Abstract
The role of subclonal TP53 mutations, defined by a variant allele frequency of <20%, has not been addressed in acute myeloid leukemia yet. We, therefore, analyzed their prognostic value in a cohort of 1,537 patients with newly diagnosed disease, prospectively treated within three trials of the “German-Austrian Acute Myeloid Leukemia Study Group”. Mutational analysis was performed by targeted deep sequencing and patients with TP53 mutations were categorized by their variant allele frequency into groups with frequencies >40%, 20%-40% and <20%. A total of 108 TP53 mutations were found in 98 patients (6.4%). Among these, 61 patients had variant allele frequencies >40%, 19 had variant allele frequencies between 20%-40% and 18 had frequencies <20%. Compared to specimens with clonal TP53 mutations, those with subclonal ones showed significantly fewer complex karyotypes and chromosomal losses. In either TP53-mutated group, patients experienced significantly fewer complete responses (P<0.001) and had worse overall and event-free survival rates (P<0.0001). In Cox regression analyses adjusting for age, white blood cell count, cytogenetic risk and type of acute myeloid leukemia, the adverse prognostic effect of TP53 mutations remained significant for all TP53-mutated subgroups. These data suggest that subclonal TP53 mutations are a novel prognostic parameter in acute myeloid leukemia and emphasize the usefulness of next-generation sequencing technologies for risk stratification in this disorder. The study was registered at ClinicalTrials.gov with number NCT00146120.
Introduction
Acute myeloid leukemia (AML) is an aggressive malignancy with an annual, age-adjusted incidence of 3.5 cases per 100,000 adults, rising to 15-20 cases per 100,000 above the age of 60 years.1 It, therefore, contributes substantially to morbidity and mortality of the elderly. The pathogenesis of AML represents a multistep process involving mutagenesis, epigenetic dysregulation and formation of copy number aberrations. During the process of leukemogenesis, initiating mutations affect hematopoietic stem and progenitor cells, giving rise to preleukemic/leukemic stem cells and, ultimately, frank leukemia. Virtually every AML genome is characterized by a number of subclones of variable size. These subclones may have different pathobiological properties as well as responses to antileukemic treatments.2–4
Aberrations of the TP53 tumor suppressor gene have been described in a variable number of cases of AML. They are present in less than 10% of cases of de novo AML, whereas their rates in therapy-related AML and erythroid leukemias exceed 20% and 90%, respectively.5–7 These aberrations include gene mutations, most of which are located within the DNA binding domain of the gene, and/or deletions of different sizes affecting the TP53 locus on chromosome 17p13. Although the majority of TP53 aberrations are somatically acquired, constituting an early leukemogenic event, germline mutations are increasingly being recognized, predominantly in patients with therapy-related AML.8,9 Aberrations of TP53 are associated with an exceedingly adverse prognosis as demonstrated by several independent reports.5,10,11 Recently, it was shown that TP53 mutations and deletions encompassing the TP53 locus have a different prognostic impact in AML, with only mutations but not deletions significantly influencing survival of these patients.12 As a consequence, testing for TP53 mutations has been introduced into the 2017 recommendations of the European LeukemiaNet.13
However, in the studies performed so far, TP53 mutations were assessed as a dichotomous variable only. With the advent of next-generation sequencing technologies, mutational subclones can now be detected with high sensitivity. Here, we aimed to investigate the clinical characteristics associated with subclonal TP53 mutations and their prognostic impact in a large cohort of AML patients prospectively treated within studies of the “German-Austrian AML Study Group” (AMLSG).
Methods
The study was approved by the ethics committees of the University of Ulm, Germany, and the Medical University of Graz, Austria and conducted in accordance with REMARK guidelines (“REporting recommendations for tumor MARKer prognostic studies”).14
Study participants
Data from a total of 1537 intensively treated AML patients enrolled in three prospective, multicenter, clinical trials of the AMLSG were analyzed.15–17 Details of these studies as well as a list of AMLSG investigators and centers are provided in the Online Supplementary Appendix.
Sequence analysis
Genetic profiling of a total of 1,537 diagnostic AML specimens using a targeted sequencing approach with 111 genes associated with myeloid neoplasms was reported previously for this combined cohort.2 Sequencing data were deposited in the European Genome-Phenome Archive (www.ebi.ac.uk/ega, accession number EGAS00001000275) and retrieved for the present study. Methodological details are provided in the Online Supplementary Methods together with information on ultradeep sequencing used for the analysis of selected, sequential patients’ samples.
Statistical analysis
The study was designed to assess differences in overall survival between AML patients exhibiting a TP53 wild-type status and those with subclonal TP53 mutations. Based on data from Papaemmanuil et al.2 an absolute reduction of 35% in 3-year overall survival compared to that of patients with a TP53 wild-type status should be detectable using a sample size of 574 patients assuming an overall survival rate of 55% for TP53 wild-type patients and a frequency of 5% for TP53 muted subclones with a variant allele frequency (VAF) as low as 5% (power=90%; α=0.05).
The main outcome parameters assessed were overall survival and event-free survival, as defined by the European LeukemiaNet.13 We determined median survival times and estimated 3-year survival rates along with their 95% confidence intervals (95% CI). Survival rates of patients with a TP53 wild-type status and patients with TP53 mutations were plotted using the Kaplan-Meier method and compared by the log-rank test. In addition, TP53-mutated patients were further categorized according to their VAF (>40%, 20%-40%, <20%). This categorization into three different groups according to the TP53 VAF was based on different biological features regarding concomitant chromosomal aberrations as outlined in the “Results” section. It also allowed a comparison with previous reports on the impact of TP53 VAF in patients with myelodysplastic syndromes.18,19 Univariable and multivariable Cox regression analyses were performed to identify relevant prognostic factors. We assessed age, white blood cell count, cytogenetic risk group and type of AML (de novo, secondary or therapy-related) in addition to the TP53 status. Hazard ratios (HR) are presented along with their 95% CI. To compare patients’ characteristics among the groups defined by their TP53 status and VAF, we performed a Fisher exact test for categorical parameters and the Kruskal-Wallis or Mann-Whitney-U tests, respectively, for continuous parameters. If the overall test between the three TP53 VAF groups showed statistically significant differences (α=0.05), post-hoc tests were performed. Due to the multiple groups tested, we employed a Bonferroni correction and considered a P-value of <0.017 as statistically significant. All statistical analyses were conducted using R version 3.4.4 (https://www.r-project.org).
Results
Characteristics of TP53-mutated subclones in acute myeloid leukemia
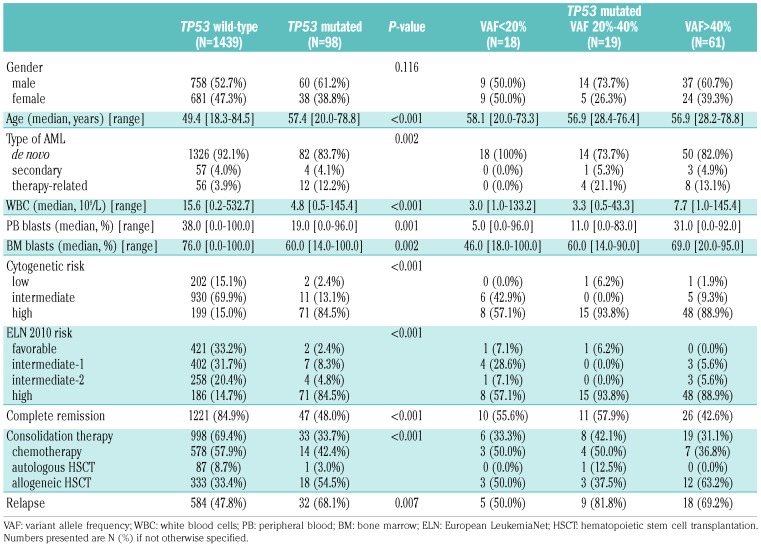
A total of 1,537 patients, enrolled in the AMLSG trials HD98A, HD98B and 07-04, were evaluated in this study. Their clinical characteristics are shown in Table 1. Of those patients, 1,408 (91.6%) had de novo AML, 61 (4.0%) had secondary AML following myelodysplastic syndromes and 68 (4.4%) had therapy-related AML. The median follow-up of all patients was 25 months (range, 0 - 219.6).
Table 1.
Patients’ characteristics.
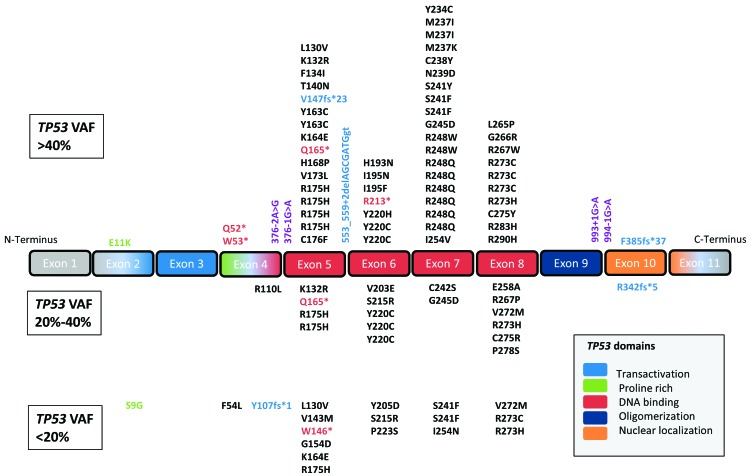
When analyzing diagnostic AML specimens, 108 pathogenic TP53 mutations were found in 98 (6.4%) patients. Seven patients exhibited two TP53 mutations each and one patient had four. A detailed presentation of the mutations detected in this cohort is given in Online Supplementary Table S1. When categorizing the 98 patients with TP53 mutations according to their maximum VAF, we found 61 (62.2%) with a VAF >40% representing the major AML clone, 19 (19.4%) with a VAF between 20% and 40% and 18 (18.4%) with a VAF <20%, this last group representing subclones. The vast majority of TP53 mutations in all groups were missense mutations located in the DNA binding domain of the gene. Neither type of the mutation nor its location showed a statistically significant difference among the three TP53-mutated subgroups (P=0.279 for mutation type and P=0.687 for mutation location) (Figure 1). As compared to AML patients with TP53 wild-type, those with clonal and subclonal TP53 mutations showed significantly lower white blood cell counts as well as peripheral blood and bone marrow blasts (Table 1).
Figure 1.
Distribution of 108 TP53 mutations found in diagnostic specimens of 98/1537 patients with acute myeloid leukemia. Top panel: TP53 mutations with a variant allele frequency (VAF) of >40%; middle panel: mutations with a VAF of 20%-40%; lower panel: mutations with a VAF <20%. Missense mutations are marked in black, nonsense in red, insertions/deletions in blue and essential splice site mutations in purple. Despite different VAF, the vast majority of TP53 mutations are missense mutations located within the DNA binding domain of the gene.
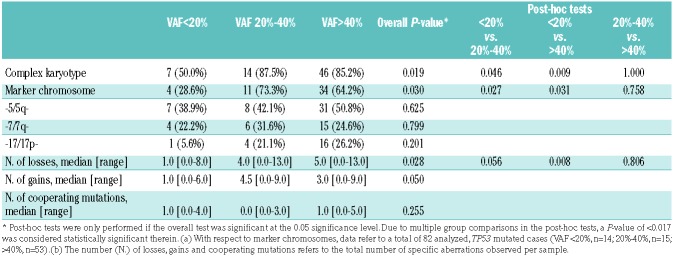
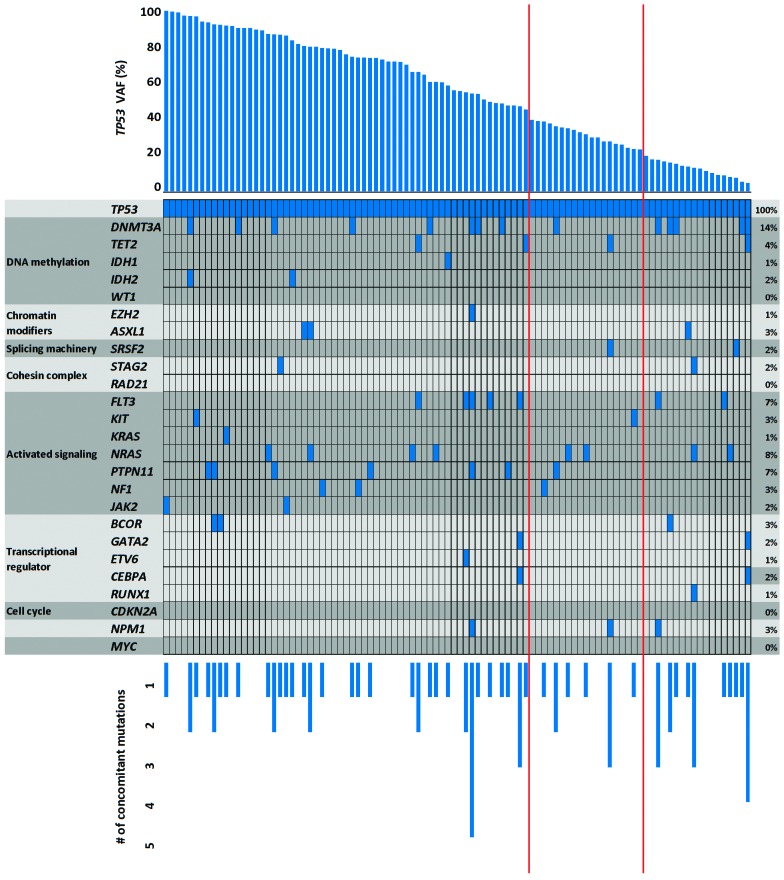
It is well documented that clonal TP53 mutations are highly associated with complex karyotypes as well as marker chromosomes arising from chromothripsis.20,21 As shown in Table 2, a significant association with complex karyotypes was observed for TP53 mutations with a VAF of >40% and between 20% and 40%, but not for subclonal ones with a VAF of <20%. There was also a statistically significant difference between TP53 wild-type and TP53 mutated cases with respect to marker chromosomes (2.9% versus 59.8%; P<0.001), however, no significant difference was observed among the three TP53-mutated subgroups. Furthermore, in the group with subclonal TP53 mutations, a significantly lower number of chromosomal losses was detected as compared to the number in the group with clonal TP53 mutations. The frequency of the chromosomal aberrations -5/5q-, -7/7q- and -17/17p- did not differ significantly among the three TP53-mutated groups, nor did the total number of chromosomal gains. The number of cooperating driver mutations was low with zero to five mutations detected per sample and did not differ significantly between the TP53-mutated groups. Mutations in DNMT3A (14/98), NRAS (8/98), FLT3 (7/98) and PTPN11 (7/98) were the most frequent cooperating mutations (Figure 2).
Table 2.
Chromosomal aberrations and cooperating gene mutations found in 98 acute myeloid leukemia patients with TP53 mutations according to their variant allele frequency.
Figure 2.
Concurrent gene mutations in 98 TP53-mutated cases of acute myeloid leukemia divided by functional classes. Of those specimens with two or more TP53 mutations, only the one with the highest variant allele frequency (VAF) is listed. Each column represents an individual patient with the total number of cooperating events stated at the bottom. Each colored box represents a mutation of the gene listed at the left. The red vertical lines indicate the 40% and 20% cut-offs with respect to TP53 VAF. #: number.
Impact of TP53-mutated subclones on survival of patients with acute myeloid leukemia
Of 1,537 patients undergoing induction therapy, 1,268 (82.5%) achieved complete remission. The complete remission rate was significantly inferior in the TP53-mutated cohort (48.0% versus 84.9% for TP53 wild-type patients, P<0.001) but equally distributed among the VAF-based groups (TP53 VAF >40%, 42.6%; 20%-40%, 57.9%; <20%, 55.6%; P=0.424) (Online Supplementary Figure S1). A total of 1,031 patients (67.1%) underwent consolidation therapy and of those, 439 (42.6%) received hematopoietic stem cell transplantation (351 allogeneic and 88 autologous; 42.1% of consolidated TP53 wild-type patients and 57.6% of TP53-mutated patients) (Table 1). The relapse rate was significantly higher in the TP53-mutated cohort (68.1% versus 47.8% for TP53 wild-type patients, P=0.007) but did not differ significantly among the TP53-mutated subgroups (TP53 VAF >40%, 69.2%; 20%-40%, 81.8%; <20%, 50.0%; P=0.336) (Table 1). Within the TP53-mutated cohort, patients who underwent allogeneic hematopoietic stem cell transplantation had a significantly better event-free survival than those given chemotherapy or autologous hematopoietic stem cell transplantation [HR, 0.25 (95% CI: 0.11-0.58); P=0.001]. No significant difference was observed for overall survival [HR, 0.47 (95% CI: 0.22-1.01); P=0.054].
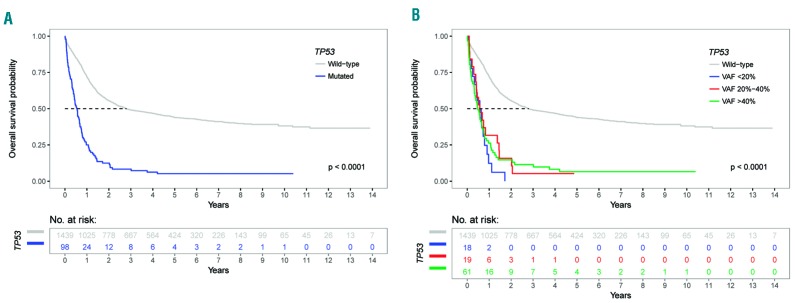
The estimated median overall survival for the entire cohort was 28.1 months (95% CI: 24.3-33.5) but differed substantially between TP53 wild-type and TP53-mutated patients [33.6 months (95% CI: 28.4-45.0) versus 6.5 months (95% CI: 5.0-8.2)]. The median overall survival was short in all TP53-mutated subgroups (TP53 VAF >40%, 5.8 months; 20%-40%, 6.9 months; <20%, 6.9 months). The estimated 3-year overall survival rate for the entire cohort was 46.5% (95% CI: 44.0-49.1) with notable differences between TP53 wild-type and TP53-mutated patients [49.1% (95% CI: 46.5-51.8) versus 8.3% (95% CI: 4.3-16.2)]. Again, 3-year overall survival rates were low in each of the TP53-mutated groups (TP53 VAF >40%, 11.5%; 20%-40%, 5.3%; <20%, 0%) (Figure 3A,B, Online Supplementary Table S2).
Figure 3.
Kaplan-Meier analysis of overall survival in 1,537 patients with acute myeloid leukemia stratified by TP53 mutational status. (A) Overall survival: TP53 wild-type patients versus TP53-mutated patients. (B) Overall survival: TP53 wild-type patients versus patients in the three groups with the defined variant allele frequencies of mutated TP53.
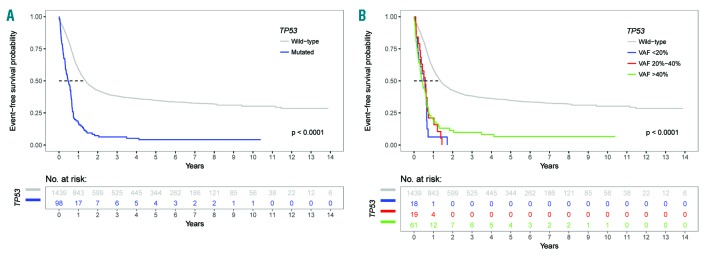
The estimated median event-free survival for the entire cohort was 15.0 months (95% CI: 13.6-16.5) with that for TP53 wild-type patients being 16.5 months (95% CI: 15.0-18.2) and that for TP53-mutated patients being 5.7 months (95% CI: 4.3-7.4). The median event-free survival was short in all TP53-mutated subgroups (TP53 VAF >40%, 5.2 months; 20%-40%, 6.9 months; <20%, 6.5 months). The estimated 3-year event-free survival rate for the entire cohort was 36.3% (95% CI: 33.9-38.8) with a pronounced difference between TP53 wild-type and TP53-mutated patients [38.3% (35.9-40.9) versus 6.3% (2.9-13.6)]. As for overall survival, 3-year event-free survival rates were low in all TP53-mutated subgroups (TP53 VAF >40%, 9.8%; 20%-40%, 0%; <20%, 0%).(Figure 4A,B, Online Supplementary Table S3).
Figure 4.
Kaplan-Meier analysis of event-free survival in 1537 patients with acute myeloid leukemia stratified by TP53 mutational status. (A) Event-free survival: TP53 wild-type patients versus TP53-mutated patients. (B) TP53 wild-type patients versus patients in the three groups with the defined variant allele frequencies of mutated TP53.
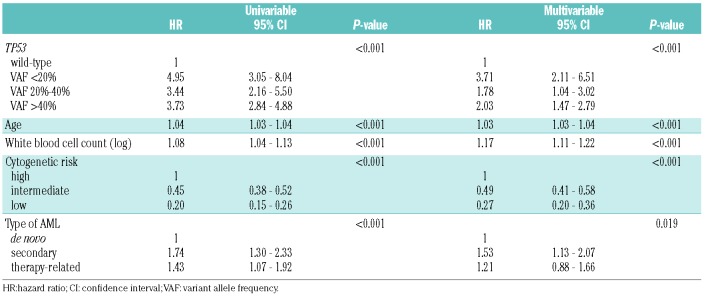
Table 3 and Online Supplementary Table S4 present the results of the Cox regression analyses assessing the impact of TP53 mutational status on overall and event-free survival. The models contain the different VAF groups, age, white blood cell count, cytogenetic risk group and type of AML as predictors. Whereas all of the factors assessed showed a significant impact on both outcome parameters in the initial univariable analysis, type of AML did not remain significant for event-free survival in the multivariable model. Importantly, each of the TP53-mutated groups showed significantly worse overall and event-free survival compared to the TP53 wild-type group. For overall survival, the HR (95% CI), for the comparison with TP53 wild-type patients, was 2.03 (1.47-2.79) for patients with a TP53 VAF >40%, 1.78 (1.04-3.02) for those with a VAF of 20%-40% and 3.71 (2.11-6.51) for those with a VAF <20%. For event-free survival, the estimates were 1.89 (1.38-2.59) for patients with a TP53 VAF >40%, 2.27 (1.35-3.80) for those with a VAF of 20%-40% and 3.57 (2.04-6.26) for those with a VAF <20%.
Table 3.
Univariable and multivariable Cox regression analysis for overall survival.
Case study on longitudinal TP53 mutational assessment
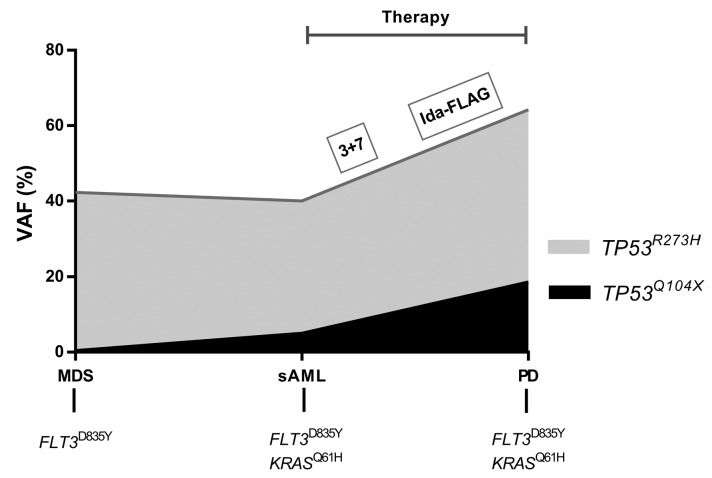
In one subject with secondary AML treated at the Medical University of Graz, Austria, outside a clinical trial, serial assessment of bone marrow specimens by error-corrected ultradeep sequencing was performed.22,23 This 68-year old male presented with myelodysplastic syndrome and an International Prognostic Scoring System risk score of “intermediate 2”. Within 2 months after diagnosis, the patient’s disease transformed into secondary AML and he was then treated with daunorubicin and cytarabine (3+7) as well as Ida-FLAG as a salvage regimen. Nevertheless, the patient died of resistant disease 4 months after diagnosis. Analysis of the diagnostic sample revealed the clonal TP53 p.273H mutation with a VAF of 42.3% and a subclonal TP53 Q104X mutation with a VAF of 0.4%. Both TP53-mutated clones expanded slightly in the course of transformation to secondary AML which was also characterized by a novel KRAS Q61H mutation. However, during two courses of intensive chemotherapy, both TP53 mutations substantially expanded to a clone size of 64.2% and 18.6%, respectively. (Figure 5 and Online Supplementary Table S5).
Figure 5.
Longitudinal mutational analyses of a patient with secondary acute myeloid leukemia showing a clonal (R273H) and a subclonal (Q104X) TP53 mutation. VAF: variant allele frequency; MDS, myelodysplastic syndrome: sAML: secondary acute myeloid leukemia; PD, progressive diesease. Bottom line: cooperating mutations.
Discussion
In this study, we analyzed a large cohort of intensively treated AML patients focusing on biological and clinical characteristics associated with subclonal TP53 mutations. We found that these aberrations represent a substantial proportion of TP53-mutated AML. Similarly to clonal TP53 mutations, subclonal ones are mainly missense mutations and are associated with an adverse outcome as evidenced by significantly inferior complete remission, overall survival and event-free survival rates.
Results similar to those presented here have been obtained for patients with chronic lymphocytic leukemia in whom those with TP53-mutated subclones with a median VAF as low as 2.1% showed comparable clinical phenotypes and survival as poor as those with clonal mutations.24,25 Interestingly, divergent data have been published for patients with myelodysplastic syndromes.18,19 Analyzing two independent cohorts of 219 and 150 patients, also including a few cases with secondary AML, Sallman et al. stratified TP53 VAF into the same categories as defined here (>40%, 20%-40%, <20%). Importantly, there was a significant difference in overall survival between patients exhibiting TP53 mutant VAF of >40% and <20%. Whereas myelodysplastic syndrome patients with a TP53 VAF >40% had a median overall survival of 124 days, overall survival was not reached in patients with a VAF <20%. In contrast, Goel et al. investigated two cohorts of 81 and 1,117 patients and did not find any significant difference with respect to TP53 VAF and their negative prognostic impact on overall survival. It was argued that one of the reasons for these discrepancies might have been the small number of TP53-mutated patients investigated. In addition, early therapeutic intervention leading to clonal expansion of a drug-resistant clone may also be relevant to these differences in survival.26 In our sufficiently powered study of 1,537 patients predominantly with de novo AML, 98 had TP53-mutated clones of various sizes. We provide evidence here that small TP53-mutated subclones share biological characteristics with clonal ones, such as mutation type, location of the mutation within TP53 domains and cooperating mutations. We also demonstrate that even TP53-mutated subclones, defined by a VAF <20%, have a statistically significant negative prognostic impact with respect to complete remission rate, overall survival and event-free survival. These findings may have implications for TP53 screening methods and future risk stratification in AML as classical Sanger sequencing with a detection limit of mutant clones set at 20% VAF should be replaced by high sensitivity next-generation sequencing approaches. Furthermore, when confirmed by others, subclonal TP53 mutations should be incorporated into future risk classification for AML.
The mechanisms by which mutant p53 mediates resistance to cytotoxic treatments are poorly understood and possibly involve novel gain-of-function properties. TP53 mutations affect preleukemic stem cells in AML and it could be shown in vitro and in vivo that genotoxic stress leads to expansion of murine hematopoietic stem and progenitor cells exhibiting either p53 haploinsufficiency or expressing a heterozygous TP53 mutation.27–29 These data are in line with the AML case presented here, in which substantial expansion of both a clonal and a subclonal TP53 mutation was observed following high-dose chemotherapy but not during the transition from myelodysplastic syndrome to frank leukemia. Similar clinical data have recently been published for patients with lymphomas showing clonal hematopoiesis including TP53 mutations at diagnosis or before undergoing autologous hematopoietic stem cell transplantation. These individuals were at increased risk of clonal expansion and, ultimately, development of therapy-related myeloid neoplasms following intensive, lymphoma-specific chemo- and radiotherapies.30–33
In our study we were able to demonstrate TP53-mutated clones with a VAF as low as 4.66% (Online Supplementary Table S1). However, recently developed next-generation sequencing techniques such as error-corrected ultradeep sequencing allow an unambiguous identificantion of mutant alleles with a frequency as low as 0.1%.34 In chronic lymphocytic leukemia, it has been demonstrated that even very small TP53-mutated clones constitute an adverse prognostic parameter.24,25 Given the mechanism of selection of such clones following genotoxic therapies as outlined above, it is very likely that a similar effect may also be operational in AML. Another issue is related to the assessment of VAF using clinical specimens. Specification of VAF refers either to mononuclear cell fractions or with a correction to represent exclusively neoplastic cells. However, it must be taken into account that mutations affecting preleukemic stem cells in AML are being propagated into mature blood cells.35 By analyzing highly purified T-lymphocytes of diagnostic AML specimens with TP53 mutations, we were able to demonstrate the specific aberration being present in up to 20% of these mature blood cells.27 Thus, analyzing mononuclear cell fractions may be the more comprehensive approach to assess VAF in AML and, possibly, other myeloid disorders. Finally, the diagnostic specimens in this study were derived from both bone marrow and peripheral blood (Online Supplementary Methods) raising the issue of comparability of these sources. Recently, Duncavage et al. demonstrated that the mutational landscape is conserved in peripheral blood of AML and myelodysplastic syndrome patients at diagnosis as well as during treatment.36 Similar data were obtained for TP53 mutations in patients with myelodysplastic syndromes.37
In this study we investigated the prognostic impact of subclonal TP53 mutations in intensively treated AML patients. Results may be somewhat different when assessing AML patients with TP53 mutations treated with hypomethylating agents such as decitabine. Although a substantial impact on overall survival could not be shown with that approach, similar response rates as those of AML patients with TP53 wild-type have been reported recently.38 It might, therefore, be necessary to prospectively address the impact of subclonal TP53 mutations in such an AML cohort.
In conclusion, we have shown that subclonal TP53 mutations represent an adverse prognostic marker in AML, independently of the clone size. Our data further corroborate the use of next-generation sequencing technologies for risk stratification of patients with AML. Future work will assess the clonal architecture of these specimens and investigate whether TP53-mutated subclones exhibit preleukemic/leukemic stem cell properties which may explain their persistence following cytotoxic therapies.
Supplementary Material
Acknowledgments
We thank Ms. Daniela Weber, Ulm, Germany, for excellent assistance and Dr. Karl Kashofer, Graz, Austria, for providing DNA for sequencing analysis.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/3/516
References
- 1.Dores GM, Devesa SS, Curtis RE, Linet MS, Morton LM. Acute leukemia incidence and patient survival among children and adults in the United States, 2001–2007. Blood. 2012;119(1)34–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23)2209–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas D, Majeti R. Biology and relevance of human acute myeloid leukemia stem cells. Blood. 2017;129(12)1577–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirsch P, Zhang Y, Tang R, et al. Genetic hierarchy and temporal variegation in the clonal history of acute myeloid leukaemia. Nat Commun. 2016;712475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rucker FG, Schlenk RF, Bullinger L, et al. TP53 alterations in acute myeloid leukemia with complex karyotype correlate with specific copy number alterations, monosomal karyotype, and dismal outcome. Blood. 2012;119(9)2114–2121 [DOI] [PubMed] [Google Scholar]
- 6.Andersen MK, Christiansen DH, Pedersen-Bjergaard J. Centromeric breakage and highly rearranged chromosome derivatives associated with mutations of TP53 are common in therapy-related MDS and AML after therapy with alkylating agents: an M-FISH study. Genes Chromosomes Cancer. 2005;42(4)358–371 [DOI] [PubMed] [Google Scholar]
- 7.Montalban-Bravo G, Benton CB, Wang SA, et al. More than 1 TP53 abnormality is a dominant characteristic of pure erythroid leukemia. Blood. 2017;129(18)2584–2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulz E, Valentin A, Ulz P, et al. Germline mutations in the DNA damage response genes BRCA1, BRCA2, BARD1 and TP53 in patients with therapy related myeloid neoplasms. J Med Genet. 2012;497422–428 [DOI] [PubMed] [Google Scholar]
- 9.Zebisch A, Lal R, Muller M, et al. Acute myeloid leukemia with TP53 germ line mutations. Blood. 2016;128(18)2270–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Middeke JM, Herold S, Rucker-Braun E, et al. TP53 mutation in patients with high-risk acute myeloid leukaemia treated with allogeneic haematopoietic stem cell transplantation. Br J Haematol. 2016;172(6)914–922 [DOI] [PubMed] [Google Scholar]
- 11.Grossmann V, Schnittger S, Kohlmann A, et al. A novel hierarchical prognostic model of AML solely based on molecular mutations. Blood. 2012;120(15)2963–2972 [DOI] [PubMed] [Google Scholar]
- 12.Stengel A, Kern W, Haferlach T, Meggendorfer M, Fasan A, Haferlach C. The impact of TP53 mutations and TP53 deletions on survival varies between AML, ALL, MDS and CLL: an analysis of 3307 cases. Leukemia. 2017;31(3)705–711 [DOI] [PubMed] [Google Scholar]
- 13.Dohner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4)424–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McShane LM, Altman DG, Sauerbrei W, et al. REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res Treat. 2006;100(2)229–235 [DOI] [PubMed] [Google Scholar]
- 15.Schlenk RF, Dohner K, Krauter J, et al. Mutations and treatment outcome in cytogenetically normal acute myeloid leukemia. N Engl J Med. 2008;358(18)1909–1918 [DOI] [PubMed] [Google Scholar]
- 16.Schlenk RF, Frohling S, Hartmann F, et al. Phase III study of all-trans retinoic acid in previously untreated patients 61 years or older with acute myeloid leukemia. Leukemia. 2004;18(11)1798–1803 [DOI] [PubMed] [Google Scholar]
- 17.Schlenk RF, Dohner K, Mack S, et al. Prospective evaluation of allogeneic hematopoietic stem-cell transplantation from matched related and matched unrelated donors in younger adults with high-risk acute myeloid leukemia: German-Austrian trial AMLHD98A. J Clin Oncol. 2010;28(30)4642–4648 [DOI] [PubMed] [Google Scholar]
- 18.Sallman DA, Komrokji R, Vaupel C, et al. Impact of TP53 mutation variant allele frequency on phenotype and outcomes in myelodysplastic syndromes. Leukemia. 2016;30(3)666–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goel S, Hall J, Pradhan K, et al. High prevalence and allele burden-independent prognostic importance of p53 mutations in an inner-city MDS/AML cohort. Leukemia. 2016;30(8)1793–1795 [DOI] [PubMed] [Google Scholar]
- 20.Rausch T, Jones DT, Zapatka M, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148(1-2)59–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bochtler T, Granzow M, Stolzel F, et al. Marker chromosomes can arise from chromothripsis and predict adverse prognosis in acute myeloid leukemia. Blood. 2017;129(10)1333–1342 [DOI] [PubMed] [Google Scholar]
- 22.Wolfler A, Erkeland SJ, Bodner C, et al. A functional single-nucleotide polymorphism of the G-CSF receptor gene predisposes individuals to high-risk myelodysplastic syndrome. Blood. 2005;105(9)3731–3736 [DOI] [PubMed] [Google Scholar]
- 23.Kinde I, Wu J, Papadopoulos N, Kinzler KW, Vogelstein B. Detection and quantification of rare mutations with massively parallel sequencing. Proc Natl Acad Sci U S A. 2011;108(23)9530–9535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malcikova J, Stano-Kozubik K, Tichy B, et al. Detailed analysis of therapy-driven clonal evolution of TP53 mutations in chronic lymphocytic leukemia. Leukemia. 2015;29(4)877–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rossi D, Khiabanian H, Spina V, et al. Clinical impact of small TP53 mutated sub-clones in chronic lymphocytic leukemia. Blood. 2014;123(14)2139–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sallman DA, Komrokji R, List A, Padron E. Reply to Goel et al. ‘TP53 mutation allele-burden and disease outcome in MDS/AML’. Leukemia. 2017;31(3)767–768 [DOI] [PubMed] [Google Scholar]
- 27.Lal R, Lind K, Heitzer E, et al. Somatic TP53 mutations characterize preleukemic stem cells in acute myeloid leukemia. Blood. 2017;129(18)2587–2591 [DOI] [PubMed] [Google Scholar]
- 28.Wong TN, Ramsingh G, Young AL, et al. Role of TP53 mutations in the origin and evolution of therapy-related acute myeloid leukaemia. Nature. 2015;518(7540)552–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, Gao R, Yao C, et al. Genotoxic stresses promote clonal expansion of hematopoietic stem cells expressing mutant p53. Leukemia. 2018;32(3)850–854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulz E, Kashofer K, Heitzer E, et al. Preexisting TP53 mutation in therapy-related acute myeloid leukemia. Ann Hematol. 2015;94(3)527–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gillis NK, Ball M, Zhang Q, et al. Clonal haemopoiesis and therapy-related myeloid malignancies in elderly patients: a proof-of-concept, case-control study. Lancet Oncol. 2017;18(1)112–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi K, Wang F, Kantarjian H, et al. Preleukaemic clonal haemopoiesis and risk of therapy-related myeloid neoplasms: a case-control study. Lancet Oncol. 2017;18(1)100–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibson CJ, Lindsley RC, Tchekmedyian V, et al. Clonal hematopoiesis associated with adverse outcomes after autologous stem-cell transplantation for lymphoma. J Clin Oncol. 2017: JCO2016716712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stahlberg A, Krzyzanowski PM, Egyud M, Filges S, Stein L, Godfrey TE. Simple multiplexed PCR-based barcoding of DNA for ultrasensitive mutation detection by next-generation sequencing. Nat Protoc. 2017;12(4)664–682 [DOI] [PubMed] [Google Scholar]
- 35.Reinisch A, Chan SM, Thomas D, Majeti R. Biology and clinical relevance of acute myeloid leukemia stem cells. Semin Hematol. 2015;52(3)150–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duncavage EJ, Uy GL, Petti AA, et al. Mutational landscape and response are conserved in peripheral blood of AML and MDS patients during decitabine therapy. Blood. 2017;129(10)1397–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Belickova M, Vesela J, Jonasova A, et al. TP53 mutation variant allele frequency is a potential predictor for clinical outcome of patients with lower-risk myelodysplastic syndromes. Oncotarget. 2016;7(24)36266–36279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welch JS, Petti AA, Miller CA, et al. TP53 and decitabine in acute myeloid leukemia and myelodysplastic syndromes. N Engl J Med. 2016;375(21)2023–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.