Abstract
Heart formation involves a complex series of tissue rearrangements, during which regions of the developing organ expand, bend, converge, and protrude in order to create the specific shapes of important cardiac components. Much of this morphogenesis takes place while cardiac function is underway, with blood flowing through the rapidly contracting chambers. Fluid forces are therefore likely to influence the regulation of cardiac morphogenesis, but it is not yet clear how these biomechanical cues direct specific cellular behaviors. In recent years, the optical accessibility and genetic amenability of zebrafish embryos have facilitated unique opportunities to integrate the analysis of flow parameters with the molecular and cellular dynamics underlying cardiogenesis. Consequently, we are making progress toward a comprehensive view of the biomechanical regulation of cardiac chamber emergence, atrioventricular canal differentiation, and ventricular trabeculation. In this review, we highlight a series of studies in zebrafish that have provided new insight into how cardiac function can shape cardiac morphology, with a particular focus on how hemodynamics can impact cardiac cell behavior. Over the long term, this knowledge will undoubtedly guide our consideration of the potential causes of congenital heart disease.
Keywords: heart development, blood flow, cardiac chambers, atrioventricular canal, trabeculation
1. Introduction
Biomechanical forces are important regulators of embryonic morphogenesis. As early as gastrulation, circumferential contractions and flow-frictional mechanisms drive the spreading of the enveloping cell layer over the zebrafish yolk (Behrndt et al., 2012). Compressing and stretching the apical surface of cells induces apoptosis and proliferation, respectively, to shape the overall structure of the Drosophila wing (Diaz de la Loza & Thompson, 2017). The periodic spacing of feather follicles in chick is governed by competing interactions between cellular contractility and substrate stiffness (Shyer et al., 2017). In many contexts, physical cues provoke specific cellular behaviors that ultimately give rise to tissue morphology.
Forces are especially crucial during cardiac development, since blood flow and cardiac contractility produce distinct physical inputs while heart formation is underway (Freund et al., 2012; Haack & Abdelilah-Seyfried, 2016). It is particularly interesting to consider how flow-induced forces create spatiotemporally heterogeneous patterns that provide regionalized cues within the developing organ. Fluid frictional forces, for example, induce shear stress that deforms cells in the direction of blood flow. Blood pressure, on the other hand, causes cells to stretch circumferentially. Finally, the direction of blood flow can itself administer distinct mechanical cues to cells: reversing or retrograde flows, for instance, are thought to activate specific downstream pathways. Together, these fluid forces sculpt the heart, in a biological manifestation of “form follows function”.
How do fluid forces prompt cardiac cells to enlarge, elongate, divide, converge, and protrude as they create the specific architecture of the embryonic heart? Despite awareness of several mechanosensitive pathways that operate in endothelial cells (Baratchi et al., 2017), we do not yet fully understand the cellular and molecular mechanisms that drive cardiac morphogenesis in response to hemodynamic cues. To gain insights into these mechanisms, we need to be able to integrate in vivo assessment of fluid forces, high-resolution analysis of cell behaviors, and precise manipulation of mechanosensitive genes. The zebrafish embryo serves as an ideal model organism in this regard: its optical clarity allows real-time visualization of fluid dynamics as well as live imaging of morphogenesis at cellular resolution, and its genetic tractability permits both classical genetic screens and cutting-edge genome editing (Collins & Stainier, 2016; Li et al., 2016). Importantly, zebrafish offer the unique advantage that they can survive throughout embryogenesis without convective oxygen, allowing the analysis of cardiovascular defects without confounding lethality. Finally, although the zebrafish heart is anatomically simpler than the mammalian heart, the mechanosensory pathways operating during cardiac development appear to be highly conserved (Goddard et al., 2017). Together, these benefits of zebrafish have facilitated a number of investigations into the ways in which function influences form in the developing heart.
Here, we review a number of recent advances in our understanding of the impact of fluid forces on cardiac morphogenesis, with a particular focus on work performed in zebrafish. We first outline several commonly used techniques for quantification of flow parameters in the zebrafish vasculature and heart. We then highlight a series of studies that have illuminated how biomechanical inputs regulate the characteristic curvatures of the cardiac chambers, the distinctive features of the atrioventricular canal, and the elaborate network of the ventricular trabeculae. Overall, these insights emphasize the important connections between the mechanical, cellular, and molecular pathways that collaborate to construct the embryonic heart. Importantly, these studies may provide guidance for understanding the etiology of cardiac birth defects in humans, since they imply that fluctuations in blood flow in utero could have significant consequences on cardiac morphogenesis.
2. Examination of fluid forces in zebrafish
2.1. Techniques for quantification of fluid forces in the vasculature
To evaluate the influence of fluid forces on cardiac development, it is essential to be able to quantify these forces in vivo. Parameters such as shear stress and retrograde flow can then be correlated with patterns of cell behavior in order to create models for how mechanics instruct morphology. A number of techniques have been found to be useful for the assessment of fluid forces in the zebrafish heart, and several of these were first developed in the context of the zebrafish vasculature, where the size and simplicity of the major vessels facilitate live imaging of blood flow.
Digital motion analysis was an initial approach employed to visualize blood flow in zebrafish vessels (Schwerte & Pelster, 2000). Using a simple dissecting microscope and a CCD camera for video imaging, “difference images” can be created to track the motion of erythrocytes in consecutive video frames. Analysis of these images allows assessment of erythrocyte distribution, erythrocyte velocity and vessel diameter (Schwerte et al., 2003). Although easy to perform, a drawback of two-dimensional video microscopy is that slight variations in embryo orientation can significantly affect results; to some extent, this can be overcome by embedding larvae in tubular molds that are rotated to allow imaging from multiple angles (Bagatto & Burggren, 2006). Additionally, the use of high-speed cameras to follow erythrocytes expressing fluorescent reporter transgenes can substantially enhance the accuracy of cell tracking techniques (Fig. 1A-C) (Watkins et al., 2012).
Figure 1: Analysis of blood flow in the zebrafish.
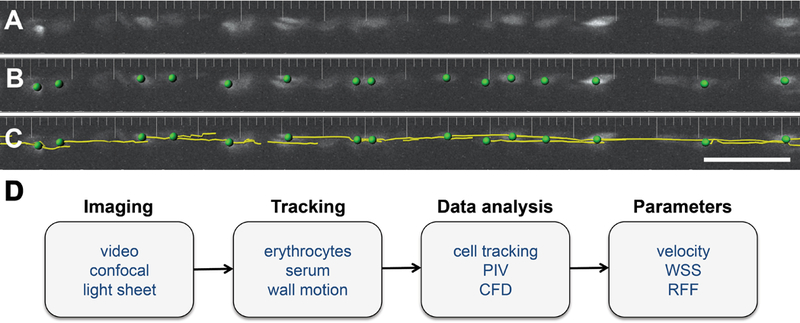
(A) Labeled erythrocytes in the dorsal aorta of an embryo carrying the transgene Tg(gata1:dsRed) are visualized at a single timepoint. (B) Image segmentation identifies individual erythrocytes (green dots). (C) A tracking algorithm reveals the trajectories (yellow lines) of the erythrocytes, allowing measurement of their displacement and estimation of their velocity. (D) Workflow options for analysis of blood flow in zebrafish. PIV, particle image velocimetry; CFD, computational fluid dynamics; WSS, wall shear stress; RFF, retrograde flow fraction. Panels A-C adapted from Watkins et al., 2012.
With the advent of particle image velocimetry (PIV), methods for evaluating flow parameters became even more rigorous. Unlike cell tracking techniques that follow only individual features, PIV averages velocity fields of sub-regions or “interrogation windows” within an image with a high signal-to-noise ratio (Yalcin et al., 2017). This information can be translated into estimates of wall shear stress when coupled with local area measurements, which can be obtained by simultaneously imaging vessel wall motion (Chen et al., 2011). PIV can be performed with both bright-field and fluorescent images, although confocal imaging of fluorescent dyes or transgene-expressing erythrocytes greatly facilitates subsequent image segmentation.
While PIV measurements can be used to estimate the distribution of physical forces, methods such as optical tweezing can directly measure the forces experienced by erythrocytes in vivo (Anton et al., 2013). In this technique, forces administered by a focused laser beam are used to hold an erythrocyte in position, such that the amount of force necessary allows assessment of the cell’s mechanical milieu. Optical tweezing can be especially useful at branch points in the vasculature, where sudden changes in the direction of erythrocyte movement can be difficult to track.
Additional techniques have been utilized to image blood flow in the zebrafish vasculature, including fluorescence correlation microscopy (Pan et al., 2007) and optical coherent tomography (Iftimia et al., 2008). Fluorescence correlation microscopy utilizes the inherent autofluorescence of erythrocytes and blood vessels to determine erythrocyte velocity, sidestepping the need to introduce fluorescent tracers. Similarly, optical coherent tomography measures backscattered and back-reflected light to image fluid flow in real time. Although these techniques are beneficial, they require specialized instruments that may not be readily accessible at all research institutions. Consequently, video imaging and confocal imaging, followed by cell tracking or PIV, remain the prevalent methods for assessing flow parameters in zebrafish vessels.
2.2. Techniques for analysis of flow parameters in the heart
The embryonic zebrafish heart is a morphologically elaborate structure that functions to propel blood from the atrium into the ventricle before finally supplying circulation to the aortic vessels. Owing to its particular architecture, imaging blood flow in the heart presents unique technical challenges distinct from those encountered in the vasculature. For example, the dimensions of the cardiac chambers provide many opportunities for erythrocytes to move in and out of the plane of focus. The motion of the chambers creates further complexity, since the chamber wall has to be properly segmented during image analysis. Moreover, the structure of the heart, coupled with the sequential contraction of the chambers, produces complex flow patterns featuring vortical and reversing flows. Nevertheless, techniques developed for the zebrafish vasculature have been successfully adapted for use in the embryonic heart.
As in the vasculature, PIV has been the method of choice to quantify wall shear stress and other flow parameters during cardiac development. Early applications of this technique used a fluorescent contrast agent to visualize serum motion and demonstrated vortical patterns of flow within the heart (Hove et al., 2003). Although still used widely, PIV has certain drawbacks when applied in the heart. For example, when utilizing video imaging to collect data for PIV analysis, three-dimensional information has to be projected in two dimensions, which results in underestimation of the distance traveled by erythrocytes (Hove et al., 2003). Techniques such as defocusing digital particle image velocimetry, which employ microscopes with two or more apertures to detect out-of-focus particles, can enable more precise tracking of intracardiac flows (Lu et al., 2008). More recently, the depth sectioning capabilities of light sheet microscopy have also been exploited for PIV analysis (Zickus & Taylor, 2018). Another complication associated with PIV measurements in the heart is the interference of cardiac wall motion with erythrocyte tracking, especially in bright-field images. To overcome this, a local signal-to-noise ratio can be used to achieve “cardiac-phase filtering”, which effectively removes artifacts from cardiac wall motion post-image acquisition (Jamison et al., 2013). Since this method identifies the position of the cardiac wall, it enables assessment of wall shear stress in addition to more accurate PIV calculations.
The development of faster imaging techniques has also been valuable for following the complex patterns of erythrocyte motion within the heart. Using a confocal laser slit scanning system, which executes line-wise illumination for faster imaging, significant retrograde flows were observed in the zebrafish heart, both during atrial contraction back into the sinus venosus, and during ventricular contraction back into the atrium (Liebling et al., 2006). Subsequent analysis with high-speed bright-field video microscopy confirmed that the region between the ventricle and the atrium, the atrioventricular canal, experiences a heightened retrograde flow fraction, which is defined as the fraction of the cardiac cycle exhibiting flow reversal (Vermot et al., 2009). These studies have motivated an interest in retrograde flow as a critical signal for atrioventricular canal differentiation, which we will discuss further in a subsequent section.
The emergence of additional strategies for image analysis, such as computational fluid dynamics (CFD), marks a new era for assessment of fluid forces in the zebrafish heart (Yalcin et al., 2017). In computational fluid dynamics, flow parameters are inferred from tracking both chamber wall motions and inlet velocities, which bypasses the requirement for high-resolution imaging as in PIV. Use of computational fluid dynamics in the zebrafish heart has allowed correlation of certain flow characteristics with specific morphogenetic processes (Lee et al., 2013). For example, mean peak shear stress increases significantly during chamber emergence and atrioventricular canal differentiation, suggesting an influence of shear forces on these steps of heart morphogenesis.
Altogether, several options are readily available for analysis of flow parameters in the zebrafish heart (Fig. 1D). Using video, confocal, or light sheet imaging to track erythrocytes, serum, or chamber walls, investigators can use cell tracking, PIV, or computational fluid dynamics approaches to assess erythrocyte velocity, wall shear stress, and retrograde flow fraction. This variety of strategies opens the door to connecting fluid forces to the regulation of a number of specific processes during cardiac morphogenesis, as discussed in the following sections.
3. Hemodynamics and cardiovascular morphogenesis
3.1. Fluid forces regulate multiple aspects of vessel development
When considering how fluid forces influence cardiac morphology, it is instructive to reflect upon our understanding of the influence of flow on vessel development. The impact of physical forces on endothelial cells in the vasculature is especially relevant to the potential roles of such forces on the endocardium, the specialized inner layer of endothelium that lines the muscular myocardial layer of the heart. Since the biomechanical regulation of vascular morphogenesis has been reviewed elsewhere (Baratchi et al., 2017; Boselli et al., 2015), this section will simply provide a few recent perspectives on how processes like lumen growth and vessel remodeling respond to fluid forces in zebrafish.
Blood flow has been shown to regulate vessel diameter through its influence on several types of endothelial cell behaviors. For example, Endoglin, a TGF-β receptor, modulates endothelial cell shape in response to hemodynamic cues, thereby restricting the diameters of the dorsal aorta and posterior cardinal vein (Sugden et al., 2017). In the caudal vein, shear forces lead to an increase in endothelial cell number via mechanosensation by Pkd2/Trpp2, a calcium channel present on endothelial cilia (Goetz et al., 2014). In the cranial vasculature, flow-induced expression of the TGF-β receptor Alk1 regulates the migration of endothelial cells in the direction opposite to blood flow, thereby limiting vessel caliber (Corti et al., 2011; Rochon et al., 2016). Finally, Yap1, a Hippo pathway effector, localizes to the nucleus in a flow-dependent manner in the dorsal longitudinal anastomotic vessel, where it regulates the maintenance of lumen size (Nakajima et al., 2017).
Fluid forces are also critical for the morphogenetic processes that are involved in vessel remodeling. For instance, flow regulates the apical membrane invagination that is required for successful tube formation during vessel fusion (Herwig et al., 2011). Blood flow is also important for the establishment of planar cell polarity in endothelial cells (Kwon et al., 2016), which could influence vessel regression, as has been predicted in mice (Franco et al., 2015). Indeed, vessel regression in the zebrafish eye (Kochhan et al., 2013) and midbrain (Chen et al., 2012) have been shown to occur in response to blood flow, potentially via flow-mediated cell rearrangement and migration.
Together, these studies underscore the importance of fluid forces during multiple aspects of vascular morphogenesis and provide inspiration for the ways in which blood flow could influence cell shape, cell number, migration, and polarity during heart development. In the following sections, we will examine the impact of cardiac function and fluid forces on cell behavior during three essential phases of cardiac morphogenesis: chamber emergence, atrioventricular canal differentiation, and ventricular trabeculation.
3.2. Chamber emergence requires hemodynamic inputs
In the early embryo, the primitive heart is a simple, relatively linear cylinder positioned at the embryonic midline. As development proceeds, the heart tube enlarges and transforms into a morphologically distinct series of cardiac chambers, each expanding into its characteristic curvatures (Collins & Stainier, 2016). Since chamber emergence takes place while the heart is beating and the blood is flowing, biomechanical inputs generated by cardiac function have the potential to influence this process. Indeed, surgical obstruction of blood flow into the embryonic chick heart results in aberrant chamber morphology (Broekhuizen et al., 1999). Mouse mutants lacking atrial contractility due to a mutation in atrial myosin light chain 2a exhibit abnormal ventricular morphogenesis (Huang et al., 2003), further supporting a connection between blood flow and cardiac chamber development. Taking advantage of the opportunities for examining chamber emergence in the context of the zebrafish embryo, a number of studies have delved deeper into the cell behaviors that shape the chambers in response to biomechanical cues.
In zebrafish, as the ventricle and atrium emerge from the linear heart tube, distinct convex and concave surfaces form the outer and inner curvatures of each chamber. These tissue shape changes are associated with regional patterns of cell shape change: in the ventricle, for example, outer curvature cardiomocytes enlarge and elongate during chamber emergence, while inner curvature cardiomyocytes remain relatively small and round (Auman et al., 2007). Analysis of cardiomyocyte morphology in mutant embryos has suggested that cell shape change at the ventricular outer curvature depends upon inputs generated by cardiac function. In weak atrium (amhc) mutants, which lack atrial contractility and therefore have reduced blood flow into the ventricle, outer curvature cardiomyocytes are smaller and rounder than in the wild-type ventricle (Fig. 2A,B) (Auman et al., 2007); these cells also exhibit diminished myofibril maturation (Lin et al., 2012). In contrast, half-hearted (vmhc) mutants, which lack ventricular contractility, display excessively enlarged and elongated cells in the ventricular outer curvature (Fig. 2C,D) (Auman et al., 2007). These studies suggest that forces produced by both blood flow and contractility modulate cardiomyocyte shape and size and therefore contribute to the regulation of chamber emergence.
Figure 2: Cardiac function modulates cardiomyocyte dimensions in the ventricle.
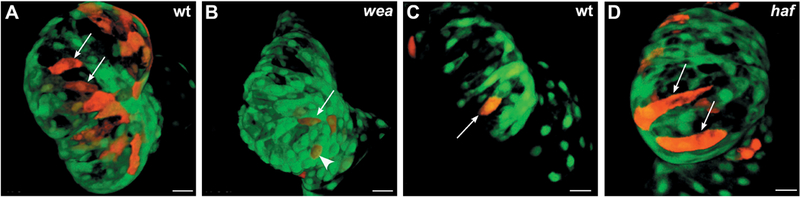
Confocal projections show the ventricle in embryos carrying the transgene Tg(myl7:egfp), with mosaic expression of Tg(myl7:dsRedt4). (A,C) At 52 hours post-fertilization (hpf), cardiomyocytes in the outer curvature of the ventricle (arrows) have acquired a characteristic size and an elongated morphology. (B) In weak atrium (wea) mutants, blood flow into the ventricle is reduced, and the cardiomyocytes in the outer curvature are abnormally small (arrow) and circular (arrowhead). (D) In half-hearted (haf) mutants, the ventricle is non-contractile, and the outer curvature cardiomyocytes are abnormally large and distended (arrows). Images adapted from Auman et al., 2007.
In conjunction with the outward expansion of the myocardium, the inner endocardial layer of the heart also grows as the chambers emerge. Cellular proliferation drives this endocardial expansion (Fig. 3A,E,F), while local differences in endocardial cell morphologies correlate with the curved chamber contours (Dietrich et al., 2014). Interestingly, in conditions where fluid forces are reduced, proliferation of the ventricular endocardium is markedly decreased (Fig. 3B-F) (Dietrich et al., 2014). Endocardial cells also fail to acquire normal morphologies under these circumstances. Together, these results suggest a model in which fluid forces trigger proliferation of endocardial cells, causing an overall enlargement of the endocardium that provokes cell shape changes within the myocardium, thereby facilitating synchronous expansion of both cardiac layers.
Figure 3: Fluid forces influence endocardial cell number during chamber emergence.
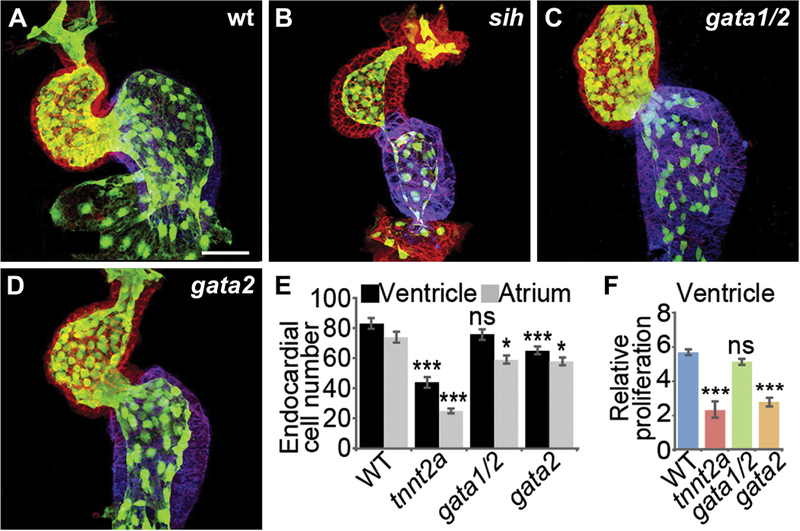
(A-D) Endocardial cells (green) are indicated by expression of the transgene Tg(kdrl:GFP); localization of Amhc (blue) and actin (red) are also shown. Whereas wild-type embryos have ~80 and ~70 endocardial cells in the ventricle and the atrium, respectively (A,E), the number of endocardial cells in both chambers is significantly reduced in silent heart (sih; tnnt2a) mutants (B,E), in which the heart is noncontractile. gata1/2 morphants, which have diminished shear forces due to their reduced hematocrit, display fewer endocardial cells in the atrium (C,E). In contrast, gata2 morphants, which have been shown to have reduced retrograde flow in addition to diminished shear forces, display fewer endocardial cells in both chambers (D,E), similar to sih mutants. (F) Consistent with this, endocardial proliferation in the ventricle is significantly reduced in sih mutants and gata2 morphants, but not in gata1/2 morphants. Images adapted from Dietrich et al., 2014.
Which molecular pathways might drive chamber emergence in response to fluid forces? Exposure of endothelial cells to shear stress in vitro induces the expression of KLF2, which encodes a mechanoresponsive transcription factor (Dekker et al., 2002), and the expression of the zebrafish homolog klf2a also appears to be flow-responsive (Vermot et al., 2009). The klf2 pathway is thought to contribute to the regulation of chamber emergence, since klf2a morphants exhibit aberrant endocardial morphology in the ventricle (Dietrich et al., 2014). Micro-RNAs are also interesting candidates for mechanoresponsive functions, since they can be regulated rapidly in response to flow (Banjo et al., 2013). Indeed, function-dependent expression of miR-143 in both the myocardium and the endocardium impacts the process of ventricular emergence (Miyasaka et al., 2011). In the myocardium, miR-143 targets adducin3, which regulates cytoskeletal actin dynamics and can thereby influence cardiomyocyte morphology (Deacon et al., 2010). In the endocardium, miR-143 limits retinoic acid signaling by directly targeting raldh2 and rxrab (Miyasaka et al., 2011). Intriguingly, miR-143 morphants exhibit gaps in the ventricular endocardium (Miyasaka et al., 2011), hinting at defects in endocardial cell number. In future studies, it will be valuable to identify not only additional mechanosensitive genes that are relevant to chamber emergence but also additional downstream effectors responsible for executing the patterns of proliferation in the endocardium and cell shape change in the myocardium.
3.3. Retrograde flow drives atrioventricular canal differentiation
As the cardiac chambers emerge, the junction between the atrium and the ventricle constricts to form the atrioventricular canal (AVC). Within this region, distinct differentiation pathways create the endocardial cushions, specialized structures that will subsequently remodel to create the atrioventricular valve. Endocardial cushion formation has been examined with cellular precision in zebrafish, owing to the optical accessibility of the embryo. At early stages, endocardial cells accumulate both through proliferation and via convergence to the AVC region (Fig. 4A-C) (Boselli et al., 2017; Steed et al., 2016), while also acquiring distinctive cuboidal morphologies (Beis et al., 2005; Steed et al., 2016). Following this, the endocardial cells invaginate as a sheet into the extracellular matrix, ultimately forming valve leaflets (Pestel et al., 2016).
Figure 4: Cardiac function regulates endocardial convergence at the AVC.
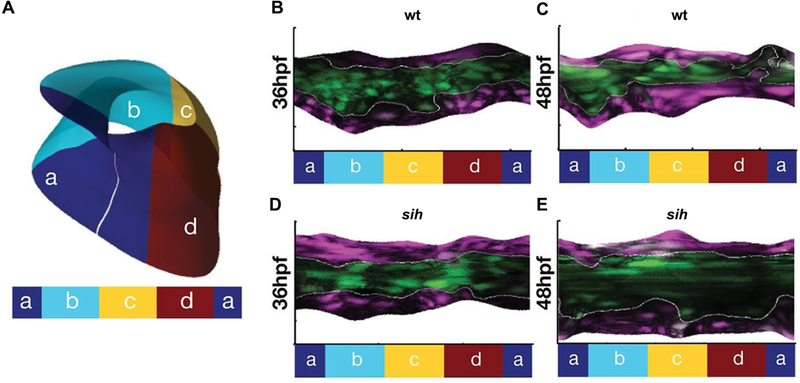
(A) The AVC endocardium can be subdivided into four regions: superior (a), exterior (b), inferior (c) and interior (d). (B-E) Representations of the unfolded endocardium in Tg(fli:kaede) embryos in which the AVC endocardium has been labeled via photoconversion. In wild-type embryos, the AVC endocardial cells converge between 36 hpf (B) and 48 hpf (C). In contrast, in sih mutants, the AVC endocardium widens between 36 hpf (D) and 48 hpf (E). Images adapted from Boselli et al., 2017.
Early work in chick had shown that impediment of mechanical forces in the heart, via surgical alteration, can result in abnormal morphology of the atrioventricular valve (Sedmera et al., 1999). Following this, several studies in zebrafish have indicated that cardiac function provides essential cues for endocardial cushion formation. Endocardial cushions fail to form in silent heart mutants and cardiofunk mutants, both of which have defects in cardiac contractility (Bartman et al., 2004; Beis et al., 2005). Similarly, physical occlusion of blood flow into the zebrafish heart impairs atrioventricular valve development (Hove et al., 2003).
The oscillatory component of wall shear stress, or retrograde flow, is known to be particularly elevated within the AVC (Heckel et al., 2015), suggesting that the magnitude and the direction of fluid forces could influence AVC differentiation. To examine the effects of these types of mechanical inputs, the phenotypes generated by knocking down gata1 and gata2, genes involved in hematopoiesis, were examined (Vermot et al., 2009). Since both genes contribute to erythrocyte development, knockdown of either should reduce shear forces in the heart. In addition, gata2 morphants display a reduction in retrograde flow fraction (RFF) in the AVC, whereas gata1 morphants display an increased RFF. Furthermore, gata2 morphants fail to develop valve leaflets, whereas gata1 morphants appear strikingly normal. Likewise, valve formation fails in weak atrium mutants, which also exhibit a reduced RFF in the AVC (Kalogirou et al., 2014). Notably, the valve defects in gata2 morphants are preceded by a failure to accumulate endocardial cells at the AVC, as well as irregular cellular morphologies (Vermot et al., 2009). The former is consistent with the observation that endocardial convergence at the AVC fails in silent heart mutants (Fig. 4B-E) (Boselli et al., 2017). Together, these findings imply that retrograde flow, and not shear, is the driving force for atrioventricular valve formation, potentially through its influence on cellular rearrangement during AVC differentiation.
Investigation of the molecular pathways triggered by retrograde flow in the AVC has primarily centered on klf2a. Expression of klf2a in the AVC endocardium is reduced in gata2 morphants (Heckel et al., 2015; Vermot et al., 2009), and klf2a morphants resemble gata2 morphants in that their AVC endocardial cells are reduced in number as well as abnormally elongated and flat (Vermot et al., 2009). Moreover, the calcium channels Trpp2 and Trpv4 appear to regulate klf2a expression in response to oscillatory flow patterns within the AVC endocardium (Heckel et al., 2015). Downstream effectors of Klf2 in the AVC endocardium include the extracellular matrix component Fibronectin: fibronectin1b morphants have defective cell clustering at the AVC, similar to klf2a morphants and gata2 morphants (Steed et al., 2016). In addition, wnt9b acts a paracrine factor downstream of klf2a during AVC differentiation (Goddard et al., 2017).
Given the importance of klf2a to mechanosensation at the AVC, it is reasonable to imagine that the levels of klf2a expression must be carefully controlled. In accordance with this, the cerebral cavernous malformation (CCM) proteins Ccm1 and Ccm2 have been shown to restrict klf2a expression, with Ccm2 acting in a manner independent of cardiac function (Renz et al., 2015). Interestingly, the expression of Heg1, which acts to stabilize Ccm1, appears to be dependent on both cardiac function and klf2a, suggesting that klf2a acts in a negative feedback loop to influence its own expression (Donat et al., 2018). Altogether, studies of the factors upstream and downstream of klf2a place it as a key node in a retrograde flow-regulated pathway that drives AVC differentiation.
When considering additional factors that may act in or parallel to the klf2a pathway in this context, it is again interesting to examine the possible roles of micro-RNAs. For example, miR-21 is expressed in the AVC in a function-dependent manner, and miR-21 morphants fail to exhibit normal atrioventricular valve formation (Banjo et al., 2013). miR-21 is thought to activate the MAP kinase cascade by targeting sprouty2, suggesting a possible mechanism for its influence on endocardial cell behavior during AVC differentiation. Further work will be necessary to evaluate whether these and/or other players interface with the klf2a pathway in executing the biomechanical regulation of AVC differentiation by retrograde flow.
3.4. Functional regulation of ventricular trabeculation
Following cardiac chamber emergence, the architecture of the ventricle becomes more elaborate through the formation of trabeculae – myocardial ridges that extend into the ventricular lumen. Analyses in zebrafish have shown that trabeculation is driven by the directional migration of cells that delaminate from the compact layer of the myocardium (Liu et al., 2010; Staudt et al., 2014). This delamination process involves constriction of the abluminal cardiomyocyte surface and is dependent upon the proper apicobasal polarization of the myocardial tissue (Jimenez-Amilburu et al., 2016; Staudt et al., 2014). In a noncontractile zebrafish heart, trabeculation fails: in silent heart morphants, for example, cardiomyocytes still extend protrusions into the ventricular lumen, but these protrusions frequently retract and stable trabeculae do not form (Staudt et al., 2014). These findings indicate that cardiac function is important for regulating trabeculation, consistent with prior observations in chick, where ventricular afterload has been implicated in regulating the thickening of the compact and trabecular layers (Sedmera et al., 1999).
Do fluid forces play a key role in controlling the cell behaviors that underlie trabeculation? Indeed, reduction of blood flow in both weak atrium mutants and gata1 morphants inhibits trabeculation (Fig. 5A,C,D) (Lee et al., 2016; Peshkovsky et al., 2011; Staudt et al., 2014; Vedula et al., 2017). Furthermore, studies in weak atrium mutants and morphants suggest that both the initial formation of luminal protrusions and their progression into stable trabeculae could be sensitive to the patterns of blood flow through the ventricle (Peshkovsky et al., 2011; Staudt et al., 2014). Flow parameters at the onset of ventricular trabeculation have not yet been carefully examined; however, the oscillatory shear index has been shown to be higher in trabecular grooves and lower in trabecular ridges at 4 days post-fertilization, after mature trabeculae are in place, and this distinct pattern is perturbed when trabeculation is inhibited (Fig. 5) (Vedula et al., 2017). In future studies, it will be valuable to investigate flow patterns in the ventricle at earlier stages in order to discern whether parameters like the oscillatory shear index can predict where trabecular protrusions will form.
Figure 5: Interplay between fluid forces and ventricular trabeculation.
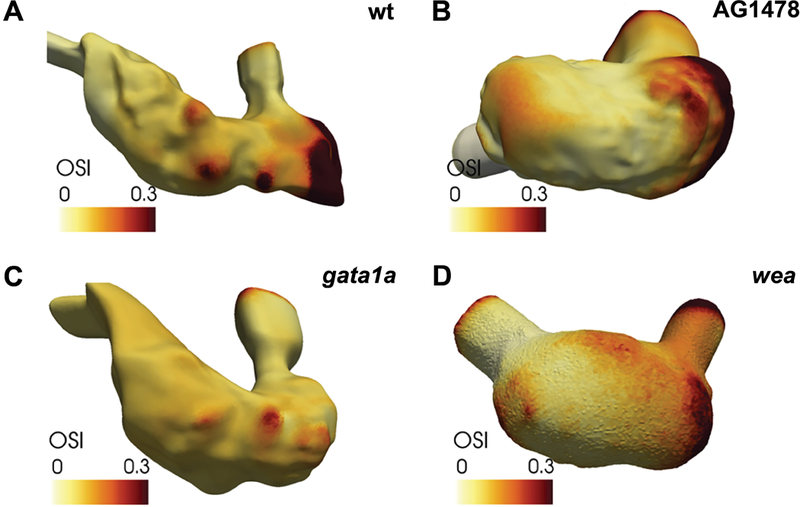
Representations of the inner surface of the ventricle, overlaid with a depiction of the oscillatory shear index (OSI) at 4 dpf. (A) In wild-type, the OSI is relatively high in trabecular grooves and relatively low in trabecular ridges. (B) In AG1478-treated embryos, the Neuregulin signaling pathway is inhibited, resulting in reduced trabeculation and a smoother inner surface of the ventricle. A similar reduction of trabeculation is observed in gata1a morphants (C) and wea mutants (D). Images adapted from Vedula et al., 2017.
Several signaling pathways that are known to be important regulators of trabeculation are also responsive to biomechanical cues. For example, the Neuregulin signaling pathway, operating through the Erbb2 receptor tyrosine kinase, is required for trabeculation (Liu et al., 2010), and expression of both erbb2 (Lee et al., 2016) and its ligand nrg2a (Rasouli & Stainier, 2017) have been shown to be dependent on cardiac function. Function-mediated Notch signaling activity in the endocardium has also been implicated in trabeculation, potentially via its role in regulating expression of the Erbb2 ligand nrg1 (Samsa et al., 2015), as has been suggested in mice (Grego-Bessa et al., 2007). Wwtr1, a Hippo pathway effector that localizes to the nucleus in response to function, is an essential regulator of the compact wall architecture supporting trabeculation, where it can also influence myocardial Notch signaling (Lai et al., 2018). Furthermore, Erbb2 signaling can direct Wwtr1 localization (Lai et al., 2018) and Notch signaling in the myocardium (Jimenez-Amilburu et al., 2016). Finally, cardiac function, Erbb2, Wwtr1, and Notch have all independently been shown to be important for the relocalization of N-cadherin in delaminating cardiomyocytes (Cherian et al., 2016; Han et al., 2016; Lai et al., 2018). Collectively, these data signify complex spatiotemporal interactions between cardiac function and the Erbb2, Notch, and Hippo signaling pathways during the modulation of trabeculation. Ongoing studies will continue to illuminate the precise nature of these interactions, while also deciphering how these pathways instruct cell behaviors as trabeculae form.
4. Summary and future directions
Recent studies in zebrafish have amply demonstrated the broad utility of this model organism for analysis of the biomechanical regulation of cardiac development. The exceptional optical access to the developing heart, coupled with a variety of options for manipulation of gene function, has illuminated multiple morphogenetic processes that are dependent upon cardiac function. Moreover, scenarios in which blood flow is altered have provided intriguing evidence linking fluid forces to several key steps in cardiac maturation. Finally, further genetic analyses have created an emerging framework of the mechanoresponsive pathways that orchestrate important patterns of cell behavior in the embryonic heart (Fig. 6).
Figure 6: Mechanoresponsive pathways regulating cardiac development in zebrafish.
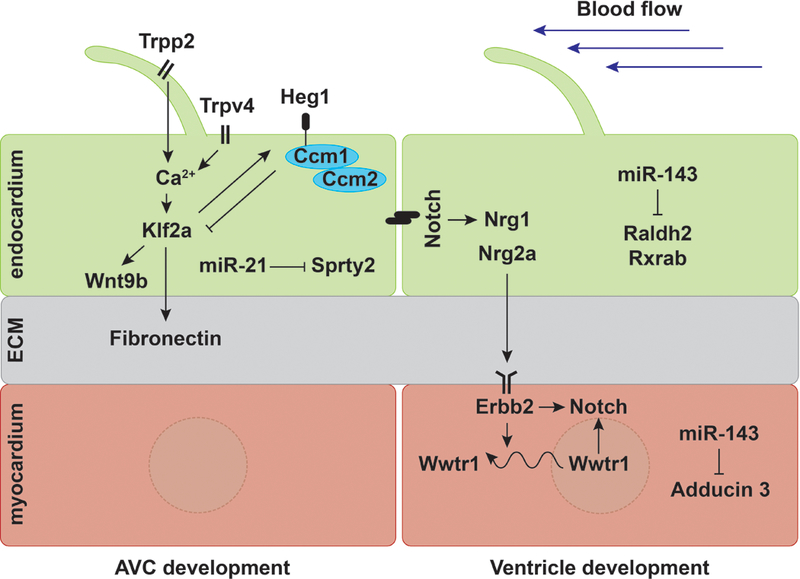
During AVC development, oscillatory flow activates mechanosensitive calcium channels Trpp2 and Trpv4 in the endocardium, which then promote klf2a expression (Heckel et al., 2015). In addition, flow-induced Heg1 represses klf2a expression in conjunction with the CCM complex, establishing a feedback loop for klf2a (Donat et al., 2018; Renz et al., 2015). Klf2a, in turn, regulates wnt9b expression in the endocardium (Goddard et al., 2017) and Fibronectin deposition in the extracellular matrix (ECM) (Steed et al., 2016). At the same time, flow-induced miR-21 represses receptor tyrosine kinase signaling by targeting sprty2 (Banjo et al., 2013). Meanwhile, during ventricle emergence, function-dependent miR-143 inhibits retinoic acid signaling in the endocardium (Miyasaka et al., 2011) and targets Adducin3 in the myocardium (Deacon et al., 2010). Mechanical forces also activate Notch signaling in the endocardium, which further induces expression of the Erbb2 ligand nrg1 (Samsa et al., 2015). During trabeculation, cardiac function regulates nrg2a (Rasouli and Stainier, 2017) and erbb2 expression (Lee et al., 2016), as well as Wwtr1 localization (Lai et al., 2018). The Nrg/Erbb2 pathway can further affect Notch signaling in cardiomyocytes (Jiménez-Amilburu et al., 2016) and regulate Wwtr1 localization, which can also activate Notch signaling (Lai et al., 2018).
Despite these recent advances, there are still substantial challenges inherent to studying the relationship between fluid forces and cardiac morphogenesis. Importantly, common strategies for interfering with cardiac function often alter several biomechanical variables simultaneously. Inhibition of cardiac contractility, for example, changes both the stretch of cardiac tissues and the various forces associated with blood flow. In addition, manipulations that alter circulation while still maintaining contractility can perturb both the velocity and the direction of flow. Hence, most conclusions reached thus far are based on correlating particular functional scenarios with interesting phenotypes, rather than demonstrating which specific types of fluid forces cause which cellular outcomes. To evaluate the effects of distinct biomechanical cues, it will be instrumental for future studies to employ techniques that administer specific forces in a targeted fashion. Applications of single molecule spectroscopy could be adapted for this purpose: optical tweezing, for instance, could be used to manipulate erythrocyte velocity or direction in a confined region (Anton et al., 2013). Similarly, magnetic tweezers could be employed to stretch cells coated with ligand-conjugated magnetic beads, where the specificity of the ligand determines spatial control (Marjoram et al., 2016). These types of directed manipulations will help to delineate causative relationships between distinct fluid forces and specific cellular responses.
Even with experiments that uncouple the effects of specific flow parameters, limitations remain in our ability to interpret how particular forces exert their impact on both the endocardium and the myocardium. These two layers of the heart appear to respond to fluid forces in a harmonious fashion, yet it is unknown how they coordinate their morphogenesis. Does the pressure created by blood flow induce stretch of both the endocardium and the myocardium simultaneously? Or do fluid forces act primarily on the endocardium, with signals relayed to the myocardium only secondarily? Does the ECM that resides between the endocardium and myocardium transmit biomechanical cues between the layers, in addition to regulating biochemical cross-talk between them? To address these questions, it will be exciting for future studies to employ tools like tissue-specific, FRET-based tension sensors (Lagendijk et al., 2017) or tissue-embedded, force-sensitive oil microdroplets (Campas et al., 2014) to elucidate the precise spatiotemporal dynamics of cardiac mechanosensation.
Finally, we cannot resolve the mechanical interactions between the endocardium and the myocardium without knowing the full roster of molecular pathways responding to forces in each of the two layers. Interesting candidates for future exploration include Piezo1, a mechanosensitive calcium channel (Ranade et al., 2014), Yap1, a nuclear target of Hippo signaling (Nakajima et al., 2017), TGF-β receptors like Endoglin (Sugden et al., 2017), and junctional components like VE-cadherin (Baratchi et al., 2017), all of which have been shown to have mechanoresponsive roles in the endothelium. It will also be important to consider possible functional overlap between paralogues of genes that have already been implicated in cardiac mechanobiology, especially since discrepancies between mutant and morphant phenotypes suggest compensatory interactions between related factors (Novodvorsky et al., 2015).
Altogether, the body of work examining the influence of fluid forces during zebrafish heart development has opened up a number of exciting avenues for further investigation into the mechanosensitive pathways that control essential morphogenetic mechanisms. Going forward, we foresee that future studies in this area will actively bridge the gap between biomechanical inputs and cellular outcomes. By measuring and manipulating specific flow parameters and monitoring their impacts, it will be feasible to connect the fluid mechanics, signal transduction, downstream effector genes, and cellular dynamics that are responsible for shaping essential features of the developing heart. Ultimately, a mechanistic understanding of the roles of fluid forces during cardiac morphogenesis is likely to provide a valuable foundation for new insights regarding how hemodynamic conditions in utero affect the incidence of congenital heart disease.
References
- Anton H, Harlepp S, Ramspacher C, Wu D, Monduc F, Bhat S, et al. (2013). Pulse Propagation by a Capacitive Mechanism Drives Embryonic Blood Flow. Development, 140, 4426–4434. [DOI] [PubMed] [Google Scholar]
- Auman HJ, Coleman H, Riley HE, Olale F, Tsai HJ, Yelon D (2007). Functional Modulation of Cardiac Form through Regionally Confined Cell Shape Changes. PLoS Biology, 5, e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagatto B, Burggren W (2006). A Three-Dimensional Functional Assessment of Heart and Vessel Development in the Larva of the Zebrafish (Danio Rerio). Physiological and Biochemical Zoology, 79, 194–201. [DOI] [PubMed] [Google Scholar]
- Banjo T, Grajcarek J, Yoshino D, Osada H, Miyasaka KY, Kida YS, et al. (2013). Haemodynamically Dependent Valvulogenesis of Zebrafish Heart Is Mediated by Flow-Dependent Expression of Mir-21. Nature Communications, 4, 1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baratchi S, Khoshmanesh K, Woodman OL, Potocnik S, Peter K, McIntyre P (2017). Molecular Sensors of Blood Flow in Endothelial Cells. Trends in Molecular Medicine, 23, 850–868. [DOI] [PubMed] [Google Scholar]
- Bartman T, Walsh EC, Wen KK, McKane M, Ren J, Alexander J, et al. (2004). Early Myocardial Function Affects Endocardial Cushion Development in Zebrafish. PLoS Biology, 2, e129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrndt M, Salbreux G, Campinho P, Hauschild R, Oswald F, Roensch J, et al. (2012). Forces Driving Epithelial Spreading in Zebrafish Gastrulation. Science, 338, 257–260. [DOI] [PubMed] [Google Scholar]
- Beis D, Bartman T, Jin SW, Scott IC, D’Amico LA, Ober EA, et al. (2005). Genetic and Cellular Analyses of Zebrafish Atrioventricular Cushion and Valve Development. Development, 132, 4193–4204. [DOI] [PubMed] [Google Scholar]
- Boselli F, Freund JB, Vermot J (2015). Blood Flow Mechanics in Cardiovascular Development. Cellular and Molecular Life Sciences, 72, 2545–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boselli F, Steed E, Freund JB, Vermot J (2017). Anisotropic Shear Stress Patterns Predict the Orientation of Convergent Tissue Movements in the Embryonic Heart. Development, 144, 4322–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekhuizen ML, Hogers B, DeRuiter MC, Poelmann RE, Gittenberger-de Groot AC, Wladimiroff JW (1999). Altered Hemodynamics in Chick Embryos after Extraembryonic Venous Obstruction. Ultrasound in Obstetrics & Gynecology, 13, 437–445. [DOI] [PubMed] [Google Scholar]
- Campas O, Mammoto T, Hasso S, Sperling RA, O’Connell D, Bischof AG, et al. (2014). Quantifying Cell-Generated Mechanical Forces within Living Embryonic Tissues. Nat Methods, 11, 183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Patrick MJ, Corti P, Kowalski W, Roman BL, Pekkan K (2011). Analysis of Early Embryonic Great-Vessel Microcirculation in Zebrafish Using High-Speed Confocal Mupiv. Biorheology, 48, 305–321. [DOI] [PubMed] [Google Scholar]
- Chen Q, Jiang L, Li C, Hu D, Bu JW, Cai D, et al. (2012). Haemodynamics-Driven Developmental Pruning of Brain Vasculature in Zebrafish. PLoS Biology, 10, e1001374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherian AV, Fukuda R, Augustine SM, Maischein HM, Stainier DY (2016). N-Cadherin Relocalization During Cardiac Trabeculation. Proceedings of the National Academy of Sciences of the United States of America, 113, 7569–7574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MM, Stainier DY (2016). Organ Function as a Modulator of Organ Formation: Lessons from Zebrafish. Current Topics in Developmental Biology, 117, 417–433. [DOI] [PubMed] [Google Scholar]
- Corti P, Young S, Chen CY, Patrick MJ, Rochon ER, Pekkan K, et al. (2011). Interaction between Alk1 and Blood Flow in the Development of Arteriovenous Malformations. Development, 138, 1573–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deacon DC, Nevis KR, Cashman TJ, Zhou Y, Zhao L, Washko D, et al. (2010). The Mir-143-Adducin3 Pathway Is Essential for Cardiac Chamber Morphogenesis. Development, 137, 1887–1896. [DOI] [PubMed] [Google Scholar]
- Dekker RJ, van Soest S, Fontijn RD, Salamanca S, de Groot PG, VanBavel E, et al. (2002). Prolonged Fluid Shear Stress Induces a Distinct Set of Endothelial Cell Genes, Most Specifically Lung Kruppel-Like Factor (Klf2). Blood, 100, 1689–1698. [DOI] [PubMed] [Google Scholar]
- Diaz de la Loza M.C., Thompson BJ (2017). Forces Shaping the Drosophila Wing. Mechanisms of Development, 144, 23–32. [DOI] [PubMed] [Google Scholar]
- Dietrich AC, Lombardo VA, Veerkamp J, Priller F, Abdelilah-Seyfried S (2014). Blood Flow and Bmp Signaling Control Endocardial Chamber Morphogenesis. Developmental Cell, 30, 367–377. [DOI] [PubMed] [Google Scholar]
- Donat S, Lourenco M, Paolini A, Otten C, Renz M, Abdelilah-Seyfried S (2018). Heg1 and Ccm1/2 Proteins Control Endocardial Mechanosensitivity During Zebrafish Valvulogenesis. eLife, 7, e28939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco CA, Jones ML, Bernabeu MO, Geudens I, Mathivet T, Rosa A, et al. (2015). Dynamic Endothelial Cell Rearrangements Drive Developmental Vessel Regression. PLoS Biology, 13, e1002125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund JB, Goetz JG, Hill KL, Vermot J (2012). Fluid Flows and Forces in Development: Functions, Features and Biophysical Principles. Development, 139, 1229–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard LM, Duchemin AL, Ramalingan H, Wu B, Chen M, Bamezai S, et al. (2017). Hemodynamic Forces Sculpt Developing Heart Valves through a Klf2-Wnt9b Paracrine Signaling Axis. Developmental Cell, 43, 274–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz JG, Steed E, Ferreira RR, Roth S, Ramspacher C, Boselli F, et al. (2014). Endothelial Cilia Mediate Low Flow Sensing During Zebrafish Vascular Development. Cell Reports, 6, 799–808. [DOI] [PubMed] [Google Scholar]
- Grego-Bessa J, Luna-Zurita L, del Monte G, Bolos V, Melgar P, Arandilla A, et al. (2007). Notch Signaling Is Essential for Ventricular Chamber Development. Developmental Cell, 12, 415–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haack T, Abdelilah-Seyfried S (2016). The Force Within: Endocardial Development, Mechanotransduction and Signalling During Cardiac Morphogenesis. Development, 143, 373–386. [DOI] [PubMed] [Google Scholar]
- Han P, Bloomekatz J, Ren J, Zhang R, Grinstein JD, Zhao L, et al. (2016). Coordinating Cardiomyocyte Interactions to Direct Ventricular Chamber Morphogenesis. Nature, 534, 700–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckel E, Boselli F, Roth S, Krudewig A, Belting HG, Charvin G, et al. (2015). Oscillatory Flow Modulates Mechanosensitive Klf2a Expression through Trpv4 and Trpp2 During Heart Valve Development. Current Biology, 25, 1354–1361. [DOI] [PubMed] [Google Scholar]
- Herwig L, Blum Y, Krudewig A, Ellertsdottir E, Lenard A, Belting HG, et al. (2011). Distinct Cellular Mechanisms of Blood Vessel Fusion in the Zebrafish Embryo. Current Biology, 21, 1942–1948. [DOI] [PubMed] [Google Scholar]
- Hove JR, Koster RW, Forouhar AS, Acevedo-Bolton G, Fraser SE, Gharib M (2003). Intracardiac Fluid Forces Are an Essential Epigenetic Factor for Embryonic Cardiogenesis. Nature, 421, 172–177. [DOI] [PubMed] [Google Scholar]
- Huang C, Sheikh F, Hollander M, Cai C, Becker D, Chu PH, et al. (2003). Embryonic Atrial Function Is Essential for Mouse Embryogenesis, Cardiac Morphogenesis and Angiogenesis. Development, 130, 6111–6119. [DOI] [PubMed] [Google Scholar]
- Iftimia NV, Hammer DX, Ferguson RD, Mujat M, Vu D, Ferrante AA (2008). Dual-Beam Fourier Domain Optical Doppler Tomography of Zebrafish. Optics Express, 16, 13624–13636. [DOI] [PubMed] [Google Scholar]
- Jamison RA, Samarage CR, Bryson-Richardson RJ, Fouras A (2013). In Vivo Wall Shear Measurements within the Developing Zebrafish Heart. PLoS One, 8, e75722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Amilburu V, Rasouli SJ, Staudt DW, Nakajima H, Chiba A, Mochizuki N, et al. (2016). In Vivo Visualization of Cardiomyocyte Apicobasal Polarity Reveals Epithelial to Mesenchymal-Like Transition During Cardiac Trabeculation. Cell Reports, 17, 2687–2699. [DOI] [PubMed] [Google Scholar]
- Kalogirou S, Malissovas N, Moro E, Argenton F, Stainier DY, Beis D (2014). Intracardiac Flow Dynamics Regulate Atrioventricular Valve Morphogenesis. Cardiovascular Research, 104, 49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochhan E, Lenard A, Ellertsdottir E, Herwig L, Affolter M, Belting HG, et al. (2013). Blood Flow Changes Coincide with Cellular Rearrangements During Blood Vessel Pruning in Zebrafish Embryos. PLoS One, 8, e75060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon HB, Wang S, Helker CS, Rasouli SJ, Maischein HM, Offermanns S, et al. (2016). In Vivo Modulation of Endothelial Polarization by Apelin Receptor Signalling. Nature Communications, 7, 11805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagendijk AK, Gomez GA, Baek S, Hesselson D, Hughes WE, Paterson S, et al. (2017). Live Imaging Molecular Changes in Junctional Tension Upon Ve-Cadherin in Zebrafish. Nature Communications, 8, 1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JKH, Collins MM, Uribe V, Jimenez-Amilburu V, Gunther S, Maischein HM, et al. (2018). The Hippo Pathway Effector Wwtr1 Regulates Cardiac Wall Maturation in Zebrafish. Development, 145. [DOI] [PubMed] [Google Scholar]
- Lee J, Fei P, Sevag Packard RR, Kang H, Xu H, Baek KI, et al. (2016). 4-Dimensional Light-Sheet Microscopy to Elucidate Shear Stress Modulation of Cardiac Trabeculation. The Journal of Clinical Investigation, 126, 3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Moghadam ME, Kung E, Cao H, Beebe T, Miller Y, et al. (2013). Moving Domain Computational Fluid Dynamics to Interface with an Embryonic Model of Cardiac Morphogenesis. PLoS One, 8, e72924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Zhao L, Page-McCaw PS, Chen W (2016). Zebrafish Genome Engineering Using the Crispr-Cas9 System. Trends in Genetics, 32, 815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebling M, Forouhar AS, Wolleschensky R, Zimmermann B, Ankerhold R, Fraser SE, et al. (2006). Rapid Three-Dimensional Imaging and Analysis of the Beating Embryonic Heart Reveals Functional Changes During Development. Developmental Dynamics, 235, 2940–2948. [DOI] [PubMed] [Google Scholar]
- Lin YF, Swinburne I, Yelon D (2012). Multiple Influences of Blood Flow on Cardiomyocyte Hypertrophy in the Embryonic Zebrafish Heart. Developmental Biology, 362, 242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Bressan M, Hassel D, Huisken J, Staudt D, Kikuchi K, et al. (2010). A Dual Role for Erbb2 Signaling in Cardiac Trabeculation. Development, 137, 3867–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Pereira F, Fraser SE, Gharib M (2008). Three-Dimensional Real-Time Imaging of Cardiac Cell Motions in Living Embryos. Journal of Biomedical Optics, 13, 014006. [DOI] [PubMed] [Google Scholar]
- Marjoram RJ, Guilluy C, Burridge K (2016). Using Magnets and Magnetic Beads to Dissect Signaling Pathways Activated by Mechanical Tension Applied to Cells. Methods, 94, 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasaka KY, Kida YS, Banjo T, Ueki Y, Nagayama K, Matsumoto T, et al. (2011). Heartbeat Regulates Cardiogenesis by Suppressing Retinoic Acid Signaling Via Expression of Mir-143. Mechanisms of Development, 128, 18–28. [DOI] [PubMed] [Google Scholar]
- Nakajima H, Yamamoto K, Agarwala S, Terai K, Fukui H, Fukuhara S, et al. (2017). Flow-Dependent Endothelial Yap Regulation Contributes to Vessel Maintenance. Developmental Cell, 40, 523–536 e526. [DOI] [PubMed] [Google Scholar]
- Novodvorsky P, Watson O, Gray C, Wilkinson RN, Reeve S, Smythe C, et al. (2015). Klf2ash317 Mutant Zebrafish Do Not Recapitulate Morpholino-Induced Vascular and Haematopoietic Phenotypes. PLoS One, 10, e0141611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Yu H, Shi X, Korzh V, Wohland T (2007). Characterization of Flow Direction in Microchannels and Zebrafish Blood Vessels by Scanning Fluorescence Correlation Spectroscopy. Journal of Biomedical Optics, 12, 014034. [DOI] [PubMed] [Google Scholar]
- Peshkovsky C, Totong R, Yelon D (2011). Dependence of Cardiac Trabeculation on Neuregulin Signaling and Blood Flow in Zebrafish. Developmental Dynamics, 240, 446–456. [DOI] [PubMed] [Google Scholar]
- Pestel J, Ramadass R, Gauvrit S, Helker C, Herzog W, Stainier DY (2016). Real-Time 3d Visualization of Cellular Rearrangements During Cardiac Valve Formation. Development, 143, 2217–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranade SS, Qiu Z, Woo SH, Hur SS, Murthy SE, Cahalan SM, et al. (2014). Piezo1, a Mechanically Activated Ion Channel, Is Required for Vascular Development in Mice. Proceedings of the National Academy of Sciences of the United States of America, 111, 10347–10352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasouli SJ, Stainier DYR (2017). Regulation of Cardiomyocyte Behavior in Zebrafish Trabeculation by Neuregulin 2a Signaling. Nature Communications, 8, 15281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renz M, Otten C, Faurobert E, Rudolph F, Zhu Y, Boulday G, et al. (2015). Regulation of Beta1 Integrin-Klf2-Mediated Angiogenesis by Ccm Proteins. Developmental Cell, 32, 181–190. [DOI] [PubMed] [Google Scholar]
- Rochon ER, Menon PG, Roman BL (2016). Alk1 Controls Arterial Endothelial Cell Migration in Lumenized Vessels. Development, 143, 2593–2602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsa LA, Givens C, Tzima E, Stainier DY, Qian L, Liu J (2015). Cardiac Contraction Activates Endocardial Notch Signaling to Modulate Chamber Maturation in Zebrafish. Development, 142, 4080–4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwerte T, Pelster B (2000). Digital Motion Analysis as a Tool for Analysing the Shape and Performance of the Circulatory System in Transparent Animals. The Journal of Experimental Biology, 203, 1659–1669. [DOI] [PubMed] [Google Scholar]
- Schwerte T, Uberbacher D, Pelster B (2003). Non-Invasive Imaging of Blood Cell Concentration and Blood Distribution in Zebrafish Danio Rerio Incubated in Hypoxic Conditions in Vivo. The Journal of Experimental Biology, 206, 1299–1307. [DOI] [PubMed] [Google Scholar]
- Sedmera D, Pexieder T, Rychterova V, Hu N, Clark EB (1999). Remodeling of Chick Embryonic Ventricular Myoarchitecture under Experimentally Changed Loading Conditions. The Anatomical Record, 254, 238–252. [DOI] [PubMed] [Google Scholar]
- Shyer AE, Rodrigues AR, Schroeder GG, Kassianidou E, Kumar S, Harland RM (2017). Emergent Cellular Self-Organization and Mechanosensation Initiate Follicle Pattern in the Avian Skin. Science, 357, 811–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt DW, Liu J, Thorn KS, Stuurman N, Liebling M, Stainier DY (2014). High-Resolution Imaging of Cardiomyocyte Behavior Reveals Two Distinct Steps in Ventricular Trabeculation. Development, 141, 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed E, Faggianelli N, Roth S, Ramspacher C, Concordet JP, Vermot J (2016). Klf2a Couples Mechanotransduction and Zebrafish Valve Morphogenesis through Fibronectin Synthesis. Nature Communications, 7, 11646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden WW, Meissner R, Aegerter-Wilmsen T, Tsaryk R, Leonard EV, Bussmann J, et al. (2017). Endoglin Controls Blood Vessel Diameter through Endothelial Cell Shape Changes in Response to Haemodynamic Cues. Nature Cell Biology, 19, 653–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedula V, Lee J, Xu H, Kuo CJ, Hsiai TK, Marsden AL (2017). A Method to Quantify Mechanobiologic Forces During Zebrafish Cardiac Development Using 4-D Light Sheet Imaging and Computational Modeling. PLoS Computational Biology, 13, e1005828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermot J, Forouhar AS, Liebling M, Wu D, Plummer D, Gharib M, et al. (2009). Reversing Blood Flows Act through Klf2a to Ensure Normal Valvulogenesis in the Developing Heart. PLoS Biology, 7, e1000246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins SC, Maniar S, Mosher M, Roman BL, Tsang M, St Croix CM (2012). High Resolution Imaging of Vascular Function in Zebrafish. PLoS One, 7, e44018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalcin HC, Amindari A, Butcher JT, Althani A, Yacoub M (2017). Heart Function and Hemodynamic Analysis for Zebrafish Embryos. Developmental Dynamics, 246, 868–880. [DOI] [PubMed] [Google Scholar]
- Zickus V, Taylor JM (2018). 3d + Time Blood Flow Mapping Using Spim-Micropiv in the Developing Zebrafish Heart. Biomedical Optics Express, 9, 2418–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]


