–14 (in various D-V orientations) was calculated. The ImageJ “Remove Outliers” function was used to remove non-specific FISH staining aggregates in images for display purposes.
For the data in Figure 5B,C, maximum-projected z-stacks of images were collected for FISH images of tll, hkb, and mist in NC12–14 embryos. These maximum-projected images were normalized to non-specific background staining, background subtracted, and analyzed in MATLAB. A mask of each embryo was created by k-means clustering the intensity image, and taking the lowest bin to be background. Morphological closing and the clearing of small structures was performed on this mask to heal minor discontinuities. The posterior-most portion of the mask was eroded to exclude germ cells, so that only the epithelial monolayer around the embryo was subjected to further analysis. The intensity of RNA staining in 9 μm wide contour around the edge around the embryo was calculated to avoid autofluorescence in the yolk. Intensity was totaled in each contour bin and normalized to the endogenous pattern. Due to the punctate expression of mist in pre-gastrulation embryos, puncta were segmented using 2-D median filtering (10 μm2) and the number of puncta was counted, rather than using the intensity of mist staining directly. To obtain the dose response curves the middle dorsal surface bins of the contour (16% of the total contour length) were averaged and the average across all embryos was normalized to the average endogenous peak value.
Analysis of gastrulation phenotypes
From DIC movies of development, embryos were classified into three groups: global contraction, expanded PMG, and normal. Embryos were considered to exhibit “global contraction” when at least the posterior-most 40% of the embryo underwent contraction and the germ cells did not invaginate. “Expanded PMG” was classified by a lack of germband extension, or germ bands that retracted or stagnated immediately after PMG invagination, which was typically exaggerated in size (See Figure S3). Gastrulation was deemed “Normal” when both PMG invagination and germband extension occurred normally. Embryos which presented defects prior to gastrulation or were unfertilized were excluded from counts. For the bar graphs of Figures 3B and 4, embryos were included for analysis as long as light stimulus began at −140 min or later and was completed by −5 mins, where 0 min is taken as the time of gastrulation.
To confirm that short durations of light still had effects on embryos, such as those observed with GOF Mek mutations, we assayed cuticle phenotypes in response to a range of optogenetic stimuli and compared with mutants (Figure S4A & B). Indeed, optogenetic activation of Erk for as little as 10 minutes is capable of disrupting segmental patterning enough to induce cuticle fusions as are observed in the weaker GOF mutants, even though gastrulation is not significantly perturbed in either case. However, long durations of Erk result in a loss of segments entirely as we observed previously (Johnson, et al., 2017), presumably do to the corresponding disruption of gastrulation. This is similar to mutants which activate Erk to higher levels, consistent with a model where the cumulative load of Erk dictates fates.
Analysis of OptoSOS-tll phenotypes
For the analysis of cuticle phenotypes in Figure S4C, cuticles for each condition were classified into 3 groups, those which lacked segments, those which had 1–7 segments and lacked filzkörper (tll), and all others. The left hand side of the plot contains the expected values for tll, TorD4021 and TorD4021 in the presence of a tll LOF mutation. These values were calculated based on the assumption that Tll embryos should make up 25% of the population, with 4% of all embryos being unfertilized, there will be a small amount of empty cuticles. Likewise one would expect that this population would be the same population which does not lose all segments in the presence of TorD4021, with the remaining embryos having no segments.
Comparison of models fits to data of gene expression vs light duration
For the model fits shown in Figure 5B-C, we compared fits to both linear and ultrasensitive models by computing the sum-of-squared error of the best fit curves in each case. The results are presented in Figure S5E-F. For the ultrasensitive model, we used the model
where y is the measured gene expression, a and b are the amplitude and offset of the gene expression intensities, t represents the duration of light stimulus, t1/2 is the duration at which a half-maximal response is observed, and n is the Hill coefficient, representing the steepness of the response. For all fits we fixed n = 10, which we observed led to an excellent match for the switchlike reponses of mist and hkb gene expression. Our linear model was simply y = +at b, with a and b as the slope and offset and t the duration.
All fitting was performed in MATLAB using the lsqcurvefit function and a sum-of-squared-error (SSE) objective function. Example fits in the case of hkb gene expression are shown in Figure S5E, showing that a linear and first-order saturating model (which is identical to the ultrasensitive model but for n = 1) fit relatively poorly, whereas the ultrasensitive model fits well. The SSE for all linear and ultrasensitive fits is presented in Figure S5F, and the results from the best-fit model in each case is presented in bold.
DATA AND SOFTWARE AVAILABILITY
Analysis scripts are available at https://github.com/h-e-j/Devcell2019. Raw image data is available upon request.
Supplementary Material
Movie S1, Related to Figure 1. Time-lapse movie of sqh-GFP during normal gastrulation in an OptoSOS embryo that was only imaged for GFP fluorescence during cellularization and gastrulation, after the sensitive window for light-dependent contractility had ended.
Movie S2, Related to Figure 1. Time-lapse movie of sqh-GFP in an OptoSOS embryo that was subjected to continuous blue light illumination for 2 h prior to imaging.
Movie S3, Related to Figure 1. Time-lapse movie of sqh-GFP in an OptoSOS embryo which has been illuminated only in the indicated region (red stripe) for 1 h prior to the start of the movie.
Movie S4, Related to Figure 2. Time-lapse movies in which an OptoSOS embryo (top) and an OptoSOS / bcdE1 embryo (bottom) were each subjected to constant global blue light illumination. Images were captured using DIC imaging through a 750 nm long-pass filter.
Movie S5, Related to Figure 2. Time-lapse movie of an OptoSOS / bcdE1 embryo (bottom) subjected to constant global blue light illumination. The embryo ruptures in response Erk-triggered tissue contractility. Images were captured using DIC imaging through a 750 nm longpass filter.
Movie S6, Related to Figure 3. Comparison of gastrulation in embryos from TorD4021/+ mothers vs OptoSOS embryos exposed to 45 minutes of blue light prior to gastrulation. Images were captured using DIC imaging through a 750 nm long-pass filter.
Highlights.
Optogenetic Erk activation drives multiple differentiation and morphogenetic events
Light-activated Erk is sufficient to switch cells between endoderm and ectoderm fates
The total dose of Erk activity, not amplitude or duration, dictates cell responses
Target gene responses to Erk range from gradual accumulation to sharp activation
Acknowledgements
We thank all members of the Toettcher lab for helpful comments, as well as Stas Shvartsman and Eric Wieschaus for their suggestions and insights throughout the study. Yuji Yamazaki kindly provided the mist RNA probe. HEJ was supported by NIH Ruth Kirschstein fellowship F32GM119297. This work was also supported by NIH grant DP2EB024247 and NSF CAREER Award 1750663 (to J.E.T.). We also thank Dr. Gary Laevsky and the Molecular Biology Microscopy Core, which is a Nikon Center of Excellence, for microscopy support.
Footnotes
Declaration of Interests
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asokan SB, Johnson HE, Rahman A, King SJ, Rotty JD, Lebedeva IP, Haugh JM, Bear JE (2015). Mesenchymal chemotaxis requires selective inactivation of Myosin II at the leading edge via a non-canonical PLCγ/PKCα pathway. Developmental Cell 31, 747–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JM, Capobianco AJ, Doyle HJ, Finney RE, McMahon M, Robbins SM, Samuels ML, and Vetter M (1994). Proto-oncogenes and plasticity in cell signaling. Cold Spring Harbor Symposia on Quantitative Biology 59, 165–71. [DOI] [PubMed] [Google Scholar]
- Bugaj LJ, Sabnis AJ, Mitchell A, Garbarino JE, Toettcher JE, Bivona TG, and Lim WA (2018). Cancer mutations and targeted drugs can disrupt dynamic signal encoding by the Ras-Erk pathway. Science 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen FB, Budgett DM, Sun Y, Malpas S, McCormick D, and Freestone PS (2017). Pulse-Width Modulation of Optogenetic Photo-Stimulation Intensity for Application to Full-Implantable Light Sources. IEEE Transactions on Biomedical Circuits and Systems 11, 28–34. [DOI] [PubMed] [Google Scholar]
- Coppey M, Boettiger AN, Berezhkovskii AM, and Shvartsman SY (2008). Nuclear trapping shapes the terminal gradient in the Drosophila embryo. Current Biology 18, 915–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EA, Basu AS, and Bayer TS (2013). Programming microbes using pulse width modulation of optical signals. Journal of Molecular Biology 425, 4161–6. [DOI] [PubMed] [Google Scholar]
- Dawes-Hoang RE, Parmar KM, Christiansen AE, Phelps CB, Brand AH, and Wieschaus EF (2005). folded gastrulation, cell shape change and the control of myosin localization. Development 132, 4165–78. [DOI] [PubMed] [Google Scholar]
- de las Heras JM, and Casanova J (2006). Spatially distinct downregulation of Capicua repression and tailless activation by the Torso RTK pathway in the Drosophila embryo. Mech Dev 123, 481–6. [DOI] [PubMed] [Google Scholar]
- De Renzis S, Elemento O, Tavazoie S, and Wieschaus EF (2007). Unmasking activation of the zygotic genome using chromosomal deletions in the Drosophila embryo. PLoS biology 5, e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dine E, Gil AA, Uribe G, Brangwynne CP, and Toettcher JE (2018). Protein Phase Separation Provides Long-Term Memory of Transient Spatial Stimuli. Cell Systems 6, 655–63 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W, Siegel V, and Nusslein-Volhard C (1990). Autonomous determination of anterior structures in the early Drosophila embryo by the bicoid morphogen. Development 109, 811–20. [DOI] [PubMed] [Google Scholar]
- Furriols M, Sprenger F, and Casanova J (1996). Variation in the number of activated torso receptors correlates with differential gene expression. Development 122, 2313–7. [DOI] [PubMed] [Google Scholar]
- Gillies TE, Pargett M, Minguet M, Davies AE, and Albeck JG (2017). Linear Integration of ERK Activity Predominates over Persistence Detection in Fra-1 Regulation. Cell Systems 5, 549–563 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal Y, Jindal GA, Pelliccia JL, Yamaya K, Yeung E, Futran AS, Burdine RD, Schupbach T, and Shvartsman SY (2017). Divergent effects of intrinsically active MEK variants on developmental Ras signaling. Nature Genetics 49, 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm O, Sanchez VZ, Kim Y, Casanova J, Shvartsman SY, Wieschaus E (2012). Torso RTK controls Capicua degradation by changing its subcellular localization. Development 139, 3962–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grusch M, Schelch K, Riedler R, Reichhart E, Differ C, Berger W, Ingles-Prieto A, and Janovjak H (2014). Spatio-temporally precise activation of engineered receptor tyrosine kinases by light. The EMBO Journal 33, 1713–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi G, Barry JD, Huber W, and De Renzis S (2015). An Optogenetic Method to Modulate Cell Contractility during Tissue Morphogenesis. Developmental Cell 35, 646–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB, Dyson S, and St Johnston D (1998). Cells’ perception of position in a concentration gradient. Cell 95, 159–62. [DOI] [PubMed] [Google Scholar]
- Hannanta-Anan P, and Chow BY (2016). Optogenetic Control of Calcium Oscillation Waveform Defines NFAT as an Integrator of Calcium Load. Cell Systems 2, 283–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A, Amourda C, Zhang S, Tolwinski NS, and Saunders TE (2017). Decoding temporal interpretation of the morphogen Bicoid in the early Drosophila embryo. eLife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C, and Wieschaus E (2000). Regulated expression of nullo is required for the formation of distinct apical and basal adherens junctions in the Drosophila blastoderm. The Journal of Cell Biology 150, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imayoshi I, Isomura A, Harima Y, Kawaguchi K, Kori H, Miyachi H, Fujiwara T, Ishidate F, and Kageyama R (2013). Oscillatory control of factors determining multipotency and fate in mouse neural progenitors. Science 342, 1203–8. [DOI] [PubMed] [Google Scholar]
- Izquierdo E, Quinkler T, and De Renzis S (2018). Guided morphogenesis through optogenetic activation of Rho signalling during early Drosophila embryogenesis. Nature communications 9, 2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez G, Guichet A, Ephrussi A, and Casanova J (2000). Relief of gene repression by torso RTK signaling: role of capicua in Drosophila terminal and dorsoventral patterning. Genes & development 14, 224–31. [PMC free article] [PubMed] [Google Scholar]
- Johnson HE, Goyal Y, Pannucci NL, Schupbach T, Shvartsman SY, and Toettcher JE (2017). The Spatiotemporal Limits of Developmental Erk Signaling. Developmental Cell 40, 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N, Kim JM, Lee M, Kim CY, Chang KY, and Heo WD (2014). Spatiotemporal control of fibroblast growth factor receptor signals by blue light. Chemistry & biology 21, 90312. [DOI] [PubMed] [Google Scholar]
- Klingler M, Erdelyi M, Szabad J, and Nusslein-Volhard C (1988). Function of torso in determining the terminal anlagen of the Drosophila embryo. Nature 335, 275–7. [DOI] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E (2004). Multiplex detection of RNA expression in Drosophila embryos. Science 305, 846. [DOI] [PubMed] [Google Scholar]
- Lim B, Dsilva CJ, Levario TJ, Lu H, Schupbach T, Kevrekidis IG, and Shvartsman SY (2015). Dynamics of Inductive ERK Signaling in the Drosophila Embryo. Current Biology 25, 1784–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim B, Samper N, Lu H, Rushlow C, Jimenez G, and Shvartsman SY (2013). Kinetics of gene derepression by ERK signaling. Proceedings of the National Academy of Sciences of the United States of America 110, 10330–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, and Alon U (2003). Structure and function of the feed-forward loop network motif. Proceedings of the National Academy of Sciences of the United States of America 100, 11980–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AJ, Peters KA, Peifer M, and Rogers SL (2013). Regulation of epithelial morphogenesis by the G protein-coupled receptor mist and its ligand fog. Science signaling 6, ra98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CJ (1995). Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell 80, 179–85. [DOI] [PubMed] [Google Scholar]
- Martin AC, Kaschube M, and Wieschaus EF (2009). Pulsed contractions of an actin-myosin network drive apical constriction. Nature 457, 495–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettetal JT, Muzzey D, Gomez-Uribe C, and van Oudenaarden A (2008). The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science 319, 482–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy LO, MacKeigan JP, and Blenis J (2004). A network of immediate early gene products propagates subtle differences in mitogen-activated protein kinase signal amplitude and duration. Molecular and cellular biology 24, 144–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakakuki T, Birtwistle MR, Saeki Y, Yumoto N, Ide K, Nagashima T, Brusch L, Ogunnaike BA, Okada-Hatakeyama M, and Kholodenko BN (2010). Ligand-specific c-Fos expression emerges from the spatiotemporal control of ErbB network dynamics. Cell 141, 884–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M, Inoue Y, and Hayashi S (2007). A wave of EGFR signaling determines cell alignment and intercalation in the Drosophila tracheal placode. Development 134, 4273–82. [DOI] [PubMed] [Google Scholar]
- Purvis JE, and Lahav G (2013). Encoding and decoding cellular information through signaling dynamics. Cell 152, 945–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi N, Averbukh I, Haskel-Ittah M, Degani N, Schejter ED, Barkai N, and Shilo BZ (2016). A WntD-Dependent Integral Feedback Loop Attenuates Variability in Drosophila Toll Signaling. Developmental Cell 36, 401–14. [DOI] [PubMed] [Google Scholar]
- Reuter R, and Leptin M (1994). Interacting functions of snail, twist and huckebein during the early development of germ layers in Drosophila. Development 120, 1137–50. [DOI] [PubMed] [Google Scholar]
- Royou A, Sullivan W, and Karess R (2002). Cortical recruitment of nonmuscle myosin II in early syncytial Drosophila embryos: its role in nuclear axial expansion and its regulation by Cdc2 activity. The Journal of Cell Biology 158, 127–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder MD, Pearce M, Fak J, Fan H, Unnerstall U, Emberly E, Rajewsky N, Siggia ED, and Gaul U (2004). Transcriptional control in the segmentation gene network of Drosophila. PLoS Biology 2, E271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schupbach T, and Wieschaus E (1986). Maternal-effect mutations altering the anterior-posterior pattern of the Drosophila embryo. Roux Arch Dev Biol 195, 302–317. [DOI] [PubMed] [Google Scholar]
- Schweitzer R, Shaharabany M, Seger R, and Shilo BZ (1995). Secreted Spitz triggers the DER signaling pathway and is a limiting component in embryonic ventral ectoderm determination. Genes & Development 9, 1518–29. [DOI] [PubMed] [Google Scholar]
- Toettcher JE, Weiner OD, and Lim WA (2013). Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson MZ, Ravindran PT, Lim WA, and Toettcher JE (2017). Tracing Information Flow from Erk to Target Gene Induction Reveals Mechanisms of Dynamic and Combinatorial Control. Molecular Cell 67, 757–769 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Movie S1, Related to Figure 1. Time-lapse movie of sqh-GFP during normal gastrulation in an OptoSOS embryo that was only imaged for GFP fluorescence during cellularization and gastrulation, after the sensitive window for light-dependent contractility had ended.
Movie S2, Related to Figure 1. Time-lapse movie of sqh-GFP in an OptoSOS embryo that was subjected to continuous blue light illumination for 2 h prior to imaging.
Movie S3, Related to Figure 1. Time-lapse movie of sqh-GFP in an OptoSOS embryo which has been illuminated only in the indicated region (red stripe) for 1 h prior to the start of the movie.
Movie S4, Related to Figure 2. Time-lapse movies in which an OptoSOS embryo (top) and an OptoSOS / bcdE1 embryo (bottom) were each subjected to constant global blue light illumination. Images were captured using DIC imaging through a 750 nm long-pass filter.
Movie S5, Related to Figure 2. Time-lapse movie of an OptoSOS / bcdE1 embryo (bottom) subjected to constant global blue light illumination. The embryo ruptures in response Erk-triggered tissue contractility. Images were captured using DIC imaging through a 750 nm longpass filter.
Movie S6, Related to Figure 3. Comparison of gastrulation in embryos from TorD4021/+ mothers vs OptoSOS embryos exposed to 45 minutes of blue light prior to gastrulation. Images were captured using DIC imaging through a 750 nm long-pass filter.
Data Availability Statement
Analysis scripts are available at https://github.com/h-e-j/Devcell2019. Raw image data is available upon request.