Abstract
The RUNX1 transcription factor has recently been shown to be obligatory for normal development. RUNX1 controls expression of genes essential for proper development in many cell lineages and tissues including blood, bone, cartilage, hair follicles and mammary glands. Compromised RUNX1 regulation is associated with many cancers. We highlight in this review evidences for RUNX1 control in both invertebrate and mammalian development, and recent novel findings of perturbed RUNX1 control in breast cancer that has implications for other solid tumors. As RUNX1 is essential for definitive hematopoiesis, RUNX1 mutations in hematopoietic lineage cells have been implicated in the etiology of several leukemias. Studies of solid tumors have revealed context-dependent function for RUNX1 as either an oncogene or a tumor suppressor. These RUNX1 functions have been reported for breast, prostate, lung, and skin cancers that are related to cancer subtypes and/or different stages of tumor development. Growing evidence suggests that RUNX1 suppresses aggressiveness in most breast cancer subtypes particularly in the early stage of tumorigenesis. Several studies have identified RUNX1 suppression of the breast cancer epithelial to mesenchymal transition (EMT). Most recently, RUNX1 repression of cancer stem cells and tumorsphere formation was reported for breast cancer. It is anticipated that these new discoveries of the context-dependent diversity of RUNX1 functions will lead to innovative therapeutic strategies for intervention of cancer and other abnormalities of normal tissues.
Keywords: RUNX1, breast cancer, hematopoiesis, mammary gland development, leukemia, cancer
1. Introduction:
The RUNX family of transcription factors have essential regulatory functions in cell differentiation, proliferation and tissue growth (Reviewed in (Coffman, 2003)). RUNX1 was historically designated Acute Myeloid Leukemia 1 protein (AML1), Core-Binding Factor Subunit α2 (CBFA2) or Polyomavirus Enhancer-Binding Protein 2 Alpha B Subunit (PEBP2αB) (Y. Ito, 2004). In mammals, there are three members of the RUNX family (RUNX1, RUNX2 and RUNX3), and each factor displays distinct tissue-restricted expression and lineage-specific roles. RUNX1 is required for hematopoiesis (Yzaguirre, de Bruijn, & Speck, 2017); RUNX2 is essential for osteogenesis (Komori, 2017); and RUNX3 is crucial for the development of dorsal root ganglia neurons and T-lymphocytes, as well as the gut system (Ebihara, Seo, & Taniuchi, 2017; Inoue et al., 2002). Deletion of any Runx gene is lethal in mice, underscoring their essential functions in development. For example, RUNX1 loss causes embryonic lethality in mice by embryonic day 12.5 (E12.5) due to major defects in the formation of the fetal liver and hemorrhaging in the central nervous system (Okuda, van Deursen, Hiebert, Grosveld, & Downing, 1996; Sood, Kamikubo, & Liu, 2017). Mice bearing a homozygous mutation in Runx2 are born without a mineralized skeleton and die just after birth due to an inability to breathe, presumably caused by complete lack of ossification (Otto et al., 1997).
It has been more than two decades since RUNX1 was first implicated in leukemia after it was cloned from Moloney murine leukemia virus and polyomavirus (Miyoshi et al., 1991; E. Ogawa et al., 1993; S. Wang et al., 1993). Since then, more than 50 RUNX1 mutations, including translocations and point mutations, have been identified as causative factors in multiple leukemias that include AML, Acute lymphoblastic leukemia (ALL), Familial platelet disorder with predisposition to myeloid leukemia (FPDMM) and Myelodysplastic syndrome (MDS) (Sood et al., 2017). Later, it was discovered that RUNX1 is required for definitive hematopoiesis and the formation of hematopoietic stem cells (HSCs) during embryogenesis (Okuda et al., 1996; Q. Wang et al., 1996). More recently it has become clear that intact RUNX1 is required for far more than hematopoiesis. Its importance in epithelial tissues such as the skin and mammary gland has been established (Osorio, Lilja, & Tumbar, 2011; Rooney et al., 2017; van Bragt, Hu, Xie, & Li, 2014). Its perturbation results in abnormal activities that are associated with solid tumors including breast, prostate, ovarian and skin (Hong et al., 2018; Hong et al., 2017; Keita et al., 2013; Takayama et al., 2015; van Bragt et al., 2014).
In this review, we discuss the conserved function of RUNX1 homology in development of invertebrates and the involvement of RUNX1 in mammalian tissue development. We comprehensively describe the molecular properties of RUNX1 and its involvement in normal tissue development and cancer, with an emphasis on mammary gland development and breast cancer.
2. Molecular properties of RUNX factors
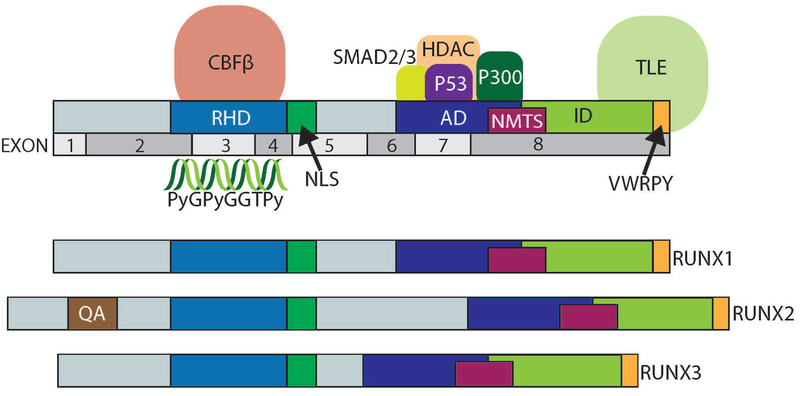
In mammals, each of the Runx genes is transcribed from two promoters, a distal P1 promoter and a proximal P2 promoter (Bangsow et al., 2001; Drissi et al., 2000; Fujiwara et al., 1999; Ghozi, Bernstein, Negreanu, Levanon, & Groner, 1996). RUNX proteins including Runx1 are defined by their shared, highly conserved, N-terminal Runt domain consisting of 128 amino acids (E. Ogawa et al., 1993) (Fig 1). The Runt Homology Domain (RHD) binds to a consensus DNA sequence (PyGPyGGTPy;Py- cytosine or thymine) (Melnikova, Crute, Wang, & Speck, 1993; Eiko Ogawa et al., 1993). Although RUNX binding sites are frequent throughout the genome, the RUNX factors occupy only a fraction of these sites (Pencovich, Jaschek, Tanay, & Groner, 2011; H. Wu et al., 2017). The mechanism defining which RUNX binding sites are targeted for occupancy is not fully elucidated. Despite this, the subnuclear localization and binding of specific cofactors requires several specific domains within the protein. A nuclear localization signal at the C-terminal end of the RHD, is essential for localizing to the nucleus (Fig 1) (Kanno et al., 1998). All RUNX family members have a conserved C-terminal region, containing a nuclear matrix-targeting signal (NMTS) (S. Kaleem Zaidi et al., 2001; Zeng et al., 1998). The NMTS in RUNX proteins is a 30–35 amino acid sequence, responsible for localization to distinct sub-nuclear sites for specific gene regulation (Stein et al., 2007; S. Kaleem Zaidi et al., 2001; Zeng et al., 1998; Zeng et al., 1997). The NMTS organizes multiple complexes of RUNX proteins with different classes of co-regulatory factors, including SMAD2 and SMAD3 (Fig 1) (Sayyed K. Zaidi et al., 2002). RUNX proteins also have a conserved C-terminal domain that contains motifs for protein-protein interactions with numerous classes of factors, such as P300, HDACs and P53 (Aronson, Fisher, Blechman, Caudy, & Gergen, 1997; Chuang, Ito, & Ito, 2013; Javed et al., 2000; Lian et al., 2004; Westendorf, 2006; Sayyed K. Zaidi et al., 2005). The VWRPY motif at the carboxy-terminus of RUNX factors binds to TLE, a critical transcriptional repressor that plays essential roles during development in multiple cellular pathways such as WNT (Fig 1) (G. Chen & Courey, 2000; Chodaparambil et al., 2014). These complex and dynamic interactions between RUNX and cofactors are highly dependent on the cellular-context (Aronson et al., 1997; Chuang et al., 2013; Coffman, 2003; Durst & Hiebert, 2004; Yoshiaki Ito, Bae, & Chuang, 2015; Javed et al., 2000), and support its engagement in the parameters of biological control that include epigenetic modification, chromatin remodeling, and hormone regulation (Coffman, 2003). In addition to the above-mentioned cofactors, RUNX protein function is dependent on binding to Core Binding Factor Beta (CBF-β) to the Runt Homology Domain (RHD), which facilitates the conformational changes of RUNX proteins and increases specificity and affinity of RUNX binding to target genes (Adya, Castilla, & Liu, 2000; Crute, Lewis, Wu, Bushweller, & Speck, 1996; Golling, Li, Pepling, Stebbins, & Gergen, 1996; Huang et al., 1998; Kagoshima et al., 2007; Y.-Y. Tang et al., 2000). Deletion of CBF-β in mice results in defects in hematopoietic and skeletal cells, reflecting the functional properties of RUNX proteins (Miller et al., 2002).
Figure 1. Protein domains within RUNX factors.
The protein domains within RUNX1 are diagrammed including the Runt Homology Domain (RHD, DNA and CBF-β binding), Nuclear Localization Signal (NLS), Nuclear Matrix Targeting Signal (NMTS), Activation Domain (AD), Inhibitory Domain (ID), and VWRPY domains. The exons of RUNX1 that correspond to these domains are also shown. Diagrams of the three RUNX factors are displayed. RUNX2 also contains a polyglutamate-alanine rich QA domain.
Together, the unique and conserved properties of the RUNX transcription factors support gene expression for normal development in a context-dependent manner and their dysregulation results in disease phenotypes which will be discussed in following sections.
3. Conserved Functions of RUNX Factors in Development
Evolutionarily, Runx genes have been identified in all metazoans and unexpectedly in the unicellular amoeboid halozoan Capsaspora owczarzaki, suggesting the RUNX family is involved in fundamental biological processes (Sebé-Pedrós, de Mendoza, Lang, Degnan, & Ruiz-Trillo, 2011). The roles of Runx genes have been intensively studied in invertebrate animal models including Drosophila melanogaster (Dm), Strongylocentrotus purpuratus (Sp) and Caenorhabditis elegans (Ce). Because RUNX function is highly context-dependent and partially redundant in vertebrates, invertebrate animal models with simple genetic backgrounds have facilitated identification of ancestral functions for RUNX proteins. The mechanistic insights obtained from these models have informed RUNX functions that may be applicable in mammals, including humans.
In the fruit fly, Drosophila melanogaster, there are four Runx genes (Bao & Friedrich, 2008; Rennert, Coffman, Mushegian, & Robertson, 2003). The most extensively studied RUNX family member is runt, which was identified for its function in development. DmRunt is one of the five pair-rule genes that regulate the spatial expression of other pair-rule and segment polarity genes (J. Peter Gergen & Wieschaus, 1985; Nusslein-Volhard & Wieschaus, 1980). Deletion of DmRunt results in the loss of larval segments and consequently, smaller than wild-type flies (J. Peter Gergen & Wieschaus, 1985). In addition, DmRunt plays a role in sex determination and neurogenesis (Canon & Banerjee, 2000; Duffy & Gergen, 1991; Duffy, Kania, & Gergen, 1991; J. P. Gergen & Butler, 1988; Kania, Bonner, Duffy, & Gergen, 1990). Another RUNX family member studied in Drosophila is lozenge (lz), which is required for eye development and hematopoiesis (Canon & Banerjee, 2000). The functions of two other Runx genes, CG34145 (RunxA) and CG42267 (RunxB) remain unclear. However, it has been shown that RunxB is involved in the control of cell survival (Boutros et al., 2004).
Caenorhabditis elegans only expresses a single RUNX homolog, termed rnt-1 which regulates the balance between proliferation/self-renewal and differentiation during development of the lateral neuroectodermal seam cells (Hughes & Woollard, 2017; Kagoshima et al., 2005; Nimmo, Antebi, & Woollard, 2005; Xia, Zhang, Huang, Sun, & Zhang, 2007). The seam cells are multi-potent stem-like cells formed during C. elegans embryogenesis (Sulston & Horvitz, 1977). Rnt-1 is expressed in seam cells during embryogenesis and throughout larval development and functions to regulate their division (Braun & Woollard, 2009). Consequently, in rnt-1 mutant worms, the number of seam cells is reduced from 16 to an average of 13 per worm (Kagoshima et al., 2005; Nimmo et al., 2005). Importantly, overexpression of rnt-1 leads to hyper-proliferation and expansion of seam cells (Kagoshima et al., 2007; Kagoshima et al., 2005). As a result, worms with rnt-1 overexpression develop massive hyperplasia leading to a tumor-like appearance of the seam cell tissue, which normally forms a straight line of cells at each side of the worm (Kagoshima et al., 2007).
There are two RUNX genes in sea urchin S. purpuratus, but only one, SpRunt-1, is expressed (Braun & Woollard, 2009). SpRunt-1 is expressed in various tissues during embryogenesis and transiently in adult coelomocytes upon presenting a challenge to their immune system (Coffman, Kirchhamer, Harrington, & Davidson, 1996; Fernandez-Guerra et al., 2006; Pancer, Rast, & Davidson, 1999; Robertson, Dickey, McCarthy, & Coffman, 2002). During embryogenesis, spRunt-1 regulates the expression of transcription factors and other markers of terminal differentiation in all major tissues (Robertson, Coluccio, Knowlton, Dickey-Sims, & Coffman, 2008). SpRunt-1 activates the WNT signaling pathway to positively regulate cell proliferation (Minokawa, Wikramanayake, & Davidson, 2005; Robertson et al., 2008).
The role of RUNX genes as master regulators that specify lineage was further studied in more complex vertebrate animal models. RUNX1 is expressed in hematopoietic progenitors in Zebrafish and Xenopus where it controls stem cell differentiation (Burns, Traver, Mayhall, Shepard, & Zon, 2005; Kalev-Zylinska et al., 2002; Tracey, Pepling, Horb, Thomsen, & Gergen, 1998). In Xenopus, RUNX1 is required for Rohon/Beard neuron development, which mediates touch response in the larval stage (Park, Hong, Weaver, Rosocha, & Saint-Jeannet, 2012). And in Zebrafish, RUNX1 is essential for blood and vessel development (Kalev-Zylinska et al., 2002).
In summary, the key function of RUNX factors is fine-tuning the balance between proliferation and differentiation. This function is retained and conserved across the animal kingdom in species separated by millions of years of evolution, highlighting the significance of RUNX factors during development.
4. Functions of RUNX1 in Mammalian Development
In mammals, all RUNX proteins play essential roles in both normal developmental processes and disease. RUNX1 is essential for hematopoiesis (Okuda et al., 1996), RUNX2 is required for osteoblast maturation (Otto et al., 1997), and RUNX3 is involved in gastrointestinal development, neurogenesis of the dorsal root ganglia and T-cell differentiation (Inoue et al., 2002; Levanon et al., 2002; Q.-L. Li et al., 2002). The concept of fundamental core mechanism(s) for RUNX protein function in development has been posited, however no single common mechanism that governs the development of different tissues has been identified (Yoshiaki Ito et al., 2015). Instead, RUNX proteins utilize multiple spatiotemporal processes to regulate development of different tissues depending on tissue type or stage.
Runx1 and Hematopoiesis
RUNX1 is widely considered the master regulator of developmental hematopoiesis (Okuda et al., 1996; Yzaguirre et al., 2017), which begins with primitive hematopoiesis. At this stage, a limited number of blood lineages (erythrocyte progenitors, erythrocyte/ megakaryocyte progenitors and primitive macrophages) that sustain early embryonic development are produced primarily from the yolk sac (Ferkowicz & Yoder, 2005; Palis, Robertson, Kennedy, Wall, & Keller, 1999; Tober et al., 2007; Tracey et al., 1998; Xu et al., 2001). RUNX1 is expressed in the mesodermal masses in the yolk sac, and in the primitive hematopoietic cells, with the exception of primitive erythrocyte progenitors (Georges Lacaud et al., 2002; North et al., 1999). Although RUNX1 is not required for primitive hematopoiesis, all three primitive hematopoietic lineages are affected by its absence. Without normal RUNX1 function, primitive macrophages are absent (Georges Lacaud et al., 2002; Z. Li, Chen, Stacy, & Speck, 2005), the number of megakaryocytes is reduced (Potts et al., 2014), and primitive erythrocytes are abnormal in function with decreased expression of the erythroid marker Ter118 and the transcription factors Klf1 and Gata1 (Castilla et al., 1996; Yokomizo et al., 2008). Taken together, these findings support a role for RUNX1 in the initial stages of hematopoiesis.
Following primitive hematopoiesis, endothelial cells undergo a process designated definitive hematopoiesis, which constitutes the second and third waves of blood development (Yzaguirre et al., 2017). During this stage, the earliest hematopoietic stem cells (HSCs) are formed at embryonic day E10.5 at the aorta–gonad–mesonephros (AGM) region (B. Chen et al., 2014). HSCs have capacity for long-term repopulation and can produce any of the hematopoietic lineages (Bryder, Rossi, & Weissman, 2006). They are derived from a subset of epithelial cells termed the hemogenic endothelium (HE), a component of the interior lining of blood vessels in embryos (Swiers, Rode, Azzoni, & de Bruijn, 2013). HE cells are a transitional population that undergoes endothelial-to-hematopoietic transition (EHT) to form hematopoietic progenitors and stem cells (Kissa & Herbomel, 2010). RUNX1 is indispensable for definitive hematopoiesis and a critical transcription factor regulating these processes by suppressing the endothelial transcriptional program and initiating the hematopoietic program (M. J. Chen, Yokomizo, Zeigler, Dzierzak, & Speck, 2009; de Bruijn & Dzierzak, 2017; Lancrin et al., 2012; North et al., 1999; Yokomizo et al., 2001). In the absence of RUNX1, no definitive HSCs are formed (Okuda et al., 1996; Q. Wang et al., 1996). In Runx1 heterozygous mutant embryos, definitive hematopoiesis is suppressed and the spatial and temporal development of HSCs is altered (Cai et al., 2000; Mukouyama et al., 2000; Q. Wang et al., 1996). For example, in Runx1+/− embryos, HSCs are abundant in the AGM region and prematurely active in yolk sacs at E10 (Cai et al., 2000). Depletion of RUNX1 in specific tissues or developmental stages in mice demonstrated that RUNX1 expression is required specifically in endothelial cells for de novo generation of HSCs, but is not essential for their renewal or survival thereafter (M. J. Chen et al., 2009; Yzaguirre et al., 2017). Even so, RUNX1 is required for lineage-specific differentiation and homeostasis; for example, RUNX1 is necessary for megakaryocytic maturation and differentiation of B-cells and T-cells in mouse bone marrow (Ichikawa et al., 2004; Niebuhr et al., 2013; Seo, Ikawa, Kawamoto, & Taniuchi, 2012). A C-terminal truncation of the RUNX1 ablating the nuclear matrix targeting signal (NMTS) and activation and inhibitory cofactor binding domains of RUNX1 resulted in a lack of HSC function and deregulation of various hematopoietic markers (Dowdy et al., 2010). Strikingly, a single point mutation in the NMTS of RUNX1 did not appear to affect HSC emergence, but resulted in defects in multiple differentiated hematopoietic lineages.
Runx1 and development of tissues
Runx1 may function in embryogenesis at an even earlier stage than hematopoiesis. In human embryonic stem cells (hESCs), RUNX1 is transiently expressed during early mesendodermal differentiation, which starts at E5.5 (Lu Wang & Chen, 2016). In fact, RUNX1 is the first RUNX member that is expressed upon retinoic acid-induced differentiation of hESCs and promotes an epithelial to mesenchymal transition in a transforming growth factor beta (TGF-β)- dependent manner (J. J. VanOudenhove et al., 2017; Jennifer J. VanOudenhove et al., 2016). In addition to its role in defining hematopoietic lineages, Runx1 is involved in the development of other tissues including hair follicles, bone, cartilage, the nervous system, mammary glands and muscles (Hoi et al., 2010; Kanaykina et al., 2010; Lian et al., 2003; Osorio et al., 2008; Sokol et al., 2015; Umansky et al., 2015; van Bragt et al., 2014; Yamashiro, Åberg, Levanon, Groner, & Thesleff, 2002).
It has been well documented that RUNX1 modulates the developmental activation and proliferation of hair follicle cells (Osorio et al., 2008). The formation of hair follicle stem cells requires constant interaction between epithelial and mesenchymal cells, both of which require RUNX1 expression (Osorio et al., 2008; Raveh et al., 2006; Sennett & Rendl, 2012). In epithelial cells, depletion of RUNX1 delays the formation of hair follicles due to lack of hair follicle stem cell emergence (Osorio et al., 2008; Osorio et al., 2011). However, the function of RUNX1 in this cell type appears dispensable, as the defects are overcome with time (Osorio et al., 2011). In RUNX1-deficient mice, mesenchymal cells still mature into hair follicles, but are not functional and form enormous sebaceous cysts that do not contain the bulb and bulge region at the first hair cycle (Osorio et al., 2011). RUNX1 is also crucial for regulating the hair cycle at the transition into adult skin homeostasis.
Importantly, Runx1 contributes to skeletal development by regulating mesenchymal stem cell (MSC) that differentiate into chondrocytes (Lian et al., 2003) and bone lineage cells. Because a global knockout of Runx1 in mice results in embryonic lethality (Okuda et al., 1996; Sood et al., 2017), other types of mouse models revealed a critical role for RUNX1 in early development of the skeleton. For example, characterizing mice expressing Runx1-lacZ identified that RUNX1 is robustly expressed in periosteum, perichondrium, calvarial sutures, trachea, lung, thyroid and skin, implicating Runx1 in organ development (Lian et al., 2003; Miller et al., 2002). A mesenchymal cell-specific conditional knockout (Dermo1-Cre) mouse deleting CBFβ had underdeveloped larynx, tracheal cartilage, and severe skeletal deformities. These mice exhibited defective intramembranous and endochondral bone with delayed growth plate maturation of chondrocytes and impaired osteoblast differentiation (M. Wu et al., 2014). In a different model, conditional ablation of RUNX1 in MSCs using Prx1-Cre delayed chondrogenesis during fracture healing (Soung et al., 2012).
5. Abnormal RUNX1 Activities and Cancer
As discussed, RUNX1 is involved in the development of multiple tissues, and the precise regulation and integrity of RUNX1 is necessary for normal function. Deregulation of RUNX1 is a major contributing factor in many types of cancers.
RUNX1 (AML1) was first cloned in 1991 based on its location at a breakpoint on chromosome 21 in leukemia (Miyoshi et al., 1991). Later it was discovered that a RUNX1 fusion protein, RUNX1-ETO (AML1-ETO), is encoded by a translocation between chromosomes 8 and 21 (t8:21) (Erickson et al., 1992; Miyoshi et al., 1993; Miyoshi et al., 1991). This aberration is the most common genetic alteration in acute myeloid leukemia (AML) (Lin, Mulloy, & Goyama, 2017; Sood et al., 2017), which has a younger onset and relatively good prognosis (Lin et al., 2017). The RUNX1-ETO fusion protein contains the N-terminal 177 amino acids of RUNX1, including the entire Runt Homology DNA-binding domain, fused in frame with almost the entire ETO protein. ETO contains four evolutionarily conserved domains termed nervy homology regions (NHR) that mediate homodimerization of RUNX1-ETO (Davis, McGhee, & Meyers, 2003; Kwok, Zeisig, Qiu, Dong, & So, 2009; Liu et al., 2006; Yan, Ahn, Hiebert, & Zhang, 2009). Like RUNX1, RUNX1-ETO regulates gene expression by forming complexes with diverse coregulatory proteins but gains the ability to interact with aberrant partners. For example, RUNX1-ETO forms a repressive complex with nuclear receptor co-repressor (NCOR1), histone deacetylase (HDAC1), and SIN3A/HDAC at the ETO NHR domain (Amann et al., 2001; Davis et al., 2003; Gelmetti et al., 1998; Lin et al., 2017; Lutterbach et al., 1998; J. Wang, Hoshino, Redner, Kajigaya, & Liu, 1998). RUNX1-ETO also interacts with E proteins through the NHR domain to inhibit E-protein-induced transcriptional activation (Zhang, Kalkum, Yamamura, Chait, & Roeder, 2004). Oligomerization of RUNX1-ETO is now considered to be the dominant mechanism by which RUNX1-ETO interferes with the RUNX1 transcriptome (Minucci et al., 2000). Dominant-negative inhibition of the native RUNX1 function may therefore be the central mechanism for RUNX1-ETO induced leukemogenesis (Goyama & Mulloy, 2011).
In addition to the 8;21 translocation, more than 50 other chromosomal translocations affect RUNX1. Most of them are related to leukemia, but only about half of the partner genes have been identified among these translocations (Etienne De Braekeleer 2011). Other common translocations include t(12;21) in pediatric acute lymphoblastic leukemia (ALL), known as TEL-RUNX1 (Jamil, Theil, Kahwash, Ruymann, & Klopfenstein, 2000); and t(3:21) in therapy-related AML and myelodysplastic syndrome (MDS), known as RUNX1-MECOM (Yang et al., 2012). RUNX1 somatic point mutations are detected in approximately 3% of pediatric and 15% of adult AML patients (Sood et al., 2017). RUNX1 is also one of the most frequently mutated genes in MDS and ALL, about 10% and 25% respectively (Bejar et al., 2011; Grossmann et al., 2011; Haferlach et al., 2014; Mullighan, 2012; Papaemmanuil et al., 2013; Speck & Gilliland, 2002). In adult AML, RUNX1 mutations are associated with older age and worse prognosis (Gaidzik et al., 2016). These leukemic cells, with defects in differentiation due to mutated RUNX1, generally have a growth advantage over hematopoietic progenitor cells (Greif et al., 2012; Mendler et al., 2012; Schuback, Arceci, & Meshinchi, 2013; Skokowa et al., 2014; J.-L. Tang et al., 2009). Since RUNX1 is a key regulator of hematopoiesis by maintaining a proper balance between proliferation and differentiation, the well-known mutations in RUNX1 in some populations of AML, MDS, and ALL patients are therefore a major factor resulting in leukemogenesis. The high frequency loss-of-function somatic point mutations or translocations in hematologic malignancies has led several companies including Invitae and NEO Genomics to provide screening for RUNX1 mutations in leukemia patients to evaluate prognosis and select therapeutic strategies.
Beyond its impact on leukemia, RUNX1 is either over- or under-expressed in many solid tumors, implying that RUNX1 either promotes or represses epithelial cancers depending on the cellular context (Scheitz & Tumbar, 2013). For example, RUNX1 is identified as a tumor promoter in ovarian and skin cancers and exhibits tumor suppressor activity in lung and prostate cancers (Keita et al., 2013; Ramsey et al., 2017; Scheitz, Lee, McDermitt, & Tumbar, 2012; Takayama et al., 2015). Notably, RUNX1 was reported to promote an epithelial to mesenchymal transition (EMT) in renal carcinoma (Zhou et al., 2018) and is elevated upon EMT in endometrial cancer (Alonso-Alconada et al., 2014). However RUNX1 is clearly a suppressor of breast cancer EMT (see section 7). Together, these examples highlight the context-dependent functions of RUNX1 related to specific tumor tissues. The involvement of RUNX1 in skin cancer was first discovered in a chemically-induced mouse model. Loss of RUNX1 significantly decreases the number of skin tumors formed (Hoi et al., 2010). Using lineage tracing, it has been shown that the RUNX1-expressing hair follicle stem cells are the origin of these chemically induced skin tumors (Scheitz et al., 2012). Mechanistically, RUNX1 maintains an active/phosphorylated form of the oncogene STAT3 by repression of Suppressor of cytokine signaling 3/4 (SOCS3/4) in skin cancer and thus increases cell survival, proliferation and invasion (Scheitz et al., 2012).
6. RUNX1 in Mammary Gland development
The mammalian mammary gland is a highly dynamic organ that undergoes profound physiological changes in structure and function during the reproductive cycle and pregnancy (Hennighausen & Robinson, 2005; Richert, Schwertfeger, Ryder, & Anderson, 2000; Watson & Khaled, 2008). The development of the mouse mammary gland starts at puberty when the embryonic epithelial placode transforms into a branched network of collecting ducts and tubes, which consist of two distinct types of cell lineages: the inner layer of the luminal lineage (including ductal and alveolar luminal cells) and the outer layer of the basal lineage (myoepithelial cells) (Muschler & Streuli, 2010). During pregnancy, increased progesterone and prolactin levels result in greater branching and formation of mature lobuloalveolar units that contain terminally differentiated cells for milk production (Hennighausen, Robinson, Wagner, & Liu, 1997). Post-partum milk is released by contraction of ductal and lobular myoepithelial cells (Haaksma, Schwartz, & Tomasek, 2011). Following lactation, the mammary gland returns to a virgin-like state through involution, with the death of epithelial cells and extensive tissue remodeling (Inman, Robertson, Mott, & Bissell, 2015; Richert et al., 2000; Watson & Khaled, 2008).
RUNX1 is differentially expressed during physiological stages of mammary gland development, and exhibits a spatiotemporal expression pattern. The highest levels are observed in virgin and early-pregnant glands and decrease in late pregnancy and during lactation (McDonald et al., 2014; Rooney et al., 2017; van Bragt et al., 2014). Compared with cells of the luminal lineage, RUNX1 is expressed at higher levels in basal progenitor cells (McDonald et al., 2014; van Bragt et al., 2014). As RUNX1 expression is lost from differentiated alveolar luminal cells, it has been speculated that a reduction in RUNX1 expression is necessary for milk production and secretion (van Bragt et al., 2014). Other than its expression pattern, the role of RUNX1 in the regulation of mammary development and in the normal mammary gland is minimally understood.
The following studies offer insight into functional activities of RUNX1 in specific cell populations. In normal-like basal MCF10A cells, RUNX1 is essential for three-dimensional growth in Matrigel (Lixin Wang, Brugge, & Janes, 2011). Furthermore, without RUNX1, mammary stem cells cannot exit the bipotent state and differentiate into mature lobules and ducts (Sokol et al., 2015). In vivo, deletion of Runx1 specifically in the mouse mammary gland reduces the proportion of luminal cells; in particular, loss of Runx1 results in a deficit in mature estrogen receptor (ER) positive luminal cells (van Bragt et al., 2014). There are relatively few studies devoted to determining the role of RUNX1 in the basal lineage of myoepithelial cells, even though RUNX1 is expressed at a higher level in this subpopulation compared with luminal cells (van Bragt et al., 2014). Interestingly, Runx1 conditional knockout mice have defects in myoepithelial cell contraction and milk ejection, which is released by contraction of ductal and lobular myoepithelial cells (Haaksma et al., 2011). Most of the Runx1 conditional null pups die within 24 hours after birth with no observed milk spots (van Bragt et al., 2014). It is noteworthy that smooth muscle contraction is among the top down-regulated pathways in embryonic stem cells with Runx1 depletion (Jennifer J. VanOudenhove et al., 2016). These data reveal a potential role for RUNX1 in maintaining the normal phenotype of basal myoepithelial cells.
7. RUNX1 and Breast Cancer
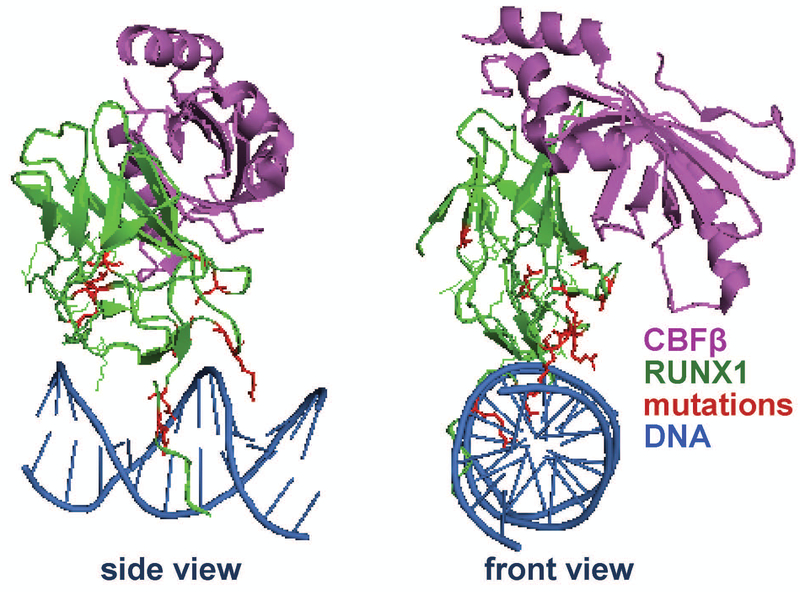
In recent years, growing evidence has indicated that RUNX1 is involved in breast cancer. RUNX1 was initially identified as a potential tumor suppressor, as it was down-regulated among a 17-gene signature associated with metastasis in adenocarcinomas including breast cancer (Ramaswamy, Ross, Lander, & Golub, 2003). The expression of RUNX1 was later shown to decrease when comparing normal mammary tissue to breast cancer, with a greater decrease in higher-grade tumors (Kadota et al., 2010). In a tissue microarray study, RUNX1 intensity was decreased in breast cancer tumors compared with normal mammary tissues (Browne et al., 2015). Moreover, sequencing of breast cancer patient samples then identified that 6% of all invasive breast cancers and 10% of invasive lobular breast cancers have an alteration in the RUNX1 gene (Ciriello et al., 2015; Rooney et al., 2017). Both whole genome and whole exome sequencing have identified point mutations and deletions of RUNX1 in luminal and basal breast cancers (Banerji et al., 2012; Ellis et al., 2012; Network, 2012). In these studies, RUNX1 is a frequently mutated gene along with other well-known tumor suppressors and oncogenes (including PTEN, CDH1, TP53, PIK3CA) which have been extensively investigated in breast cancer (Bertheau et al.; Kechagioglou et al., 2014; Maeirah Afzal & Ezharul Hoque, 2016; Mukohara, 2015). Interestingly, many of RUNX1 mutations, including point mutations, frame-shift mutations, and deletions, are located at the interface between the Runt Homology Domain (DNA-binding domain) and DNA, suggesting that the mutants cannot bind properly to RUNX1 target genes (Fig 2). Mutations were also identified in the RUNX1 binding partner CBF-β (Network, 2012). Thus, it is possible that loss of RUNX1 function(s) by disrupting RUNX1-DNA binding or through loss of RUNX1/CBF-β interaction promotes tumorigenesis in the mammary gland. Recently, two studies independently identified RUNX1 mutations as driver mutations in breast cancer, thus strongly suggesting that RUNX1 loss promotes breast cancer progression (Kas et al., 2017; Pereira et al., 2016).
Figure 2. Mutations in the Runt Homology Domain of RUNX1 in breast cancer.
Ribbon representation showing CBF-β in purple, DNA binding Runt Homology Domain in green, and DNA in blue. RUNX1 mutations identified in breast cancer patients are shown in red. For clarity, the structure is shown in two different orientations (front and side), rotated 90 degrees relative to one another. The image was rendered from the Public Data Base (PDB) code 1H9D (Bravo, Li, Speck, & Warren, 2001). The locations of the mutations are in the DNA binding domain suggesting RUNX1 loses its putative function in breast tumors.
Molecular mechanisms underlying RUNX1 tumor suppressor activity remain unclear and require further investigation in cell lines, mouse models, and human patient samples. Multiple studies using cell lines and mouse models have been carried out to identify RUNX1 function in breast cancer. In normal mammary epithelial cells, loss of RUNX1 in a 3D Matrigel assay resulted in hyper-proliferation and abnormal morphogenesis, which required normal FOXO1 function (Lixin Wang et al., 2011). In another study, conditional knockout of Runx1 in mouse mammary epithelial cells reduced the proportion of ER+ luminal cells, but did not result in mammary tumors (van Bragt et al., 2014). However, loss of TP53 or RB1 rescued this phenotype and resulted in hyper-proliferation of Runx1-deficient ER+ luminal cells. It has been suggested that Mice harboring a double mutation (Runx1/TP53 or Runx1/RB1) could eventually develop breast cancer (van Bragt et al., 2014). Further exploration using double-knockout mice to examine whether these mice develop abnormal mammary hyperplasias or tumors will be necessary to understand the role of RUNX1 in breast cancer initiation and progression. Recent work from the Frenkel lab has demonstrated that loss of RUNX1 in Luminal A breast cancer cells facilitates estrogen-induced WNT signaling by suppressing the scaffold protein AXIN1 (Chimge et al., 2016). Therefore, along with genetic data, growing evidence in cell lines and mouse models establishes the concept that RUNX1 has tumor suppressor activity in breast cancer, especially in the luminal subtype.
Recently, we have reported that loss of RUNX1 in both mammary epithelial and breast cancer cells causes activation of an epithelial-to-mesenchymal transition (EMT) and cancer stem cell (CSC) phenotypes (Hong et al., 2018; Hong et al., 2017). In normal mammary epithelial MCF10A cells, depletion of RUNX1 changes the morphology of cells from epithelial-like to mesenchymal-like, and loss of RUNX1 initiates EMT in both normal epithelial and breast cancer cells (Hong et al., 2017) . RUNX1 expression was lost upon induction of EMT by two different methods, suggesting that reduction of RUNX1 expression is essential for initiation of EMT in these cells. Mechanistically, RUNX1 functions through both exogenous TGF-β-dependent and -independent signaling cascades. As EMT is associated with CSC, which are an important component of tumor growth, cancer stem cell properties were examined upon altering RUNX1 expression (Hong et al., 2018). The results demonstrated that RUNX1 suppresses tumorsphere formation efficiency and the cancer stem cell subpopulation. These RUNX1 activities are likely partially through the negative regulation of ZEB1, an oncogene promoting EMT and CSC. Furthermore, ectopic RUNX1 expression reduces migration and invasion in vitro and tumor growth in vivo. Independently, the Ito laboratory determined that RUNX1 suppresses tumorigenicity, stemness, and migration properties through a different mechanism by inhibiting YAP-mediated signaling. In breast cancer patient samples, high expression of YAP correlated with EMT and stemness gene signatures. In contrast, when RUNX1/RUNX3 were also highly expressed, EMT and CSC gene signatures were less enriched(Kulkarni et al., 2018). Thus studies from the two laboratories are consistent with the concept that RUNX1 is a potent suppressor of EMT and CSC in normal mammary epithelial. These studies also show that RUNX1 has the capability of exerting its inhibitory function through multiple downstream targets. RUNX1 inhibition of the cancer stem cell phenotype highlights the potential of RUNX1 for intervention of aggressive breast cancer.
In contrast, a few studies indicate that RUNX1 functions as an oncogene in breast cancer. In particular, triple negative breast cancer was correlated with high RUNX1 expression and poor prognostic outcome (Ferrari et al., 2014). Although studies of the relationship between RUNX1 and hormones are minimal, it is apparent that RUNX1 is integral for estrogen signaling (Chimge et al., 2016; Stender et al., 2010; van Bragt et al., 2014). RUNX1 inhibition in the triple negative MDA-MB-231 late stage breast cancer cell line, showed a less aggressive phenotype with decreased proliferation, migration and invasion in vitro (Recouvreux et al., 2016). Furthermore, in the MMTV-PyMT mouse model, RUNX1 expression is positively correlated with advanced disease (Browne et al., 2015). The discrepancy between the protective effects of RUNX1 in normal mammary epithelial MCF10A cells compared to the oncogenic activity of RUNX1 in late-stage triple negative breast cancer cells may be due to the lack of the hormone control and/or the cellular heterogeneities in breast cancer, which encompasses a diverse group of subtypes. These subtypes have different cellular origins (luminal versus basal) and molecular alterations (e.g., hormonal status including ER, PR, and HER2) (Eroles, Bosch, Alejandro Pérez-Fidalgo, & Lluch, 2012). In the luminal subtype of breast cancer, it has been well-accepted that RUNX1 has tumor suppressor activities (Chimge et al., 2016; van Bragt et al., 2014). On the other hand, in the basal-like subtype, RUNX1 may have a dual function depending on the stage of breast cancer. In normal mammary myoepithelial cells, loss of RUNX1 disrupts the normal function (i.e., ability to contract and eject milk (van Bragt et al., 2014)). However, in late-stage triple-negative breast cancer, RUNX1 is linked to fast proliferation and a more aggressive phenotype (Recouvreux et al., 2016). The molecular signatures of normal basal cells, early stage basal cancer and late stage basal cancer cells are significantly different (Toft & Cryns, 2011). Due to the distinct cellular context and gene expression patterns, RUNX1 may form complexes with different co-activator or co-repressor proteins. This differential binding of co-regulatory factors may differentially regulate the same subset of genes and convert its activity from being anti-aggressive to tumor promoting. Alternatively, these RUNX1 complexes may be targeted to entirely new subsets of genes.
8. Conclusion:
Although RUNX1 has long been identified for its major impact on hematologic malignancies, advances in NGS approaches have revealed its role in solid tumors. The importance of RUNX1 in breast cancer progression is highlighted by its inclusion among breast cancer driver mutations in several such studies. Despite this progress, knowledge regarding the function of RUNX1 in cancers, especially in breast cancer, is far from complete. The context-dependent role of RUNX1 as either an oncogene or a tumor suppressor indicates the complexity of signaling cascades that intersect with RUNX1 pathway. Furthermore, it remains a challenge to integrate the genomic data obtained from patients with molecular data from cell lines and animal models. Future investigation into complex mechanisms by which RUNX1 functions at different stages of breast cancer onset and progression can potentially translate into targeted therapies with great benefit for prevention and screening.
Acknowledgements:
This work was supported by NIH grants NCI P01 CA082834 (G.S.S., J.L.S.), R01 CA139322 (G.S.S.), R37 DE012528 (J.B.L.), NCI 1F32 CA220935 (A.J.F., G.S.S, J.L.S.), U01 CA196383 (J.L.S) and the Charlotte Perelman Fund for Cancer Research (G.S.S.). The authors would like to thank Caitlin Maloney for editorial assistance.
References
- Adya N, Castilla LH, & Liu PP (2000). Function of CBFβ/Bro proteins. Seminars in Cell & Developmental Biology, 11(5), 361–368. doi: 10.1006/scdb.2000.0189 [DOI] [PubMed] [Google Scholar]
- Alonso-Alconada L, Muinelo-Romay L, Madissoo K, Diaz-Lopez A, Krakstad C, Trovik J, … Consortium E (2014). Molecular profiling of circulating tumor cells links plasticity to the metastatic process in endometrial cancer. Mol Cancer, 13, 223. doi: 10.1186/1476-4598-13-223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann JM, Nip J, Strom DK, Lutterbach B, Harada H, Lenny N, … Hiebert SW (2001). ETO, a Target of t(8;21) in Acute Leukemia, Makes Distinct Contacts with Multiple Histone Deacetylases and Binds mSin3A through Its Oligomerization Domain. Molecular and Cellular Biology, 21(19), 6470–6483. doi: 10.1128/MCB.21.19.6470-6483.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson BD, Fisher AL, Blechman K, Caudy M, & Gergen JP (1997). Groucho-dependent and -independent repression activities of Runt domain proteins. Molecular and Cellular Biology, 17(9), 5581–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, … Meyerson M (2012). Sequence analysis of mutations and translocations across breast cancer subtypes. Nature, 486(7403), 405–409. doi:http://www.nature.com/nature/journal/v486/n7403/abs/nature11154.html#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangsow C, Rubins N, Glusman G, Bernstein Y, Negreanu V, Goldenberg D, … Groner Y (2001). The RUNX3 gene – sequence, structure and regulated expression. Gene, 279(2), 221–232. doi: 10.1016/S0378-1119(01)00760-0 [DOI] [PubMed] [Google Scholar]
- Bao R, & Friedrich M (2008). Conserved cluster organization of insect Runx genes. Development Genes and Evolution, 218(10), 567. doi: 10.1007/s00427-008-0244-x [DOI] [PubMed] [Google Scholar]
- Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, … Ebert BL (2011). Clinical Effect of Point Mutations in Myelodysplastic Syndromes. The New England journal of medicine, 364(26), 2496–2506. doi: 10.1056/NEJMoa1013343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertheau P, Lehmann-Che J, Varna M, Dumay A, Poirot B, Porcher R, … de Thé H (2013). p53 in breast cancer subtypes and new insights into response to chemotherapy. The Breast, 22, S27–S29. doi: 10.1016/j.breast.2013.07.005 [DOI] [PubMed] [Google Scholar]
- Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, … Perrimon N (2004). Genome-Wide RNAi Analysis of Growth and Viability in <em>Drosophila</em> Cells. Science, 303(5659), 832. [DOI] [PubMed] [Google Scholar]
- Braun T, & Woollard A (2009). RUNX factors in development: Lessons from invertebrate model systems. Blood Cells, Molecules, and Diseases, 43(1), 43–48. doi: 10.1016/j.bcmd.2009.05.001 [DOI] [PubMed] [Google Scholar]
- Bravo J, Li Z, Speck NA, & Warren AJ (2001). The leukemia-associated AML1 (Runx1)-CBF[beta] complex functions as a DNA-induced molecular clamp. Nat Struct Mol Biol, 8(4), 371–378. [DOI] [PubMed] [Google Scholar]
- Browne G, Taipaleenmäki H, Bishop NM, Madasu SC, Shaw LM, van Wijnen AJ, … Lian JB (2015). Runx1 is associated with breast cancer progression in MMTV-PyMT transgenic mice and its depletion in vitro inhibits migration and invasion. Journal of Cellular Physiology, 230(10), 2522–2532. doi: 10.1002/jcp.24989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryder D, Rossi DJ, & Weissman IL (2006). Hematopoietic Stem Cells : The Paradigmatic Tissue-Specific Stem Cell. The American Journal of Pathology, 169(2), 338–346. doi: 10.2353/ajpath.2006.060312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CE, Traver D, Mayhall E, Shepard JL, & Zon LI (2005). Hematopoietic stem cell fate is established by the Notch–Runx pathway. Genes & Development, 19(19), 2331–2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, de Bruijn M, Ma X, Dortland B, Luteijn T, Downing JR, & Dzierzak E (2000). Haploinsufficiency of AML1 Affects the Temporal and Spatial Generation of Hematopoietic Stem Cells in the Mouse Embryo. Immunity, 13(4), 423–431. doi: 10.1016/S1074-7613(00)00042-X [DOI] [PubMed] [Google Scholar]
- Canon J, & Banerjee U (2000). Runt and Lozenge function in Drosophila development. Seminars in Cell & Developmental Biology, 11(5), 327–336. doi: 10.1006/scdb.2000.0185 [DOI] [PubMed] [Google Scholar]
- Castilla LH, Wijmenga C, Wang Q, Stacy T, Speck NA, Eckhaus M, … Liu PP (1996). Failure of Embryonic Hematopoiesis andLethal Hemorrhages in Mouse Embryos Heterozygousfor a Knocked-In Leukemia Gene CBFB–MYH11. Cell, 87(4), 687–696. doi: 10.1016/S0092-8674(00)81388-4 [DOI] [PubMed] [Google Scholar]
- Chen B, Mao B, Huang S, Zhou Y, Tsuji K, & Ma F (2014). Human Embryonic Stem Cell-Derived Primitive and Definitive Hematopoiesis In Atwood CS & Meethal SV (Eds.), Pluripotent Stem Cell Biology - Advances in Mechanisms, Methods and Models (pp. Ch. 04). Rijeka: InTech. [Google Scholar]
- Chen G, & Courey AJ (2000). Groucho/TLE family proteins and transcriptional repression. Gene, 249(1), 1–16. doi: 10.1016/S0378-1119(00)00161-X [DOI] [PubMed] [Google Scholar]
- Chen MJ, Yokomizo T, Zeigler BM, Dzierzak E, & Speck NA (2009). Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature, 457(7231), 887–891. doi:http://www.nature.com/nature/journal/v457/n7231/suppinfo/nature07619_S1.html [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimge N-O, Little GH, Baniwal SK, Adisetiyo H, Xie Y, Zhang T, … Frenkel B (2016). RUNX1 prevents oestrogen-mediated AXIN1 suppression and β-catenin activation in ER-positive breast cancer. Nature Communications, 7, 10751. doi:10.1038/ncomms10751 10.1038/ncomms10751http://www.nature.com/articles/ncomms10751#supplementary-informationhttp://www.nature.com/articles/ncomms10751#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodaparambil JV, Pate KT, Hepler MRD, Tsai BP, Muthurajan UM, Luger K, … Weis WI (2014). Molecular functions of the TLE tetramerization domain in Wnt target gene repression. The EMBO Journal, 33(7), 719–731. doi: 10.1002/embj.201387188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang LSH, Ito K, & Ito Y (2013). RUNX family: Regulation and diversification of roles through interacting proteins. International Journal of Cancer, 132(6), 1260–1271. doi: 10.1002/ijc.27964 [DOI] [PubMed] [Google Scholar]
- Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, … Perou CM (2015). Comprehensive molecular portraits of invasive lobular breast cancer. Cell, 163(2), 506–519. doi: 10.1016/j.cell.2015.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman JA (2003). Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biology International, 27(4), 315–324. doi: 10.1016/S1065-6995(03)00018-0 [DOI] [PubMed] [Google Scholar]
- Coffman JA, Kirchhamer CV, Harrington MG, & Davidson EH (1996). SpRunt-1, a New Member of the Runt Domain Family of Transcription Factors, Is a Positive Regulator of the Aboral Ectoderm-SpecificCyIIIAGene in Sea Urchin Embryos. Developmental Biology, 174(1), 43–54. doi: 10.1006/dbio.1996.0050 [DOI] [PubMed] [Google Scholar]
- Crute BE, Lewis AF, Wu Z, Bushweller JH, & Speck NA (1996). Biochemical and Biophysical Properties of the Core-binding Factor α2 (AML1) DNA-binding Domain. Journal of Biological Chemistry, 271(42), 26251–26260. [DOI] [PubMed] [Google Scholar]
- Davis JN, McGhee L, & Meyers S (2003). The ETO (MTG8) gene family. Gene, 303, 1–10. doi: 10.1016/S0378-1119(02)01172-1 [DOI] [PubMed] [Google Scholar]
- de Bruijn M, & Dzierzak E (2017). Runx transcription factors in the development and function of the definitive hematopoietic system. Blood, 129(15), 2061. [DOI] [PubMed] [Google Scholar]
- Dowdy CR, Xie R, Frederick D, Hussain S, Zaidi SK, Vradii D, … Stein GS (2010). Definitive hematopoiesis requires Runx1 C-terminal-mediated subnuclear targeting and transactivation. Human Molecular Genetics, 19(6), 1048–1057. doi: 10.1093/hmg/ddp568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drissi H, Luc Q, Shakoori R, Chuva De Sousa Lopes S, Choi J-Y, Terry A, … Stein GS (2000). Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. Journal of Cellular Physiology, 184(3), 341–350. doi: [DOI] [PubMed] [Google Scholar]
- Duffy JB, & Gergen JP (1991). The Drosophila segmentation gene runt acts as a position-specific numerator element necessary for the uniform expression of the sex-determining gene Sex-lethal. Genes & Development, 5(12a), 2176–2187. [DOI] [PubMed] [Google Scholar]
- Duffy JB, Kania MA, & Gergen JP (1991). Expression and function of the Drosophila gene runt in early stages of neural development. Development, 113(4), 1223. [DOI] [PubMed] [Google Scholar]
- Durst KL, & Hiebert SW (2004). Role of RUNX family members in transcriptional repression and gene silencing. Oncogene, 23(24), 4220–4224. [DOI] [PubMed] [Google Scholar]
- Ebihara T, Seo W, & Taniuchi I (2017). Roles of RUNX Complexes in Immune Cell Development In Groner Y, Ito Y, Liu P, Neil JC, Speck NA, & van Wijnen A (Eds.), RUNX Proteins in Development and Cancer (pp. 395–413). Singapore: Springer Singapore. [DOI] [PubMed] [Google Scholar]
- Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, … Mardis ER (2012). Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature, 486(7403), 353–360. doi:http://www.nature.com/nature/journal/v486/n7403/abs/nature11143.html#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson P, Gao J, Chang KS, Look T, Whisenant E, Raimondi S, … Drabkin H (1992). Identification of breakpoints in t(8;21) acute myelogenous leukemia and isolation of a fusion transcript, AML1/ETO, with similarity to Drosophila segmentation gene, runt. Blood, 80(7), 1825. [PubMed] [Google Scholar]
- Eroles P, Bosch A, Alejandro Pérez-Fidalgo J, & Lluch A (2012). Molecular biology in breast cancer: Intrinsic subtypes and signaling pathways. Cancer Treatment Reviews, 38(6), 698–707. doi: 10.1016/j.ctrv.2011.11.005 [DOI] [PubMed] [Google Scholar]
- De Braekeleer Etienne, D.-G. N, Morel Frédéric, Le Bris Marie-Josée, Férec Claude & De Braekeleer Marc. (2011). RUNX1 translocations and fusion genes in malignant hemopathies. Future Oncol, 7(1), 77–91. doi: 10.2217/fon.10.158 [DOI] [PubMed] [Google Scholar]
- Ferkowicz MJ, & Yoder MC (2005). Blood island formation: longstanding observations and modern interpretations. Experimental Hematology, 33(9), 1041–1047. doi: 10.1016/j.exphem.2005.06.006 [DOI] [PubMed] [Google Scholar]
- Fernandez-Guerra A, Aze A, Morales J, Mulner-Lorillon O, Cosson B, Cormier P, … Genevière A-M (2006). The genomic repertoire for cell cycle control and DNA metabolism in S. purpuratus. Developmental Biology, 300(1), 238–251. doi: 10.1016/j.ydbio.2006.09.012 [DOI] [PubMed] [Google Scholar]
- Ferrari N, Mohammed ZMA, Nixon C, Mason SM, Mallon E, McMillan DC, … Blyth K (2014). Expression of RUNX1 Correlates with Poor Patient Prognosis in Triple Negative Breast Cancer. PLoS One, 9(6), e100759. doi: 10.1371/journal.pone.0100759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Tagashira S, Harada H, Ogawa S, Katsumata T, Nakatsuka M, … Takada H (1999). Isolation and characterization of the distal promoter region of mouse Cbfa11Sequences presented in this article have been submitted to the DDBJ database and appear under the accession number AB013129.1. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression, 1446(3), 265–272. doi: 10.1016/S0167-4781(99)00113-X [DOI] [PubMed] [Google Scholar]
- Gaidzik VI, Teleanu V, Papaemmanuil E, Weber D, Paschka P, Hahn J, … Döhner H (2016). RUNX1 mutations in acute myeloid leukemia are associated with distinct clinico-pathologic and genetic features. Leukemia, 30, 2160. doi:10.1038/leu.2016.126 10.1038/leu.2016.126https://www.nature.com/articles/leu2016126#supplementary-informationhttps://www.nature.com/articles/leu2016126#supplementary-information [DOI] [PubMed] [Google Scholar]
- Gelmetti V, Zhang J, Fanelli M, Minucci S, Pelicci PG, & Lazar MA (1998). Aberrant Recruitment of the Nuclear Receptor Corepressor-Histone Deacetylase Complex by the Acute Myeloid Leukemia Fusion Partner ETO. Molecular and Cellular Biology, 18(12), 7185–7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacaud Georges, Gore Lia, Kennedy Marion, Kouskoff Valerie, Kingsley Paul, Hogan Christopher, … Keller G (2002). Runx1 is essential for hematopoietic commitment at the hemangioblast stage of development in vitro. Blood, 100(2), 458–466. doi: 10.1182/blood-2001-12-0321 [DOI] [PubMed] [Google Scholar]
- Gergen JP, & Butler BA (1988). Isolation of the Drosophila segmentation gene runt and analysis of its expression during embryogenesis. Genes & Development, 2(9), 1179–1193. [DOI] [PubMed] [Google Scholar]
- Gergen JP, & Wieschaus EF (1985). The localized requirements for a gene affecting segmentation in Drosophila: Analysis of larvae mosaic for runt. Developmental Biology, 109(2), 321–335. doi: 10.1016/0012-1606(85)90459-2 [DOI] [PubMed] [Google Scholar]
- Ghozi MC, Bernstein Y, Negreanu V, Levanon D, & Groner Y (1996). Expression of the human acute myeloid leukemia gene AML1 is regulated by two promoter regions. Proceedings of the National Academy of Sciences of the United States of America, 93(5), 1935–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golling G, Li L, Pepling M, Stebbins M, & Gergen JP (1996). Drosophila homologs of the proto-oncogene product PEBP2/CBF beta regulate the DNA-binding properties of Runt. Molecular and Cellular Biology, 16(3), 932–942. doi: 10.1128/mcb.16.3.932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyama S, & Mulloy JC (2011). Molecular pathogenesis of core binding factor leukemia: current knowledge and future prospects. International Journal of Hematology, 94(2), 126–133. doi: 10.1007/s12185-011-0858-z [DOI] [PubMed] [Google Scholar]
- Greif PA, Konstandin NP, Metzeler KH, Herold T, Pasalic Z, Ksienzyk B, … Bohlander SK (2012). RUNX1 mutations in cytogenetically normal acute myeloid leukemia are associated with a poor prognosis and up-regulation of lymphoid genes. Haematologica, 97(12), 1909–1915. doi: 10.3324/haematol.2012.064667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann V, Kern W, Harbich S, Alpermann T, Jeromin S, Schnittger S, … Kohlmann A (2011). Prognostic relevance of RUNX1 mutations in T-cell acute lymphoblastic leukemia. Haematologica, 96(12), 1874–1877. doi: 10.3324/haematol.2011.043919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaksma CJ, Schwartz RJ, & Tomasek JJ (2011). Myoepithelial Cell Contraction and Milk Ejection Are Impaired in Mammary Glands of Mice Lacking Smooth Muscle Alpha-Actin1. Biology of Reproduction, 85(1), 13–21. doi: 10.1095/biolreprod.110.090639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, … Ogawa S (2014). Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia, 28(2), 241–247. doi: 10.1038/leu.2013.336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennighausen L, & Robinson GW (2005). Information networks in the mammary gland. Nat Rev Mol Cell Biol, 6(9), 715–725. [DOI] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW, Wagner K-U, & Liu X (1997). Developing a Mammary Gland is a Stāt Affair. Journal of Mammary Gland Biology and Neoplasia, 2(4), 365–372. doi: 10.1023/A:1026347313096 [DOI] [PubMed] [Google Scholar]
- Hoi CSL, Lee SE, Lu S-Y, McDermitt DJ, Osorio KM, Piskun CM, … Tumbar T (2010). Runx1 Directly Promotes Proliferation of Hair Follicle Stem Cells and Epithelial Tumor Formation in Mouse Skin. Molecular and Cellular Biology, 30(10), 2518–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D, Fritz AJ, Finstad KH, Fitzgerald MP, Weinheimer A, Viens AL, … Stein GS (2018). Suppression of Breast Cancer Stem Cells and Tumor Growth by the RUNX1 Transcription Factor. Molecular Cancer Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong D, Messier TL, Tye CE, Dobson JR, Fritz AJ, Sikora KR, … Stein GS (2017). Runx1 stabilizes the mammary epithelial cell phenotype and prevents epithelial to mesenchymal transition. Oncotarget, 8(11), 17610–17627. doi: 10.18632/oncotarget.15381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Crute BE, Sun C, Tang Y-Y, Kelley JJ, Lewis AF, … Bushweller JH (1998). Overexpression, Purification, and Biophysical Characterization of the Heterodimerization Domain of the Core-binding Factor β Subunit. Journal of Biological Chemistry, 273(4), 2480–2487. [DOI] [PubMed] [Google Scholar]
- Hughes S, & Woollard A (2017). RUNX in Invertebrates In Groner Y, Ito Y, Liu P, Neil JC, Speck NA, & van Wijnen A (Eds.), RUNX Proteins in Development and Cancer (pp. 3–18). Singapore: Springer Singapore. [Google Scholar]
- Ichikawa M, Asai T, Saito T, Yamamoto G, Seo S, Yamazaki I, … Kurokawa M (2004). AML-1 is required for megakaryocytic maturation and lymphocytic differentiation, but not for maintenance of hematopoietic stem cells in adult hematopoiesis. Nat Med, 10(3), 299–304. [DOI] [PubMed] [Google Scholar]
- Inman JL, Robertson C, Mott JD, & Bissell MJ (2015). Mammary gland development: cell fate specification, stem cells and the microenvironment. Development, 142(6), 1028. [DOI] [PubMed] [Google Scholar]
- Inoue K. i., Ozaki S, Shiga T, Ito K, Masuda T, Okado N, … Ito Y (2002). Runx3 controls the axonal projection of proprioceptive dorsal root ganglion neurons. Nat Neurosci, 5(10), 946–954. doi:http://www.nature.com/neuro/journal/v5/n10/suppinfo/nn925_S1.html [DOI] [PubMed] [Google Scholar]
- Ito Y (2004). Oncogenic potential of the RUNX gene family: ‘overview’. . Oncogene, 23(24), 4198–4208. [DOI] [PubMed] [Google Scholar]
- Ito Y, Bae S-C, & Chuang LSH (2015). The RUNX family: developmental regulators in cancer. Nat Rev Cancer, 15(2), 81–95. doi:10.1038/nrc3877 10.1038/nrc3877http://www.nature.com/nrc/journal/v15/n2/abs/nrc3877.html#supplementary-informationhttp://www.nature.com/nrc/journal/v15/n2/abs/nrc3877.html#supplementary-information [DOI] [PubMed] [Google Scholar]
- Jamil A, Theil KS, Kahwash S, Ruymann FB, & Klopfenstein KJ (2000). TEL/AML-1 fusion gene. Cancer Genet Cytogenet, 122(2), 73–78. doi: 10.1016/S0165-4608(00)00272-7 [DOI] [PubMed] [Google Scholar]
- Javed A, Guo B, Hiebert S, Choi JY, Green J, Zhao SC, … Stein GS (2000). Groucho/TLE/R-esp proteins associate with the nuclear matrix and repress RUNX (CBF(alpha)/AML/PEBP2(alpha)) dependent activation of tissue-specific gene transcription. J Cell Sci, 113(12), 2221. [DOI] [PubMed] [Google Scholar]
- Kadota M, Yang HH, Gomez B, Sato M, Clifford RJ, Meerzaman D, … Lee MP (2010). Delineating Genetic Alterations for Tumor Progression in the MCF10A Series of Breast Cancer Cell Lines. PLoS One, 5(2), e9201. doi: 10.1371/journal.pone.0009201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagoshima H, Nimmo R, Saad N, Tanaka J, Miwa Y, Mitani S, … Woollard A (2007). The C. elegans CBFβ homologue BRO-1 interacts with the Runx factor, RNT-1, to promote stem cell proliferation and self-renewal. Development, 134(21), 3905–3915. doi: 10.1242/dev.008276 [DOI] [PubMed] [Google Scholar]
- Kagoshima H, Sawa H, Mitani S, Bürglin TR, Shigesada K, & Kohara Y (2005). The C. elegans RUNX transcription factor RNT-1/MAB-2 is required for asymmetrical cell division of the T blast cell. Developmental Biology, 287(2), 262–273. doi: 10.1016/j.ydbio.2005.08.034 [DOI] [PubMed] [Google Scholar]
- Kalev-Zylinska ML, Horsfield JA, Flores MVC, Postlethwait JH, Vitas MR, Baas AM, … Crosier KE (2002). Runx1 is required for zebrafish blood and vessel development and expression of a human RUNX1-CBF2T1 transgene advances a model for studies of leukemogenesis. Development, 129(8), 2015. [DOI] [PubMed] [Google Scholar]
- Kanaykina N, Abelson K, King D, Liakhovitskaia A, Schreiner S, Wegner M, & Kozlova EN (2010). In vitro and in vivo effects on neural crest stem cell differentiation by conditional activation of Runx1 short isoform and its effect on neuropathic pain behavior. Upsala Journal of Medical Sciences, 115(1), 56–64. doi: 10.3109/03009730903572065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania MA, Bonner AS, Duffy JB, & Gergen JP (1990). The Drosophila segmentation gene runt encodes a novel nuclear regulatory protein that is also expressed in the developing nervous system. Genes & Development, 4(10), 1701–1713. [DOI] [PubMed] [Google Scholar]
- Kanno T, Kanno Y, Chen L-F, Ogawa E, Kim W-Y, & Ito Y (1998). Intrinsic Transcriptional Activation-Inhibition Domains of the Polyomavirus Enhancer Binding Protein 2/Core Binding Factor α Subunit Revealed in the Presence of the β Subunit. Molecular and Cellular Biology, 18(5), 2444–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kas SM, de Ruiter JR, Schipper K, Annunziato S, Schut E, Klarenbeek S, … Jonkers J (2017). Insertional mutagenesis identifies drivers of a novel oncogenic pathway in invasive lobular breast carcinoma. Nat Genet, 49(8), 1219–1230. doi:10.1038/ng.3905 10.1038/ng.3905http://www.nature.com/ng/journal/v49/n8/abs/ng.3905.html#supplementary-informationhttp://www.nature.com/ng/journal/v49/n8/abs/ng.3905.html#supplementary-information [DOI] [PubMed] [Google Scholar]
- Kechagioglou P, Papi RM, Provatopoulou X, Kalogera E, Papadimitriou E, Grigoropoulos P, … Gounaris A (2014). Tumor Suppressor PTEN in Breast Cancer: Heterozygosity, Mutations and Protein Expression. Anticancer Research, 34(3), 1387–1400. [PubMed] [Google Scholar]
- Keita M, Bachvarova M, Morin C, Plante M, Gregoire J, Renaud M-C, … Bachvarov D (2013). The RUNX1 transcription factor is expressed in serous epithelial ovarian carcinoma and contributes to cell proliferation, migration and invasion. Cell Cycle, 12(6), 972–986. doi: 10.4161/cc.23963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kissa K, & Herbomel P (2010). Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature, 464(7285), 112–115. doi:http://www.nature.com/nature/journal/v464/n7285/suppinfo/nature08761_S1.html [DOI] [PubMed] [Google Scholar]
- Komori T (2017). Roles of Runx2 in Skeletal Development In Groner Y, Ito Y, Liu P, Neil JC, Speck NA, & van Wijnen A (Eds.), RUNX Proteins in Development and Cancer (pp. 83–93). Singapore: Springer Singapore. [Google Scholar]
- Kulkarni M, Tan TZ, Syed Sulaiman NB, Lamar JM, Bansal P, Cui J, … Ito Y (2018). RUNX1 and RUNX3 protect against YAP-mediated EMT, stem-ness and shorter survival outcomes in breast cancer. Oncotarget, 9(18), 14175–14192. doi: 10.18632/oncotarget.24419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok C, Zeisig BB, Qiu J, Dong S, & So CWE (2009). Transforming activity of AML1-ETO is independent of CBFβ and ETO interaction but requires formation of homo-oligomeric complexes. Proceedings of the National Academy of Sciences of the United States of America, 106(8), 2853–2858. doi: 10.1073/pnas.0810558106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancrin C, Mazan M, Stefanska M, Patel R, Lichtinger M, Costa G, … Lacaud G (2012). GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood, 120(2), 314. [DOI] [PubMed] [Google Scholar]
- Levanon D, Bettoun D, Harris-Cerruti C, Woolf E, Negreanu V, Eilam R, … Groner Y (2002). The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. The EMBO Journal, 21(13), 3454–3463. doi: 10.1093/emboj/cdf370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q-L, Ito K, Sakakura C, Fukamachi H, Inoue K. i., Chi X-Z, … Ito Y (2002). Causal Relationship between the Loss of RUNX3 Expression and Gastric Cancer. Cell, 109(1), 113–124. doi: 10.1016/S0092-8674(02)00690-6 [DOI] [PubMed] [Google Scholar]
- Li Z, Chen MJ, Stacy T, & Speck NA (2005). Runx1 function in hematopoiesis is required in cells that express Tek. Blood, 107(1), 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian JB, Balint E, Javed A, Drissi H, Vitti R, Quinlan EJ, … Stein GS (2003). Runx1/AML1 hematopoietic transcription factor contributes to skeletal development in vivo. Journal of Cellular Physiology, 196(2), 301–311. doi: 10.1002/jcp.10316 [DOI] [PubMed] [Google Scholar]
- Lian JB, Javed A, Zaidi SK, Lengner C, Montecino M, van Wijnen AJ, … Stein G (2004). Regulatory Controls for Osteoblast Growth and Differentiation: Role of Runx/Cbfa/AML Factors. 14(1&2), 42. doi: 10.1615/CritRevEukaryotGeneExpr.v14.i12.10 [DOI] [PubMed] [Google Scholar]
- Lin S, Mulloy JC, & Goyama S (2017). RUNX1-ETO Leukemia In Groner Y, Ito Y, Liu P, Neil JC, Speck NA, & van Wijnen A (Eds.), RUNX Proteins in Development and Cancer (pp. 151–173). Singapore: Springer Singapore. [Google Scholar]
- Liu Y, Cheney MD, Gaudet JJ, Chruszcz M, Lukasik SM, Sugiyama D, … Bushweller JH (2006). The tetramer structure of the Nervy homology two domain, NHR2, is critical for AML1/ETO’s activity. Cancer Cell, 9(4), 249–260. doi: 10.1016/j.ccr.2006.03.012 [DOI] [PubMed] [Google Scholar]
- Lutterbach B, Westendorf JJ, Linggi B, Patten A, Moniwa M, Davie JR, … Hiebert SW (1998). ETO, a Target of t(8;21) in Acute Leukemia, Interacts with the N-CoR and mSin3 Corepressors. Molecular and Cellular Biology, 18(12), 7176–7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeirah Afzal A, & Ezharul Hoque C (2016). Cadherins: The Superfamily Critically Involved in Breast Cancer. Current Pharmaceutical Design, 22(5), 616–638. doi: 10.2174/138161282205160127095338 [DOI] [PubMed] [Google Scholar]
- McDonald L, Ferrari N, Terry A, Bell M, Mohammed ZM, Orange C, … Blyth K (2014). RUNX2 correlates with subtype-specific breast cancer in a human tissue microarray, and ectopic expression of <em>Runx2</em> perturbs differentiation in the mouse mammary gland. Disease Models & Mechanisms, 7(5), 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikova IN, Crute BE, Wang S, & Speck NA (1993). Sequence specificity of the core-binding factor. Journal of Virology, 67(4), 2408–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendler JH, Maharry K, Radmacher MD, Mrózek K, Becker H, Metzeler KH, … Bloomfield CD (2012). RUNX1 Mutations Are Associated With Poor Outcome in Younger and Older Patients With Cytogenetically Normal Acute Myeloid Leukemia and With Distinct Gene and MicroRNA Expression Signatures. Journal of Clinical Oncology, 30(25), 3109–3118. doi: 10.1200/JCO.2011.40.6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J, Horner A, Stacy T, Lowrey C, Lian JB, Stein G, … Speck NA (2002). The core-binding factor β subunit is required for bone formation and hematopoietic maturation. Nature Genetics, 32, 645. doi: 10.1038/ng1049 [DOI] [PubMed] [Google Scholar]
- Minokawa T, Wikramanayake AH, & Davidson EH (2005). cis-Regulatory inputs of the wnt8 gene in the sea urchin endomesoderm network. Developmental Biology, 288(2), 545–558. doi: 10.1016/j.ydbio.2005.09.047 [DOI] [PubMed] [Google Scholar]
- Minucci S, Maccarana M, Cioce M, De Luca P, Gelmetti V, Segalla S, … Pelicci PG (2000). Oligomerization of RAR and AML1 transcription factors as a novel mechanism of oncogenic activation. Mol Cell, 5(5), 811–820. [DOI] [PubMed] [Google Scholar]
- Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, … Ohki M (1993). The t(8;21) translocation in acute myeloid leukemia results in production of an AML1-MTG8 fusion transcript. The EMBO Journal, 12(7), 2715–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, & Ohki M (1991). t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proceedings of the National Academy of Sciences of the United States of America, 88(23), 10431–10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukohara T (2015). PI3K mutations in breast cancer: prognostic and therapeutic implications. Breast Cancer : Targets and Therapy, 7, 111–123. doi: 10.2147/BCTT.S60696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukouyama Y. s., Chiba N, Hara T, Okada H, Ito Y, Kanamaru R, … Watanabe T (2000). The AML1 Transcription Factor Functions to Develop and Maintain Hematogenic Precursor Cells in the Embryonic Aorta–Gonad–Mesonephros Region. Developmental Biology, 220(1), 27–36. doi: 10.1006/dbio.2000.9617 [DOI] [PubMed] [Google Scholar]
- Mullighan CG (2012). Molecular genetics of B-precursor acute lymphoblastic leukemia. The Journal of Clinical Investigation, 122(10), 3407–3415. doi: 10.1172/JCI61203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschler J, & Streuli CH (2010). Cell–Matrix Interactions in Mammary Gland Development and Breast Cancer. Cold Spring Harbor Perspectives in Biology, 2(10), a003202. doi: 10.1101/cshperspect.a003202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Network TCGA (2012). Comprehensive molecular portraits of human breast tumours. Nature, 490(7418), 61–70. doi:http://www.nature.com/nature/journal/v490/n7418/abs/nature11412.html#supplementary-information [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebuhr B, Kriebitzsch N, Fischer M, Behrens K, Günther T, Alawi M, … Stocking C (2013). Runx1 is essential at two stages of early murine B-cell development. Blood, 122(3), 413. [DOI] [PubMed] [Google Scholar]
- Nimmo R, Antebi A, & Woollard A (2005). mab-2 encodes RNT-1, a C. elegans Runx homologue essential for controlling cell proliferation in a stem cell-like developmental lineage. Development, 132(22), 5043. [DOI] [PubMed] [Google Scholar]
- North T, Gu TL, Stacy T, Wang Q, Howard L, Binder M, … Speck NA (1999). Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development, 126(11), 2563. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, & Wieschaus E (1980). Mutations affecting segment number and polarity in Drosophila. Nature, 287(5785), 795–801. [DOI] [PubMed] [Google Scholar]
- Ogawa E, Inuzuka M, Maruyama M, Satake M, Naito-Fujimoto M, Ito Y, & Shigesada K (1993). Molecular Cloning and Characterization of PEBP2β, the Heterodimeric Partner of a Novel Drosophila runt-Related DNA Binding Protein PEBP2α. Virology, 194(1), 314–331. doi: 10.1006/viro.1993.1262 [DOI] [PubMed] [Google Scholar]
- Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, … Ito Y (1993). PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proceedings of the National Academy of Sciences of the United States of America, 90(14), 6859–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, & Downing JR (1996). AML1, the Target of Multiple Chromosomal Translocations in Human Leukemia, Is Essential for Normal Fetal Liver Hematopoiesis. Cell, 84(2), 321–330. doi: 10.1016/S0092-8674(00)80986-1 [DOI] [PubMed] [Google Scholar]
- Osorio KM, Lee SE, McDermitt DJ, Waghmare SK, Zhang YV, Woo HN, & Tumbar T (2008). Runx1 modulates developmental, but not injury-driven, hair follicle stem cell activation. Development, 135(6), 1059. [DOI] [PubMed] [Google Scholar]
- Osorio KM, Lilja KC, & Tumbar T (2011). Runx1 modulates adult hair follicle stem cell emergence and maintenance from distinct embryonic skin compartments. The Journal of Cell Biology, 193(1), 235–250. doi: 10.1083/jcb.201006068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, … Owen MJ (1997). Cbfa1, a Candidate Gene for Cleidocranial Dysplasia Syndrome, Is Essential for Osteoblast Differentiation and Bone Development. Cell, 89(5), 765–771. doi: 10.1016/S0092-8674(00)80259-7 [DOI] [PubMed] [Google Scholar]
- Palis J, Robertson S, Kennedy M, Wall C, & Keller G (1999). Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development, 126(22), 5073. [DOI] [PubMed] [Google Scholar]
- Pancer Z, Rast JP, & Davidson EH (1999). Origins of immunity: transcription factors and homologues of effector genes of the vertebrate immune system expressed in sea urchin coelomocytes. Immunogenetics, 49(9), 773–786. doi: 10.1007/s002510050551 [DOI] [PubMed] [Google Scholar]
- Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, … Campbell PJ (2013). Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood, 122(22), 3616–3627. doi: 10.1182/blood-2013-08-518886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B-Y, Hong C-S, Weaver JR, Rosocha EM, & Saint-Jeannet J-P (2012). Xaml1/Runx1 is required for the specification of Rohon-Beard sensory neurons in Xenopus. Developmental Biology, 362(1), 65–75. doi: 10.1016/j.ydbio.2011.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pencovich N, Jaschek R, Tanay A, & Groner Y (2011). Dynamic combinatorial interactions of RUNX1 and cooperating partners regulates megakaryocytic differentiation in cell line models. Blood, 117(1), e1. [DOI] [PubMed] [Google Scholar]
- Pereira B, Chin S-F, Rueda OM, Vollan H-KM, Provenzano E, Bardwell HA, … Caldas C (2016). The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nature Communications, 7, 11479. doi: 10.1038/ncomms11479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts KS, Sargeant TJ, Markham JF, Shi W, Biben C, Josefsson EC, … Taoudi S (2014). A lineage of diploid platelet-forming cells precedes polyploid megakaryocyte formation in the mouse embryo. Blood, 124(17), 2725. [DOI] [PubMed] [Google Scholar]
- Ramaswamy S, Ross KN, Lander ES, & Golub TR (2003). A molecular signature of metastasis in primary solid tumors. Nat Genet, 33(1), 49–54. doi:http://www.nature.com/ng/journal/v33/n1/suppinfo/ng1060_S1.html [DOI] [PubMed] [Google Scholar]
- Ramsey J, Butnor K, Peng Z, Leclair T, van der Velden J, Stein G, … Kinsey CM (2017). Loss of RUNX1 is associated with aggressive lung adenocarcinomas. Journal of Cellular Physiology, 233(4), 3487–3497. doi: 10.1002/jcp.26201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveh E, Cohen S, Levanon D, Negreanu V, Groner Y, & Gat U (2006). Dynamic expression of Runx1 in skin affects hair structure. Mechanisms of Development, 123(11), 842–850. doi: 10.1016/j.mod.2006.08.002 [DOI] [PubMed] [Google Scholar]
- Recouvreux MS, Grasso EN, Echeverria PC, Rocha-Viegas L, Castilla LH, Schere-Levy C, … Rubinstein N (2016). RUNX1 and FOXP3 interplay regulates expression of breast cancer related genes. Oncotarget, 7(6), 6552–6565. doi: 10.18632/oncotarget.6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennert J, Coffman JA, Mushegian AR, & Robertson AJ (2003). The evolution of Runx genes I. A comparative study of sequences from phylogenetically diverse model organisms. BMC Evolutionary Biology, 3(1), 4. doi: 10.1186/1471-2148-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richert MM, Schwertfeger KL, Ryder JW, & Anderson SM (2000). An Atlas of Mouse Mammary Gland Development. Journal of Mammary Gland Biology and Neoplasia, 5(2), 227–241. doi: 10.1023/A:1026499523505 [DOI] [PubMed] [Google Scholar]
- Robertson AJ, Coluccio A, Knowlton P, Dickey-Sims C, & Coffman JA (2008). Runx Expression Is Mitogenic and Mutually Linked to Wnt Activity in Blastula-Stage Sea Urchin Embryos. PLoS One, 3(11), e3770. doi: 10.1371/journal.pone.0003770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson AJ, Dickey CE, McCarthy JJ, & Coffman JA (2002). The expression of SpRunt during sea urchin embryogenesis. Mechanisms of Development, 117(1), 327–330. doi: 10.1016/S0925-4773(02)00201-0 [DOI] [PubMed] [Google Scholar]
- Rooney N, Riggio AI, Mendoza-Villanueva D, Shore P, Cameron ER, & Blyth K (2017). Runx Genes in Breast Cancer and the Mammary Lineage In Groner Y, Ito Y, Liu P, Neil JC, Speck NA, & van Wijnen A (Eds.), RUNX Proteins in Development and Cancer (pp. 353–368). Singapore: Springer Singapore. [DOI] [PubMed] [Google Scholar]
- Scheitz CJF, Lee TS, McDermitt DJ, & Tumbar T (2012). Defining a tissue stem cell‐driven Runx1/Stat3 signalling axis in epithelial cancer. The EMBO Journal, 31(21), 4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheitz CJF, & Tumbar T (2013). New insights into the role of Runx1 in epithelial stem cell biology and pathology. Journal of Cellular Biochemistry, 114(5), 985–993. doi: 10.1002/jcb.24453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuback HL, Arceci RJ, & Meshinchi S (2013). Somatic Characterization of Pediatric Acute Myeloid Leukemia Using Next-Generation Sequencing. Seminars in Hematology, 50(4), 325–332. doi: 10.1053/j.seminhematol.2013.09.003 [DOI] [PubMed] [Google Scholar]
- Sebé-Pedrós A, de Mendoza A, Lang BF, Degnan BM, & Ruiz-Trillo I (2011). Unexpected Repertoire of Metazoan Transcription Factors in the Unicellular Holozoan Capsaspora owczarzaki. Molecular Biology and Evolution, 28(3), 1241–1254. doi: 10.1093/molbev/msq309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sennett R, & Rendl M (2012). Mesenchymal-epithelial interactions during hair follicle morphogenesis and cycling. Seminars in Cell & Developmental Biology, 23(8), 917–927. doi: 10.1016/j.semcdb.2012.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo W, Ikawa T, Kawamoto H, & Taniuchi I (2012). Runx1–Cbfβ facilitates early B lymphocyte development by regulating expression of Ebf1. The Journal of Experimental Medicine, 209(7), 1255–1262. doi: 10.1084/jem.20112745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skokowa J, Steinemann D, Katsman-Kuipers JE, Zeidler C, Klimenkova O, Klimiankou M, … Welte K (2014). Cooperativity of RUNX1 and CSF3R mutations in severe congenital neutropenia: a unique pathway in myeloid leukemogenesis. Blood, 123(14), 2229. [DOI] [PubMed] [Google Scholar]
- Sokol ES, Sanduja S, Jin DX, Miller DH, Mathis RA, & Gupta PB (2015). Perturbation-Expression Analysis Identifies RUNX1 as a Regulator of Human Mammary Stem Cell Differentiation. PLoS Comput Biol, 11(4), e1004161. doi: 10.1371/journal.pcbi.1004161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood R, Kamikubo Y, & Liu P (2017). Role of RUNX1 in hematological malignancies. Blood. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung DY, Talebian L, Matheny CJ, Guzzo R, Speck ME, Lieberman JR, … Drissi H (2012). Runx1 dose-dependently regulates endochondral ossification during skeletal development and fracture healing. Journal of Bone and Mineral Research, 27(7), 1585–1597. doi: 10.1002/jbmr.1601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speck NA, & Gilliland DG (2002). Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer, 2(7), 502–513. [DOI] [PubMed] [Google Scholar]
- Stein GS, Lian JB, van Wijnen AJ, Stein JL, Javed A, Montecino M, … Young D (2007). Organization of transcriptional regulatory machinery in nuclear microenvironments: implications for biological control and cancer. Adv Enzyme Regul, 47, 242–250. doi: 10.1016/j.advenzreg.2006.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stender JD, Kim K, Charn TH, Komm B, Chang KCN, Kraus WL, … Katzenellenbogen BS (2010). Genome-Wide Analysis of Estrogen Receptor α DNA Binding and Tethering Mechanisms Identifies Runx1 as a Novel Tethering Factor in Receptor-Mediated Transcriptional Activation. Molecular and Cellular Biology, 30(16), 3943–3955. doi: 10.1128/MCB.00118-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, & Horvitz HR (1977). Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Developmental Biology, 56(1), 110–156. doi: 10.1016/0012-1606(77)90158-0 [DOI] [PubMed] [Google Scholar]
- Swiers G, Rode C, Azzoni E, & de Bruijn MFTR (2013). A short history of hemogenic endothelium. Blood Cells Mol Dis, 51(4), 206–212. doi: 10.1016/j.bcmd.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K. i., Suzuki T, Tsutsumi S, Fujimura T, Urano T, Takahashi S, … Inoue S (2015). RUNX1, an androgen- and EZH2-regulated gene, has differential roles in AR-dependent and -independent prostate cancer. Oncotarget, 6(4), 2263–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J-L, Hou H-A, Chen C-Y, Liu C-Y, Chou W-C, Tseng M-H, … Tien H-F (2009). AML1/RUNX1 mutations in 470 adult patients with de novo acute myeloid leukemia: prognostic implication and interaction with other gene alterations. Blood, 114(26), 5352. [DOI] [PubMed] [Google Scholar]
- Tang Y-Y, Crute BE, Kelley JJ, Huang X, Yan J, Shi J, … Bushweller JH (2000). Biophysical characterization of interactions between the core binding factor α and β subunits and DNA. FEBS Lett, 470(2), 167–172. doi: 10.1016/S0014-5793(00)01312-0 [DOI] [PubMed] [Google Scholar]
- Tober J, Koniski A, McGrath KE, Vemishetti R, Emerson R, de Mesy-Bentley KKL, … Palis J (2007). The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hematopoiesis. Blood, 109(4), 1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toft DJ, & Cryns VL (2011). Minireview: Basal-Like Breast Cancer: From Molecular Profiles to Targeted Therapies. Molecular Endocrinology, 25(2), 199–211. doi: 10.1210/me.2010-0164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tracey WD, Pepling ME, Horb ME, Thomsen GH, & Gergen JP (1998). A Xenopus homologue of aml-1 reveals unexpected patterning mechanisms leading to the formation of embryonic blood. Development, 125(8), 1371. [DOI] [PubMed] [Google Scholar]
- Umansky KB, Gruenbaum-Cohen Y, Tsoory M, Feldmesser E, Goldenberg D, Brenner O, & Groner Y (2015). Runx1 Transcription Factor Is Required for Myoblasts Proliferation during Muscle Regeneration. PLoS Genet, 11(8), e1005457. doi: 10.1371/journal.pgen.1005457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bragt MPA, Hu X, Xie Y, & Li Z (2014). RUNX1, a transcription factor mutated in breast cancer, controls the fate of ER-positive mammary luminal cells. Elife, 3. doi: 10.7554/eLife.03881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanOudenhove JJ, Medina R, Ghule PN, Lian JB, Stein JL, Zaidi SK, & Stein G (2017). Precocious Phenotypic Transcription-Factor Expression during Early Development. Journal of Cellular Biochemistry, 118(5), 953–958. doi: 10.1002/jcb.25723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanOudenhove Jennifer J., Medina R, Ghule Prachi N., Lian Jane B., Stein Janet L., Zaidi Sayyed K., & Stein Gary S. (2016). Transient RUNX1 Expression during Early Mesendodermal Differentiation of hESCs Promotes Epithelial to Mesenchymal Transition through TGFB2 Signaling. Stem Cell Reports, 7(5), 884–896. doi: 10.1016/j.stemcr.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Hoshino T, Redner RL, Kajigaya S, & Liu JM (1998). ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proceedings of the National Academy of Sciences of the United States of America, 95(18), 10860–10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brugge JS, & Janes KA (2011). Intersection of FOXO- and RUNX1-mediated gene expression programs in single breast epithelial cells during morphogenesis and tumor progression. Proceedings of the National Academy of Sciences of the United States of America, 108(40), E803–E812. doi: 10.1073/pnas.1103423108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, & Chen Y-G (2016). Signaling Control of Differentiation of Embryonic Stem Cells toward Mesendoderm. Journal of Molecular Biology, 428(7), 1409–1422. doi: 10.1016/j.jmb.2015.06.013 [DOI] [PubMed] [Google Scholar]
- Wang Q, Stacy T, Binder M, Marin-Padilla M, Sharpe AH, & Speck NA (1996). Disruption of the Cbfa2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proceedings of the National Academy of Sciences of the United States of America, 93(8), 3444–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wang Q, Crute BE, Melnikova IN, Keller SR, & Speck NA (1993). Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Molecular and Cellular Biology, 13(6), 3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CJ, & Khaled WT (2008). Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development, 135(6), 995. [DOI] [PubMed] [Google Scholar]
- Westendorf JJ (2006). Transcriptional co-repressors of Runx2. Journal of Cellular Biochemistry, 98(1), 54–64. doi: 10.1002/jcb.20805 [DOI] [PubMed] [Google Scholar]
- Wu H, Gordon JAR, Whitfield TW, Tai PWL, van Wijnen AJ, Stein JL, … Lian JB (2017). Chromatin dynamics regulate mesenchymal stem cell lineage specification and differentiation to osteogenesis. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms, 1860(4), 438–449. doi: 10.1016/j.bbagrm.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Li C, Zhu G, Wang Y, Jules J, Lu Y, … Chen W (2014). Deletion of Core-binding factor β (Cbfβ) in mesenchymal progenitor cells provides new insights into Cbfβ/Runxs complex function in cartilage and bone development. Bone, 65, 49–59. doi: 10.1016/j.bone.2014.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Zhang Y, Huang X, Sun Y, & Zhang H (2007). The C. elegans CBFβ homolog, BRO-1, regulates the proliferation, differentiation and specification of the stem cell-like seam cell lineages. Developmental Biology, 309(2), 259–272. doi: 10.1016/j.ydbio.2007.07.020 [DOI] [PubMed] [Google Scholar]
- Xu M. j., Matsuoka S, Yang F-C, Ebihara Y, Manabe A, Tanaka R, … Tsuji K (2001). Evidence for the presence of murine primitive megakarycytopoiesis in the early yolk sac. Blood, 97(7), 2016. [DOI] [PubMed] [Google Scholar]
- Yamashiro T, Åberg T, Levanon D, Groner Y, & Thesleff I (2002). Expression of Runx1, −2 and −3 during tooth, palate and craniofacial bone development. Mechanisms of Development, 119, S107–S110. doi: 10.1016/S0925-4773(03)00101-1 [DOI] [PubMed] [Google Scholar]
- Yan M, Ahn E-Y, Hiebert SW, & Zhang D-E (2009). RUNX1/AML1 DNA-binding domain and ETO/MTG8 NHR2-dimerization domain are critical to AML1-ETO9a leukemogenesis. Blood, 113(4), 883–886. doi: 10.1182/blood-2008-04-153742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JJ, Cho SY, Suh J-T, Lee HJ, Lee W-I, Yoon H-J, … Park TS (2012). Detection of RUNX1-MECOM Fusion Gene and t(3;21) in a Very Elderly Patient Having Acute Myeloid Leukemia with Myelodysplasia-Related Changes. Annals of Laboratory Medicine, 32(5), 362–365. doi: 10.3343/alm.2012.32.5.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomizo T, Hasegawa K, Ishitobi H, Osato M, Ema M, Ito Y, … Takahashi S (2008). Runx1 is involved in primitive erythropoiesis in the mouse. Blood, 111(8), 4075. [DOI] [PubMed] [Google Scholar]
- Yokomizo T, Ogawa M, Osato M, Kanno T, Yoshida H, Fujimoto T, … Ito Y (2001). Requirement of Runx1/AML1/PEBP2αB for the generation of haematopoietic cells from endothelial cells. Genes to Cells, 6(1), 13–23. doi: 10.1046/j.1365-2443.2001.00393.x [DOI] [PubMed] [Google Scholar]
- Yzaguirre AD, de Bruijn MFTR, & Speck NA (2017). The Role of Runx1 in Embryonic Blood Cell Formation In Groner Y, Ito Y, Liu P, Neil JC, Speck NA, & van Wijnen A (Eds.), RUNX Proteins in Development and Cancer (pp. 47–64). Singapore: Springer Singapore. [Google Scholar]
- Zaidi SK, Javed A, Choi J-Y, van Wijnen AJ, Stein JL, Lian JB, & Stein GS (2001). A specific targeting signal directs Runx2/Cbfa1 to subnuclear domains and contributes to transactivation of the osteocalcin gene. J Cell Sci, 114(17), 3093. [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Sullivan AJ, van Wijnen AJ, Stein JL, Stein GS, & Lian JB (2002). Integration of Runx and Smad regulatory signals at transcriptionally active subnuclear sites. Proceedings of the National Academy of Sciences of the United States of America, 99(12), 8048–8053. doi: 10.1073/pnas.112664499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Choi J-Y, Pratap J, Javed A, Montecino M, … Stein GS (2005). The dynamic organization of gene-regulatory machinery in nuclear microenvironments. EMBO Reports, 6(2), 128–133. doi: 10.1038/sj.embor.7400337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, McNeil S, Pockwinse S, Nickerson J, Shopland L, Lawrence JB, … Stein GS (1998). Intranuclear targeting of AML/CBFα regulatory factors to nuclear matrix-associated transcriptional domains. Proceedings of the National Academy of Sciences of the United States of America, 95(4), 1585–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, van Wijnen AJ, Stein JL, Meyers S, Sun W, Shopland L, … Hiebert SW (1997). Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBF-α transcription factors. Proceedings of the National Academy of Sciences of the United States of America, 94(13), 6746–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kalkum M, Yamamura S, Chait BT, & Roeder RG (2004). E Protein Silencing by the Leukemogenic AML1-ETO Fusion Protein. Science, 305(5688), 1286. [DOI] [PubMed] [Google Scholar]
- Zhou T, Luo M, Cai W, Zhou S, Feng D, Xu C, & Wang H (2018). Runt-Related Transcription Factor 1 (RUNX1) Promotes TGF-beta-Induced Renal Tubular Epithelial-to-Mesenchymal Transition (EMT) and Renal Fibrosis through the PI3K Subunit p110delta. EBioMedicine, 31, 217–225. doi: 10.1016/j.ebiom.2018.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]