Abstract
Objective
To determine the putative protective relationship of educational attainment on Alzheimer disease (AD) risk using Mendelian randomization and to test the hypothesis that by using genetic regions surrounding individually associated single nucleotide polymorphisms (SNPs) as the instrumental variable, we can identify genes that contribute to the relationship.
Methods
We performed Mendelian randomization using genome-wide association study summary statistics from studies of educational attainment and AD in two stages. Our instrumental variable comprised (1) 1,271 SNPs significantly associated with educational attainment and (2) individual 2-Mb regions surrounding the genome-wide significant SNPs.
Results
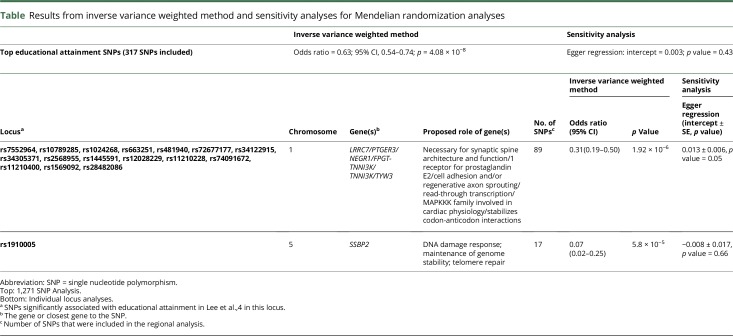
A causal inverse relationship between educational attainment and AD was identified by the 1,271 SNPs (odds ratio = 0.63; 95% confidence interval, 0.54–0.74; p = 4.08 x 10−8). Analysis of individual loci identified 2 regions that significantly replicated the causal relationship. Genes within these regions included LRRC2, SSBP2, and NEGR1; the latter a regulator of neuronal growth.
Conclusions
Educational attainment is an important protective factor for AD. Genomic regions that significantly paralleled the overall causal relationship contain genes expressed in neurons or involved in the regulation of neuronal development.
Education has consistently been identified as an important antecedent factor in Alzheimer disease (AD), whereby advanced educational attainment is thought to reduce AD risk.1,2 In addition, educational attainment and AD may be genetically related based on the genome-wide correlation of −0.31 (p = 4 × 10−4).3 However, confounding factors that affect educational attainment such as socioeconomic status, nutrition, and ethnicity blur the relationship.
Mendelian randomization limits confounding by using instrumental variables associated with a risk factor to establish effects on the outcome. The largest (n = 1,131,881) genome-wide association study (GWAS) of educational attainment4 identified 1,271 significantly associated single nucleotide polymorphisms (SNPs), and a separate 2013 GWAS from the International Genomics of Alzheimer's Project (IGAP) provided AD SNP association data. The data created an opportunity to examine the role of educational attainment in AD through Mendelian randomization.
In GWAS analyses, the significantly associated tag SNPs may not be the causal SNP.5 When pleiotropic effects and heterogeneity remain minimal, the instrumental variable composed of the index and all associated SNPs in a region improves the validity of the relationship,6 explains a greater percentage of variation in the phenotype, and reduces heterogeneous effects of using fewer SNPs.7
In addition to testing the overall hypothesis that educational attainment has a protective effect on AD risk, we tested the hypothesis that a subset of genetic loci related to educational attainment might individually significantly reflect the overall protective relationship with AD. To maximize detection, we included independent SNPs in a 2-Mb region surrounding each of the 1,271 SNPs significantly associated with educational attainment.3
Methods
We used available summary statistics from the analysis of all discovery data excluding the 23andMe cohort in the largest GWAS meta-analysis of educational attainment3 (n = 766,345). Summary statistics from the IGAP GWAS of AD,8 consisting of 54,162 individuals and 7,055,881 analyzed SNPs, were used for the outcome. There is a small overlap of samples used from the educational attainment and AD GWAS. Sample overlap in a 2-sample Mendelian randomization can cause bias in results; however, because of the large sample size of the educational attainment GWAS and minimal overlap of the sample, no appreciable bias was expected. The study population in both GWASs were of European descent.
SNP selection
TwoSampleMR, an R package that performs Mendelian randomization using data from MR-Base,9 was used in R (R v.3.3.1) to perform all analyses. Mendelian randomization was performed using two different schemes for selecting SNPs for the instrumental variable:
1,271 SNP analysis: the instrumental variables consisted of the 1,271 approximately independent SNPs (all with p < 5 × 10−8) significantly associated with educational attainment and were used to test for causality to AD. Independence for SNPs in the analysis was established using linkage disequilibrium (LD) clumping (r2 ≥ 0.001 within a 10,000-kb window).
Individual locus analyses: Mendelian randomization was performed independently on each of the 1,271 loci using SNPs within 1-Mb upstream and downstream of individual SNPs. We used more liberal inclusion criteria including SNPs associated with educational attainment at p < 0.01 and clumped at r2 ≥ 0.1 within a 250-kb window.
Of the 1,271 independent loci, SNPs found within a 2-Mb region of another were merged together, resulting in 441 independent loci.
Mendelian randomization analyses
The strictest LD threshold was applied for the first analysis, using default settings in MR-Base; for analysis 2, we allowed for more SNPs with still minimal LD to be included in the analysis, as we hypothesized that this would strengthen the per-loci analysis. Those SNPs remaining after LD clumping were queried within the IGAP summary statistics, if they were not present in that data set, genetic proxies were found, and finally SNPs for which no proxies could be found were excluded. Next, SNPs were harmonized for the effect allele between the 2 GWAS data sets or removed if harmonization predictions were inconclusive. For the joint 1,271 SNP analysis, 387 remained after LD clumping, and 13 SNPs were neither found in the IGAP nor had appropriate proxies and thus removed. Finally, 57 SNPs were removed because of low confidence that the effect allele of the exposure corresponded to the same allele in the outcome. Therefore, a total of 317 SNPs were included in the initial joint SNP analysis. For the regional analyses, SNPs were similarly removed if not found in the IGAP or if allele harmonization failed. The inverse variance weighted (IVW) method was used to calculate odds ratios (ORs). Results are reported as the OR (±95% confidence interval [CI]) of AD risk per SD increase in educational attainment in each test.
Sensitivity analyses
Mendelian randomization requires that that the instrumental variables meet 3 requirements: it must be associated with the risk factor, not associated with any confounder of the risk factor or outcome, and is only associated with the outcome through the risk factor. Sensitivity analyses following the initial analysis provide confidence that assumptions of the instrumental variables were not broken. In addition to the IVW, the weighted median method was used to measure causality and provided consistent results with IVW when at least 50% of the instrumental variables were valid. Next, the intercept of the Mendelian randomization–Egger test was used to determine potential horizontal pleiotropy or an effect of the instrumental variables on a phenotype other than the outcome. As the intercept neared zero in the mendelian randomization–Egger test, horizontal pleiotropy was reduced. The Steiger test of directionality was used to confirm directionality of the effect, i.e., that the SNPs first affected educational attainment and subsequently that AD risk was affected through educational attainment. Finally, the radial regression analyses were performed for the inverse variance methods, which identify any significant SNP outliers using the Cochran's Q-statistic.10
Results
Using the 1,271 SNPs associated with educational attainment, we detected statistically significant evidence for causation between educational attainment and AD such that an increase of 4.2 years of educational attainment was associated with a 37% reduction in AD risk (OR—scaled per SD, 4.2 years = 0.63; 95% CI, 0.54–0.74; p = 4.08 × 10−8).
In the per-loci analyses of the 1,271 regions, 2 independent SNP regions demonstrated a statistically significant inverse relationship between educational attainment and AD risk (Bonferroni-corrected threshold p < 1.2 × 10−4). These regions include the neuronal growth regulator precursor (NEGR1) gene, leucine-rich repeat containing 7 (LRCC7) gene, and prostaglandin E receptor 3 (PTGER3) gene (table 1).
Table.
Results from inverse variance weighted method and sensitivity analyses for Mendelian randomization analyses
Sensitivity analyses were performed for all Mendelian randomization analyses. There was no indication of pleiotropy, reverse causality, or heterogeneity, and the weighted median method showed consistent results with IVW overall in the significant analyses.
Discussion
Consistent with earlier reports, we found an inverse relationship of educational attainment with AD.11–13 In addition, we also identified regions that individually significantly replicated the causal relationship of education on AD, several of which were found to contain genes expressed in neurons or involved in regulation of neuronal development. For example, one of the SNPs is within the LRCC7 gene, which is crucial to dendritic spine architecture and function and may be involved in bipolar disorder.14 Another SNP within the intronic region of the NEGR1 gene is also highly expressed in neurons and has been found to be associated with major depressive disorder.15 An SNP was found in the PTGER3 gene, which is thought to be involved in the modulation of neurotransmitter release in neurons.
One limitation of using the Mendelian randomization method for outcomes with other strong nongenetic factors risk factors is that the CIs tend to be large. However, this outcome is preferable to the bias that is inherent in epidemiologic studies, which unavoidably include confounders.16 Taken together, the results presented here, along with earlier reports, establish a putative causal relationship between educational attainment and AD.
The key next steps are to replicate these findings in diverse ethnic groups with more variable educational experiences and to identify and validate specific variants within these loci that account for the association.
Acknowledgments
The authors thank the International Genomics of Alzheimer's Project and Lee et al. for providing summary statistics for this project.
Glossary
- AD
Alzheimer disease
- CI
confidence interval
- GWAS
genome-wide association study
- IGAP
International Genomics of Alzheimer's Project
- IVW
inverse variance weighted
- LD
linkage disequilibrium
- OR
odds ratio
- SNP
single nucleotide polymorphism
Author contributions
All authors of the study contributed to the study design, study analysis, and writing and editing.
Study funding
This work was supported by grants from the National Institute on Aging from the NIH (U01AG032984 and RO1AG041797) and the National Center for Advancing Translational Sciences (TL1TR001875).
Disclosure
N.S. Raghavan reports no disclosures. B. Vardarajan has served as a consultant for BISC Global. R. Mayeux has served as a consultant for the Alzheimer's Disease Center at Rush University Medical School and has received research support from the National Institute on Aging. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/NG.
References
- 1.Caamano-Isorna F, Corral M, Montes-Martinez A, Takkouche B. Education and dementia: a meta-analytic study. Neuroepidemiology 2006;26:226–232. [DOI] [PubMed] [Google Scholar]
- 2.Barnes DE, Yaffe K. The projected effect of risk factor reduction on Alzheimer's disease prevalence. Lancet Neurol 2011;10:819–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okbay A, Beauchamp JP, Fontana MA, et al. . Genome-wide association study identifies 74 loci associated with educational attainment. Nature 2016;533:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JJ, Wedow R, Okbay A, et al. . Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat Genet 2018;50:1112–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaid DJ, Chen W, Larson NB. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat Rev Genet 2018;19:491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgess S, Bowden J, Fall T, Ingelsson E, Thompson SG. Sensitivity analyses for Robust causal inference from mendelian randomization analyses with multiple genetic variants. Epidemiology 2017;28:30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol 2011;40:740–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. . Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet 2013;45:1452–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hemani G, Zheng J, Wade KH, et al. . MR-base: a platform for systematic causal inference across the phenome using billions of genetic associations. bioRxiv 2016. [Google Scholar]
- 10.Bowden J, Spiller W, Del Greco MF, et al. . Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the radial plot and radial regression. Int J Epidemiol 2018;47:1264–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anderson E, Wade KH, Hemani G, et al. . The causal effect of educational attainment on Alzheimer's disease: a two-sample Mendelian randomization study. bioRxiv 2017. [Google Scholar]
- 12.Larsson SC, Traylor M, Malik R, Dichgans M, Burgess S, Markus HS. Modifiable pathways in Alzheimer's disease: mendelian randomisation analysis. BMJ 2017;359:j5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Østergaard SD, Mukherjee S, Sharp SJ, et al. . Associations between potentially modifiable risk factors and Alzheimer disease: a mendelian randomization study. PloS Med 2015;12:e1001841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiorentino A, Sharp SI, Kandaswamy R, Gurling HM, Bass NJ, McQuillin A. Genetic variant analysis of the putative regulatory regions of the LRRC7 gene in bipolar disorder. Psychiatr Genet 2016;26:99–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hyde CL, Nagle MW, Tian C, et al. . Identification of 15 genetic loci associated with risk of major depression in individuals of European descent. Nat Genet 2016;48:1031–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bautista LE, Smeeth L, Hingorani AD, Casas JP. Estimation of bias in nongenetic observational studies using mendelian triangulation. Ann Epidemiol 2006;16:675–680. [DOI] [PubMed] [Google Scholar]