Abstract
Background/purpose
Oral submucous fibrosis (OSF) is a potentially malignant disorder of oral squamous cell carcinoma (SCC). In this study, we obtained the genetic expression signatures of OSF and SCC by microarray analysis.
Materials and methods
Five patients with clinically evident OSF, five patients with SCC who also had existing OSF, and four normal volunteers who did not have a history of chewing betel quids were recruited. Biopsy specimens were obtained with an approved Institutional Review Board protocol. Total RNA from OSF or SCC was isolated and hybridized to a Human Oligo 1A (V2) Microarray (G4110B) chip against normal control RNA that was pooled from the four healthy volunteers.
Results
We found similar, but distinct genetic expression signatures for OSF and SCC. At the hierarchical clustering analysis, 24 known genes (23 upregulated and 1 downregulated) in OSF were differentially expressed consistently in all participants. Among the genes, XRCC5 was cloned and transfected into oral cancer GNM cells. The results demonstrated that the overexpression of XRCC5 increased the resistance of GNM cells to low-density X-ray irradiation and promoted the cell growth rate.
Conclusion
The distinct but similar genetic expression signatures seen in OSF and SCC suggested that this expression may be used as a supplemental diagnostic tool in pathology practice. This preliminary study showed that the XRCC5 gene promoted GNM cell growth and conferred resistance to low-density X-ray irradiation. Further studies on the effect of XRCC5 in oral cancer cells are in progress.
Keywords: genetic signature, microarray, oral submucous fibrosis, SCC, XRCC5/Ku80
Introduction
Oral submucous fibrosis (OSF) is a progressively scarring chronic disease of the oral mucosa. It is characterized by increasing mucosal rigidity that is caused by the proliferation of fibroelastic tissue and the deposition of dense fibrous connective tissue in the superficial submucosa resulting from increased collagen synthesis,1, 2 decreased collagenase activity,3 or both. Oral submucous fibrosis is frequently associated with the habitual chewing of betel quid, which is a prevalent habit in Southeast Asia.4 The disease affects 0.2–1.2% of the urban population that visits dental clinics in India.5 A recent study on OSF showed that, of the 2 million people in Taiwan who habitually chew betel quid, 85.4% will develop OSF6 and one-third of these patients may eventually develop squamous cell carcinoma (SCC). Therefore, OSF is a potentially malignant disorder of the oral mucosa that may develop into oral squamous cell carcinoma (SCC).1
Despite its potential for malignant transformation, it is difficult to predict on the basis clinical and histopathologic examinations alone whether OSF will develop into a malignancy. Previous studies of OSF on the molecular mechanism of premalignant transformation of the oral mucosa have identified some molecules such as COX-2,7 type I plasminogen activator inhibitor,8 p53,9, 10 keratinocyte growth factor-1,11 interleukin-6,12 tissue inhibitor of metalloprotease 1,13 and adenomatous polyposis coli.10 However, these studies were insufficient in discriminating OSF from SCC.
This study used a microarray analysis to identify potential biomarkers that can predict the malignant transformation of OSF. The results support the hypothesis that every cancerous or precancerous lesion may possess a specific genetic expression signature that can be distinguished from others after hierarchical clustering analysis.
Materials and methods
Case recruitment
Oral mucosal specimens were collected, as described by other researchers, from five patients with clinically evident OSF.14, 15 Clinical signs and symptoms include trismus, marble-like pallor on the buccal mucosa and a progressive stiffness of subepithelial tissue, and a grayish white oral mucosa.1 All patients came to the Oral Surgery Clinic at the Chung Shan Medical University Hospital (Taichung City, Taiwan) for diagnosis and treatment. A review of their social habits showed all patients were smokers and areca nut chewers. Specimens of SCC were also collected from five patients who were undergoing surgical removal of the tumors at the same hospital. All five patients had SCC in the presence of clinically evident OSF and were all smokers and areca nut chewers. Normal tissues were obtained from the posterior mucobuccal fold of four healthy volunteers who did not have any history of chewing betel quid and were undergoing mandibular third molar extraction. All patients and volunteers were male and between the age of 30 and 55 years. The study protocol was approved by the Institutional Review Board of the Chung Shan Medical University Hospital. All patients with OSF or SCC and the healthy volunteers were provided with research information and informed consent was obtained from everyone before the study. All specimens were immediately frozen in –80°C until further use. The RNA extracted from the four healthy volunteers was pooled as one normal control for all microarray hybridization test, which is described later.
RNA extraction
The RNA extraction, labeling, and microarray analysis were performed in cooperation with DigiGenomics Co., Ltd. (Taipei county, Taiwan). In brief, the collected tissue samples were pulverized into fine powder in liquid nitrogen-filled mortar. They were then homogenized in TRI REAGENT (Molecular Research Center, Inc., Cincinnati, OH, USA). The total RNA was extracted in accordance with the manufacturer's protocol. To remove residual genomic DNA, 50 μg or less of total RNA was treated with Ambion RNase-free DNase I (Ambion, Austin, TX, USA) in accordance with the manufacturer's protocol. The enzyme was then removed by phenol/chloroform extraction. The RNA was recovered by ethanol precipitation. The integrity of RNA was checked in 0.9% agarose run in tris-acetate-EDTA (TAE) buffer to ensure that the RNA was of good quality for subsequent labeling steps.
Labeling and hybridization
The total RNA (1 μg) from each tissue sample was reverse transcribed into cDNA, further transcribed in vitro into cRNA, and labeled with CyDye by using the MessageAmp aRNA kit (Ambion, Austin, TX, USA) in accordance with the manufacturer's protocol. The cRNA obtained from the RNA sample of the normal tissue was labeled with Cy3 and was the reference sample. The cRNA obtained from the RNA sample of the diseased tissue was labeled with Cy5 and was the experimental sample. The labeled cRNA of the reference and experimental sample was purified to remove uncoupled CyDyes, combined in equal amounts, and mixed with 2× hybridization buffer (based on the manufacturer's protocol) before hybridization onto Human Oligo 1A (V2) Microarray (G4110B) (Agilent Technologies, Inc., Santa Clara, CA, USA). The array contains approximately 22,000 oligonucleotides, representing more than 20,000 genes. Conditions and procedures for hybridization and washing followed the Agilent 60-mer oligo microarray processing protocol (Agilent Technologies, Inc., Santa Clara, CA, USA).
Microarray image analysis, data normalization, and data analysis
Microarray images were acquired by using the GenePix 4000B scanner (Axon Instruments, Union City, CA, USA). The image was analyzed by using GenePix Pro 5.1 software (Molecular Devices Corporation, Downingtown, PA, USA). Individual microarray data was normalized by using Expressionist Pro Refiner software (Genedata AG, Basel, Switzerland). A filter procedure was applied to eliminate nongene features such as positive controls, negative controls, and blanks. Defective features were masked before data processing. Background correction was applied to genes with an S/N ratio over 2. Nonlinear normalization (i.e., Lowess) was performed globally with the smoothness factor set at 0.3.
Expression profiling was analyzed by using Expressionist Pro Analyst software (Genedata AG, Basel, Switzerland). All microarrays were normalized before comparison by using median intensity set at a fixed value and with the reference value set at 1. Features were excluded from analysis when the S/N ratio was less than 2.0.
To find common upregulated and downregulated genes among the samples, a threshold of expression ratio greater than 1.5 or less than 0.67 is the set criterion for screening significant genes. Genes passing this criterion in more than 50% of microarray experiments are chosen for subsequent hierarchical clustering.
Hierarchical clustering analysis
The hierarchical clustering analysis was performed, based on complete linkage and Manhattan (L1) distance. Clusters of genes showing opposite expression profiles between two distinct stages were chosen.
Cloning the full length XRCC5 cDNA
Total RNA from normal oral tissue was isolated from healthy volunteers. Reverse transcriptase polymerase chain reaction (RT-PCR) was performed with a primer pair that can amplify the full length of XRCC5 cDNA. The size of RT-PCR product was verified with electrophoresis (∼2.2 kb).
The same RT-PCR reaction was performed with the same primer pair and the resultant reaction solution was ligated to a pcDNA3.1 vector by using the TOPO TA cloning kit (Life Technologies, Grand Island, NY, USA). The reaction was transformed to TOP10' competent cells. After culturing on agarose plate, positive colonies were selected and the plasmid DNA was prepared by using the QIAprep Spin Miniprep kit (Qiagen, Germantown, MD, USA). The XRCC5 cDNA sequence was verified by a sequencing procedure.
Cell culture
GNM cells were originally derived from a 63-year-old female patient who was diagnosed as having gingival SCC with neck lymph node metastasis.16 Transfected GNM cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1% antibiotics.
Transient transfection
Oral cancer GNM cells were plated at 1 × 106 cell density per well in six-well dishes. Two milliters of DMEM with 10% FBS was added to the wells and lipofectamine 2000/DNA mixture was added into each well in accordance with the manufacturer's instructions. In brief, 4 μg plasmid DNA (XRCC5 or pcDNA) was diluted in 250 μL DMEM, mixed with 10 μL Lipofectamine 2000, and diluted in 250 μL DMEM at room temperature for 2–5 minutes to form a DNA-Lipofectamine 2000 complex. Culture media were removed and cells were washed with PBS twice, followed by 1500 μL 10% FBS DMEM. The DNA-Lipofectamine2000 complex was then gradually inserted into the wells. The dishes were gently rocked and incubated in 5% CO2-containing chamber for 24 hours. Cells were collected 24 hours, 48 hours, and 72 hours after transfection.
Results
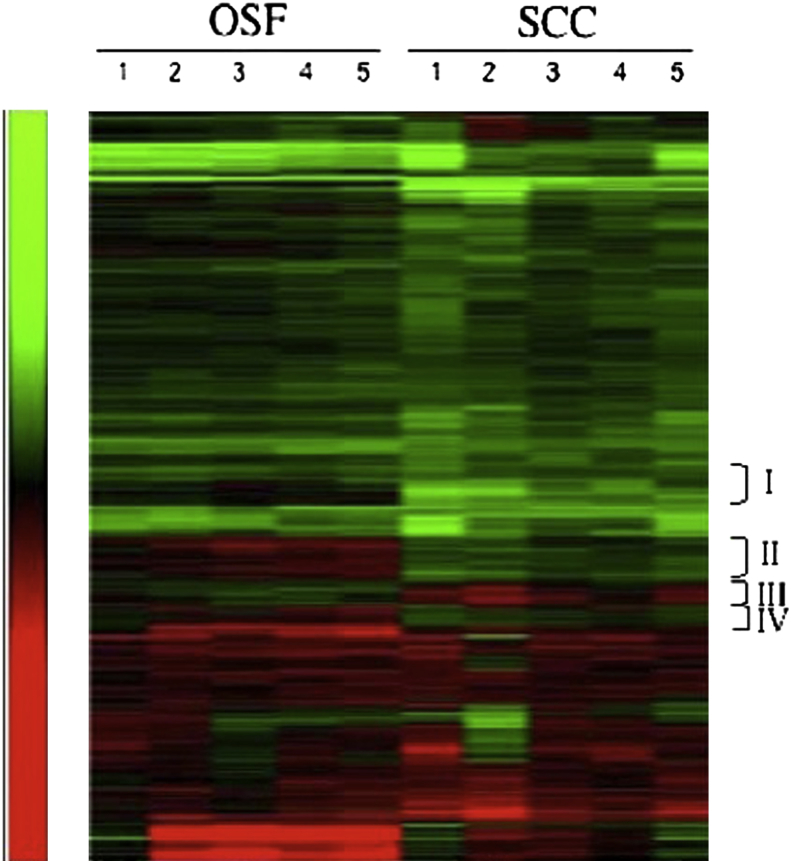
With a cut-off threshold of 1.5-fold (upregulated) or 0.67-fold (downregulated) in at least 50% of the comparisons, we found 1015 differentially expressed genes (approximately 5% of the total genes). The hierarchical analysis provided genetic expression signatures of OSF and SCC, which were distinct, but very similar. (Fig. 1) The hierarchical clustering analysis identified four clusters (Group I to IV in Fig. 1) that could discriminate OSF from SCC. Among these, 24 known genes (23 upregulated and 1 downregulated) in OSF were consistently expressed in all five cases of OSF in comparison to all cases of SCC. This therefore provided the most discriminating power (Table 1). Among the 24 differentially expressed genes, downregulation of XRCC5 in SCC in comparison to OSF was confirmed by quantitative RT-PCR and was cloned for further studies.
Figure 1.
Genetic expression signatures of OSF and SCC. Groups I, II, III and IV represented gene clusters that can most discriminate OSF from SCC, among which, 24 known potential genomic biomarkers were selected.
Table 1.
Genomic biomarkers identified in cDNA microarray that may predict malignant transformation of OSF.
| Assession # | Gene ID | Fold change |
|---|---|---|
| AF098269 | Procollagen C-endopeptidase enhancer 2 | 2.1 – up |
| AF329483 | Testis-specific serine kinase 6 | 2.4 – down |
| AK001592 | β-carotene 15,15'-monooxygenase 1 | 2.3 – up |
| AK024892 | SH3 domain binding glutamic acid-rich protein like | 1.6 – up |
| AK026775 | Regulator of G-protein signaling 5 | 3.4 – up |
| AK098393 | Glycine amidinotransferase | 3.1 – up |
| AL833232 | Hemicentin | 2.6 – up |
| BC002580 | Zine finger protein 593 | 3.0 – up |
| BC014418 | Superoxide dismutase 3 | 1.5 – up |
| BCO17396 | Prostatic binding protein | 1.9 – up |
| BC022279 | Leukocyte immunoglobulin-like receptor | 1.7 – up |
| BC033721 | SPARC-like 1 | 4.4 – up |
| BC035782 | Tyrosine kinase, non-receptor 1 | 3.7 – up |
| D12485 | Ectonucleotide pyrophosphatase/phosphodiesterase 1 | 3.4 – up |
| D67031 | Adducin 3 | 2.6 – up |
| M30938 | XRCC5 | 2.2 – up |
| NM000385 | Aquaporin 1 | 3.9 – up |
| NM002404 | Microfibrillar-associated protein 4 | 3.9 – up |
| NM019113 | Fibroblast growth factor 21 | 2.6 – up |
| NM032280 | Zine finger, CCHC domain containing 9 | 3.1 – up |
| NM133267 | Homeobox protein GSH-2 | 1.5 – up |
| NM152407 | GrpE-like 2, mitochondrial | 1.7 – up |
| U02570 | Rho GTPase activating protein 1 | 2.8 – up |
| Z70293 | Chemokine ligand 14 | 2.5 –up |
OSF = oral submucous fibrosis.
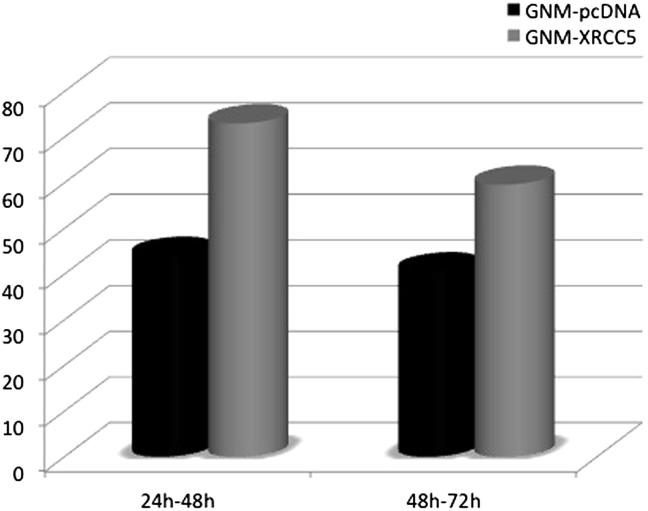
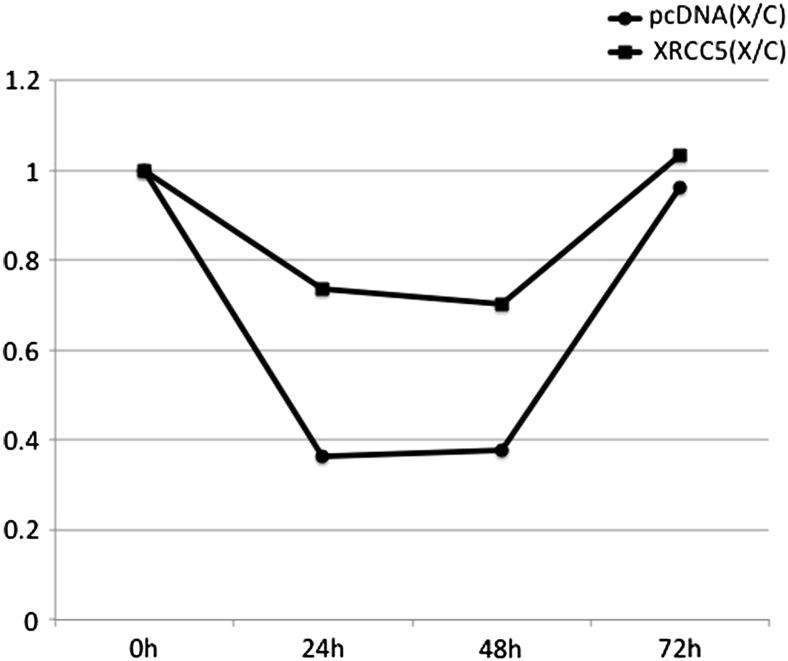
To study if the cell growth was influenced by the overexpression of XRCC5, GNM cells were transiently transfected with pcDNA3.1 or with XRCC5. The cells were harvested at 24 hours, 48 hours, and 72 hours after transfection. The cell growth rate was calculated as the ratio of the cell number difference to the cell count at the earlier time point. The results showed that the growth rate of XRCC5-transfected GNM cells was higher than the growth rate of the control group (Fig. 2). When the transfected cells were irradiated with 70 kvp at 10 mA (Minray intraoral X-ray unit, Soredex, Shenzhen, China) at a distance of 2 cm for 3 seconds (approximately 0.015 mSv), the growth rate was calculated as the ratio of the X-ray irradiated group to the nonirradiated control group. The results showed that fewer XRCC5-transfected cells died from X-ray irradiation and they grew faster at 24 hours and 48 hours after irradiation (Fig. 3).
Figure 2.
The proliferation of XRCC5 transfected GNM cells with X-ray irradiation. Data are presented as ratios of X-ray irradiated to non-irradiated groups.
Figure 3.
The growth rate observed between 24–48h and 48–72h with X-ray irradiation.
Discussion
We have reported the genetic expression signature from the epithelium of OSF by using laser capture microdissection and a cDNA microarray analysis.17 To demonstrate the usefulness of the genetic expression signature in distinguishing diseases, five cases each of OSF and SCC were included in the microarray analysis. The genetic expression signatures of 5 OSF cases were very similar to each another, as were the genetic expression signatures in the SCC group (Fig. 1). It is also very interesting that these two diseases showed different genetic expression signatures, but with certain degrees of similarity. This is different from the cases of synovial sarcoma and fibrosarcoma which show completely different genetic expression signatures were demonstrated between OSF and SCC.18 An interpretation of this finding is that OSF is a potentially malignant disorder of SCC, but synovial sarcoma and fibrosarcoma are totally unrelated diseases. Therefore, we hypothesized that every disease or tumor possesses a unique genetic expression signature that, in the future, may be used as a molecular diagnostic tool. However, to do so, it is important to realize that one single normal tissue group should be used as control for all hybridizations, and more disease entities should be included for examination.
With the rapid progression in molecular biology research, modern technology such as the Affymetrix array (Santa Clara, CA, USA) may survey thousands of genes at one time to identify potential biomarkers. However, our cDNA microarray has identified 24 known genes that were consistently and differentially expressed in all five OSF cases, compared to the five cases of SCC. Limited cases were included in this study, although a similar approach has been applied in different expression array-based studies with a limited case number.19, 20, 21, 22
It would be ideal to use normal tissue from the same patient for microarray analysis, as reported by Shieh et al.13, 23 In another study on oral SCC, cDNA microarray analysis used normal tissue from the same patient as the control for the analysis.24 However, it is impractical to obtain clinically and biologically normal tissue from patients with OSF or SCC. As other scientists have shown, peritumor normal tissue may not be totally normal in that cancer cells nay influence the gene expression of the adjacent tissue.24 To eliminate the possible contamination of the tissue samples, we decided to use normal tissues from healthy volunteers who did not have any history of chewing betel quids. We pooled their RNAs as one normal control for microarray analysis to minimize possible variations.
X-ray cross-complementing (XRCC) genes were discovered mainly through their role in protecting mammalian cells from damage caused by ionizing radiation. They are also important in genetic stability. There are two main pathways in eukaryotic cells for repairing double strand DNA breakage—namely, nonhomologous end joining and homologous recombination (HR). Nonhomologous end joining provides a mechanism for the repair of double strand DNA breakage throughout the cell cycle. However, it is particularly important during the G0, G1, and early S phases in mitotic cells and is mediated by the XRCC5, XRCC6 and XRCC7 genes. The DNA repair protein Ku acts as a heterodimer of the two 70-kDa (Ku70) and 80-kDa (Ku80) subunits and binds to DNA ends, nicks, or single- to double-strand transition.25, 26, 27 It serves as a DNA-binding component of the DNA-dependent protein kinase (DNA-PK) that phosphorylates certain chromatin-bound proteins in vitro. The XRCC5 gene encodes Ku80 and forms a heterodimer with Ku70 and functions as DNA end-binding at the double strand breakage site. Ku binds to the end of the DNA double-strand breakage and appears to stabilize the binding of the DNA-PKCs to DNA.28, 29, 30, 31, 32, 33, 34, 35
No data have reported the association of XRCC5 gene in OSF and/or in the subsequent development of SCC. However, a recent study has demonstrated frequent chromosomal breakage at the 1cen-1q12 region in the mucosa cells of betel nuts chewers.36 It has also been shown that the suppression of Ku70 increases radiosensitivity in one human lung carcinoma cell line.37 It is thus possible that the upregulation of the XRCC5 gene in the OSF patients in our study reflects a repair mechanism of DNA breakage, whereas the decreased expression of the XRCC5 gene in SCC possibly signifies an increased potential for malignant transformation of OSF to SCC.
The effects of X-ray irradiation on GNM cells transfected with the XRCC5 gene were preliminarily studied. However, only dental periapical irradiation was tested in this preliminary study, which emits relative low radiation. The XRCC5-transfected GNM cells were interestingly more resistant to low-dose X-ray irradiation and had enhanced cell growth. Higher doses of irradiation should be tested to understand more about the effects of XRCC5 on the cells.
In summary, hierarchical clustering analysis showed distinct, but similar genetic expression signatures of OSF and SCC. The genetic expression signature in OSF and SCC and other cancers17, 18 renders this analysis a possible diagnostic tool in the future. Preliminary studies also showed that XRCC5 gene overexpression enhanced GNM cell proliferation and provided more resistance to low-dose X-ray irradiation. Further studies on the effects of XRCC5 will be performed in the future.
References
- 1.Neville B.W., Damm D.D., Allen C.M., Bouquot J.E. W.B. Saunders; Philadelphia, PA: 2002. Oral and Maxillofacial Pathology. [Google Scholar]
- 2.Meghji S., Scutt A., Harvey W., Canniff J.P. An in-vitro comparison of human fibroblasts from normal and oral submucous fibrosis tissue. Arch Oral Biol. 1987;32:213–215. doi: 10.1016/0003-9969(87)90138-5. [DOI] [PubMed] [Google Scholar]
- 3.Shieh T.Y., Yang J.F. Collagenase activity in oral submucous fibrosis. Proc Natl Sci Counc Repub China B. 1992;16:106–110. [PubMed] [Google Scholar]
- 4.Sinor P.N., Gupta P.C., Murti P.R. A case-control study of oral submucous fibrosis with special reference to the etiologic role of areca nut. J Oral Pathol Med. 1990;19:94–98. doi: 10.1111/j.1600-0714.1990.tb00804.x. [DOI] [PubMed] [Google Scholar]
- 5.Jayanthi V., Probert C.S., Sher K.S., Mayberry J.F. Oral submucosal fibrosis—a preventable disease. Gut. 1992;33:4–6. doi: 10.1136/gut.33.1.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee C.H., Ko Y.C., Huang H.L. The precancer risk of betel quid chewing, tobacco use and alcohol consumption in oral leukoplakia and oral submucous fibrosis in southern Taiwan. Br J Cancer. 2003;88:366–372. doi: 10.1038/sj.bjc.6600727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsai C.H., Chou M.Y., Chang Y.C. The up-regulation of cyclooxygenase-2 expression in human buccal mucosal fibroblasts by arecoline: a possible role in the pathogenesis of oral submucous fibrosis. J Oral Pathol Med. 2003;32:146–153. doi: 10.1034/j.1600-0714.2003.00004.x. [DOI] [PubMed] [Google Scholar]
- 8.Yang S.F., Hsieh Y.S., Tsai C.H., Chou M.Y., Chang Y.C. The upregulation of type I plasminogen activator inhibitor in oral submucous fibrosis. Oral Oncol. 2003;39:367–372. doi: 10.1016/s1368-8375(02)00123-9. [DOI] [PubMed] [Google Scholar]
- 9.Chiang C.P., Lang M.J., Liu B.Y. Expression of p53 protein in oral submucous fibrosis, oral epithelial hyperkeratosis, and oral epithelial dysplasia. J Formos Med Assoc. 2003;99:229–234. [PubMed] [Google Scholar]
- 10.Liao P.H., Lee T.L., Yang L.C., Yang S.H., Chen S.L., Chou M.Y. Adenomatous polyposis coli gene mutation and decreased wild-type p53 protein expression in oral submucous fibrosis: a preliminary investigation. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2001;92:202–207. doi: 10.1067/moe.2001.116816. [DOI] [PubMed] [Google Scholar]
- 11.Tsai C.H., Yang S.F., Chen Y.J., Chou M.Y., Chang Y.C. Raised keratinocyte growth factor-1 expression in oral submucous fibrosis in vivo and upregulated by arecoline in human buccal mucosal fibroblasts in vitro. J Oral Pathol Med. 2005;34:100–105. doi: 10.1111/j.1600-0714.2004.00288.x. [DOI] [PubMed] [Google Scholar]
- 12.Tsai C.H., Yang S.F., Chen Y.J., Chu S.C., Hsieh Y.S., Chang Y.C. Regulation of interleukin-6 expression by arecoline in human buccal mucosal fibroblasts is related to intracellular glutathione levels. Oral Dis. 2004;10:360–364. doi: 10.1111/j.1601-0825.2004.01041.x. [DOI] [PubMed] [Google Scholar]
- 13.Shieh D.H., Chiang L.C., Shieh T.Y. Augmented mRNA expression of tissue inhibitor of metalloproteinase-1 in buccal mucosal fibroblasts by arecoline and safrole as a possible pathogenesis for oral submucous fibrosis. Oral Oncol. 2003;39:728–735. doi: 10.1016/s1368-8375(03)00101-5. [DOI] [PubMed] [Google Scholar]
- 14.Tadakamadla J., Kumar S., Mamatha G. Evaluation of serum copper and iron levels among oral submucous fibrosis patients. Med Oral Patol Oral Cir Bucal. 2001 doi: 10.4317/medoral.17083. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 15.Patidar K., Parwani R., Wanjari S. Correlation of salivary and serum IgG, IgA levels with total protein in oral submucous fibrosis. J Oral Sci. 2011;53:97–102. doi: 10.2334/josnusd.53.97. [DOI] [PubMed] [Google Scholar]
- 16.Gin H.-S. Hu-Bei Medical University; WuHan: 1997. Human Oral SCC with Neck Lymph Node Metastasis and Tongue SCC Cell Lines—Establishment and Biological Characterization. PhD dissertation. [Google Scholar]
- 17.Huang Y.-F., Hansen M.F., Yang H.-W. Determining the genetic signature of oral submucous fibrosis by the use of laser capture microdissection and a cDNA microarray. J Dent Sci. 2006;1:66–73. [Google Scholar]
- 18.Allander S.V., Illei P.B., Chen Y. Expression profiling of synovial sarcoma by cDNA microarrays: association of ERBB2, IGFBP2, and ELF3 with epithelial differentiation. Am J Pathol. 2002;161:1587–1595. doi: 10.1016/S0002-9440(10)64437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alevizos I., Mahadevappa M., Zhang X. Oral cancer in vivo gene expression profiling assisted by laser capture microdissection and microarray analysis. Oncogene. 2001;20:6196–6204. doi: 10.1038/sj.onc.1204685. [DOI] [PubMed] [Google Scholar]
- 20.Al Moustafa A.E., Alaoui-Jamali M.A., Batist G. Identification of genes associated with head and neck carcinogenesis by cDNA microarray comparison between matched primary normal epithelial and squamous carcinoma cells. Oncogene. 2002;21:2634–2640. doi: 10.1038/sj.onc.1205351. [DOI] [PubMed] [Google Scholar]
- 21.Mendell J.T., Sharifi N.A., Meyers J.L., Martinez-Murillo F., Dietz H.C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat Genet. 2004;36:1238. doi: 10.1038/ng1429. [DOI] [PubMed] [Google Scholar]
- 22.Horiuchi K., Umetani M., Minami T. Wilms' tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proc Natl Acad Sci USA. 2006;103:17278–17283. doi: 10.1073/pnas.0608357103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shieh D.H., Chiang L.C., Lee C.H., Yang Y.H., Shieh T.Y. Effects of arecoline, safrole, and nicotine on collagen phagocytosis by human buccal mucosal fibroblasts as a possible mechanism for oral submucous fibrosis in Taiwan. J Oral Pathol Med. 2004;33:581–587. doi: 10.1111/j.1600-0714.2004.00229.x. [DOI] [PubMed] [Google Scholar]
- 24.Tomioka H., Morita K.-I., Hasegawa S., Omura K. Gene expression analysis by cDNA microarray in oral SCC. J Oral Pathol Med. 2006;35:206–211. doi: 10.1111/j.1600-0714.2006.00410.x. [DOI] [PubMed] [Google Scholar]
- 25.Mimori T., Hardin J.A. Mechanism of interaction between ku protein and DNA. J Biol Chem. 1986;261:10375–10379. [PubMed] [Google Scholar]
- 26.Mimori T., Hardin J.A., Steitz J.A. Characterization of the DNA-binding protein antigen ku recognized by autoantibodies from patients with rheumatic disorders. J Biol Chem. 1986;261:2274–2278. [PubMed] [Google Scholar]
- 27.Rathmell W.K., Chu G. Involvement of the ku autoantigen in the cellular response to DNA double-strand breaks. Proc Natl Acad Sci USA. 1994;91:7623–7627. doi: 10.1073/pnas.91.16.7623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jackson S.P. DNA-dependent protein kinase. Int J Biochem Cell Biol. 1997;29:935–938. doi: 10.1016/s1357-2725(97)00006-x. [DOI] [PubMed] [Google Scholar]
- 29.Jeggo P.A. DNA-PK: at the cross-roads of biochemistry and genetics. Mutat Res. 1997;384:1–14. doi: 10.1016/s0921-8777(97)00009-8. [DOI] [PubMed] [Google Scholar]
- 30.Smith G.C., Jackson S.P. The DNA-dependent protein kinase. Genes Dev. 1999;13:916–934. doi: 10.1101/gad.13.8.916. [DOI] [PubMed] [Google Scholar]
- 31.Dvir A., Peterson S.R., Knuth M.W. Ku autoantigen is the regulatory component of a template-associated protein kinase that phosphorylates RNA polymerase II. Proc Natl Acad Sci USA. 1992;89:11920–11924. doi: 10.1073/pnas.89.24.11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gottlieb T.M., Jackson S.P. The DNA-dependent protein kinase: requirement for DNA ends and association with ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 33.Blier P.R., Griffith A.J., Craft J. Binding of ku protein to DNA. Measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem. 1993;268:7594–7601. [PubMed] [Google Scholar]
- 34.Jin S., Inoue S., Weaver D.T. Functions of the DNA dependent protein kinase. Cancer Surv. 1997;29:221–261. [PubMed] [Google Scholar]
- 35.Smith G.C., Divecha N., Lakin N.D. DNA-dependent protein kinase and related proteins. Biochem Soc Symp. 1999;64:91–104. [PubMed] [Google Scholar]
- 36.Rupa D.S., Eastmond D.A. Chromosomal alterations affecting the 1cen-1q12 region in buccal mucosal cells of betel quid chewers detected using multicolor fluorescence in situ hybridization. Carcinogenesis. 1997;18:2347–2351. doi: 10.1093/carcin/18.12.2347. [DOI] [PubMed] [Google Scholar]
- 37.Omori S., Takiguchi Y., Suda A. Suppression of a DNA double-strand break repair gene, Ku70, increases radio- and chemosensitivity in a human lung carcinoma cell line. DNA Repair (Amst) 2002;1:299–310. doi: 10.1016/s1568-7864(02)00006-x. [DOI] [PubMed] [Google Scholar]