Abstract
The process of documentation is one of the most important parts of electronic health records (EHR). It is time-consuming, and up until now, available documentation procedures have not been able to overcome this type of EHR limitations. Thus, entering information into EHR still has remained a challenge. In this study, by applying the trigram language model, we presented a method to predict the next words while typing free texts. It is hypothesized that using this system may save typing time of free text. The words prediction model introduced in this research was trained and tested on the free texts regarding to colonoscopy, transesophageal echocardiogram, and anterior-cervical-decompression. Required time of typing for each of the above-mentioned reports calculated and compared with manual typing of the same words. It is revealed that 33.36% reduction in typing time and 73.53% reduction in keystroke. The designed system reduced the time of typing free text which might be an approach for EHRs improvement in terms of documentation.
Keywords: Electronic health record, Data capture, Natural language processing, Word prediction, N-gram, Trigram model, Free-text, Data entry
Introduction
Patient-centralized functional modules in a typical electronic health records (EHR) system contain both structured and unstructured data [1]. EHRs provide several methods for documentation, including structured and unstructured data [2, 3] stored in clinical records [4]. There are several benefits for EHR regarding structured data application [5]. That is, the structured fields provide the opportunity for reusing and analyzing data quickly through a top-down menus and decrease the time of typing [6]. They create a structure for assessing documents, and emphasize the need for some data elements in order to meet the quality and monitoring objectives [7]. Despite the positive aspects of structured documentation in EHR, there are still concerns regarding to patients’ information connectivity and unstructured clinical notes are still the preferred form of inter-practitioner communication about patients [8]. According to a survey [9] the physicians do not trust the information recorded in EHR and prefer verbal communication to collect patient’s information [10].
Due to variety of clinicians with diverse specialties; they have personal preferences and there are varied document requirements [11]. Two general methods for data entry are known including electronic interface systems for structured data and Manual data entry [12], which is mainly unstructured data.
Often clinical notes appear as dictated and unstructured in EHR. The ability to explain what is expressed by natural language is considered as one of the benefits of free texts [13]. Clinical professionals spend most of their time investigating this part of the electronic health records [14].
Unstructured data or narrative data are not easily analyzed and cannot be linked to the structured records, and entering these data are time-consuming and illegible as well [2]. To utilize the effectiveness of clinical records, both of the structured and unstructured data are necessary [11]. In electronic health records, a great part of the data stored is unstructured [15]. Another key role of free texts is their acting as a strong communication tool between physicians and nurses [2, 16].
It is reported that nurses and physicians believe on more documentation time using EHR [17, 18] Despite efforts made to structure data entry into EHR there are many problems about the patients’ history because of structured method data collection. Hence, in many cases, in order to transmit supplementary information to all parties involved in the treatment process (physicians, nurses, radiologists, physiotherapists, psychologists, etc.), it is better to use free texts in order to transfer the data of diagnoses, treatment methods, and results [19]. To provide reliable information and early diagnosis, different technologies are used to collect and process data [20].
In spite of different data entry ways of information registry for EHR including dictation [21], speech recognition [22, 23], handwriting recognition [11], scanning paper reports [11], template [24], typing [25] and combined approach, none of them have been known as the inclusive and fast way for data entry in EHR [12, 25].
Many attempts have been made to investigate the difficulties in clinical data entry in order to promote acceptance and quality-in-use of clinical information systems [26].
The documentation process in the EHR is known as a time-consuming process.
Recent studies showed the limitation of increasing the time of entering data into EHR is now considered as one of the reasons for the low acceptance rate of this system [27]. In [13], it is found that a considerable part (58%) of the entry time of electronic health records data is spent for entering free texts. Personnel’s waste of time and dissatisfaction by 44% put EHR’s usefulness under question. Therefore, it is necessary to develop an exact mechanism assisting physicians, nurses, and other staffs to register free texts on EHR in a time saving method.
In recent years, natural language processing (NLP) has been recognized as a viable alternative for data entry [28]. NLP area, which performs tasks such text prediction through understanding and interpretation of texts and speeches, became popular [21].
We propose to examine the utilization of text prediction applied in free text entry in EHR. Text prediction is the task of suggesting the next word, letters, phrase or sentence while the user is typing. A language model is needed to improve prediction [29]. Text prediction tools as an assisted data entry function aim to save time and effort by reducing the number of keystrokes needed and to improve text quality by preventing misspellings [30, 31].
In this paper, we explore the tasks of word prediction, where a system displays a list of suggestions for the next word when users start typing free text entry in EHR.
Related work
The application of text prediction systems is diverse in the clinical domain. These systems serve as an intermediate tool for producing reports such radiology report [32] and discharge summary [33] to save time and effort by reducing the number of keystrokes needed.
In work [34] an autocomplete tool has been developed helping physicians at the time of prescribing medication to select the correct drug name. Term selection strategies in this work are Edit Distances, Radix Tree, Inverted Files. This tool has been tested in the context of the medical prescription of the HUG. The tool helps to select the most appropriate term by ranking the possible results in a clever manner. Experimental evaluation shows promising results and indicates the tool ease the terminology manipulations.
The N-gram language model has been used to improve the text input rate in work [35]. Results show that by reducing the number of phrases that Mentally Disabled Huntington’s Patients can type, text input rate has improved.
The study [31] evaluates the multi-prefix matching algorithm, which propose terms whose words’ prefixes own all words in the letters typed by the user. The evaluation results indicate that the use of this algorithm leads to a decrease in keystrokes (saves an additional 4.7 keystrokes on average) and it can be used to search and retrieve terms from the SNOMED-CT medical dictionary.
The study [36] used word prediction to improve the efficiency and quality of the input of structured data. In this work, prediction lists were used to recognize the word being typed. The results of this study on the public database of Morbidity and Mortality show 13.0% time reduction and 3.9% increase of response accuracy in a data entry function.
Gong and coworkers proposed two predictive text functions, which are connected to narrative comment field widely used in patient safety reporting systems. The results show 87.1% for keystroke savings and increase of 70.5% in text generation rate [26].
The study [37] presents a solution to the word prediction and completion tasks. Their system best recall value is 71.28% for word completion and best value Keystroke savings value is 44.81% for word completion on the test set.
The results of a review of word prediction systems in the non-medical domain in 2015 [38] on different languages indicate that the use of the word prediction system based on (2–6) -GRAM for English is 37.4% for the Spanish language 51.9% and for Swedish 42.7% savings on keystroke.
Methodology
There are a variety of approaches to predict the next word used in natural language modeling from the early 1980s.
These methods can be classified into three groups [39]:
Statistical word prediction is based on the Markov assumption, in which only n-1 last words influence the next word. This is also called the N-gram Markov model.
Knowledge-based modeling, which systems that use statistical modeling to predict, often examine the terms in grammatical terms in order to predict. For example, Syntactic Prediction suggests a grammatically consistent word for parts of speech tags and phrase structures also in semantic prediction tries to predict words that are syntactically and semantically true.
Heuristic modeling is a compatibility model that adapts the system to the user and provides appropriate predictions. In this modeling, the frequency tags attached to the words contained in the corpus as the user constructs the sentences.
N-gram
One of the first NLP approaches in the computer science field is the N-gram model. N-gram is a method of examining a sequence of n items available in a text or voice utilized in order to predict the following word [18] based on their applications, items can be phonemes, syllables, letters, words, and base pairs. N-grams are generally collected from text or speech corpuses [40]. This is a potential predicting model which measures the probability of the occurrence of a word after a sequence of n-1 words based on Markov chain model [16, 24].
N-gram equation
N-grams are named according to their size. Unigram refers to an N-gram with the size of one, Bigram refers to an N-gram with the size of two, and trigram refers to an N-gram with the size of three. Larger N-grams are named as four gram [41]. In this study, the trigram language model was applied.
| 1 |
Trigram equation
For example, in the sentence “The patient was transferred to the operating room”, the N-gram calculations will be as follows:
Unigram: “The”, “patient”, “was”, “transferred”, “to”, “the”, “operating”, “room”
| 2 |
Bigram equation
Bigram: In this model, the words are considered in pairs (formula 2):
“The patient”, “patient was”, “was transferred”, “transferred to”, “to the“, “the operating”, “operating room”
Trigram: By using formula 1, the words are produced as three consecutive words:
“The patient was”, “patient was transferred”, “was transferred to”, “transferred to the”, “to the operating”, “the operating room”
Generation of tri-gram
By employing trigram linguistic model, we can follow a three-word sequence and anticipate the following possible words. To meet this end, we need to assess all the sentences available in the corpuses collected in Sect. 3.3 For example, the following sentence is selected from the corpus belonging to the scope of transesophageal-echocardiogram:
“Localizing x-ray verified the marker to be right at the C3–4 interspace.”
Since our proposed model is trigram, after each pair of words, we should examine what word has been viewed. According to Table 1, the trigram calculated on the sentence above is produced as follows:
Table 1.
Sample of calculations for trigram linguistic model
| Word-A | Word-B | Predict word |
|---|---|---|
| Localizing | X-ray | Verified |
| X-ray | Verified | The |
| Verified | The | Marker |
| The | Marker | To |
| Marker | To | Be |
| To | Be | Right |
| Be | Right | At |
| Right | at | The |
| At | The | C3-4 |
| The | C3-4 | Interspace |
According to this model of trigram, when two pairs of words of “localizing” and “x-ray” follow each other, the following word will be “verified”. This operation will be performed on all statements available in each of the corpuses collected in Sect. 3.3. Thus, due to the corpus comprehensives, it is highly possible that after two pairs of words consideration, some words would be available as suggestions.
Data
In this study, the available textual reports of anterior-cervical-decompression, colonoscopy, and transesophageal-echocardiogram were applied. The notes included admission and discharge, outpatient clinical encounter, clinic visit and etc.). In order to create a standard corpus for each field, we collected and integrated the reports. These three clinical reports were randomly retrieved from two different non-government clinics and used for training and testing. This corpus was containing 1,509,716 words and the test corpus containing 23,356 words.
Suggesting best tri-gram
During system development, we faced problem of providing the best candidate to user. For instance, if the user typed two words of “Anterior” and “cervical” trigrams should be examined based on the collected corpus of the two pairs of typed words. In following examples, there are two pairs of “cervical” and “Anterior” which have been frequently repeated in the corpus:
Anterior cervical discectomy at C5–C6 for neural decompression.
Anterior cervical instrumentation at C5–C6 for stabilization by Uniplate construction at C5–C6.
Anterior cervical discectomy at C5–C6 and C6–C7 for neural decompression.
According to the statements assessed in the corpus and the two pairs of typed words, the following trigrams can be considered:
Anterior cervical discectomy
Anterior cervical instrumentation
Anterior cervical discectomy
At this stage, bearing in mind the corpus comprehensiveness, a large number of trigrams are produced among which we must select the best and offer it to the users. To solve this problem at the production stage of the trigrams, the frequency of the occurrence of words after the two pairs of typed words is also counted in the corpus. The words, which have the highest frequency after the two pairs of words typed, can have a greater opportunity to be selected by the user for display. The following figure demonstrations a part of the calculations on the corpus where the trigrams and their frequency are also shown (Fig. 1).
Fig. 1.
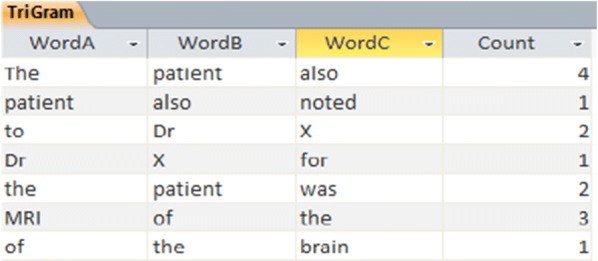
Sample of calculations of trigram linguistic model with frequency of words in the database
Thus, after performing the calculations, the trigrams with the highest probability will be offered to the user. In Fig. 2, after receiving the two word pairs of “the” and “patient”, the system proposes the words of “was”, “remained”, “in”, “tolerated”, and “with” which have the highest probability. Here, the system is designed based on five words having the highest probability which can be changed based on more or less words.
Fig. 2.
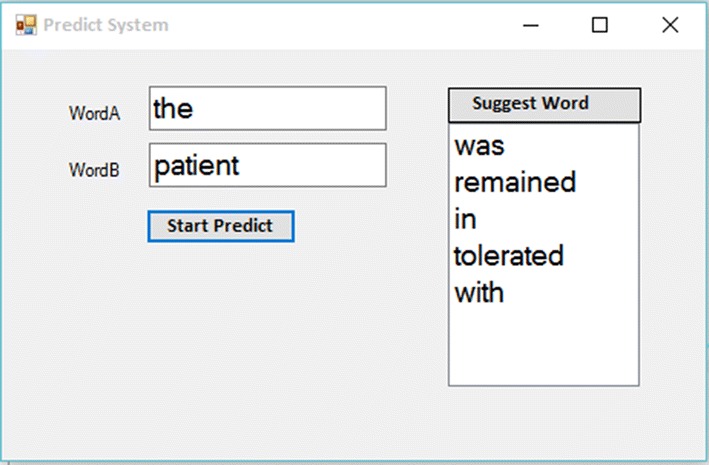
Sample of the suggestions of word-predicting model to users
Optimizing tri-gram model process
N-gram calculations are always included issues with high run-time order. Therefore, in order to have an acceptable performance, we are forced to convert the calculations from an online mode to an Offline one. For this purpose, as the process of trigram calculations is done once, and then the generated database is employed repeatedly. Hence, instead of constantly carrying out the calculations process on the corpus with the run-time order of O(n), the generated database is repeatedly used. In this process, after the production of the database based on corpus, it will no longer be required to do the calculations. In other words, all the possible calculations are conducted on the corpus only once, and then the database becomes the reference of all calculations. Figure 3 demonstrates a part of the calculations performed on the corpus (Sect. 3.3) which leads to the production of the database.
Fig. 3.
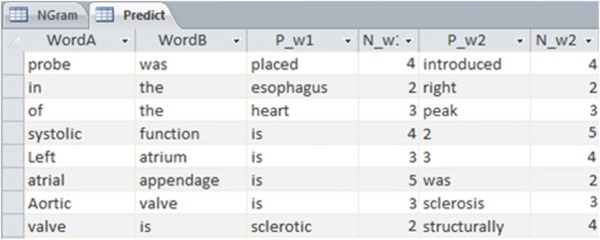
A part of the offline database showing pre-calculations of the trigram linguistic model
The search is conducted based on the two pairs of words typed by the user; in addition, by finding a record, all the words, which are placed after these two words, will be accessible along with their frequency number. For example, if the user has first entered the word of “aortic” and then “valve”, the system will offer words, presented in Table 2, to the user. In point of fact, these words are exactly five words which have come into the corpus after the term “aortic valve” and have the highest frequency compared to the other words. At the final step by using trigram Eq. (1), probability of each five candidates is calculated and sort based on the highest probability.
Table 2.
Sample of the words suggested to the user at the time of typing
| Predict Word | Frequent | Probability |
|---|---|---|
| Is | 3 | 0.036 |
| Sclerosis | 1 | 0.012 |
| Reveals | 2 | 0.019 |
| Planimetry | 1 | 0.006 |
| Area | 4 | 0.051 |
Result
In the present study, to assess the implementation of this system, we received help from 150 employees working in the field of healthcare such as nurse, physician and medical transcriber. For each of the selected fields, some texts were set aside as the test texts offered to the volunteers for typing. This process was carried out in two stages including using the given system and Microsoft Office Word for typing the sample free text. We assigned typing the sample text by either developed system or Microsoft Office Word to a user randomly with no intentional consequences. The average time was compared for two methods.
The typing speed in colonoscopy reports, anterior-cervical-decompression reports, and transesophageal-echocardiogram improved by 28.20, 35.82, and 36.06%, respectively. These results can be seen in Table 3.
Table 3.
Compares the speed of typing in the model presented compared to the manual typing of clinical reports
| Domain | Manual type time | Proposed type system time | Improve (%) |
|---|---|---|---|
| Colonoscopy | 3′:44″ | 2′:47″ | 28.20 |
| Anterior-cervical-decompression | 3′:26″ | 2′:11″ | 35.82 |
| Transesophageal-echocardiogram | 4′:26″ | 2′:50″ | 36.06 |
Experiment shows precision increases, until number of suggest words lower than 6 and bigger than 5 words. The recall tends to get maximum 93% for 5–6 number of suggest words and will decrease for bigger window size (Fig. 4).
Fig. 4.
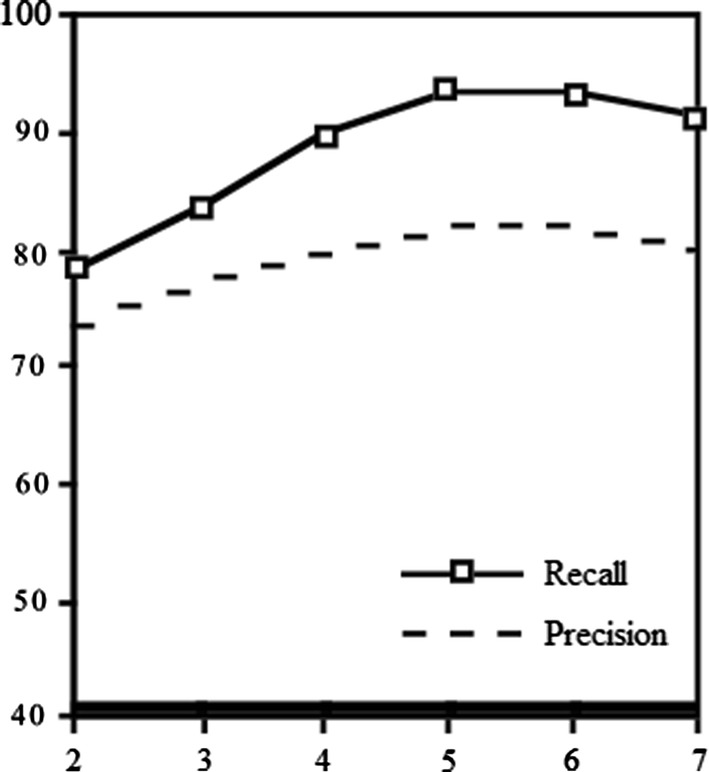
Precision and recall’s suggestion words
In the flowing table, different numbers of suggestion words’ recalls and precisions are compared. As it is shown, precisions and recall percentages depend on window size (Table 4).
Table 4.
Compares the speed of typing in the model presented compared to the manual typing of clinical reports
| Window size | Recall (%) | Precision (%) |
|---|---|---|
| 2 | 79 | 73 |
| 3 | 84 | 78 |
| 4 | 91 | 80 |
| 5 | 93 | 82 |
| 6 | 93 | 82 |
| 7 | 91 | 81 |
As Fig. 5 specifies, the speed of typing in the three reports of the free texts has improved on average by 33.36% via applying the proposed model.
Fig. 5.
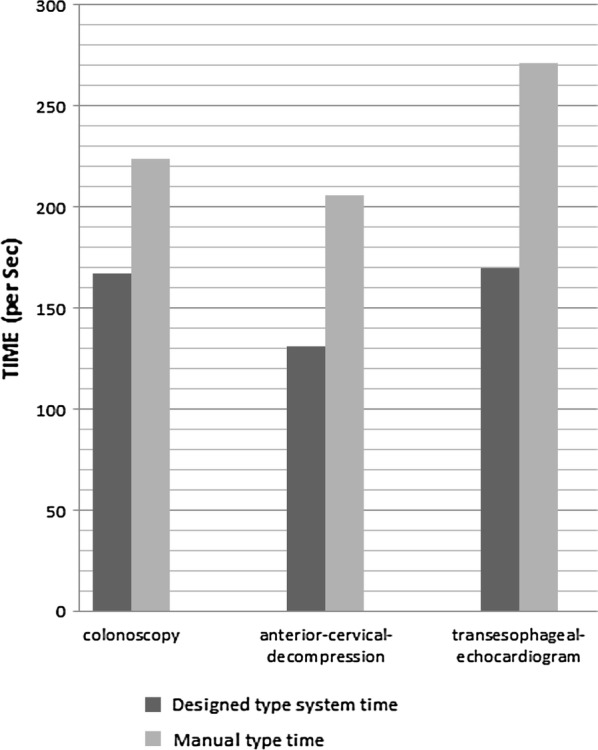
Compares the speed of typing in the proposed model compared with the manual typing of clinical reports
Discussion
Despite time-consuming of entering free text, these data play a very important role in health information collection in the electronic health record. Although clinicians have a time limitation, they are expected to report well in a timely manner [42]. Therefore, the use of systems that improve the efficiency of clinicians and medical typist in data entry seems to be necessary. Text prediction tools aim to save time and effort by reducing the number of keystrokes needed and to improve text quality by misspellings prevention.
Our text prediction system is based on N-gram language model. We examined three measures associated with the efficiency in terms of time reduction, keystrokes and text generation rates. Our system can significantly reduce the number of keystrokes necessary to type narrative texts. Therefore, the typing duration time of free texts decreased (33.36% on average) compared with similar studies [36].
In order to evaluate the effectiveness of the proposed system according to the keystroke measurement, 150 reports were randomly selected out of the corpus. The proposed system reduced the average number of keystrokes per report from 359 (without proposed system) to 95. Based on Table 5, the system presented 73.53% reduction in keystroke, which is promising in comparison with the reviewed works.
Table 5.
Comparion of keystroks saving(%)
Also, the proposed system performance was evaluated from the point of view of the text generation rate. According to the text generation rate, this rate on colonoscopy free texts were 0.57 (word per second), text generation rates of trans esophageal echocardiogram were 0.56 and anterior-cervical-decompression text generation rates were 0.73. On average, 0.61 words were generated on all three free text categories.
Conclusion
In this paper, we developed a system accelerating the free text entry into EHR based on the trigram language model. The designed system was tested on three sets of free texts, and the time of typing these texts was measured separately for all of the three sets. During the survey, we observed a decline in the time of typing in the three fields. By using the model presented in this study, the time of typing all of the unstructured clinical texts entered in EHR might be improved Based on our knowledge.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shah S, et al. Neural networks for mining the associations between diseases and symptoms in clinical notes. Health Inf Sci Syst. 2019;7(1):1. doi: 10.1007/s13755-018-0062-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finn C. Narrative nursing notes in the electronic health record: a key communication tool. Online J Nurs Inform. 2015;19(2):3. [Google Scholar]
- 3.Pollard SE, et al. How physicians document outpatient visit notes in an electronic health record. Int J Med Inform. 2013;82(1):39–46. doi: 10.1016/j.ijmedinf.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 4.Shah AD, Martinez C, Hemingway H. The freetext matching algorithm: a computer program to extract diagnoses and causes of death from unstructured text in electronic health records. BMC Med Inform Decis Mak. 2012;12(1):88. doi: 10.1186/1472-6947-12-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannon J, Lucci S. Transcription and EHRs: benefits of a blended approach. J AHIMA. 2010;81(2):36–40. [PubMed] [Google Scholar]
- 6.Abdelhak M, Grostick S, Hanken MA. Health Information: Management of a Strategic Resource. St. Louis: Elsevier; 2014. [Google Scholar]
- 7.Rosenbloom ST, et al. Generating clinical notes for electronic health record systems. Appl Clin Inform. 2010;1(3):232. doi: 10.4338/ACI-2010-03-RA-0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W, et al. A genetic algorithm enabled ensemble for unsupervised medical term extraction from clinical letters. Health Inf Sci Syst. 2015;3(1):5. doi: 10.1186/s13755-015-0013-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hall JM, Powell J. Understanding the person through narrative. Nurs Res Prac. 2011;2011:293837. doi: 10.1155/2011/293837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chou D. Health IT and patient safety: building safer systems for better care. JAMA. 2012;308(21):2282. [Google Scholar]
- 11.Hoyt RE, Yoshihashi, AK. Health informatics: practical guide for healthcare and information technology professionals. 2014: Lulu.com.
- 12.Shortliffe EH, Cimino JJ. Biomedical informatics: computer applications in health care and biomedicine. London: Springer; 2013. [Google Scholar]
- 13.Rule A, et al. Validating free-text order entry for a note-centric EHR. In: AMIA annual symposium proceedings. 2015. American Medical Informatics Association. [PMC free article] [PubMed]
- 14.Collins SA, et al. Relationship between nursing documentation and patients’ mortality. Am J Crit Care. 2013;22(4):306–313. doi: 10.4037/ajcc2013426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou L, et al. How many medication orders are entered through free-text in EHRs? A study on hypoglycemic agents. In: AMIA annual symposium proceedings. 2012. American Medical Informatics Association. [PMC free article] [PubMed]
- 16.Rosenbloom ST, et al. Data from clinical notes: a perspective on the tension between structure and flexible documentation. J Am Med Inform Assoc. 2011;18(2):181–186. doi: 10.1136/jamia.2010.007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han H, Lopp L. Writing and reading in the electronic health record: an entirely new world. Med Edu Online. 2013;18(1):18634. doi: 10.3402/meo.v18i0.18634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hripcsak G, et al. Use of electronic clinical documentation: time spent and team interactions. J Am Med Inform Assoc. 2011;18(2):112–117. doi: 10.1136/jamia.2010.008441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Embi PJ, et al. Computerized provider documentation: findings and implications of a multisite study of clinicians and administrators. J Am Med Inform Assoc. 2013;20(4):718–726. doi: 10.1136/amiajnl-2012-000946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yazdani A, et al. Scalable architecture for telemonitoring chronic diseases in order to support the CDSSs in a common platform. Acta Inform Med. 2018;26(3):195–200. doi: 10.5455/aim.2018.26.195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaufman DR, et al. Natural language processing-enabled and conventional data capture methods for input to electronic health records: a comparative usability study. JMIR Med Inform. 2016;4(4):e35. doi: 10.2196/medinform.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoyt R, Yoshihashi A. Lessons learned from implementation of voice recognition for documentation in the military electronic health record system. Perspectives in health information management/AHIMA, American Health Information Management Association, 2010. 7 (Winter). [PMC free article] [PubMed]
- 23.Johnson M, et al. A systematic review of speech recognition technology in health care. BMC Med Inform Decis Mak. 2014;14(1):94. doi: 10.1186/1472-6947-14-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sequeira KS. Electronic health records documentation: nurses attitudes and preferences in a given hospital. 2010, RGUHS.
- 25.dela Cruz JE, et al. Typed versus voice recognition for data entry in electronic health records: emergency physician time use and interruptions. West J Emerg Med. 2014;15(4):541. doi: 10.5811/westjem.2014.3.19658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gong Y, Hua L, Wang S. Leveraging user’s performance in reporting patient safety events by utilizing text prediction in narrative data entry. Comput Methods Programs Biomed. 2016;131:181–189. doi: 10.1016/j.cmpb.2016.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sas J, Poreba T. Optimal acoustic model complexity selection in polish medical speech recognition. J Med Inform Technol. 2011;17:115–122. [Google Scholar]
- 28.Paulett JM, Langlotz CP. Improving language models for radiology speech recognition. J Biomed Inform. 2009;42(1):53–58. doi: 10.1016/j.jbi.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Lin C-H, et al. Comparison of a semi-automatic annotation tool and a natural language processing application for the generation of clinical statement entries. J Am Med Inform Assoc. 2014;22(1):132–142. doi: 10.1136/amiajnl-2014-002991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sevenster M, Aleksovski Z. SNOMED CT saves keystrokes: quantifying semantic autocompletion. In: AMIA annual symposium proceedings. 2010. American Medical Informatics Association. [PMC free article] [PubMed]
- 31.Sevenster M, van Ommering R, Qian Y. Algorithmic and user study of an autocompletion algorithm on a large medical vocabulary. J Biomed Inform. 2012;45(1):107–119. doi: 10.1016/j.jbi.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 32.Eng J, Eisner JM. Informatics in radiology (info RAD) radiology report entry with automatic phrase completion driven by language modeling. Radiographics. 2004;24(5):1493–1501. doi: 10.1148/rg.245035197. [DOI] [PubMed] [Google Scholar]
- 33.Chen C-H, et al. Design and implementation of web-based discharge summary note based on service-oriented architecture. J Med Syst. 2012;36(1):335–345. doi: 10.1007/s10916-010-9479-y. [DOI] [PubMed] [Google Scholar]
- 34.Ehrler F, Lovis C. Supporting drug prescription through autocompletion. Stud Health Technol Inform. 2013;186:120–124. [PubMed] [Google Scholar]
- 35.Gelšvartas J, Simutis R, Maskeliūnas R. User adaptive text predictor for mentally disabled Huntington’s patients. Comput Intell Neurosci. 2016;2016:2. doi: 10.1155/2016/3054258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hua L, Wang S, Gong Y. Text prediction on structured data entry in healthcare. Appl Clin Inform. 2014;5(01):249–263. doi: 10.4338/ACI-2013-11-RA-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spithourakis GP, Petersen SE, Riedel S. Clinical text prediction with numerically grounded conditional language models. arXiv preprint arXiv:1610.06370, 2016.
- 38.Makkar R, Kaur M, Sharma DV. Word prediction systems: a survey. Adv Comput Sci Inform Technol. 2015;2(2):177–180. [Google Scholar]
- 39.Ghayoomi M, Momtazi S. An overview on the existing language models for prediction systems as writing assistant tools. In: IEEE international conference on systems, man and cybernetics, 2009. SMC 2009. IEEE.
- 40.Sarker A, Gonzalez G. Portable automatic text classification for adverse drug reaction detection via multi-corpus training. J Biomed Inform. 2015;53:196–207. doi: 10.1016/j.jbi.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Golkar A, et al. Word sense disambiguation based on number of lexical’s senses weighting in conceptual density. Int J Artif Intell Mechatron. 2014;3(1):12–15. [Google Scholar]
- 42.Poissant L, et al. The impact of electronic health records on time efficiency of physicians and nurses: a systematic review. J Am Med Inform Assoc. 2005;12(5):505–516. doi: 10.1197/jamia.M1700. [DOI] [PMC free article] [PubMed] [Google Scholar]


