Abstract
We utilized a system for sequence-specific RNA base editing via Adenosine Deaminases acting on RNA (ADAR) enzymes with associated ADAR guide RNAs (adRNAs). We systematically engineered it to harness ADARs, and comprehensively evaluated its specificity and activity in vitro and in vivo via two mouse models of human disease. We anticipate this platform will enable tunable and reversible engineering of RNAs for diverse applications.
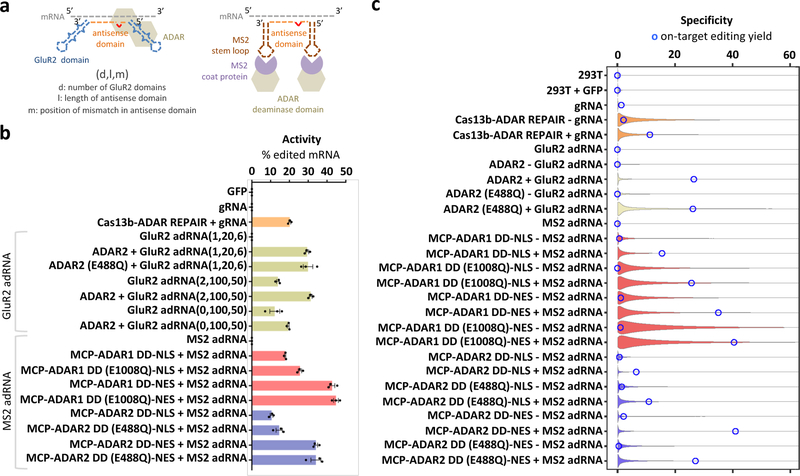
Adenosine to inosine RNA editing, a common post-transcriptional RNA modification, is catalyzed by Adenosine Deaminases acting on RNA (ADAR) enzymes1. Inosine is a deaminated form of adenosine that is biochemically recognized as guanine. Recently, multiple studies have demonstrated ADAR mediated targeted RNA editing2–9. Building on these, we engineered here two orthogonal toolsets for sequence-specific programmable RNA base editing in vitro and in vivo. Specifically, we utilized a system for targeted RNA editing via ADAR1/2 with associated ADAR guide RNAs (adRNAs) (Figure 1a). The adRNAs comprise in part a programmable antisense region that is complementary to the target RNA sequence with a mismatched cytidine opposite the target adenosine. Additionally, they bear in one version, zero, one, or two ADAR-recruiting domains engineered from the naturally occurring ADAR substrate GluR2 pre-mRNA3,4 (referred hereon as GluR2 adRNA); and in a second format, two MS2 hairpins flanking the antisense region (referred hereon as MS2 adRNA). The GluR2 adRNA was systematically optimized to enhance recruitment of exogenous and/or endogenous ADARs by evaluating multiple scaffold variants, including mutagenized scaffolds based on G-C versus A-U pairing, addition of editing inducer elements10, and antisense domain length11 and mis-match position modifications (Supplementary Figures 1, 2a–c). The latter MS2 adRNA version was in turn optimized to harness synthetic proteins comprising the deaminase domains (DD) of ADAR1 or ADAR2 fused to the MS2 Coat Protein (MCP), via systematic antisense domain length and mis-match position modifications, coupled with use of hyper-active versions of the deaminase domains, and versions bearing nuclear localization (NLS) versus export (NES) signals (Figure 1b, Supplementary Figures 3a, 3b).
Figure 1: Engineering programmable RNA editing and characterizing specificity profiles:
(a) Schematics of RNA editing via constructs utilizing the full length ADAR2 and an engineered adRNA derived from the GluR2 transcript, or MS2 Coat Protein (MCP) fusions to the ADAR1/2 deaminase domains and the corresponding MS2 hairpin bearing adRNA. (b) Comparison of RNA editing efficiency of the endogenous RAB7A transcript by different RNA editing constructs quantified by Sanger sequencing (efficiency calculated as a ratio of Sanger peak heights G/(A+G)). Experiments were carried out in HEK 293T cells. Values represent mean +/− SEM (n=3). (c) Violin plots representing distributions of A->G editing yields observed at reference sites where at least one treatment sample was found to have a significant change (Fisher’s exact test, FDR = 1%) in editing yield relative to the control sample. Blue circles indicate editing yields at the target A-site within the RAB7A transcript. Black dots represent median off-target editing yields. To better visualize the shapes of the distributions, their maximum extent along the y-axis was equalized across all plots, and were truncated at 60% yield.
We comprehensively evaluated the activity of the above two systems and also benchmarked it with the recently developed RNA editing system based on Cas13b7. Via these in vitro experiments we observed: 1) the engineered constructs were active in their ability to effect targeted RNA editing with yields comparable to the Cas13b based system (Figure 1b, Supplementary Figure 4a, Supplementary Tables 1, 2), and U6 transcribed adRNAs and chemically synthesized adRNAs were both effective formats (Supplementary Figure 4b); 2) adRNAs bearing long antisense domains, both with and without GluR2 domains, suffice to recruit exogenously expressed ADARs, and to a degree endogenous ADARs12 too to enable efficient RNA editing (Figure 1b, Supplementary Figures 2b, 2c, 4c); 3) the constructs based on the MS2 adRNAs and corresponding MCP-ADAR1/2 fusions showed the highest and most robust activity, including across a large panel of endogenous genes chosen across a spectrum of different expression levels (Figure 1b, Supplementary Figure 4c); 4) use of a NES and/or hyper-active deaminase domains in the MCP-ADAR1/2 fusions consistently yielded higher RNA editing yields at the target adenosine, but also led to a higher propensity of editing at non-targeted adenosines in the flanking sequences (Figure 1b, Supplementary Figure 5a). To further validate this, we showed that a similar promiscuity ensued also from deletion of the native NLS domain in ADAR2 (∆1–138)13 (Supplementary Figures 5b–d); and 5) these two toolsets we operationally orthogonal: specifically, we evaluated the editing efficiency of the MCP-ADAR2 deaminase domain fusion with a co-expressed MS2 adRNA or GluR2 adRNA and observed on-target editing only via the former. Conversely, we also confirmed that full-length ADAR2 was recruited by the GluR2 adRNA and not the MS2 adRNAs (Supplementary Figure 3b).
Having demonstrated robust activity of this toolset, we next investigated its specificity profiles via analysis of the transcriptome-wide off-target A->G editing effected by this system (Figure 1c). To this end, HEK 293T cells were transfected with each construct and analyzed by RNA-seq. Untransfected cells were included as controls. From each sample, we collected ~40 million uniquely aligned sequencing reads. We then used Fisher’s exact test to quantify significant changes in A->G editing yields, relative to untransfected cells, at each reference adenosine site having sufficient read coverage. The number of sites with at least one A->G editing event detected in any of the samples was computed. Of these, the number of sites with statistically significant A->G edits, at a false discovery rate (FDR) of 1%, and with fold change of at least 1.1, was found to vary over a wide range, from lowest for the MCP-ADAR2 DD-NLS construct, to highest for the MCP-ADAR1 DD (E1008Q)-NES (Supplementary Figures 6–9, Supplementary Tables 3, 4). To investigate the distribution of editing yields, we generated violin plots considering the A-sites whose editing yields changed significantly in at least one sample (Figure 1). Taken together, our RNA-seq experiments revealed that transcriptome-wide off-target edits were: 1) less prevalent in MCP-ADAR constructs with NLS than constructs with NES; 2) less prevalent in MCP-ADAR2 constructs than MCP-ADAR1 constructs; 3) less prevalent in the wild-type MCP-ADAR constructs than the E>Q hyperactive mutants (Supplementary Figure 10a, Supplementary Table 4); and 4) the off-targets were primarily due to ADAR overexpression and use of adRNAs alone resulted in least number of off-targets (Supplementary Figure 10b).
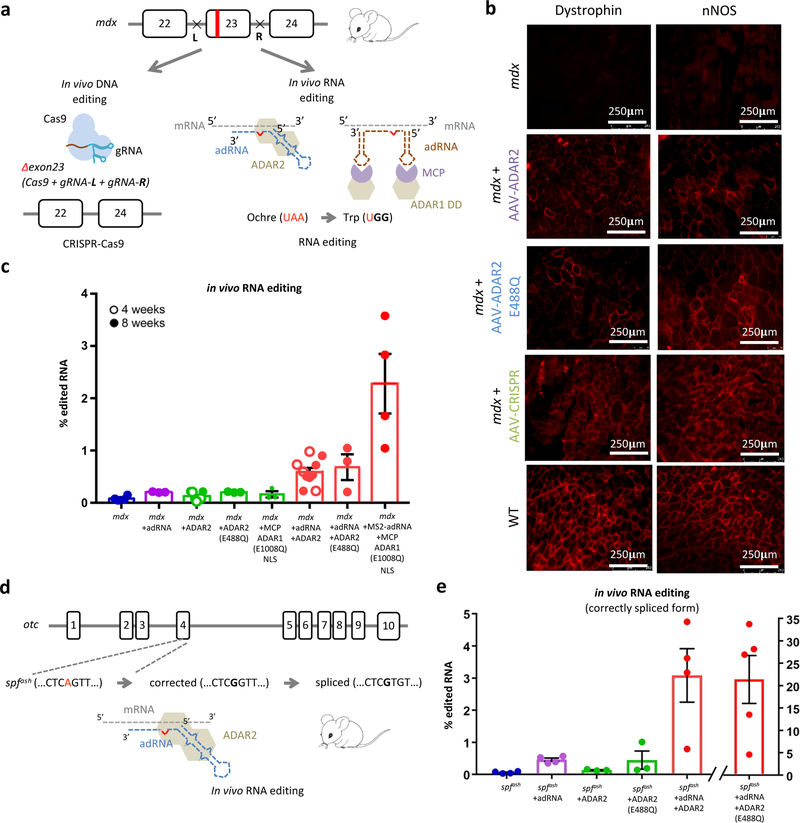
Following these in vitro studies, we next evaluated our system in in vivo RNA targeting for gene therapy applications, utilizing the adRNA cum exogenous ADAR expression construct versions, as those consistently enabled the highest in vitro RNA editing yields. We focused first on the mdx mouse model for Duchenne muscular dystrophy (DMD) which bears an ochre stop site in exon 23 of the dystrophin gene. This choice was additionally motivated by the fact that nonsense mutations in general are responsible for nearly 11% of all described gene lesions causing inheritable human disease, and close to 20% of disease-associated single base substitutions that affect the coding regions of genes14. Thus, validation of an RNA editing strategy here would have broad therapeutic application. Towards this, we first optimized RNA editing of stop codons in vitro (Supplementary Figure 11). Notably, we observed that addition of a second copy of the adRNA significantly improved the targeting efficiencies (Supplementary Figure 11c), and thus in all our in vivo studies we utilized a dual-adRNA delivery approach. We next packaged our constructs into AAV8, and injected 2E+12 vector genomes (vg)/muscle into the tibialis anterior (TA) or gastrocnemius of mdx mice. To further benchmark our approach, we concurrently also targeted the mdx mice via CRISPR-Cas9 based excision of exon 2315–17 (Figure 2a). Four or eight weeks post injection, TA and gastrocnemius muscles were collected from mdx mice, wild type mice, mice treated with adRNA targeting and non-targeting controls, and CRISPR-Cas9. Immunofluorescence staining revealed clear restoration of dystrophin expression via targeted RNA editing (Figure 2b, Supplementary Figure 12a). In addition, nNOS activity was also restored at the sarcolemma (Figure 2b, Supplementary Figure 12a). RNA editing yields (TAA->TGG/TAG/TGA) of up to 3.6%, and TAA->TGG up to 2.4% were observed in treated mice (Figure 2c, Supplementary Figure 11e). Western blots of the treated muscles confirmed the immunofluorescence observations, demonstrating 1–2.5% protein restoration. (Supplementary Figures 12b). As benchmark, muscles injected with vectors bearing CRISPR-Cas9 also expectedly led to restoration of dystrophin expression in a subset of the muscle cells (Figure 2b), with Western blots of the treated muscles confirming up to 10% protein restoration. (Supplementary Figure 12c).
Figure 2: In vivo RNA editing in mouse models of human disease:
(a) Schematic of the DNA and RNA targeting approaches to restore dystrophin expression in the mdx mouse model of Duchenne Muscular Dystrophy: (i) a dual gRNA-CRISPR based approach leading to in frame excision of exon 23 and (ii) ADAR2 and MCP-ADAR1 based editing of the ochre codon. (b) Immunofluorescence staining for dystrophin in the TA muscle shows partial restoration of expression in treated samples (intra-muscular injections of AAV8-ADAR2, AAV8-ADAR2 (E488Q), and AAV8-CRISPR). Partial restoration of nNOS localization is also seen in treated samples (scale bar: 250μm). (c) In vivo TAA->TGG/TAG/TGA RNA editing efficiencies in corresponding treated adult mdx mice. Values represent mean +/− SEM (n=4, 3, 7, 3, 3, 10, 3, 4 independent TA muscles respectively). (d) Schematic of the OTC locus in the spfash mouse model of Ornithine Transcarbamylase deficiency which have a G->A point mutation at a donor splice site in the last nucleotide of exon 4, and approach for correction of mutant OTC mRNA via ADAR2 mediated RNA editing. (e) In vivo RNA correction efficiencies in the correctly spliced OTC mRNA in the livers of treated adult spfash mice (retro-orbital injections of AAV8-ADAR2 and AAV8-ADAR2 (E488Q)). Values represent mean +/− SEM (n=4, 4, 3, 3, 4, 5 independent animals respectively).
To further confirm the efficacy of this approach, we next evaluated ADAR mediated RNA editing in an independent mouse model of human disease, the male sparse fur ash (spfash) mouse model of ornithine transcarbamylase (OTC) deficiency. The spfash mice harbor a G->A point mutation in the last nucleotide of the fourth exon of the OTC gene, which leads to OTC mRNA deficiency and production of a mutant protein18. Recent studies have demonstrated the use of CRISPR-Cas9 and homologous recombination based strategies for robust correction of this mutation in neonatal mice19. To test the effectiveness of our system in editing the point mutation in spfash OTC mRNA (Figure 2d), we initially evaluated our constructs in vitro (Supplementary Figure 13a). We next packaged our constructs into AAV8, which has high liver tropism19, and injected 2.5E+12 vg/mouse in 10–12 week old spfash mice. Three to four weeks post injection, we collected liver samples from spfash mice, wild-type litter mates, and spfash mice treated with the ADAR2 targeting and non-targeting vectors and evaluated corresponding editing efficiency via NGS. Notably, upon delivery of the adRNA and the ADAR2, we observed 0.8–4.7% edited mRNA amongst the correctly spliced OTC mRNA, and interestingly adRNA alone resulted in low but significant RNA editing yields (Figure 2e, refer Methods). Moreover, upon the delivery of the hyper-active ADAR2 mutant (E488Q), we observed a high edited fraction (4.6–33.8%) in the correctly spliced OTC mRNA (Figure 2e, Supplementary Figure 13b), 4.6–8.2% in the OTC pre-mRNA (Supplementary Figure 13c), and confirmed a reduction in the incorrectly spliced product (Supplementary Figure 13d). Western blots of the treated liver samples confirmed partial (2.5–5%) restoration of OTC protein (Supplementary Figure 13e).
Taken together, our results establish the utility of RNA-guided ADARs for in vivo RNA editing of point mutations. Moving forward, we note that, sequence preferences of the ADAR enzymes, RNA folding, intrinsic half-life, localization, translation machinery9, and resident RNA binding proteins can potentially impact accessibility and editability of target sites in the RNA, and will be important design parameters to consider for enabling efficacious targeting. For instance, in the mdx model, ADAR based RNA editing approaches have to compete with nonsense mediated decay of mutant dystrophin mRNA, and also the requirement for effecting two A->I substitutions in the context of non-ideal flanking nucleotides to eliminate the premature stop codon and potential impact on RNA stability and function. Furthermore, in the spfash model, the need to target the transient OTC pre-mRNA entails rapid target engagement and editing. Further progress will also require addressing important limitations of the system such as the off-targets induced by intrinsic enzyme-RNA binding, processivity, promiscuity, stimulation of the interferon response by the delivery modalities themselves (such as lipid, nanoparticles or viral) leading in turn to increased endogenous ADAR expression, potential of adRNAs to induce RNAi, and also off-target hybridization of the antisense domain of the adRNA which could potentially have deleterious effects20. In this regard, our studies revealed toxicity in mice systemically injected with the hyperactive ADAR mutants (Supplementary Figure 14). While we hypothesize that off-target RNA editing due to the hyper-active version might be responsible for the negative impact on mouse health, additional work needs to be performed to ascertain underlying mechanisms. These studies will be critical to aid systematic improvement of the specificity and safety of this approach. Another important consideration while considering RNA targeting for gene therapy, especially via the use of non-integrating vectors, is the necessity for periodic re-administration of the effector constructs, owing to the typically limited half-life of edited mRNAs and effectors. In this regard, compared to the CRISPR based RNA editing approaches, the RNA-guided ADAR strategy is directly human therapeutics relevant, as versions of the same solely utilize effector RNAs and human proteins. Additionally, as ADARs are widely expressed, for instance, ADAR1 across most human tissues and ADAR2 in particular in the lung and brain, endogenous recruitment of these via adRNAs bearing long-antisense domains (as demonstrated in Figures 1, 2e and Supplementary Figures 2, 4) presents a very attractive strategy for efficacious RNA editing. With progressive improvements, we thus anticipate this toolset will have broad implications for diverse basic science and therapeutic applications.
ONLINE METHODS
Vector design and construction
One or two copies of the adRNAs were cloned into an AAV vector containing a human U6 and mouse U6 promoter along with a CMV promoter driving the expression of the enzyme. To construct the GFP reporters – GFP-Amber, GFP-Ochre and GFP-Opal, three gene blocks were synthesized with ‘TAG’, ‘TAA’ and ‘TGA’ respectively replacing the Y39 residue of the wild type GFP and were cloned downstream of a CAG promoter. To construct the OTC and DMD reporters, 200 bp fragments of the spfash OTC and mdx DMD transcript bearing the target adenosine(s) to be edited were cloned downstream of the CAG promoter.
Mammalian cell culture and transfection
All HEK 293T cells were grown in Dulbecco’s Modified Eagle Medium supplemented with 10% FBS and 1% Antibiotic-Antimycotic (Thermo Fisher) in an incubator at 37 °C and 5% CO2 atmosphere. All in vitro transfection experiments were carried out in HEK 293T cells using the commercial transfection reagent Lipofectamine 2000 (Thermo Fisher). All in vitro RNA editing experiments involving a reporter were carried out in 24 well plates using 400ng of reporter plasmid and 800ng of the adRNA+enzyme plasmid. All in vitro RNA editing experiments targeting an endogenous transcript were carried out in 24 well plates using 800ng of the adRNA/Enzyme plasmid. dCas13b-ADAR2DDE488Q based RNA editing experiments were carried out using 800ng of the enzyme plasmid (Addgene #103864) as well 800 ng of the gRNA plasmid. Cells were transfected at 25–30% confluence and harvested 60 hours post transfection for quantification of editing. Chemically synthesized adRNAs (synthesized via IDT or Synthego) were transfected using Lipofectamine 3000 (Thermo Fisher) at an amount of 20 pmol/well.
Production of AAV vectors
AAV8 particles were produced using HEK 293T cells via the triple transfection method and purified via an iodixanol gradient. Confluency at transfection was about 80%. Two hours prior to transfection, DMEM supplemented with 10% FBS was added to the HEK 293T cells. Each virus was produced in 5 × 15 cm plates, where each plate was transfected with 7.5 ug of pXR-8, 7.5 of ug recombinant transfer vector, 7.5 ug of pHelper vector using PEI (1ug/uL linear PEI in 1×DPBS pH 4.5, using HCl) at a PEI:DNA mass ratio of 4:1. The mixture was incubated for 10 minutes at RT and then applied dropwise onto the cell media. The virus was harvested after 72 hours and purified using an iodixanol density gradient ultracentrifugation method. The virus was then dialyzed with 1 × PBS (pH 7.2) supplemented with 50 mM NaCl and 0.0001% of Pluronic F68 (Thermo Fisher) using 50kDA filters (Millipore), to a final volume of ~1 mL and quantified by qPCR using primers specific to the ITR region, against a standard (ATCC VR-1616).
AAV-ITR-F: 5’-CGGCCTCAGTGAGCGA-3’ and AAV-ITR-R: 5’-GGAACCCCTAGTGATGGAGTT-3’.
RNA isolation and Next Generation Sequencing library preparation
RNA from animal tissue was extracted using the RNeasy Plus Universal Mini Kit (Qiagen), according to the manufacturer’s protocol. RNA from cells was extracted using the RNeasy Mini Kit (Qiagen). cDNA was synthesized from 500ng RNA using the Protoscript II First Strand cDNA synthesis Kit (NEB). Next generation sequencing libraries were prepared as follows. Briefly, 1ul of cDNA prepared above was amplified by PCR with primers that amplify about 150 bp surrounding the sites of interest using KAPA Hifi HotStart PCR Mix (Kapa Biosystems). PCR products were purified (Qiagen PCR Purification Kit/ Gel Extraction Kit) to eliminate byproducts. Libraries were constructed with NEBNext Multiplex Oligos for Illumina kit (NEB). 10 ng of input DNA was amplified with indexing primers. Samples were then pooled and loaded on an Illumina Miseq (150bp single-end run) or Hiseq (100bp paired-end run). Data analysis was performed using CRISPResso (Pinello, L. et al. 2016). A minimum of 100,000 reads were analyzed for all in vivo experiments. RNA-seq libraries were prepared from 300ng of RNA, using the NEBNext Poly(A) mRNA magnetic isolation module and NEBNext Ultra RNA Library Prep Kit for Illumina. Samples were pooled and loaded on an Illumina Hiseq (100bp paired-end run).
Quantification of OTC mRNA editing yields in the spfash mice
The spfash mice bear three forms of OTC RNA: the pre-mRNA, the correctly spliced mRNA and an incorrectly spliced, elongated mRNA formed due to the use of a cryptic splice site 48 base pairs into intron 418. Let the total number of the correctly spliced mRNA be X, incorrectly spliced variant be Y and the pre-mRNA be Z. Xe, Ye and Ze denote the A->G edited mRNA in the three forms. The mRNA editing yield ideally would be calculated as (Xe+Ye+Ze)/(X+Y+Z). However, since it is not possible to amplify the spliced and pre-mRNA variants using the same primers, in Figure 2e, we report the fraction of edited transcripts in the correctly spliced mRNA (Xe/X) which will in turn be translated to produce the OTC protein. In addition, in Supplementary Figure 14c, we also report the fraction of edited transcripts in the pre-mRNA (Ze/Z). This fraction, upon correct splicing will contribute to formation of OTC protein. Finally, the incorrectly spliced mRNA results in the production of a protein elongated by 16 amino acids which is selectively degraded. We have not quantified the edited transcripts in this fraction of mRNA since these edited transcripts cannot be used to produce a functional protein. In Supplementary Figure 14d, bands corresponding to X and Y are shown.
Animal experiments
All animal procedures were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee (IACUC) of the University of California, San Diego. All mice were acquired from Jackson labs. AAVs were injected into the gastrocnemius or TA muscle of mdx mice (C57BL/10ScSn-Dmdmdx/J) using 2E+12 vg/muscle. AAVs were injected into spfash mice (B6EiC3Sn a/A-Otcspf-ash/J) via retro-orbital injections using 2.5E+12 vg/mouse. Mice that appeared to have a rough hair coat, moved slowly and appeared slightly hunched were termed as sick mice and euthanized.
Immunofluorescence
Harvested gastrocnemius or TA muscles were placed in molds containing OCT compound (VWR) and flash frozen in liquid nitrogen. 10 μm sections were cut onto pre-treated histological slides. Slides were fixed using 4% Paraformaldehyde. Dystrophin and nNOS were detected with rabbit polyclonal antibodies against the C-terminal domain of dystrophin (1:200, Abcam 15277) and N-terminal domain of nNOS (1:100, Immunostar 24431) respectively, followed by a donkey anti-rabbit Alexa 546 secondary antibody (1:400, Thermo Fisher).
Western Blots
Muscle biopsies from mdx mice and liver biopsies from spfash mice were fragmented in RIPA buffer (Sigma) with a proteinase inhibitor cocktail (Roche) and incubated for 1 hour on ice with intermittent vortexing. Samples were centrifuged at 15500×g for 30 min at 4°C and the supernatant was isolated and quantified with a Pierce Coomassie Plus (Bradford) assay kit (Thermo Fisher). Protein isolate was mixed with 4× Laemmli Loading buffer (Biorad) and 2-Mercaptoethanol (Biorad) and boiled at 100°C for 10 min. 100 μg total protein from muscle biopsies or 60 ug from liver biopsies was loaded into each well of a4–15% Mini Protean TGX gel (Biorad) with Tris-Glycine-SDS buffer (Biorad) and electrophoresed for 60 min at 100 V. Protein from muscle biopsies was transferred to nitrocellulose membranes overnight at 34V while that from liver biopsies was transferred at 65V for 1hour 30 minutes in a 1X tris-glycine transfer buffer containing 10% methanol and 0.1% SDS at 4°C. The blot was blocked for 1 hour in 5% milk-TBST. Blots were probed with rabbit anti-dystrophin (1:200, Abcam 15277), rabbit anti-GAPDH (1:4000, Cell Signaling 2118S), rabbit anti-OTC (1:800, Abcam 203859) and mouse anti-ADAR2 (1:150, Santa Cruz Biotechnology 73409) overnight at 4°C in 5% milk-TBST. Blots were washed with TBST and then incubated with anti-rabbit or anti-mouse horseradish peroxidase-conjugated secondary antibodies (Cell Signaling) for 1 hour in 5% milk-TBST. After washing with TBST, blots were visualized using SuperSignal West Femto Chemiluminescent Substyeild (Thermo Fisher) and X-Ray films.
Statistics and Reproducibility
In vitro experiments: All in vitro experiments were carried out once with a minimum of 3 independent replicates.
In vivo experiments: For the mdx mouse model, ADAR2 and MCP-ADAR1 (E1008Q) NLS based experiments were carried out twice. Both rounds of experiments yielded consistent RNA editing efficiencies, dystrophin immunofluorescence and dystrophin restoration as seen by western blots. ADAR2 (E488Q) and CRISPR-Cas9 based experiments were carried out once. For the spfash mouse model, all experiments were carried out twice, based on the availability of mice. RNA editing efficiencies of the OTC transcript, both the spliced and pre-mRNA were consistent in both rounds of experiments. RT-PCR and Western blots were carried out on all animals in experimental set 1.
Quantification of RNA A->G editing
RNA-seq read alignment
RNA-seq read pairs with 100 bases per read mate were aligned to the GRCh38 reference genome using STAR aligner version 2.6.0c (Dobin A et al 2013). The genome index was built using primary assembly annotations from GENCODE release 28 (GRCh38.p12). Default parameters were used to run STAR, except for the following relevant settings: readMapNumber=−1, alignSJoverhangMin=5, alignSJDBoverhangMin=1, alignEndsType=EndToEnd, outFilterMismatchNmax=10, outFilterMultimapNmax=1, outSAMunmapped=None, outSAMmultNmax=1. The reads of the resulting uniquely aligned pairs were sorted by genomic coordinate using samtools sort (Li H. et al 2009). Duplicated read pairs were marked using samtools markdup and were removed from subsequent analysis. Tallies of total, aligned, duplicated, and remaining reads (not pairs) are reported for each sample in Supplementary Table 3.
Selection of reference sites for quantification of editing yields
The assessment of sites with significant changes in A-to-G editing yields (see below) is sensitive to the number of uniquely aligned reads available for each sample. To minimize potential biases when comparing different samples in terms of significantly edited sites, the uniquely aligned reads for each HEK293T sample were down-sampled using samtools view with option -s and the down-sampling fractions reported in Supplementary Table 3. These fractions were calculated by dividing the smallest number of uniquely aligned reads among all samples by the number of uniquely aligned reads available for the sample being down-sampled. Down-sampling was not performed on the reads of the control sample, the first in Supplementary Table 3. The down-sampled reads where then processed using samtools mpileup. The output of this tool was parsed to extract the counts of each base found in the aligned reads at each A-site and T-site in the GRCh38 reference genome sequence. Insertions and deletions were ignored. Reference sites with read coverage less than 10 were omitted from downstream analysis. The number of remaining reference A- and T-sites with read coverage of at least 10 varied by ~15% across the samples listed in Supplementary Table 3. Without down-sampling, such number was found to vary by ~50%. From the reference A- and T-sites with read coverage of at least 10, a final list of total sites (A-sites and T-sites) was selected by choosing those sites that were common to all samples and for which at least one G or C was observed at a reference A- or T-site, respectively, in the aligned reads of at least one sample. The other sites, those not common to all samples or with zero observed editing events in all samples, were discarded.
Assessment of significant changes in A-to-G editing yields
To uncover significant changes in A-to-G editing yields, several pairs of control and treatment samples were considered. For each pair, the control sample was the first sample listed in in Supplementary Table 3, while the treatment sample was one of the samples shown in Figure 1. For each pair of compared samples, and for each reference A-site selected as described above, a Fisher exact test was carried out using a 2×2 contingency matrix C with entries defined as follows: C1,1 = count of bases other than G observed in the control sample, C2,1 = count of G bases observed in the control sample, C1,2 = count of bases other than G observed in the test sample, C2,2 = count of G bases observed in the test sample. A similar contigency matrix was used for each selected reference T-site, except that G was replaced with C in the above definitions. The p-values calculated for all selected reference sites and for a given comparison of samples were adjusted for multiple testing using the Benjamini-Hochberg method. A-sites and T-sites with adjusted p-values less than a false discovery rate (FDR) of 1% and with a fold change of at least 1.1 in editing yield were deemed to have a significant change in A-to-G editing yield on forward and reverse transcripts, respectively. The counts of these sites for each comparison of samples are shown as Nsig in Supplementary Figures 6–10, and are reported under the column “changed sites” in Supplementary Table 4. The total number of reference sites with a significant change in A-to-G editing yield was computed. The editing yields at these sites were used to construct the distributions shown in Figure 1. The on-target A-to-G editing yields shown as blue circles in Figure 1 and Supplementary Figures 6–10 were estimated for each sample as C2,2 / (C1,2 + C2,2) using counts observed at the intended target A-site in the RAB7A transcript. These values are reported under the column “editing yield” in Supplementary Table 4. The 1-based genomic coordinate of the intended target A-site was found to be chr3:128814202 by submitting the following sequence to BLAT after selecting reference assembly hg38:
AGCGGCAGTATTCTGTACAGTAGACACAAGAATTATGTACGCCTTTTATCA.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Ana Moreno, Lauren Hodge, Dongxin Zhao, Udit Parekh, and other members of the Mali lab for advice and help with experiments, and the Salk GT3 viral core for advice on AAV production. We also thank the UCSD IGM Genomics Center for providing NGS technologies and services. This work was generously supported by the Burroughs Wellcome Fund (1013926), and NIH grants (R01HG009285, RO1CA222826, RO1GM123313, F32DK112682).
Footnotes
COMPETING FINANCIAL INTERESTS
D.K. and P.M. have filed patents based on this work. P.M. is a scientific co-founder of Navega Therapeutics, Pretzel Therapeutics, Engine Biosciences, and Shape Therapeutics. The terms of these arrangements have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
DATA AVAILABILITY AND ACCESSION CODE AVAILABILITY STATEMEMTS
All data and code used in the manuscript, if not already accessible, is fully available upon request.
REFERENCES
- 1.Melcher T et al. A mammalian RNA editing enzyme. Nature 379, 460–464 (1996). [DOI] [PubMed] [Google Scholar]
- 2.Montiel-Gonzalez MF, Vallecillo-Viejo I, Yudowski GA & Rosenthal JJC Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proc. Natl. Acad. Sci. U. S. A 110, 18285–90 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wettengel J, Reautschnig P, Geisler S, Kahle PJ & Stafforst T Harnessing human ADAR2 for RNA repair - Recoding a PINK1 mutation rescues mitophagy. Nucleic Acids Res 45, 2797–2808 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukuda M et al. Construction of a guide-RNA for site-directed RNA mutagenesis utilising intracellular A-to-I RNA editing. Sci. Rep 7, 41478 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vallecillo-Viejo IC, Liscovitch-Brauer N, Montiel-Gonzalez MF, Eisenberg E & Rosenthal JJC Abundant off-target edits from site-directed RNA editing can be reduced by nuclear localization of the editing enzyme. RNA Biol 15, 104–114 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinnamon JR et al. Site-directed RNA repair of endogenous Mecp2 RNA in neurons. Proc. Natl. Acad. Sci. U. S. A 114, E9395–E9402 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox DBT et al. RNA editing with CRISPR-Cas13. Science 358, 1019–1027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Azad MTA, Bhakta S & Tsukahara T Site-directed RNA editing by adenosine deaminase acting on RNA for correction of the genetic code in gene therapy. Gene Ther 24, 779–786 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Vogel P et al. Efficient and precise editing of endogenous transcripts with SNAP-tagged ADARs. Nat. Methods 15, 535–538 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel C, Widmark A, Rigardt D & Öhman M Editing inducer elements increases A-to-I editing efficiency in the mammalian transcriptome. Genome Biol 18, 195 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woolf TM, Chase JM, & Stinchcomn DT Toward the therapeutic editing of mutated RNA sequences. Proc. Natl. Acad. Sci. USA 92, 8298–8302 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung H et al. Human ADAR1 Prevents Endogenous RNA from Triggering Translational Shutdown. Cell 172, 811–824.e14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desterro JMP et al. Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci 116, 1805–1818 (2003). [DOI] [PubMed] [Google Scholar]
- 14.Mort M, Ivanov D, Cooper DN & Chuzhanova NA A meta-analysis of nonsense mutations causing human genetic disease. Hum. Mutat 29, 1037–1047 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Nelson CE et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351, 403–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabebordbar M et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science (80-. ) 351, 407–411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Long C et al. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science (80-. ) 351, 400–403 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hodges PE & Rosenberg LE The spfash mouse: a missense mutation in the ornithine transcarbamylase gene also causes aberrant mRNA splicing. Proc. Natl. Acad. Sci. U. S. A 86, 4142–4146 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang Y et al. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol 34, 334–338 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med 19, 209–216 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.