Abstract
We have previously identified NOD2 genotype and inflammatory bowel diseases (IBD) phenotype, as associated with shifts in the ileal microbiome (“dysbiosis”) in a patient cohort. Here we report an integrative analysis of an expanded number of Crohn's disease (CD) related genetic defects in innate immune function (NOD2, ATG16L1, IRGM, CARD9, XBP1, ORMDL3) and composition of the ileal microbiome by combining the initial patient cohort (Batch 1, 2005–2010, n = 165) with a second consecutive patient cohort (Batch 2, 2010–2012, n = 118). These combined patient cohorts were composed of three non-overlapping phenotypes: 1.) 106 ileal CD subjects undergoing initial ileocolic resection for diseased ileum, 2.) 88 IBD colitis subjects without ileal disease (predominantly ulcerative colitis but also Crohn’s colitis and indeterminate colitis, and 3.) 89 non-IBD subjects. Significant differences (FDR < 0.05) in microbiota were observed between macroscopically disease unaffected and affected regions of resected ileum in ileal CD patients. Accordingly, analysis of the effects of genetic and clinical factors were restricted to disease unaffected regions of the ileum. Beta-diversity differed across the three disease categories by PERMANOVA (p < 0.001), whereas no significant differences in alpha diversity were noted. Using negative binomial models, we confirmed significant effects of IBD phenotype, C. difficile infection, and NOD2 genotype on ileal dysbiosis in the expanded analysis. The relative abundance of the Proteobacteria phylum was positively associated with ileal CD and colitis phenotypes, but negatively associated with NOD2R genotype. Additional associations with ORMDL3 and XBP1 were detected at the phylum/subphylum level. IBD medications, such as immunomodulators and anti-TNFα agents, may have a beneficial effect on reversing dysbiosis associated with the IBD phenotype. Exploratory analysis comparing microbial composition of the disease unaffected region of the resected ileum between 27 ileal CD patients who subsequently developed endoscopic recurrence within 6–12 months versus 34 patients who did not, suggested that microbial biomarkers in the resected specimen helped stratify patients with respect to risk of post-surgical recurrence.
Introduction
Inflammatory bowel disease (IBD) describes a group of disorders in which the intestines become inflamed. Two major types of IBD are ulcerative colitis (UC) and Crohn's disease (CD). UC is limited to the colon or large intestine. Crohn's disease, on the other hand, can involve any part of the gastrointestinal tract from the mouth to the anus. Most commonly, though, it affects the ileum or the colon or both. Abnormal host-microbial interactions and genetic susceptibility are implicated in the pathogenesis of IBD, reviewed in [1]. We previously examined the effects of the NOD2 and ATG16L1 polymorphisms on ileal microbial composition in 1) ileal CD subjects undergoing initial ileal resection, 2) IBD colitis without ileitis (predominantly ulcerative colitis) subjects and 3) subjects without inflammatory bowel disease (non-IBD) [2–4]. These previous studies identified NOD2 genotype and C. difficile infection, in addition to IBD phenotype, as associated with shifts in ileal microbiota or “dysbiosis” [2–4]. We also found that differential expression of genes involved in Paneth cell function were associated with shifts in ileal microbial composition [5]. In the current study, the effects of an expanded panel of Crohn’s disease risk alleles [6–10] that are associated with defects in innate immunity (NOD2, ATG16L1, IRGM, CARD9, XBP1, ORMDL3) on ileal microbial composition, were analyzed by combining the original patient cohort (referred to as “Batch 1 2005–2010”) with a second consecutive patient cohort collected between 2010 and 2012 (“Batch 2 2010–2012”, see Table 1”) Some of these genes are implicated in autophagy (e.g. ATG16L1, IRGM), or endoplasmic reticulum stress (e.g. XBP1, ORMDL3), and/or Paneth cell dysfunction (NOD2 ATG16L1, IRGM).
Table 1. Distribution of NOD2, ATG16L1, IRGM, CARD9 and ORMDL3 risk alleles and clinical characteristics in ileal CD, colitis and non-IBD subjects in batches 1 and 2.
The percent of subjects recruited at each of the three IBD centers with complete genotype and clinical characteristics with at least one risk allele and the percent of subjects who have at least one risk allele (see Methods) are listed.
| Ileal CD | Colitis | Non-IBD | |||||||
|---|---|---|---|---|---|---|---|---|---|
| n = 106 | n = 88 | n = 89 | |||||||
| Batch1 | Batch2 | P value | Batch1 | Batch2 | P value | Batch1 | Batch2 | P value | |
| n = 50 | n = 56 | n = 59 | n = 29 | n = 56 | n = 33 | ||||
| IBD Center | |||||||||
| Wash. U.-St. Louis | 100% | 62% | 100% | 83% | 100% | 73% | |||
| Stony Brook U. | 0% | 9% | 0% | 17% | 0% | 27% | |||
| U. of North Carolina | 0% | 29% | 0% | 0% | 0% | 0% | |||
| IBD Risk Allele | |||||||||
| NOD2R composite | 40% | 30% | 0.38 | 19% | 14% | 0.78 | 14% | 52% | <0.001 |
| ATG16L1R | 94% | 79% | 0.05 | 81% | 76% | 0.79 | 77% | 79% | 0.99 |
| IRGMR | 30% | 27% | 0.90 | 29% | 17% | 0.34 | 27% | 18% | 0.38 |
| CARD9R | 94% | 96% | 0.98 | 93% | 93% | 1 | 88% | 91% | 0.85 |
| XBP1R | 12% | 14% | 0.99 | 10% | 14% | 0.84 | 14% | 0% | 0.01 |
| ORMDL3R | 76% | 73% | 0.90 | 75% | 79% | 0.88 | 62% | 70% | 0.51 |
| Clinical covariates | |||||||||
| Male gender % | 46% | 43% | 0.91 | 53% | 55% | 1 | 41% | 39% | 0.99 |
| Caucasian race % | 94% | 82% | 0.11 | 91% | 97% | 0.55 | 89% | 94% | 0.56 |
| Median age years (range) |
33 (18–72) |
32 (17–67) |
44 (18–69) |
47 (20–72) |
61 (24–84) |
62 (17–86) |
|||
| Duration IBD years (range) |
5 (0–38) |
5 (0–35) |
5 (0–45) |
5 (0.1–30) |
NA | NA | |||
| Current smoker % | 32% | 29% | 0.90 | 10% | 3% | 0.46 | 27% | 18% | 0.38 |
| + fecal C. difficile toxin % | 0% | 5% | 0.32 | 24% | 0% | 0.01 | 0% | 6% | 0.23 |
| Colon Cancer % | 6% | 0% | 0.20 | 17% | 3% | 0.13 | 54% | 45% | 0.46 |
| Median BMI kg/m2 (range) |
24 (16–41) |
23 (14–44) |
26 (16–43) |
28 (19–36) |
28 (18–47) |
26 (20–48) |
|||
| 5-ASA % | 60% | 38% | 0.04 | 63% | 48% | 0.27 | 0% | 3% | 0.65 |
| Steroids % | 48% | 54% | 0.67 | 54% | 52% | 1 | 2% | 12% | 0.11 |
| Immunomodulators % | 48% | 30% | 0.09 | 29% | 28% | 1 | 4% | 6% | 0.98 |
| Anti-TNF alpha % | 24% | 45% | 0.04 | 31% | 52% | 0.09 | 2% | 0% | 0.96 |
The majority of patients with the ileal CD phenotype eventually undergo surgical resection of diseased ileum because of stricturing and penetrating complications [11]. Unfortunately, disease recurrence in previously disease-free segments of the ileum at the surgical anastomosis is common [12–16]. A reduced relative abundance of Faecalibacterium prausnitzii, a commensal anaerobic bacterium in the distal intestine (ileum and colon), has been a consistent feature associated with the ileal CD phenotype [1, 3,4,17–21]. This bacterial species, along with other closely related clostridial species, are key sources of the short chain fatty acid butyrate, which is the preferred energy source for enterocytes in the distal intestine, and exhibits anti-inflammatory and pro-intestinal barrier properties in experimental mouse models [22]. A diminished relative abundance of ileal F. prausnitzii at the time of resection has been associated with a higher risk of post-operative endoscopic recurrence of ileal CD six months after surgery [17,23–25]. Consequently, an additional goal of this study was to determine whether alterations in the relative abundances of specific bacterial taxa, such as F. prausnitzii, at the time of ileal resection were predictive of subsequent endoscopic recurrence. To this end, ileal microbiota in the disease unaffected region of the ileal resection were compared between a group of ileal CD subjects who subsequently developed endoscopic recurrence in the neo-terminal ileum and a group of ileal CD subjects who did not, 6–12 months after initial surgery.
Materials and methods
Patient recruitment
This study was approved by the Institutional Review Boards of Stony Brook University (IRB# 245010), Washington University-St. Louis (IRB# 201101774), and the University of North Carolina (IRB# 10–0355). Patient written consents were obtained from all study participants and assent and parental consents were obtained for children <18 years of age at their respective institutions. Coded samples stripped of all identifying information were collected from subjects in the three following categories: 1) Ileal CD patients undergoing initial ileocolic resection (ICR) of diseased ileum; 2) colitis patients without ileitis (predominantly ulcerative colitis or UC, but also Crohn’s colitis and indeterminate colitis) undergoing initial total colectomy; and 3) patients without IBD undergoing initial right or total colectomy were prospectively enrolled to donate tissue, blood and longitudinal clinical information in a consecutive fashion by the Stony Brook University GI Biobank (Batch 2, 2010–2012), the Washington University Digestive Diseases Research Core Center Biobank Core (Batch 1, 2005–2010; Batch 2, 2010–2012) and the U. of North Carolina Multidisciplinary IBD Center (Batch 2, 2011–2012) as previously described [4]. The diagnosis of ileal CD, UC, indeterminate colitis and Crohn’s colitis was made ultimately on the basis of pathological criteria (surgical resection specimen) [26–28]. A minimum of 4 ex-vivo biopsies were taken separately from the macroscopically disease unaffected proximal ileal margin (from all subjects) and from the disease affected region (from ileal CD subjects) of fresh pathological specimens using Radial Jaw4 large capacity biopsy forceps (Boston Scientific, Natick, MA), and immediately placed in RNA stabilization solution (RNAlater, Life Technologies, Grand Island, NY, USA) overnight at 4°C prior to storing at -80o C. A subset of the ileal CD patients (n = 61) underwent follow up colonoscopy within 6–12 months to assess endoscopic disease recurrence as previously described [29]. Ex-vivo research ileal biopsies, blood and longitudinal clinical information were also prospectively collected from non-IBD patients undergoing colonoscopies for colon cancer screening by the Stony Brook University GI Biobank (2010–2012) and the Washington University Digestive Diseases Research Core Center Biobank Core (2005–2012).
Additional clinical metadata on smoking, obesity, and IBD medications were obtained on samples collected by the Stony Brook University GI Biobank and the Washington University Digestive Disease Research Core Center Biobank Core and Dr. Sartor’s laboratory, by reviewing the medical records including the pathology report of the resected intestine by a gastroenterologist (EL, RBS) [4]. Preoperative mechanical bowel preparations were not routinely ordered for surgical procedures on IBD patients but were often ordered for the non-IBD patients. A smoker was defined as smoking ≥7 cigarettes a week for at least a year [12,30]. In order to assess the potentially confounding effect of obesity [31], body mass index (BMI) was also recorded. C. difficile infection, which has been associated with IBD [32], was recorded as the presence of a positive fecal C. difficile toxin B [33] within a week of the sample collection. Antibiotics have a significant effect on the microbiome [34] and all patients received preoperative surgical antibiotic prophylaxis (long-acting beta lactam 30 minutes before the initial incision was made) [35]. Patients diagnosed with C. difficile infections (predominantly colitis patients) were treated with oral vancomycin up until surgery. The few non-IBD (total <10) subjects from whom ileal research biopsies were collected during colonoscopy did not receive antibiotics prior to collection of the ileal biopsies for at least two months. Dietary information collected on the subjects revealed that none of the subjects were vegetarian and none of the subjects were on either elemental or polymeric enteral feedings [34]. Because there was no significant difference between the three institutions with respect to race and ethnicity (all predominantly non-Hispanic White/Caucasian), the Batch 2 patients were analyzed as a single cohort.
IBD genotyping
Because of their relationship to Crohn’s disease phenotype, our analysis focused on the following single nucleotide polymorphisms (SNPs), which are implicated in microbial sensing, autophagy, endoplasmic reticulum stress, and/or Paneth cell dysfunction [6–10]: 1) NOD2 risk alleles (rs2066847, rs2066884, rs2066845, rs5743289) [9,36–41], ATG16L1 (rs2241880) [42–45], IRGM (rs13361189) [46–48], 2) CARD9 (rs10870077) [49,50], 3) XBP1 (rs35873774) [51], 4) ORMDL3 (rs2872507) [52]. The subjects were categorized as 1) homozygous for both non-risk alleles NR/NR), termed NR or 2) carrying at least one risk allele (R/NR, R/R), termed R. Four major NOD2 risk alleles were combined to form two composite categories: 1) NOD2NR, subjects harboring none of the four major risk alleles 2) NOD2R, subjects harboring at least one of the four major risk alleles (i.e., NOD2R/NR + NOD2R/R). Illumina Immunochip genotyping using genomic DNA prepared from peripheral blood and/or tissue was performed on all of the subjects [53–55]. A subset of these patients had previously undergone genotyping by using the Sequenom MassArray System (Sequenom Inc., San Diego, CA) in the Washington University Sequenom Technology Core 3]. In the patients for which genotyping of the three major nonsynonymous NOD2 risk alleles, Leu1007fs (rs2066847, SNP13), R702W (rs2066884, SNP8) and G908R (rs2066845, SNP12), could not be assigned by Illumina Immunochip, genotyping for these SNPs was performed by Taqman Genotyping Assays (Life Technologies, Grand Island, NY, USA) as previously described [4]. For the ATG16L1 and IRGM genotypes, the value of a missing SNP was imputed from other tightly linked SNPs. For ATG16L1 the tightly linked SNP was rs12994997 and for IRGM genotype the SNPs were rs10065172 and rs11747270.
16S rRNA amplicon library construction and sequencing
Amplicons of the V3-V5 hypervariable regions of the bacterial 16S rRNA gene were sequenced using the 454 FLX Titanium Sequencing Platform and the same primers employed for characterizing the microbial communities in healthy individuals at different body sites including the gastrointestinal tract by the Human Microbiome Project [4, 56]. Library construction and sequencing for Batch 1 samples (2005–2010), was performed at the Genome Institute at Washington University-St. Louis. Library construction and sequencing for a small subset (n = 15) of Batch 1 samples and for all the Batch 2 (2010–2012) samples for all subjects recruited was performed in the Frank laboratory (UC Denver) and the sequencing was performed at The Centre for Applied Genomics at the Hospital for Sick Children in Toronto, Canada following the same standard operating procedures [4]. The variance in the relative abundances of phyla/subphyla taxa between duplicate libraries sequenced at the two different centers (n = 15) did not exceed the variance observed for duplicate libraries sequenced at a single center (n = 15). Clinical, genotyping, and sequencing data can be accessed through the dbGAP authorized access system (Request access to: phs000255.v2). In order to request access to any of the individual-level datasets within the controlled-access portions of the database, the Principal Investigator (PI) and the Signing Official (SO) at the investigator’s institution will need to co-sign a request for data access, which will be reviewed by an NIH Data Access Committee at the appropriate NIH Institute or Center (https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?page=login).
Sequence reads were screened for basic quality defects by the software program BARTAB [57]. All sequences were checked for chimerism with Uchime (usearch6.0.203_i86linux32) [58] using the Schloss Silva references [59]. The filtered sequences were aligned and classified with SINA (1.2.11 using the 418,497 bacterial sequences in Silva 115NR99 as reference configured to yield Silva technology [60,61]. Operational taxonomic units (OTUs) at the genera level were produced by clustering sequences with identical taxonomic assignments. Relative abundances were calculated by dividing OTU counts were normalized between samples by dividing sequence counts by the total number of high-quality 16S sequences generated per sample to calculate the average relative abundance values shown in (Table 2). Phylum/subphyla level OTU tables were generated by collapsing lower level OTUs into higher-level categories corresponding to the categories used previously [4]: 1) Actinobacteria, 2) Bacteroidetes, 3) Firmicutes/Clostridia/Ruminococcaceae (family corresponding to Clostridia Group IV), 4) Firmicutes/Clostridia/Lachnospiriceae (family corresponding to Clostridia Group XIVa), 5) Firmicutes/Clostridia/Other, 6) Firmicutes/Bacillus (class), 7) Firmicutes/Other, 8) Proteobacteria, and 9) Other taxa.
Table 2. Comparison of the relative abundances of phyla/subphyla taxa between the disease unaffected and disease affected regions of resected ileum in ileal CD subjects undergoing initial ICR.
The mean relative abundance ± standard deviation is shown for each bacterial category in Batch 1 (2005–2010) and Batch 2 (2010–2012) as well as the FDR for respectively macroscopic pathology (disease affected vs. disease unaffected) and the sample batch (Batch 1 Batch 2). A total of 101 disease affected and 111 disease unaffected samples were analyzed, of which 91 were paired samples.
| Disease affected | Disease unaffected | FDR | ||||
|---|---|---|---|---|---|---|
| Batch 1 | Batch 2 | Batch 1 | Batch 2 | Path | Batch | |
| Actinobacteria | 0.028 ±0.055 |
0.006 ±0.011 |
0.049 ±0.088 |
0.009 ±0.016 |
0.092 | <0.001 |
| Bacteroidetes | 0.314 ±0.272 |
0.447 ±0.255 |
0.292 ±0.273 |
0.356 ±0.278 |
0.346 | 0.165 |
|
Firmicutes/Clostridia/ Lachnospiraceae |
0.166 ±0.161 |
0.172 ±0.141 |
0.127 ±0.135 |
0.148 ±0.144 |
0.071 | 0.449 |
|
Firmicutes/Clostridia/ Ruminococcaceae |
0.028 ±0.039 |
0.045 ±0.074 |
0.021 ±0.031 |
0.036 ±0.08 |
0.023 | 0.473 |
|
Firmicutes/Clostridia/ Other |
0.059 ±0.121 |
0.056 ±0.127 |
0.058 ±0.136 |
0.061 ±0.141 |
0.631 | 0.449 |
| Firmicutes/Bacilli | 0.124 ±0.195 |
0.051 ±0.119 |
0.149 ±0.177) |
0.088 ±0.177 |
0.071 | <0.001 |
| Proteobacteria | 0.158 ±0.206 |
0.146 ±0.183 |
0.197 ±0.223 |
0.204 ±0.222 |
0.026 | 0.637 |
Quantitative PCR for targeted bacterial subgroups
QPCR assays were performed using established primers for total bacteria (forward, 5’- GTG STG CAY GGY TGT CGT CA-3’ and reverse 5’- ACG TCR TCC MCA CCT TCC TC-3’) [62], F. prausnitzii (forward 5’-CCC TTC AGT GCC GCA GT-3’ and reverse 5’-GTC GCA GGA TGT CAA GAC-3’) [63], C. coccoides-E. rectales subgroup (forward, 5’–CGG TAC CTG ACT AAG C-3’and reverse 5’–AGT TT(C/T) ATT CTT GCG AAC G-3’) [63]. The log2 transformation of the relative abundance of each bacterial subgroup was measured by ΔCt = Ct (threshold cycle) total bacteria−Ct subgroup. All assays were carried out in triplicate and results averaged. Plasmid quantification standards were prepared from representative clones of the target organisms to insure that the assays were conducted within the linear range and with similar slopes [64].
Statistical analysis
Patient samples with less than 100 total high-quality 16S rRNA sequence counts were excluded from the analysis. Alpha diversity indices (i.e., Chao1, Shannon complexity H) were calculated for each using the software package Explicet (v2.9.4, www.explicet.org) [65]. Beta diversity was calculated using the adonis function in the R vegan package as previously described [66,67] at the phylum/subphylum level, family and genus levels. This function uses a non-parametric multivariate analysis of variance test (PERMANOVA) [68], which was applied using Bray-Curtis, Morisita-Horn and Jaccard indices as distance measurements [69].
Because over-dispersion is often observed in microbiome sequence count data, negative binomial regression models [70, 71] were used to analyze the 16S rRNA sequence data. The OTUs (relative abundance ≥0.0001 and prevalence ≥0.01) were grouped primarily at the phylum/subphylum level as described above [4]. To identify taxa with significant differences in relative abundance between disease affected and disease unaffected regions of the resected ileum in ileal CD patients undergoing initial ICR, a generalized linear mixed model was used:
Here Yijk denotes the OTU k’s observed count for tissue j in patient i with μijk being its mean. The symbol ϕk is the dispersion parameter for the count of OTU k. The OTU k of patient i is associated with a random coefficient bik in order to assess paired disease affected and disease unaffected samples collected from the same patient i. The Batch variable refers to whether samples were collected in Batch 1 (2005–2010) or Batch 2 (2010–2012). The log total count of each patient sample is considered as an offset.
Negative binomial models were used to identify associations between OTUs and IBD phenotype (ileal CD, colitis without ileal disease, and control non-IBD), IBD genotype, and clinical co-variates (see Table 1).
For OTU k, the model is:
Yik is the observed counts for subject i on OTU k. The IBD genotypes and covariates in the model are listed in Table 1. The Batch variable refers to whether samples were collected in Batch 1 (2005–2010) or Batch 2 (2010–2012). The interactions included first-order interactions between phenotype, genotypes, clinical covariates and Batch. Stepwise variable selection based on Bayesian information criterion (BIC) was applied to generate the final model. The p value of coefficients in final model was subject to FDR correction [72], with the FDR threshold set at 0.05.
The relative abundances of F. prausnitzii and C. coccoides-E.rectales measured by qPCR were analyzed using a permutation-based linear regression model.
Results
Distribution of IBD phenotype, genotype, and clinical covariates in Batch 1 (2005–2010) and Batch 2 (2010–2012) samples
Samples were analyzed from 154 subjects in Batch 2 in addition to the 170 subjects in Batch 1 analyzed in our previous study [4]. Of the 324 total subjects, genotype, and clinical data were completed for 283 (87%) subjects. The distributions of IBD phenotype, genotype, and clinical covariates for the 283 of 324 subjects with complete datasets are summarized in Table 1. All of the Batch 1 samples and the majority (62–83%) of Batch 2 samples were collected at the Washington University–St. Louis medical center and the remaining subjects (17–38%) were recruited at the Stony Brook University and University of North Carolina medical centers (Table 1). At all three institutions, the race and ethnicity of the subjects were predominantly White/Caucasian. The proportion of subjects for each disease phenotype who had at least one of the following risk alleles: the NOD2 risk super allele (rs2066847, rs2066884, rs2066845, rs5743289), the ATG16L1T300A allele (rs2241880), the IRGM risk allele (rs13361189), the CARD9 risk allele (rs10870077), the XBP1 risk allele (rs35873774), and the ORMDL3 risk allele (rs2872507) are shown in Table 1. The high proportion of NOD2R non-IBD subjects in Batch 2 reflects preferential selection of these individuals since our previous analysis indicated that NOD2 genotype was associated with shifts in ileal microbiome composition, independent of disease phenotype.
The clinical covariates for Batch 1 and Batch 2 samples (Table 1) revealed that the median duration of IBD prior to initial surgery was 5 years for both ileal CD and colitis subjects. As previously noted [4], ileal CD subjects were younger than non-IBD patients, and active smoking was less prevalent in colitis subjects. A higher proportion of the Batch 2 ileal CD and colitis subjects received anti-TNF alpha biologics at the time of surgery than Batch 1 subjects (See Table 1). A lower proportion of the Batch 2 colitis subjects had C. difficile infections immediately prior to surgery than the Batch 1 colitis patients.
16S rRNA sequencing analysis of ileal CD, colitis and non-IBD samples
A total of 5,739,816 high-quality V3–V5 sequences (median 6024 reads/sample, IQR 2950–10314) were generated from the ileal specimens in this study. All libraries had a Goods coverage of ≥ 95% (median 98%, IQR 97.9–98.8%) at the rarefaction point of 500 sequences. All of the sequences were binned using an updated pipeline as described in Methods. Greater than 90% of the sequences could be binned into the following eight phylum/subphylum categories: 1) Actinobacteria, 2) Bacteroidetes, 3) Firmicutes/Clostridia/Ruminococcaceae, 4) Firmicutes/Clostridia/Lachnospiraceae, 5) Firmicutes/Clostridia/Other, 6) Firmicutes/Bacillus, 7) Firmicutes/Other, 8) Proteobacteria, (see Fig 1). The Firmicutes phyla were sub-divided between classes Clostridia, Bacilli and Firmicutes/Other. The Clostridia taxa were then further subdivided into Clostridia/Ruminococcaceae and Clostridia/Lachnospiraceae at the family level, corresponding to the Clostridium Group IV and XIVa phylum/subphylum categories designated when the Batch 1 samples were previously analyzed [4]. The remaining Clostridia OTUs were grouped as Clostridia/Other.
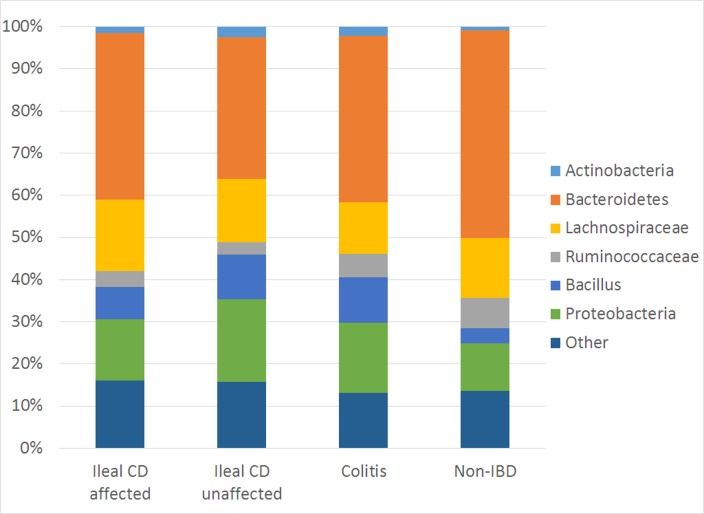
Fig 1. Phylum/Subphylum comparison of ileum-associated bacterial communities of ileal CD, colitis, and non-IBD samples.
The average relative abundances of 6 of 8 phylum/subphylum bacterial category are shown for macroscopically disease affected and macroscopically disease unaffected samples, respectively from ileal CD, and from macroscopically disease unaffected samples from colitis and control subjects. In this figure “Other” includes Firmicutes/Clostridia/Other, Firmicutes/Other and all remaining taxa.
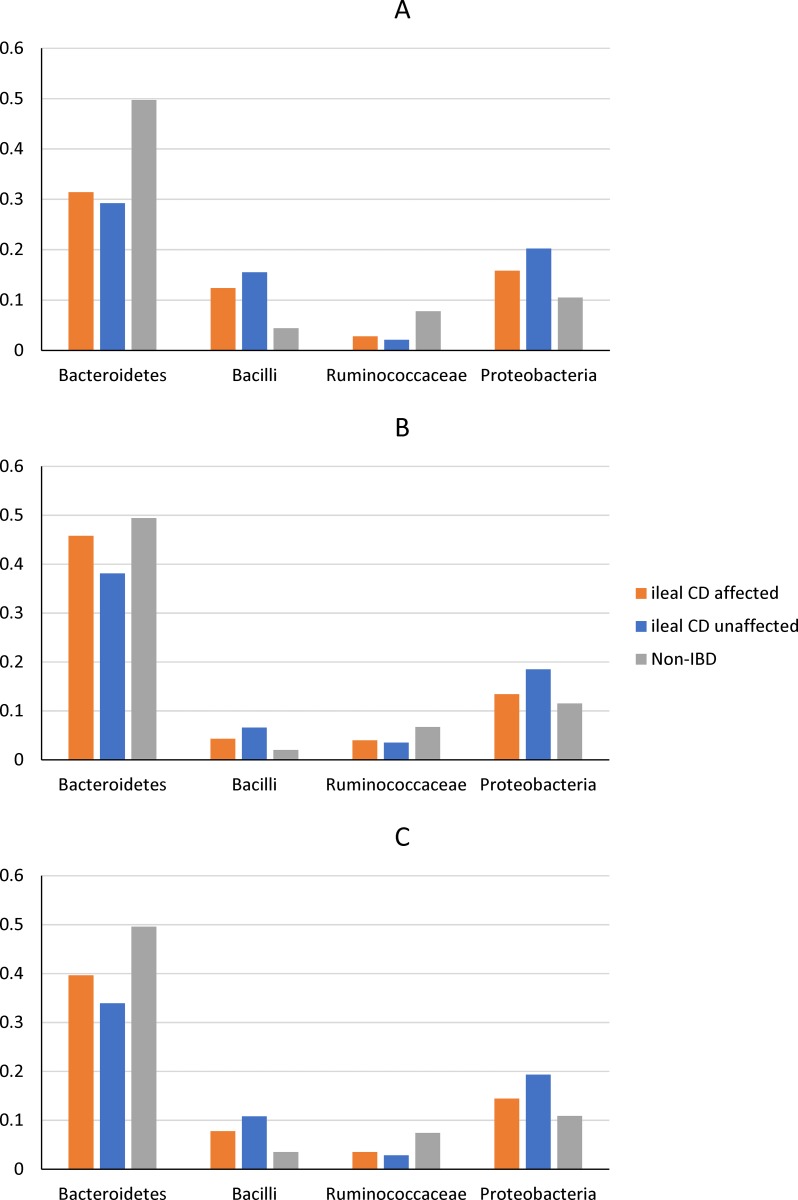
Because samples were collected from both the macroscopically disease unaffected region (proximal margin) and the disease affected region of the resected ileum, we first examined the within-subject differences in microbiota between disease unaffected and affected regions of the ileum. To this end, a generalized linear mixed effect negative binomial model was used to measure the effects of pathology (disease affected vs. disease unaffected) as well as sample batch on ileal microbiota binned at the phyla/subphyla level (see Table 2). The effect of pathology was significant for Ruminococcaceae (FDR = 0.023) and Proteobacteria (FDR = 0.026, see Table 2). As shown in Fig 2 and S1 Table, the mean relative abundance of Proteobacteria was consistently higher in the disease unaffected regions than the adjacent disease affected regions from ileal CD subjects.
Fig 2. The mean relative abundances of selected phyla/subphyla groups between disease affected ileal samples from ileal CD subjects, disease unaffected ileal samples from ileal CD subjects and disease unaffected ileal samples from non-IBD subjects.
A. Batch 1; B. Batch 2; C. Batch 1 and Batch 2 combined.
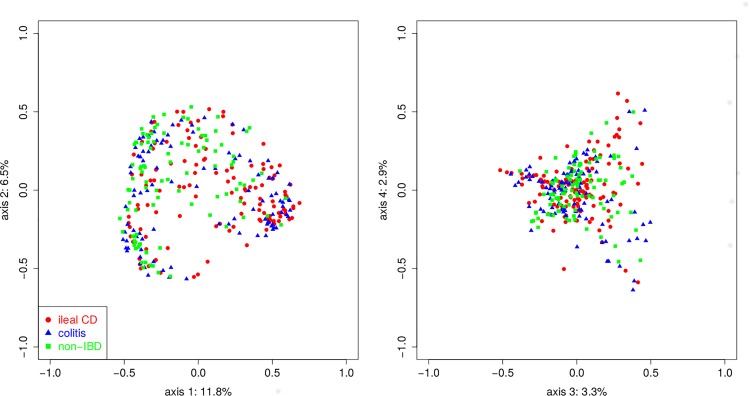
No significant differences (p < 0.05) were detected in measures of alpha diversity (complexity measured by Shannon H or richness, measured by Chao1) of the ileal microbiome in ileal CD, colitis, and non-IBD disease unaffected samples or between disease unaffected and disease affected ileal CD samples. There were significant differences in beta diversity using three different dissimilarity indices, Bray-Curtis (p < 0.001, Fig 3), Jaccard (p < 0.001), and Morasita-Horn (p <0.001) of the ileal microbiome in disease unaffected regions between the three phenotypes. Furthermore, pairwise comparisons revealed significant differences between all three phenotypes (see S2 Table). While significant differences were detected for individual phyla/subphyla categories, particularly Proteobacteria, between disease unaffected and disease affected ileal CD samples, no significant difference in beta diversity was detected using any three of the dissimilarity indices.
Fig 3. Principal coordinate analysis (PCoA) conducted on genus-level microbiome data using a dissimilarity matrix of Bray Curtis scores.
The IBD phenotypes are color coded as follows: ileal CD (designated by red circle), colitis without ileal disease (designated by red triangle), non-IBD (designated by green square). The four largest dimensions (PC1, PC2, PC3, PC4) are shown and account for 11.8%, 6.5%, 3.3% and 2.9% of the differences, respectively.
Integrated analysis of the effect of IBD phenotype, genotype and clinical covariates on phyla/subphyla bacterial categories
Because significant differences were detected between disease affected and disease unaffected samples from ileal CD subjects, analysis of the effects of IBD genotype (NOD2, ATG16L1, IRGM, CARD9, XBP1 and ORMDL3) on each phylum/subphylum category was restricted to only disease-unaffected samples from the three phenotypes (ileal CD, colitis, and non-IBD). Integrated analysis of the effects of the three IBD phenotypes, six IBD genotypes, 11 clinical covariates, sample batch, and all first order interactions, was conducted by building negative binomial models for each of the eight phyla/subphyla categories, using these factors as predictor variables (see Table 3).
Table 3. Negative binomial model results for IBD phenotype, genotype and clinical covariates at the phylum/subphylum level.
Variables with significant effects are bolded.
| Actinobacteria | Log fold change | p -value | FDR |
|---|---|---|---|
| Main effects1 | |||
| ileal CD | 2.055 | <0.0001 | <0.0001 |
| Colitis | 1.368 | <0.0001 | <0.0001 |
| Duration IBD | -0.064 | <0.0001 | <0.0001 |
| Batch | -1.194 | <0.0001 | <0.0001 |
| + C. difficile fecal toxin | -2.878 | 0.0056 | 0.0083 |
| Anti TNF alpha | -0.889 | 0.0015 | 0.0026 |
| Current smoker | -0.333 | 0.1081 | 0.1179 |
| Age | 0.001 | 0.9801 | 0.9801 |
| First order interactions | |||
| + C. difficile fecal toxin*Current smoker | 2.806 | 0.0009 | 0.0017 |
| + C. difficile fecal toxin *Age | 0.530 | 0.0124 | 0.0149 |
| Duration IBD * Anti TNF alpha | 0.008 | 0.0064 | 0.0083 |
| Firmicutes/Clostridia/Lachnospiraceae | Log fold change | p -value | FDR |
| Main effects | |||
| Immunomodulators | -0.347 | 0.0068 | 0.0068 |
| Firmicutes/Clostridia/Ruminococcaceae | Log fold change | p -value | FDR |
| Main effects | |||
| ileal CD | -0.989 | 0.0001 | 0.0002 |
| Colitis | -0.244 | 0.2714 | 0.2714 |
| Age | -0.017 | 0.0030 | 0.0032 |
| Immunomodulators | 1.879 | 0.0160 | 0.0224 |
| BMI | 0.0282 | 0.0674 | 0.0787 |
| First order interactions | |||
| Immunomodulators * BMI | -0.939 | 0.0012 | 0.0027 |
| Firmicutes/Clostridia/Other | Log fold change | p -value | FDR |
| NOD2R | 0.569 | 0.0028 | 0.0042 |
| ORMDL3R | 0.716 | 0.0001 | 0.0003 |
| XBP1R | -1.000 | 0.0001 | 0.0003 |
| + C. difficile fecal toxin | 1.365 | 0.0001 | 0.0003 |
| Steroids | 0.606 | 0.0026 | 0.0042 |
| Immunomodulators | -2.387 | 0.0027 | 0.0042 |
| BMI | -0.0416 | 0.0065 | 0.0081 |
| Colon Cancer | -0.0384 | 0.8703 | 0.8703 |
| Current smoking | -0.3546 | 0.2724 | 0.3143 |
| First order interactions | |||
| Current smoking * 5-ASA | 1.9875 | <0.0001 | 0.0002 |
| Current smoking * Steroids | -1.287 | 0.006 | 0.0081 |
| Immunomodulators * BMI | 0.1074 | 0.0002 | 0.0005 |
| Firmicutes/Bacilli | Log fold change | p-value | FDR |
| Main effects | |||
| ileal CD | 1.7384 | <0.0001 | <0.0001 |
| Colitis | 1.2696 | <0.0001 | <0.0001 |
| Batch | -0.9925 | <0.0001 | <0.0001 |
| 5-ASA | -0.8868 | 0.0049 | 0.0057 |
| Steroids | -0.3359 | 0.2659 | 0.2659 |
| First order interactions | |||
| 5-ASA*Steroids | 1.4087 | 0.0014 | 0.0019 |
| Proteobacteria | Log fold change | p-value | FDR |
| Main effects | |||
| ileal CD | 0.6767 | <0.0001 | 0.0001 |
| Colitis | 0.5257 | 0.002 | 0.0023 |
| NOD2R | -1.0788 | 0.0002 | 0.0003 |
| ORMDL3R | -0.2539 | 0.1433 | 0.1433 |
| First order interactions | |||
| NOD2R* ORMDL3R | 1.3856 | <0.0001 | 0.0001 |
The relative abundance of the Actinobacteria phylum was positively associated with both ileal CD and colitis phenotype and negatively associated with detection of C. difficile fecal toxin and anti-TNFα biologic use within 8 weeks of surgery. Significant first order interactions were observed for C. difficile fecal toxin b*age, C. difficile fecal toxin * current smoker, and IBD duration * anti-TNFα use.
The relative abundance of the Firmicutes/Clostridia/Lachnospiraceae (Clostridia Group XIVa) family was negatively associated with immunomodulatory use. The relative abundance of C. coccoides-E. rectales bacterial group determined by qPCR corresponds roughly to the relative abundance of the Clostridia/Lachnospiraceae family [4]. Univariate analysis did not detect a significant difference between NOD2R and NOD2NR ileal CD patients as previously reported for the Batch 1 samples [4]. A linear regression model (see Table 4) detected a positive association with the colitis phenotype. There was a trend towards a negative association between relative abundance of the C. coccoides-E. rectales group and immunomodulatory use and a significant association with the immunomodulator * C. difficile fecal toxin first order interactions.
Table 4. Linear regression results for disease phenotype, IBD genotype and additional clinical covariates (see Table 1) and the relative abundance of the C. coccoides-E. rectales bacterial subgroup and F. prausnitzii.
Log fold change was determined by real time qPCR. Variables with significant effects were bolded.
| C. coccoides-E-rectales bacterial subgroup | Log fold change | p-value | FDR |
|---|---|---|---|
| Main effects | |||
| ileal CD | 0.129 | 0.772 | 0.772 |
| Colitis | 1.456 | 0.002 | 0.004 |
| Immunomodulators | -0.762 | 0.095 | 0.143 |
| C. difficile fecal toxin | -1.08 | 0.290 | 0.347 |
| First order interactions | |||
| Immunomodulators* C. difficile fecal toxin | 4.71 | 0.001 | 0.004 |
| Faecalibacterium prausnitzii | Log fold change | p-value | FDR |
| Main effects | |||
| ileal CD | -1.691 | 0.001 | 0.001 |
| Colitis | -0.930 | 0.074 | 0.074 |
The relative abundance of the Firmicutes/Clostridia/Ruminococcaceae (Clostridia Group IV) family was strongly negatively associated with ileal CD phenotype and to a lesser extent with age, and positively associated with immunomodulator use. A significant first order interaction was observed for immunomodulators * BMI (see Table 3). At a more granular level, the relative abundance of the Faecalibacterium genus within the Ruminococcaceae family revealed significant negative associations with ileal CD phenotype (FDR = 0.0004) and age (FDR = 0.004), and significant positive associations with immunomodulatory use (FDR = 0.004) and BMI (FDR = 0.0006). In addition, a significant first order interaction was observed for immunomodulators * BMI (FDR = 0.0007). The negative association with ileal CD was further confirmed by measuring the relative abundance of F. prausnitzii by qPCR (FDR = 0.001, see Table 4). In addition, within the Ruminococcaceae (Clostridia Group IV) family, the relative abundance of the Subdoligranulum genus was also negative associated with the ileal CD phenotype (FDR < 0.0001) and colitis phenotype (FDR = 0.006).
Multiple associations, including IBD genotype (NOD2R, ORMDL3R, XBP1R), medications (steroids, immunomodulators), C. difficile fecal toxin and BMI were detected with the relative abundance of the remaining Clostridia taxa binned as Firmicutes/Clostridia/Other (see Table 3). Exploratory analyses did not detect associations between these three IBD genotypes with the predominant families binned within this group, such as Clostridiales/Clostridiaceae or Clostridiales/Peptostreptococcaceae.
The relative abundance of the Firmicutes/Bacilli class was positively associated with both ileal CD and colitis phenotype and negatively associated with 5-ASA (see Table 3). A significant first order interaction was observed for 5-ASA * Steroids. At a more granular level, similar associations with ileal CD (FDR = 0.04) and colitis (FDR = 0.02) phenotypes and negatively associated with 5-ASA (FDR = 0.003) were observed for the Streptococcaceae family, which is binned within this subphylum.
The relative abundance of the Proteobacteria phylum was positively associated with ileal CD and colitis phenotypes, but negatively associated with NOD2 genotype (see Table 3). Significant first order interactions were observed for NOD2R * ORMDL3R genotype and 5-ASA * anti-TNFα. Similar associations were observed for the relative abundance of the Pseudomonadaceae family and ileal CD phenotype (FDR <0.0001) and colitis phenotype (FDR = 0.0006). A similar association was observed between the relative abundance of the Pseudomonas genus and ileal CD phenotype (FDR <0.0001). In addition, significant associations were detected with XBP1R (FDR = 0.0004) and with NOD2R*XBP1R first order interactions (FDR = 0.005). Although the negative association between relative abundance of the Enterobacteriaceae family and NOD2Rgenotype did not reach significance (FDR = 0.15, FDR threshold = 0.05), a significant association was detected with first order NOD2R * IRGMR interaction. Finally, significant associations were detected between the Enterobacteriaceae family and ATG16L1R genotype (FDR = 0.03), 5-ASA (FDR = 0.035), BMI (FDR = 0.001), and 5-ASA * BMI first order interactions (FDR = 0.01).
No significant effects were detected for the relative abundance of the remaining phyla/subphyla categories including the Bacteroidetes phylum.
Microbial biomarkers of post-operative endoscopic recurrence in ileal CD subjects
Of the 124 ileal CD subjects included in this study, 61 subjects underwent postoperative ileoscopy of the neo-terminal ileum 6–12 months after surgery. The distribution of subjects for each Rutgeerts score were as follows: i0, n = 22; i1, n = 12; i2, n = 15; i3, n = 7; i4, n = 5. Therefore, 34 ileal CD subjects did not subsequently develop endoscopic recurrence (i0-i1) in the neo-terminal ileum, whereas the remaining 27 ileal CD subjects did subsequently develop endoscopic recurrence (i2-i4). Further analyses were conducted to compare the microbial composition of the disease unaffected region of the resected ileum in the 27 subjects who subsequently developed endoscopic recurrence with that of the 34 subjects who did not.
No difference in alpha- or beta-diversity at the phylum, family, or genus level, was detected in the disease unaffected specimens collected from the ileal CD subjects who did or did not subsequently develop recurrence. To identify potential microbial biomarkers of recurrence, univariate negative binomial analyses were conducted to identify taxa with significant differences in relative abundances between the ileal CD subjects who did or did not subsequently develop recurrence. No significant difference was observed when the bacteria were binned at the phyla/subphyla level. At the family level, the relative abundance of a number of bacterial taxa, including two potentially pathogenic families, Pasteurellaceae and Mycobacteriaceae, differed between ileal CD patients with and without recurrence (see Table 5). However, although the relative abundance of the Ruminococcaceae family was decreased in ileal CD patients who subsequently developed endoscopic recurrence compared to those who did not, the difference did not reach statistical significance. QPCR measurements of the relative abundance of C. Coccoides-E. rectales (i.e., members of the Lachnospiraceae family), and F. prausnitzii (i.e., an abundant species of the Ruminococcaceae family indicated significantly reduced levels of both taxa in patients with endoscopic recurrence (see Table 4). The difference between the qPCR and 16S rRNA sequence count data may reflect in part the looser correlation between qPCR and sequencing compared to the correlation between different sequencing platforms [73].
Table 5. Univariate negative binomial results (p-value and FDR) for endoscopic recurrence: family-level analysis.
Only families with FDR corrected p-values <0.05 are included.
| Ileal CD | p-value | FDR | ||
|---|---|---|---|---|
| Increased in recurrence | Recurrence Mean ±std |
No recurrence Mean ±std |
||
|
Proteobacteria/Gammaproteobacteria/ Pasteurellales/Pasteurellaceae |
0.0467 ±0.1581 |
0.0173 ±0.0664 |
1E-5 | 0.002 |
|
Proteobacteria/Alphaproteobacteria/ Rhodospirillales/ Acetobacteraceae |
0.0005 ± 0.1583 |
0.0000 ±0.0001 |
5E-5 | 0.004 |
|
Firmicutes/Clostridia/Clostridiales/ Other |
0.0354 ± 0.1405 |
0.0022 ± 0.0046 |
4E-4 | 0.016 |
|
Actinobacteria/Corynebacteriales/ Mycobacteriaceae |
0.0001 ±0.0003 |
0.0000 ±0.0003 |
0.001 | 0.026 |
| Decreased in recurrence | ||||
|
Proteobacteria/Alphaproteobacteria/ Rhizobiales/ Methylobacteriaceae |
0.0092 ± 0.023 |
0.0137 ± 0.0505 |
1E-4 | 0.006 |
|
Proteobacteria/Betaproteobacteria/ Burkholderiales/ Comamonadaceae |
0.0028 ± 0.0044 |
0.018 ± 0.0663 |
0.001 | 0.02 |
| Proteobacteria/Gammaproteobacteria/ Pseudomonadales/Moraxellaceae | 0.001 ± 0.002 |
0.0185 ± 0.0799 |
0.001 | 0.02 |
|
Proteobacteria/Betaproteobacteria/ Burkholderiales/ Alcaligenaceae |
0.004 ±0.005 |
0.018 ± 0.02 |
0.001 | 0.02 |
|
Firmicutes/Erysipelotrichi/ Erysipelotrichales/Erysipelotrichaceae |
0.0001 ± 0.0003 |
0.007 ± 0.030 |
0.002 | 0.03 |
Discussion
The current studies report findings from an integrative approach to examine the relationships between host genetic factors, clinical factors, and dysbiosis in an expanded IBD patient dataset collected from three institutions with approximately double the number of subjects than was previously reported [4]. This study focused on ileal CD phenotypes and, to reduce heterogeneity, included two non-overlapping phenotypes without ileal CD: 1) non-IBD subjects and 2) colitis subjects without evidence of ileal disease. It is important to note however that the IBD genotype profiles associated with UC/indeterminate colitis, Crohn’s colitis and ileal CD/ileocolonic CD phenotypes represent a continuum [9]. Our analysis emphasizes samples taken from macroscopically disease-unaffected regions of the ileum, because we detected significant differences in ileal microbiota between samples obtained from adjacent disease-affected regions of the ileum, and included only samples from initial ileocolic resections. This is because increased reflux of colonic content into the neo-terminal ileum, would be anticipated after surgical removal of the ileocolic valve.
This study linking IBD genotype and clinical covariates to ileal mucosal samples represents one of the larger patient cohorts and is comparable in size to a previous study, which combined ileal mucosal biopsies from three institutions in Boston, Toronto and the Netherlands [55]. The V3-V5 pyrosequencing platform used in the current study, differs from the previous study which utilized the V4 Illumina sequencing [55]. The Illumina sequencing platform is generally conducted at a greater depth of sequencing (9,55,76–79), however Illumina sequencing read lengths (V1-V2, V3-V4, V4) are shorter than the pyrosequencing read length (V3-V5),. In the current study, significant differences in α-diversity were not detected between the three phenotypes, possibly because of the lower depth of sequencing using the pyrosequencing platform.
The dimensions of the linked data sets in the current study were reduced for integrative analysis by restricting the number of distinct non overlapping IBD phenotypes to 3, binning the taxa into 8 major phyla/subphyla categories, and by restricting the IBD loci to 6 that have previously been associated with innate immunity. The analysis was also restricted to macroscopically normal appearing disease unaffected ileum at the time of initial surgical resection, in contrast to the previous study where the ileal samples analyzed were more heterogeneous [55]. Our study is also unique in that detection of fecal C. difficile toxin (primarily in colitis subjects) was included as a potentially confounding co-variate. The rationale for including this co-variate is based on our previous study of Batch 1 subjects [4], and a more recent study analyzing the effect of FMT demonstrated marked dysbiosis associated with recurrent C. difficile infections in patients with and without ulcerative colitis compared to healthy subjects and compared to ulcerative colitis patients without C. difficile infections [74]. However it remains difficult to determine whether the marked dysbiosis is related to C. difficile infection or antibiotic treatment of the infection.
This expanded study confirmed our previous report [4] linking NOD2R genotype, C. difficile infection, and IBD phenotype to ileal dysbiosis in Batch1 subjects alone, despite the addition of four additional genetic loci associated with defects in innate immunity (IRGM, CARD9, XBP1, and ORMDL3) to the analysis. In our previous study [3], NOD2R genotype was linked to the relative abundance of the Proteobacteria phylum (p = 0.016, FDR = 0.07) independent of IBD phenotype. In this current expanded study, this association now reached significance (FDR = 0.0003). The observation that while ileal CD phenotype is positively associated with the relative abundance of Proteobacteria phylum in disease unaffected ileal mucosa, NOD2R genotype is negatively associated is somewhat puzzling. It may be related to the observation that the mean relative abundance of Proteobacteria was consistently lower in disease affected ileal mucosa than in adjacent disease unaffected mucosa in ileal CD subjects. Both observations reinforce the concept that dysbiosis is detectable in macroscopically normal ileal mucosa in ileal CD subjects
Aggregate analysis at the phylum/subphylum level could mask the contribution of individual taxa within each of these groups, particularly if significant associations with phylogenetically related taxa exhibit opposing polarities. Identifying significant associations with bacterial subcategories at the family or genus level likely was limited by the reduced relative abundance of the individual taxa and the increased number of multiple comparisons. We are currently pursuing deeper sequencing and expanding the patient cohort to increase statistical power to detect differences in lower-level taxa. Nonetheless, at a more granular level, a possible effect of NOD2 genotype was detected through NOD2 * XBP1 first order interactions with the relative abundance of Pseudomonas genus, and through the NOD2*IRGM first order interaction with the relative abundance of the Enterobacteriaceae family (FDR <0.05). The NOD2*IRGM interaction may reflect, at least in part, direct interactions between the IRGM, NOD2, and ATG16L1 gene products, which have been reported to form a molecular complex that modulates autophagic reactions to microbial products [8]. NOD2R genotype was not significantly associated with lower Enterobacteriaceae (FDR = 0.15) based on our threshold of FDR = 0.05 in the current study, but the results suggest a trend. In contrast, NOD2 risk dosage has been previously correlated with higher Enterobacteriaceae (FDR = 0.11, FDR threshold = 0.25) in ileal samples collected in two of three cohorts in the previous study (Boston and the Netherlands, n = 314) [55]. In summary, while both the current and the previous IBD genotype–ileal microbiota studies [55] detect associations between NOD2 risk alleles and Proteobacteria taxa, these associations had opposite polarities. This may relate to the analysis of only macroscopically disease unaffected ileal sample obtained from initial ileal resection in three relatively distinct IBD subphenotypes (ileal CD, colitis and non-IBD) in the current study.
NOD2 and other additional IBD genotypes (ORMDL3, XBP1) were also linked to the relative abundance of Clostridia spp. not binned into either the Ruminococcaceae or Lachnospiraceae families, but we were unable to attribute this association to any of the taxa within this bacterial category. In the expanded study we could not confirm the increased abundance of C. coccoides–E. rectales group and F. prausnitzii previously detected by PCR analysis in NOD2R ileal CD subjects compared to NOD2NR ileal CD subjects in Batch 1 alone [4]
A recent analysis of the effect of NOD2 homozygotes and compound heterozygotes compared to wild type NOD2 homozygotes in CD patients in clinical remission and in non-IBD controls detected no significant differences in the fecal microbiota [75]. This may be due in part to the smaller sample size, to the asymptomatic status of the CD patients, and to compositional differences between ileal microbiota and fecal microbiota [76–78].
Associations between ileal CD phenotype were confirmed with the decreased relative abundance of the Ruminococcaceae family and increased relative abundances of the Actinobacteria phylum and the Firmicutes/Bacillus class. These analyses also confirmed similar associations between the relative abundances of F. prausntizii within the Ruminococcaceae family, the Corynebacteriaceae family in the Actinobacteria phylum, and the Streptococcaceae family in the Bacillus class.
The current study has a larger or comparable cohort size compared with previous studies on microbial predictors of post-ICR endoscopic recurrence at the time of resection [17,22–25,79]. Q-PCR analysis of F. prausnitzii abundance confirmed that there was reduced abundance of this species in subjects that subsequently went on to develop endoscopic recurrence at 6–12 months compared to those that did not develop recurrence. In addition exploratory analysis identified two potentially pathogenic taxa, Pasteurellaceae and Mycobacteriaceae, with increased abundance in subjects that subsequently developed endoscopic recurrence compared to those that did not. Atypical mycobacteria have been implicated in the pathogenesis of CD [80]. The abundance of Pasteurellaceae is increased in fecal microbiota collected from treatment-naïve new-onset CD subjects compared to non-IBD subjects [21]. Somewhat puzzling is a report from a recent study [78] that the abundance of Pasteurellaceae is increased in the resected ileum collected from patients that remain in remission compared to those that have endoscopic recurrence. It remains to be determined whether the taxa associated with endoscopic recurrence simply reflect subtle alterations in the local environment, such as changes in pH or oxidation, or whether they play a causal role in promoting recurrent inflammation in the peri-anastomotic neo-terminal ileum.
Our study likely was limited by the batch effect observed, most notably for the relative abundance of the Actinobacteria phylum and the Firmicutes/Bacilli class between Batch 1 (2005–2010) and Batch 2 (2010–2012) cohorts. Several differences between the two patient sample cohorts, Batch 1 and 2, could contribute to this effect. Despite the use of the same primers for library construction and the same pyrosequencing platforms, which generated concordant results in a preliminary duplicate analysis of a small subset of samples, some systematic technical errors may have contributed to batch effects. Another factor could be related to demographic differences between Batch 1 and Batch 2 patients, which included a higher proportion of NOD2R non-IBD patients, a higher proportion of patients treated with anti-TNFα biologics, and a lower proportion of patients treated with 5-ASA medications in Batch 2 compared to Batch 1 (see Table 1) This study was also limited with respect to assessing the effect of antibiotics, because all of the patient received antibiotics prior to the collection of the samples during surgical resection except <10 non IBD subjects, who contributed endoscopic biopsies in Batch 2, where they received no antibiotics.
In summary, the results of this integrative analysis of a large number of uniformly curated samples, confirm a significant effect of IBD phenotype, C. difficile infection, and NOD2 genotype on ileum-associated microbiota. Furthermore, we present data demonstrating that additional IBD-related genotypes, specifically alleles of ORMDL3 and XBP1, are associated with changes in the ileal microbiome at the phyla/subphyla level, particularly Proteobacteria, either as direct effects or through interactions with NOD2 or other clinical variables. As we continue to expand our systematic accrual of subjects, further associations between host, environment, and microbial factors with ileal CD phenotype and clinical outcome will emerge and help delineate the complex etiology of this disease.
Supporting information
(DOCX)
(DOCX)
Acknowledgments
The authors thank the patients who have contributed their medical information, blood and tissue samples to the Digestive Diseases Research Core Center (DDRCC) Clinical Database, the faculty of the Section of Colon and Rectal Surgery and the Division of Gastroenterology at Washington University, Stony Brook University and the University of North Carolina.
Data Availability
Clinical, genotyping, and sequencing data can be accessed through the dbGAP authorized access system (Request access to: phs000255.v2). In order to request access to any of the individual-level datasets within the controlled-access portions of the database, the Principal Investigator (PI) and the Signing Official (SO) at the investigator’s institution will need to co-sign a request for data access, which will be reviewed by an NIH Data Access Committee at the appropriate NIH Institute or Center (https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?page=login).
Funding Statement
This work was supported in part by the National Institute of Health grants P30DK52574 (N.O.D.), UH2DK083994 (E.L.), and HG005964 (D.N.F.), the Barnes-Jewish Foundation (E.L. donation from Dr. Carl Lyss and the Lenauers), The Barnes-Jewish Hospital Foundation, the MAC Crohn's and Colitis Foundation senior Research award Ref.370763, the Pakula IBD Innovation Fund at Washington University St. Louis and the Givin' It All for Guts Foundation https://givinitallforguts.org/(M.C.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sartor RB, Wu GD. Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology. 2017. February;152(2):327–339.e4. 10.1053/j.gastro.2016.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frank DN, Robertson CE, Hamm CM, Kpadeh Z, Zhang T, Chen H, et al. Disease phenotype and genotype are associated with shifts in intestinal-associated microbiota in inflammatory bowel diseases. Inflamm Bowel Dis. 2011;17:179–84. 10.1002/ibd.21339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frank DN, Zhu W, Sartor RB, Li E. Investigating the biological and clinical significance of human dysbioses. Trends Microbiol. 2011;19, 427–434. 10.1016/j.tim.2011.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li E, Hamm CM, Gulati AS, Sartor RB, Chen H, Wu X, et al. Inflammatory bowel diseases phenotype, C. difficile and NOD2 genotype are associated with shifts in human ileum associated microbial composition. PLoS One. 2012;7:e26284 10.1371/journal.pone.0026284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang T, DeSimone RA, Jiao X, Rohlf FJ, Zhu W, Gong QQ, et al. Host genes related to paneth cells and xenobiotic metabolism are associated with shifts in human ileum-associated microbial composition. PLoS One. 2012;7:e30044 10.1371/journal.pone.0030044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoefkens E, Nys K, John JM, Van Steen K, Arijs I, van der Goten J, et al. Genetic association and functional role of Crohn disease risk alleles involved in microbial sensing, autophagy, and endoplasmic reticulum (ER) stress. Autophagy. 2013;9:2046–55. 10.4161/auto.26337 [DOI] [PubMed] [Google Scholar]
- 7.Fritz T, Niederreiter L, Adolph T, Blumberg RS, Kaser A. Crohn's disease: NOD2, autophagy and ER stress converge. Gut. 2011;60:1580–8. 10.1136/gut.2009.206466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chauhan S, Mandell MA, Deretic V. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Mol Cell. 2015;58:507–21. 10.1016/j.molcel.2015.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner J, Sim W H, Ellis J A, Ong E K, Catto-Smith A G, Cameron D J, et al. Interaction of Crohn’s Disease susceptibility genes in an Australian paediatric cohort. PLoS ONE, 2010; 5: e15376 10.1371/journal.pone.0015376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleynen I, Boucher G, Jostins L, Schumm LP, Zeissig S, Ahmad T, et al. Inherited determinants of Crohn's disease and ulcerative colitis phenotypes: a genetic association study. Lancet. 2016;387:156–67. 10.1016/S0140-6736(15)00465-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strong SA, Koltun WA, Hyman NH, Buie WD; Standards Practice Task Force of The American Society of Colon and Rectal Surgeons. Practice parameters for the surgical management of Crohn’s disease. Dis Colon Rectum. 2007;50:1735–46. 10.1007/s10350-007-9012-7 [DOI] [PubMed] [Google Scholar]
- 12.Farmer RG, Whelan G, Fazio VW. Long-term follow-up of patients with CD. Relationship between the clinical pattern and prognosis. Gastroenterology. 1985;88:1818–25. [DOI] [PubMed] [Google Scholar]
- 13.Bernell O, Lapidus A, Hellers G. Risk factors for surgery and recurrence in 907 patients with primary ileocaecal CD. Br J Surg. 2000;87:1697–701. 10.1046/j.1365-2168.2000.01589.x [DOI] [PubMed] [Google Scholar]
- 14.Unkart JT, Anderson L, Li E, Miller C, Yan Y, Gu CC, et al. Risk factors for surgical recurrence after ileocolic resection of Crohn's disease. Dis Colon Rectum. 2008;51:1211–6. 10.1007/s10350-008-9348-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunningham MF, Doherty NG, Coffey JC, Burke JP, O'Connell PR. Postsurgical recurrence of ileal Crohn’s disease: an update on risk factors and intervention points to a central role for impaired host-microflora homeostasis. World J. Surg 2010; 34: 1615–26. 10.1007/s00268-010-0504-6 [DOI] [PubMed] [Google Scholar]
- 16.Manser CN, Frei P, Grandinetti T, Biedermann L, Mwinyi J, Vavricka SR, et al. Risk factors for repetitive ileocolic resection in patients with Crohn's disease: results of an observational cohort study. Inflamm Bowel Dis. 2014;20:1548–54. 10.1097/MIB.0000000000000123 [DOI] [PubMed] [Google Scholar]
- 17.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, et al. Faecalibacterium prausnitzii is an anti inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008; 105: 16731–16736. 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Willing BP, Dicksved J, Halfvarson J, Andersson AF, Lucio M, Zheng Z, et al. A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology. 2010; 139:1844–1854.e1. 10.1053/j.gastro.2010.08.049 [DOI] [PubMed] [Google Scholar]
- 19.Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 2012; 13: R79 10.1186/gb-2012-13-9-r79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tong M, Li X, Wegener Parfrey L, Ippoliti A, Wei B, Borneman J, et al. A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease. PLoS One 2013;8:e80702 10.1371/journal.pone.0080702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014;15:382–92. 10.1016/j.chom.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, et al. Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe 2015; 17: 662–671. 10.1016/j.chom.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Cruz P, Kang S, Wagner J, Buckley M, Sim WH, Prideaux L, et al. Association between specific mucosa-associated microbiota in Crohn's disease at the time of resection and subsequent disease recurrence: a pilot study. J Gastroenterol Hepatol. 2015;30:268–78. 10.1111/jgh.12694 [DOI] [PubMed] [Google Scholar]
- 24.Dey N, Soergel DA, Repo S, Brenner SE. Association of gut microbiota with post-operative clinical course in Crohn's disease. BMC Gastroenterol. 2013;13:131 10.1186/1471-230X-13-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mondot S, Lepage P, Seksik P, Allez M, Tréton X, Bouhnik Y, et al. Structural robustness of the gut mucosal microbiota is associated with Crohn's disease remission after surgery. Gut. 2015; pii: gutjnl-2015-309184. 10.1136/gutjnl-2015-30918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satsangi J, Silverberg MS, Vermeire S, Colombel JF. The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut. 2006;55:749–53 10.1136/gut.2005.082909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Assche G, Dignass A, Panes J, Beaugerie L, Karagiannis J, et al. The second European evidence-based Consensus on the diagnosis and management of Crohn's disease: Definitions and diagnosis. J Crohns Colitis. 2010;4:7–27. 10.1016/j.crohns.2009.12.003 [DOI] [PubMed] [Google Scholar]
- 28.Geboes K, Van Eyken P. Inflammatory bowel disease unclassified and indeterminate colitis: the role of the pathologist. J Clin Pathol. 2009. March;62:201–5. 10.1136/jcp.2008.059311 [DOI] [PubMed] [Google Scholar]
- 29.Rutgeerts P, Geboes K, Vantrappen G, Beyls J, Kerremans R, Hiele M. Predictability of the postoperative course of Crohn's disease. Gastroenterol. 1999; 99:956–63. [DOI] [PubMed] [Google Scholar]
- 30.Aldhous MC, Drummond HE, Anderson N, Smith LA, Arnott ID, et al. Does cigarette smoking influence the phenotype of Crohn's disease? Analysis using the Montreal classification. Am J Gastroenterol. 2007;102:577–88. 10.1111/j.1572-0241.2007.01064.x [DOI] [PubMed] [Google Scholar]
- 31.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–3. 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- 32.Reddy SS, Brandt LJ. Clostridium difficile infection and inflammatory bowel disease. J Clin Gastroenterol. 2013;47:666–71. 10.1097/MCG.0b013e31828b288a [DOI] [PubMed] [Google Scholar]
- 33.Humphries RM, Uslan DZ, Rubin Z. Performance of Clostridium difficile toxin enzyme immunoassay and nucleic acid amplification tests stratified by patient disease severity. J. Clin. Microbiol. 2013;51:869–873. 10.1128/JCM.02970-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis JD, Chen EZ, Baldassano RN, Otley AR, Griffiths AM, Lee D, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn's disease. Cell Host Microbe. 2015;18:489–500. 10.1016/j.chom.2015.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson RL, Gladman E, Barbaleskovic M. Antimicrobial prophylaxis for colorectal surgery. Cochrane Database Syst Rev. 2014. May 9;(5):CD001181 10.1002/14651858.CD001181.pub4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cézard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. 10.1038/35079107 [DOI] [PubMed] [Google Scholar]
- 37.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–6. 10.1038/35079114 [DOI] [PubMed] [Google Scholar]
- 38.Cuthbert AP, Fisher SA, Mirza MM, King K, Hampe J, Croucher PJ, et al. The contribution of NOD2 gene mutations to the risk and site of disease in inflammatory bowel disease. Gastroenterology. 2002;122:867–74. [DOI] [PubMed] [Google Scholar]
- 39.Ogura Y, Lala S, Xin W, Smith E, Dowds TA, Chen FF et al. Expression of NOD2 in Paneth cells: a possible link to Crohn's ileitis. Gut. 2003;52:1591–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen H, Lee A, Bowcock A, Zhu W, Li E, et al. Influence of Crohn's disease risk alleles and smoking on disease location. Dis Colon Rectum. 2011;54:1020–5. 10.1007/DCR.0b013e31821b94b3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Germain A, Guéant RM, Chamaillard M, Allen PB, Bresler L, Guéant JL, et al. NOD2 gene variant is a risk factor for postoperative complications in patients with Crohn's disease: A genetic association study. Surgery. 2016; March 2. [DOI] [PubMed] [Google Scholar]
- 42.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, et al. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. 10.1038/ng2032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prescott NJ, Fisher SA, Franke A, Hampe J, Onnie CM, Soars D, et al. A nonsynonymous SNP in ATG16L1 predisposes to ileal Crohn's disease and is independent of CARD15 and IBD5. Gastroenterology. 2007;132:1665–71. 10.1053/j.gastro.2007.03.034 [DOI] [PubMed] [Google Scholar]
- 44.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–63. 10.1038/nature07416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.VanDussen KL, Liu TC, Li D, Towfic F, Modiano N, Winter R, et al. Genetic variants synthesize to produce paneth cell phenotypes that define subtypes of Crohn's disease. Gastroenterology. 2014;146:200–9. 10.1053/j.gastro.2013.09.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCarroll SA, Huett A, Kuballa P, Chilewski SD, Landry A, Goyette P, et al. Deletion polymorphism upstream of IRGM associated with altered IRGM expression and Crohn's disease. Nat Genet. 2008;40:1107–12. 10.1038/ng.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberts RL, Hollis-Moffatt JE, Gearry RB, Kennedy MA, Barclay ML, et al. Confirmation of association of IRGM and NCF4 with ileal Crohn's disease in a population-based cohort. Genes Immun. 2008;9:561–5. 10.1038/gene.2008.49 [DOI] [PubMed] [Google Scholar]
- 48.Liu B, Gulati AS, Cantillana V, Henry SC, Schmidt EA, Daniell X, et al. Irgm1-deficient mice exhibit Paneth cell abnormalities and increased susceptibility to acute intestinal inflammation. Am J Physiol Gastrointest Liver Physiol. 2013;305:G573–84. 10.1152/ajpgi.00071.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsu YM, Zhang Y, You Y, Wang D, Li H, Duramad O, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat Immunol. 2007;8:198–205. 10.1038/ni1426 [DOI] [PubMed] [Google Scholar]
- 50.Zhernakova A, Festen EM, Franke L, Trynka G, van Diemen CC, Monsuur AJ, et al. Genetic analysis of innate immunity in Crohn's disease and ulcerative colitis identifies two susceptibility loci harboring CARD9 and IL18RAP. Am J Hum Genet. 2008;82:1202–10. 10.1016/j.ajhg.2008.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, Tilg H, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–56. 10.1016/j.cell.2008.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–62. 10.1038/ng.175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cortes A, Brown MA. Promise and pitfalls of the Immunochip. Arthritis Res Ther. 2011;13:101 10.1186/ar3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012. November 1;491(7422):119–24. 10.1038/nature11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knights D, Silverberg MS, Weersma RK, Gevers D, Dijkstra G, et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 2014;6:107 10.1186/s13073-014-0107-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jumpstart Consortium Human Microbiome Project Data Generation Working Group. Evaluation of 16S rDNA-based community profiling for human microbiome research. PLoS One. 2012;7:e39315 10.1371/journal.pone.0039315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frank DN. BARCRAWL and BARTAB: software tools for the design and implementation of barcoded primers for highly multiplexed DNA sequencing. BMC Bioinformatics. 2009;10:362 10.1186/1471-2105-10-362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011; 27: 2194–2200. 10.1093/bioinformatics/btr381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schloss PD, Westcott SL. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol. 2011;77:3219–26. 10.1128/AEM.02810-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pruesse E, Peplies J, Glockner FO. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–9. 28: 1823–29. 10.1093/bioinformatics/bts252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41(Database issue):D590–6. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maeda H, Fujimoto C, Haruki Y, Maeda T, Kokeguchi S, Petelin M, et al. Quantitative real-time PCR using TaqMan and SYBR Green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol Med Microbiol. 2003. 24;39:81–6. [DOI] [PubMed] [Google Scholar]
- 63.Rinttilä T, Kassinen A, Malinen E, Krogius L, Palva A. Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol. 2004;97:1166–77. 10.1111/j.1365-2672.2004.02409.x [DOI] [PubMed] [Google Scholar]
- 64.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A. 2007;104:13780–5. 10.1073/pnas.0706625104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robertson CD, Harris JK, Wagner BD, Granger D, Browne K, Tatem B, et al. (2013) Explicet: graphical user interface software for metadata driven management, analysis and visualization of microbiome data. Bioinformatics 29:3100–01. 10.1093/bioinformatics/btt526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Frank DN, Bales ES, Monks J, Jackman MJ, MacLean PS, Ir D, et al. (2015) Perilipin-2 modulates lipid absorption and microbiome responses in the mouse intestine. PLoS One 10:e0131944 10.1371/journal.pone.0131944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oksanen J, Kindt R, Legendre P, O'Hara B, Simpson GL, Solymos P, et al. (2008) Vegan: Community Ecology Package. R package version 1.15–1. http://cran.r-project.org/, http://vegan.r-forge.r-project.org. [Google Scholar]
- 68.Anderson MJ A new method for non-parametric multivariate analysis of variance. Austral Ecology 2001; 26: 32–46. 10.1111/j.1442-9993.2001.01070.pp.x [DOI] [Google Scholar]
- 69.Anderson MJ, Crist TO, Chase JM, Vellend M, Inouye BD, Freestone AL, et al. Navigating the multiple meanings of β diversity: a roadmap for the practicing ecologist. Ecol Lett. 2011; 14:19–28. 10.1111/j.1461-0248.2010.01552.x [DOI] [PubMed] [Google Scholar]
- 70.Son JS, Zheng LJ, Rowehl LM, Tian X, Zhang Y, Zhu W, et al. Comparison of fecal microbiota in children with autism spectrum disorders and neurotypical siblings in the Simons Simplex Collection. PLoS One. 2015;10:e0137725 10.1371/journal.pone.0137725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10:e1003531 10.1371/journal.pcbi.1003531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B 1995; 57: 289–300. [Google Scholar]
- 73.Wu X, Berkow K, Frank DN, Li E, Gulati AS, Zhu W. Comparative analysis of microbiome measurement platforms using latent variable structural equation modeling. BMC Bioinformatics. 2013;14:79 10.1186/1471-2105-14-79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mintz M, Khair S, Grewal S, LaComb JF, Park J, Channer B, et al. Longitudinal microbiome analysis of single donor fecal microbiota transplantation in patients with recurrent Clostridium difficile infection and/or ulcerative colitis. PLoS One. 2018;13:e0190997 10.1371/journal.pone.0190997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kennedy NA, Lamb CA, Berry SH, Walker AW, Mansfield J, Parkes M, et al. The Impact of NOD2 Variants on Fecal Microbiota in Crohn's Disease and Controls Without Gastrointestinal Disease. Inflamm Bowel Dis. 2018;24:583–592. 10.1093/ibd/izx061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen L, Wang W, Zhou R, Ng SC, Li J, Huang M, et al. Characteristics of fecal and mucosa-associated microbiota in Chinese patients with inflammatory bowel disease. Medicine (Baltimore). 2014;93(8):e51 10.1097/MD.0000000000000051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen W, Liu F, Ling Z, Tong X, Xiang C. Human intestinal lumen and mucosa-associated microbiota in patients with colorectal cancer. PLoS One. 2012;7(6):e39743 10.1371/journal.pone.0039743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wright EK, Kamm MA, Wagner J, Teo SM, Cruz P, Hamilton AL, et al. Microbial Factors Associated with Postoperative Crohn's Disease Recurrence. J Crohns Colitis. 2017;11:191–203. 10.1093/ecco-jcc/jjw136 [DOI] [PubMed] [Google Scholar]
- 79.Pascal V, Pozuelo M, Borruel N, Casellas F, Campos D, Santiago A, et al. A microbial signature for Crohn's disease. Gut. 2017;66:813–822. 10.1136/gutjnl-2016-313235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liverani E, Scaioli E, Cardamone C, Dal Monte P, Belluzzi A. Mycobacterium avium subspecies paratuberculosis in the etiology of Crohn's disease, cause or epiphenomenon? World J Gastroenterol. 2014;20:13060–70. 10.3748/wjg.v20.i36.13060 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
(DOCX)
Data Availability Statement
Clinical, genotyping, and sequencing data can be accessed through the dbGAP authorized access system (Request access to: phs000255.v2). In order to request access to any of the individual-level datasets within the controlled-access portions of the database, the Principal Investigator (PI) and the Signing Official (SO) at the investigator’s institution will need to co-sign a request for data access, which will be reviewed by an NIH Data Access Committee at the appropriate NIH Institute or Center (https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?page=login).