Abstract
Polyomavirus family consists of a highly diverse group of small DNA viruses. The founding family member (MPyV) was first discovered in the newborn mouse in the late 1950s, which induces solid tumors in a wide variety of tissue types that are the epithelial and mesenchymal origin. Later, other family members were also isolated from a number of mammalian, avian and fish species. Some of these viruses significantly contributed to our current understanding of the fundamentals of modern biology such as transcription, replication, splicing, RNA editing, and cell transformation. After the discovery of first two human polyomaviruses (JC virus [JCV] and BK virus [BKV]) in the early 1970s, there has been a rapid expansion in the number of human polyomaviruses in recent years due to the availability of the new technologies and brought the present number to 14. Some of the human polyomaviruses cause considerably serious human diseases, including progressive multifocal leukoencephalopathy, polyomavirus‐associated nephropathy, Merkel cell carcinoma, and trichodysplasia spinulosa. Emerging evidence suggests that the expression of the polyomavirus genome is more complex than previously thought. In addition to encoding universally expressed regulatory and structural proteins (LT‐Ag, Sm t‐Ag, VP1, VP2, and VP3), some polyomaviruses express additional virus‐specific regulatory proteins and microRNAs. This review summarizes the recent advances in polyomavirus genome expression with respect to the new viral proteins and microRNAs other than the universally expressed ones. In addition, a special emphasis is devoted to the recent structural and functional discoveries in the field of polyomavirus agnoprotein which is expressed only by JCV, BKV, and simian virus 40 genomes.
Keywords: Agnoprotein, BKV, dimer and oligomer formation, DNA replication, Merkel cell carcinoma polyomavirus, NMR, polyomavirus JCV, progressive multifocal leukoencephalopathy, SV40, transcription and viroporin
1 |. POLYOMAVIRUSES
The polyomavirus family was first named after the discovery of the founding family member, mouse polyomavirus, (MPyV; polyoma: means many tumors) in mouse during the late 1950s (Stewart, Eddy, & Borgese, 1958). Soon after, a second member of the family, simian virus 40 (SV40) was isolated as a covert contaminant of poliovirus vaccines prepared using macaques monkey renal cell cultures (Sweet & Hilleman, 1960). To date, many diverse family members have been isolated from a variety of mammalian, avian, and fish species including humans, monkey, cattle, rabbit, mouse, hamster, parakeet, athymic rat, and fish (Imperiale & Major, 2007; Van Doorslaer et al., 2018). The polyomaviruses were first classified as a genus of the family Papovaviridae along with papillomaviruses. However, in the year 2000, the International Committee on the Taxonomy of Viruses split the Papovaviridae into two families, Polyomaviridae and Papillomaviridae. Two of the first human polyomaviruses, JC virus (JCV) and BK virus (BKV) were isolated in 1971. JCV was isolated using a tissue culture system that was inoculated with the biopsy brain tissue sample obtained from a patient with a demyelinating disease, progressive multifocal leukoencephalopathy (PML; Padgett, Zu Rhein, Walker, Echroade, & Dessel, 1971). BKV, on the other hand, was cultivated from a kidney transplant recipient with polyomavirus‐associated nephropathy (PVAN; Gardner, Feild, Colleman, & Hulme, 1971). It is now well established that JCV is the etiologic agent of a fatal demyelinating disease of the central nervous system (CNS) known as PML. Seroepidemiological data suggest that the majority of the world’s population (70–80%) is infected with this virus. After the asymptomatic infection, JCV establishes a persistent infection in the kidneys (latent infection) in the archetype form and reactivates itself from latency under immunocompromised conditions (Berger, 2000; Berger, Kaszovitz, Post, & Dickinson, 1987; Ferenczy et al., 2012). During the reactivation period, the regulatory region of the archetype strain undergoes deletions and duplications rendering a variety of virulent strains or forms including Mad‐4 and Mad‐1 (Frisque & White, 1992). These virulent forms can travel to the CNS by an unknown mechanism; and lytically and abortively infect oligodendrocytes and astrocytes, respectively, and lead to the onset of the PML (Ferenczy et al., 2012; Frisque & White, 1992). In addition to the kidneys, the hematopoietic progenitor cells, peripheral blood B lymphocytes, and tonsillar stromal cells were also found to harbor JCV, suggesting that these sites could serve as additional sites for JCV latent infection (Monaco, Jensen, Hou, Durham, & Major, 1998). PML, a rare disease in the normal healthy population, is mostly associated with immunosuppressive conditions of acquired immunodeficiency syndrome (AIDS; Berger, 1992; Berger et al., 1992) but the treatment of AIDS patients with highly active antiretroviral therapy significantly reduced the PML cases. Recent studies suggest that JCV is also associated with another brain pathologies called “granule cell neuronopathy” (Dang, Vidal, et al., 2012; Du Pasquier et al., 2003; Soleimani‐Meigooni et al., 2017; Wuthrich, Batson, Anderson, White, & Koralnik, 2016; Wuthrich et al., 2009). More than a decade ago, PML noticeably resurfaced again in some multiple sclerosis (MS) and Crohn’s disease patients, who were treated with immunomodulatory antibodies such as natalizumab and efalizumab. These antibodies target specific cell surface molecules on T and B cells (Kleinschmidt-DeMasters & Tyler, 2005; Langer‐Gould, Atlas, Green, Bollen, & Pelletier, 2005; Van Assche et al., 2005). The rationale behind using these immunomodulatory antibodies in MS and Crohn’s disease treatment is to prevent extravasation of T cells into brain and infiltration of B cells into the layers of the skin, respectively, thereby aiming to downregulate the harmful effects of these immune cells in target organs. Efalizumab targets an integrin molecule, CD11‐ɑ, on both B and T cells and blocks their interaction with the intercellular adhesion molecules on endothelial cells and prevents their infiltration into target organs (Gottlieb et al., 2002) whereas natalizumab interacts with the α4 integrin on either B or T cells (Kleinschmidt-DeMasters & Tyler, 2005). α4 integrin is known to heterodimerize either with integrin β1 (α4β1) or with integrin β7 (α4β7) on both B and T cells. Both complexes (α4β1 and α4β7) then serve as attachment ligands for vascular cell adhesion molecules on endothelial cells, and thus prevents T‐cell extravasation into the brain. Another immunomodulatory antibody, rituximab, targets the CD20 receptor on B cells and causes their death through a complement-dependent lysis (Tobinai et al., 1998). Every one of these drug treatment cases creates a severe immunosuppression in treated people, which consequently provides an opportunity for JCV to reactivate itself from the latency and induce PML. One might think that the loss of immune surveillance inherit to these therapies would be sufficient to induce PML in every MS and Crohn’s disease. In fact, there seem to be additional unknown factors contributing to the reactivation of JCV in immunocompromised individuals. The genetic makeup of those individuals, for example, may significantly contribute to the reactivation of JCV and, therefore, to the development of PML. There are also reports indicating that the specific mutations in the JCV VP1 gene can significantly affect the outcome of the JCV infections with respect to the progression of the PML (Gorelik, Goelz, & Sandrock, 2009; Gorelik et al., 2011). Similarly, BKV which is also highly prevalent in human population latently infects kidney epithelial cells and induces a disease known as polyomavirus associated nephropathy (PVAN) most frequently seen in patients receiving highly immunosuppressive drugs. The end result would be an allograft failure when BKV is reactivated.
In addition to these two important human polyomavirus (JCV and BKV), there has been a rapid increase in the discovery of the additional human polyomaviruses in recent years, due to availability of the new technologies, such as digital transcriptome subtraction and rolling circle amplification, bringing the total number to 14 (Table 1). Note that, not every human polyomavirus is associated with a human disease. Besides JCV and BKV, two other recently discovered polyomaviruses have been known to cause diseases in humans. For example, trichodysplasia spinulosa‐associated polyomavirus (TSPyV) induces trichodysplasia spinulosa (TS; van der Meijden et al., 2015) and Merkel cell carcinoma-associated polyomavirus (MCPyV) is associated with a Merkel cell carcinoma (MCC; Feng, Shuda, Chang, & Moore, 2008). The remaining human polyomaviruses, including Karolinska Institute polyomavirus (KIPyV; Allander et al., 2007), Washington University polyomavirus (WUPyV; Gaynor et al., 2007), human polyomavirus 6 (HPyV6; Schowalter, Pastrana, Pumphrey, Moyer, & Buck, 2010), human polyomavirus 7 (HPyV7; Schowalter et al., 2010), human polyomavirus 9 (HPyV9; Scuda et al., 2011), human polyomavirus 10 (HPyV10; Buck et al., 2012), Malawi human polyomavirus (MWPyV; Siebrasse et al., 2012), MX human polyomavirus (MXPyV; Yu et al., 2012), St. Louis polyomavirus (SLPyV‐11; Lim et al., 2013; Pastrana et al., 2013), human polyomavirus 12 (HPyV12; Korup et al., 2013), and New Jersey polyomavirus (NJPyV; Mishra et al., 2014) have yet to be linked to a particular human disease. The involvement of the human polyomaviruses in the pathology of various human diseases is illustrated in Table 1.
TABLE 1.
Currently known human polyomaviruses
| Full names | Abbreviations | Source of isolation | Time of discovery | Associated disease |
|---|---|---|---|---|
| JC virus | JCPyV | Brain, PML patient | Padgett et al. (1971) | Progressive multifocal leukoencephalopathy |
| BK virus | BKPyV | Urine, kidney transplant recipient | Gardner et al. (1971) | BK virus-associated nephropathy, Hemorrhagic cystitis |
| Karolinska Institute polyomavirus | KIPyV | Nasopharyngeal tissue | Allander et al. (2007) | Respiratory disease |
| Washington University polyomavirus | WUPyV | Nasopharyngeal tissue | Gaynor et al. (2007) | Respiratory disease |
| Merkel cell polyomavirus | MCPyV | Skin lesions, Merkel cell carcinoma patient | Feng et al. (2008) | Merkel cell carcinoma |
| Human polyomavirus 6 | HPyV6 | Normal skin | Schowalter et al. (2010) | Keratoacanthoma Kimura disease |
| Human polyomavirus 7 | HPyV7 | Normal skin | Schowalter et al. (2010) | Unknown |
| Trichodysplasia spinulosa-associated polyomavirus | TSPyV | Trichodysplasia spinulosa, skin lesions | van der Meijden et al. (2015) | Trichodysplasia spinulosa |
| Human polyomavirus 9 | HPyV9 | Blood, skin, urine, kidney transplant recipient | Scuda et al. (2011) | Unknown |
| Malawi polyomavirus | MWPyV | Stool, a patient with warts, hypogammaglobulinemia, infections, and myelokathexis (WHIM) syndrome | Buck et al. (2012) | Unknown |
| Human polyomavirus 12 | HPyV12 | Resected human liver tissue | Korup et al. (2013) | Unknown |
| St. Louis polyomavirus | STPyV | Stool samples, healthy child | Lim et al. (2013), Pastrana et al. (2013) | Unknown |
| MX polyomavirus | MXPyV | Stool samples from children | Yu et al. (2012) | Unknown |
| New Jersey polyomavirus | NJPyV | Biopsy from the swollen endothelial cells, pancreatic transplant recipient | Mishra et al. (2014) | Unknown |
2 |. POLYOMAVIRUS GENOME AND ITS EXPRESSION
2.1 |. Genomic structure of polyomaviruses
All polyomaviruses have a small, circular, double‐stranded DNA genome encapsidated in icosahedral virions (Imperiale & Major, 2007; Saribas, Ozdemir, Lam, & Safak, 2010). The viral DNA is primarily made up of two major elements: The noncoding regulatory region and two coding regions (early and late; Figure 1). The regulatory region contains (a) promoter–enhancer elements and (b) the origin of DNA replication. This region regulates bidirectional expression of the coding regions and the viral DNA replication. In some of the family members, the noncoding regulatory elements of the virus display a hypervariability with respect to the recombination of the promoter–enhancer elements. For example, JCV undergoes a complicated recombination process within its regulatory region during the evolution from an archetype strain to a more virulent strain, including Mad‐1 and Mad‐4. The archetype strain, for example, contains only one 98 bp repeat in its regulatory region, but this 98 bp element is split by two unique insertional blocks, which are 23 and 66 bp in length (Safak, Gallia, & Khalili, 1999). A prototype strain of JCV (Mad‐1), in contrast, is characterized by the presence of two 98 bp tandem repeats within its regulatory region but it lacks the 23‐ and 66‐bp insertional sequences which are completely eliminated from the viral genome during the transition from the archetype to the more virulent strains. Similar to Mad‐1 strain, Mad‐4 strain of JCV contains only two 84 bp tandem repeats. Again, Mad‐4 does not have any of those 23 and 66 bp insertional elements (Safak, Gallia, & Khalili, 1999). The occurrence of the various rearrangements within the regulatory region has also been reported for the different isolates of JCV (Ferrante et al., 2003; Reid et al., 2011). The rearranged forms in the noncoding regulatory region have also been reported for BKV (Barcena‐Panero et al., 2012; Randhawa et al., 2003).
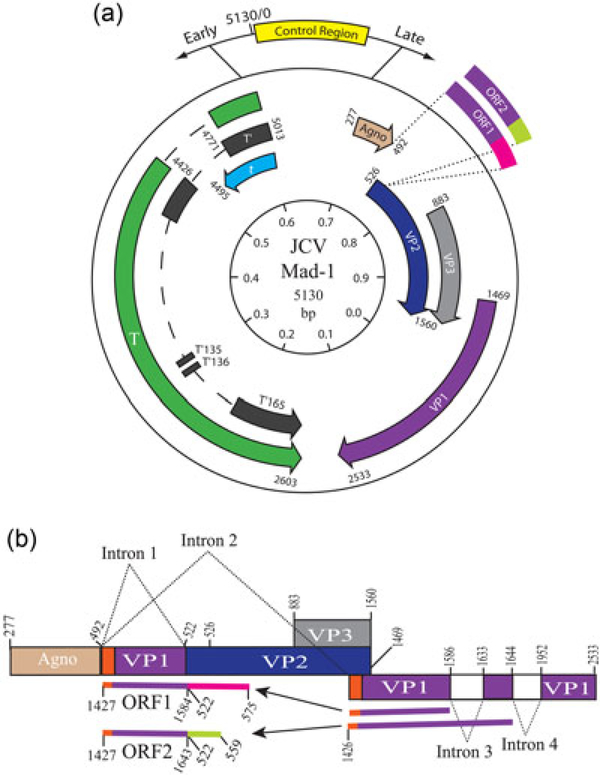
FIGURE 1.
Circular map of JC virus. (a) The genome of JC virus contains three functional regions. 1, Control region, which contains the origin of DNA replication and promoter–enhancer elements for early and late genes. 2, Early coding region, which encodes LT‐Ag, Sm t‐Ag, T’proteins (T’165, T’135, and T’136) due to cis‐alternative splicing of the early pre‐mRNA. 3, Late coding region which encodes capsid proteins (VP1, VP2, and VP3) and a regulatory protein, agnoprotein. In addition, late region also encodes two additional proteins ORF1 and ORF2 due to trans‐splicing of the late pre‐mRNA. (b) The coding regions of ORF1 and ORF2 are created by insertion of a partial coding region of VP1 in between agnoprotein and VP2 coding regions. A schematic demonstration of the mechanism of the creation of ORF1 and ORF2, open reading frames (see text for details; Saribas, DeVoto, et al., 2018). LT‐Ag: large tumor antigen; mRNA: messenger RNA; Sm t‐Ag: small tumor antigen [Color figure can be viewed at wileyonlinelibrary.com]
2.2 |. The expression of the early coding region of polyomaviruses
The coding regions of the polyomaviruses encode (a) regulatory and (b) structural proteins, and (c) regulatory microRNA (miRNA) species. The coding regions have two directions, early and late, for transcription. Transcription extends directionally from initiation sites near the origin to produce the early and late messenger RNAs (mRNAs). They are transcribed from the opposite strands of the viral genome. For the sake of simplicity, we will mainly focus on the description of the expression patterns of the human (JCV, BKV, MCPyV, and TSPyV), monkey (SV40), and mouse (MPyV) polyomavirus coding regions rather than describing it for all polyomaviruses. However, JCV coding regions will be described in more details and deviations from it for the above‐mentioned polyomaviruses will be highlighted. The JCV early coding region encodes primarily two major proteins: Large tumor antigen (LT‐Ag) and small tumor antigen (Sm t‐Ag; Figure 1). Note that LT‐Ag and Sm t‐Ag are universally expressed by all polyomaviruses from their early coding regions. In 1995, however, the discovery of the several new alternatively spliced variants of these two major proteins were also reported by Trowbridge and Frisque (1995), and they were named T’ proteins (T’165, T’136, T’135, T’147, and T’152). Significantly high number of amino acid sequences of all T’ proteins are shared with those of LT‐Ag (Prins & Frisque, 2001).
The LT‐Ag is the major regulatory protein of JCV and is required for the initiation of the replication of the viral genome and transactivation of the viral late promoter. In addition, it suppresses its own promoter by an autoregulatory loop to regulate the level of early gene expression (Safak, Gallia, Ansari, & Khalili, 1999). The precise function of Sm t‐Ag is unknown but plays regulatory roles in the viral replication cycle, where, along with the LT‐Ag, it pushes the cells into the S‐phase of the cell cycle, during which all DNA viruses including polyomaviruses replicate their DNA (Khalili, Sariyer, & Safak, 2008). The function of the T’ proteins (T’135, T’136, and T’165) in the viral life cycle was also not studied in great detail. However, recent evidence suggests that they may have roles in cell transformation through interaction with tumor suppressor proteins, p107 and p130 (Bollag, Kilpatrick, Tyagarajan, Tevethia, & Frisque, 2006).
Several other polyomaviruses also express alternatively spliced forms of proteins from their early coding regions. In addition to the expression of its Sm t‐Ag and LT‐Ag, mouse polyomavirus (MPyV) encodes two additional proteins from its early coding region, one of which is a membrane‐anchored protein termed middle T antigen, 420 aa long protein and has well‐known oncogenic properties. It binds and activates Src‐family protein tyrosine kinases in a PP2A‐dependent manner and thereby stimulates the Scr signaling pathway during cell transformation (Fluck & Schaffhausen, 2009; Ichaso & Dilworth, 2001; Zhou et al., 2011). Another one is tiny T antigen, which localizes to both cytoplasm and nucleus; and stimulates the ATPase activity of Hsp70 through its J domain (Riley, Yoo, Mda, & Folk, 1997). The monkey polyomavirus, SV40, also expresses a 17‐kDa protein from its early coding region called 17K protein. Although the function of it is not studied in detail (Rundell, Hearing, & Yang, 1980; Y. C. Yang, Hearing, & Rundell, 1979), it was reported to have oncogenic activity in both human and rodent cells (Boyapati, Wilson, Yu, & Rundell, 2003; Zerrahn, Knippschild, Winkler, & Deppert, 1993). Long after the discovery of JCV T’ proteins, Abend, Joseph, Das, Campbell‐Cecen, and Imperiale (2009) reported the expression of a 17–20‐kDa protein from the early region of BKV in 2009, which is a truncated form of BKV LT‐Ag (truncTAg). TruncT‐Ag is expressed in both BKV‐transformed and lytically infected cells; and primarily localizes to the nucleus (Abend et al., 2009) and it is believed to be involved in cell transformation (Abend et al., 2009). Another human polyomavirus, MCPyV, associated with human MCC was also reported to express two additional spliced products from its early coding region, one of which is a truncated form of LT‐Ag (a 27‐kDa protein) called the large T open reading frame (ALTO; Carter et al., 2013) and other one is called 57 kT protein (DeCaprio, 2017). ALTO appears not to be required for the replication of MCPyV in tissue culture but it was suggested to play an auxiliary role for the virus life cycle. It is likely that the early coding region of these polyomaviruses and other ones may generate additional splice variants of the early mRNA, the identity of those has yet to be discovered.
All of the above‐mentioned alternative splicing events for JCV, BKV, MCPyV, MPyV, and SV40 occur by a conventional splicing process known as cis‐splicing, a process that occurs within a single transcript between a splice donor and acceptor site (Black, 2003). Alternatively, the joining of sequences from the distinct transcripts (two different transcripts) can also take place during the splicing to diversify viral proteins. This type of splicing event is called as trans‐splicing, which was first detected in 1986 in trypanosomes and soon after in the nematode Caenorhabditis elegans (Krause & Hirsh, 1987; Sutton & Boothroyd, 1986). Such a trans‐splicing process was also reported for SV40 early large T antigen pre‐mRNA transcript (Eul, Graessmann, & Graessmann, 1996a, 1996b; Eul & Patzel, 2013). This trans‐splicing event was determined to generate a 147‐nt duplication of the SV40 LT‐Ag exon 2 sequence between nucleotides 4571‐4425, which corresponds to a duplication of amino acids 83‐131 followed by the beginning of the exon 2 again, which is a tandem repeat of this region. Duplicated region contains the binding region for pRb which is essential for cell transformation by LT‐Ag. This event generates an open reading frame (ORF) for a 100‐kDa LT‐Ag protein which is called “super LT‐Ag” due to its very high transforming activity in rodent cells (Eul & Patzel, 2013).
2.3 |. Expression of the late coding region of polyomaviruses
Contrary to the early region, the late coding regions of the polyomaviruses are known to primarily encode structural capsid proteins including VP1, VP2, and VP3. There is an exception to this trend. That is, two of the human polyomaviruses, JCV and BKV and monkey polyomavirus virus, SV40, encode a small regulatory protein from their late genome called Agnoprotein (Frisque, Bream, & Cannella, 1984; Saribas et al., 2016). However, recent discoveries show that the late coding region of one of these three viruses, JCV, also generate various alternatively spliced variants of the late premRNA (Saribas, DeVoto, et al., 2018; Shishido‐Hara et al., 2000). Some of these splice variants, in fact, encode regulatory proteins for JCV.
Among the viral structural proteins, capsid protein, VP1, forms the outer shell of the virions and plays an essential role in the attachment of the virus to the cell surface receptors and thereby facilitates the virus spread between the cells. The other two capsid proteins, VP2 and VP3, which have overlapping sequences, are located at the inner parts of the capsid and are critical for the nucleation of the viral DNA into the capsids during the encapsidation process. In addition to VP1, VP2, and VP3 capsid proteins, SV40 late mRNA encodes a novel “late protein” named “VP4” (Daniels, Sadowicz, & Daniel, 2007). Similar to VP3, the expression of VP4 is generated from a downstream AUG start codon within the VP2 transcript at aa Met228, which is approximately a 13.9‐kDa protein. Its expression was detected not only in the translation of VP2 coding sequences in reticulocyte lysates but also in SV40‐infected cells (Daniels et al., 2007). It is expressed late during the viral life cycle and is suggested to contribute to the lytic release of the SV40 virions from the infected cells (Giorda, Raghava, & Hebert, 2012; Raghava, Giorda, Romano, Heuck, & Hebert, 2011, 2013; Tange, Imai, & Nakanishi, 2011). In contrast to these findings regarding the expression of SV40 VP4, a study by Henriksen et al., failed to detect the expression of VP4 in SV40‐infected cells, perhaps technical difficulties associated with in their detection system. This group also suggests that VP4 is not required for the release of BKV from the infected cells (Henriksen, Hansen, Bruun, & Rinaldo, 2016).
As stated earlier, the genome of the polyomaviruses, particularly JCV can generate more than predicted splice variants from its late transcripts (Shishido‐Hara et al., 2000). However, their coding capacity was unknown. Recently, two novel splice variants of JCV late transcripts were discovered by our group (Saribas, DeVoto, et al., 2018), These variants are generated by an unusual splicing process called trans‐splicing rather than cis‐splicing, and contain novel ORFs, called ORF1 and ORF2. They result from (a) the trans‐splicing of two different lengths of the 5′‐short coding regions of VP1 into between the coding regions of agnoprotein and VP2 after replacing the intron which is located between Agnoprotein and VP2 coding sequences; and (b) frame‐shifts occurring within the VP2 coding sequences terminated by a stop codon (Figure 1a,b). ORF1 and ORF2 encode 58 and 72 amino acid long proteins, respectively. The expression of ORF1 is found not only in infected cells in vitro but also in PML patient samples in vivo (Saribas, DeVoto, et al., 2018). Each ORF protein shares a common coding region with VP1 but also has their own unique coding sequences at their C‐terminus. When blocked the expression of these ORFs by a mutation in the viral background, it was found that the mutant viruses replicate less efficiently than the wild‐type, suggesting that these ORFs may play important regulatory roles in the viral life cycle (Saribas, DeVoto, et al., 2018). Further analysis of the JCV late transcripts for additional splicing products revealed not only novel splice recombinants but also new ORFs for the JCV late transcripts (unpublished data). It is more likely that other human polyomaviruses also generate various recombinant splice products from the late transcripts and perhaps generate novel ORFs. A further investigation is required to identify, if any, those transcripts and novel ORFs. The generation of the novel viral proteins by polyomaviruses due to alternative splicing is summarized in Table 2.
TABLE 2.
Polyomavirus genomes generate the novel viral proteins due to alternative splicing
| Virus names | Early/late gene expression | Putative protein | No. of amino acids | Expected size (kDa) | References |
|---|---|---|---|---|---|
| TSPyV | Early | Top T | 332 | 38 | van der Meijden et al. (2015) |
| TSPyV | Early | Tiny T | 85 | 10 | van der Meijden et al. (2015) |
| TSPyV | Early | Small T* | 197 | 24 | van der Meijden et al. (2015) |
| TSPyV | Early | 21 kT | 184 | 21 | van der Meijden et al. (2015) |
| MCPyV | Early | 57 kT | 432 | 48 | Carter et al. (2013) |
| MCPyV | Early | ALTO | 248 | 27 | Carter et al. (2013) |
| MPyV | Early | Top T | 421 | 46 | Soeda et al. (1979) |
| MPyV | Early | Tiny T | 75 | 8 | Riley et al. (1997) |
| JCPyV | Early | T'165 | 165 | 18 | Prins and Frisque (2001) |
| JCPyV | Early | T'136 | 136 | 15 | Prins and Frisque (2001) |
| JCPyV | Early | T'135 | 135 | 15 | Prins and Frisque (2001) |
| JCPyV | Early | T'152 | 152 | 17 | Prins and Frisque (2001) |
| JCPyV | Early | T'147 | 147 | 16 | Prins and Frisque (2001) |
| JCPyV | Late | ORF1 | 58 | 6.4 | Saribas, Devoto et al. 2018 |
| JCPyV | Late | ORF2 | 72 | 7.9 | Saribas, Devoto et al. 2018 |
| JCPyV | Late | Agnoprotein | 71 | 7.8 | Del Valle et al. (2002) |
| SV40PyV | Early | 17 kDa | 135 | 17 | Zerrahn et al. (1993) |
| SV40PyV | Late | Agnoprotein | 62 | 6.8 | Gilbert et al. (1981) |
| BKPyV | Early | TruncTAg | ND | 17–20 | Abend et al. (2009) |
| BKPyV | Late | Agnoprotein | 68 | 7.5 | Rinaldo et al. (1998) |
Note. All polyomaviruses universally express large and small T antigens and they are not included in this table.
ND: not determined.
In addition to encoding structural proteins, several polyomaviruses also encode a small regulatory phosphoprotein from their late coding region, named agnoprotein. Among many, only JCV, BKV, and SV40 encode this protein from their late genome. Encoded agnoproteins from these three viruses are 71, 68, and 62 amino acid long, respectively (Khalili, White, Sawa, Nagashima, & Safak, 2005). They all have a highly basic amino acid composition and share a highly significant sequence homology between them (Khalili et al., 2005). JCV agnoprotein is the most studied one in details among three agnoproteins with respect to its binding partners, functional and structural properties (Saribas et al., 2016). In this review, we further described the recent developments in the structural and functional properties of agnoprotein of JCV, BKV, and SV40 in great detail as highlighted in upcoming sections below.
3 |. EXPRESSION OF miRNAS BY POLYOMAVIRUSES
MiRNAs are non‐coding–small RNA molecules that are encoded by a diverse genome of the organisms including viruses, protists, plants, and animals (He & Hannon, 2004). MiRNAs are produced in relatively longer precursor RNA transcripts by RNA polymerase II and III in the nucleus, which are subsequently cleaved into precursor miRNAs and then exported to the cytoplasm, where it is further processed to be mature and become active guide miRNAs. Mature miRNAs not only regulate the translation of transcripts by binding to the complementary sequences on the target RNA but sometimes induce their degradation as well (Iwakawa & Tomari, 2015). In their mature state, they can either reside intracellularly or can be released from cells through the extracellular vesicles including exosomes and apoptotic bodies, (Makarova et al., 2016; Turchinovich & Cho, 2014; Turchinovich, Weiz, Langheinz, & Burwinkel, 2011; Zhang et al., 2015). Such secreted miRNAs can play roles in intercellular communication with respect to gene regulation (Hannafon & Ding, 2013).
The identification and functional analysis of the first miRNA encoded by the polyomaviruses, namely for SV40 (SVmicroRNA), is reported by Sullivan, Grundhoff, Tevethia, Pipas, and Ganem (2005). SVmicroRNA accumulates late during the infection cycle and is perfectly complementary to the viral early RNA sequences, and induces early RNA degradation. In addition, it was also demonstrated that SVmicroRNA reduces the susceptibility of the infected cells to be lysed by the cytotoxic T cells (Sullivan et al., 2005). In other words, SV40 uses its own SVmicroRNA to evade immune response, thereby enhances its ability to successfully complete its life cycle. Expression of similar miRNAs from the genomes of two human polyomaviruses, JCV and BKV was also detected (Seo, Fink, O’Hara, Atwood, & Sullivan, 2008; Tian et al., 2014). It turned out that these miRNAs are also expressed late during the infection cycle and autoregulate the early gene expression of both viruses as reported for SV40 miRNA. In addition, it appears that both JCV and BKV also use these miRNA to evade both innate and adaptive immune system, perhaps to sustain their latent infection in the body. One such an example to this evasion mechanism is that JCV and BKV miRNAs were shown to target the stress‐induced ligand ULBP3, which is the protein that is recognized by natural killer cell receptor NKG2D, consequently resulting in the reduced NKG2D‐mediated killing of the infected cells by natural killer cells (Bauman et al., 2011). Another human polyomavirus, MCPyV, also reported to encode a single miRNA precursor from the late strand of its genome. This miRNA is then processed to produce two mature miRNAs designated as MCV‐miR‐M1–5p and MCV‐mir‐M1–3p (Lee et al., 2011; Seo, Chen, & Sullivan, 2009). MCV‐miR‐M1–5p sequence was found to be perfectly complementary to the coding sequences of LT‐Ag of MCPyV which attenuates its expression through which it limits the viral replication (Seo et al., 2009). In an additional study, investigators found that MCV‐miR‐M1–5p targets SP100 mRNA, and its translation product is implicated in innate immune response against dsDNA viruses including MCPyV, suggesting that MCPyV also uses its miRNA to evade host immune system (Akhbari, Tobin, Poterlowicz, Roberts, & Boyne, 2018). The founding family member of the polyomavirus family, the mouse polyomavirus (MPyV) was also found to encode a precursor miRNA from its genome (Sullivan et al., 2009), which is processed into two mature miRNAs. This miRNA like other miRNAs encoded by some other polyomaviruses targets the viral early transcripts for cleavage but is not required for their regulation of the efficiency of the viral infection cycle (Sullivan et al., 2009).
4 |. AGNOPROTEIN OF POLYOMAVIRUSES
4.1 |. Cellular distribution of polyomavirus agnoprotein
Agnoprotein is mainly a cytoplasmic protein but exhibits high accumulations in the perinuclear area of the infected cells (Ellis & Koralnik, 2015; Gerits & Moens, 2012; Nomura, Khoury, & Jay, 1983; Rinaldo & Hirsch, 2007; Saribas et al., 2016; Saribas, White, & Safak, 2012). However, a small amount of agnoprotein is also consistently detected in the nucleus by microscopy studies (Saribas et al., 2012). The nuclear localization of agnoprotein is highly consistent with the bioinformatics predictions suggesting that agnoprotein has a weak “bipartite nuclear localization signal” (BNLS; Dingwall & Laskey, 1986, 1991), which is localized toward the N‐terminal region of the protein (Saribas et al., 2012; Figure 2). Perinuclear localization of agnoprotein suggests that it may also colocalize with the endoplasmic reticulum (ER). In fact, Suzuki et al. (2010) demonstrated the colocalization of agnoprotein with ER in JCV‐infected cells using ER markers including calreticulin and BiP. The same investigators further investigated the impact of some of the critical residues of the BNLS on the distribution of agnoprotein to ER. Findings showed that although substitution of the Arg8 and Lys9 residues with Ala did not affect the targeting of the protein to ER, the conversion of the Lys22, Lys23, and Arg24 residues to Ala, Ala, and Gly, respectively, resulted in the loss of ER targeting activity. These observations suggest that some of these basic amino acid residues found within the BNLS region of agnoprotein are important for ER targeting (Suzuki et al., 2010). It is important to note that the distribution of agnoprotein to cytoplasm and nucleus suggests its diverse roles in both compartments. Similarly, the cellular distribution of BKV agnoprotein was also investigated using enhanced green fluorescent protein fusion constructs by Unterstab et al. (2010). Interestingly, these studies showed the colocalization of the agnoprotein with “lipid‐droplets (LD)” in the infected cells and the predicted that α‐helical region of BKV agnoprotein, amino acids encompassing amino acids 20 through 42 are important for this localization. The double substitution mutants, Ala25Asp and Phe39Glu within this region abrogated the targeting of the agnoprotein into the LD (Unterstab et al., 2010), suggesting that maintenance of the amino acid integrity within the α-helical region is critical for its localization to LD.
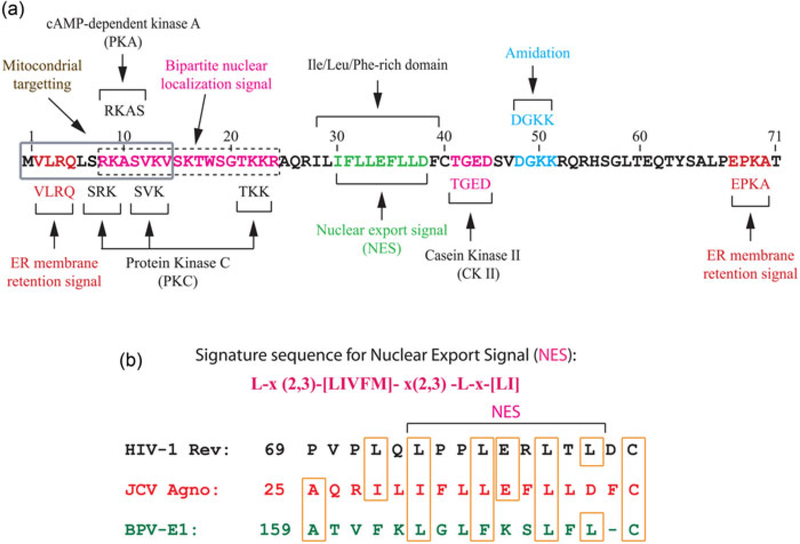
FIGURE 2.
Predicted and validated functional motifs of JCV agnoprotein. (a) Phosphorylation of Ser7, Ser11, and Thr21 was previously demonstrated (Johannessen et al., 2008; Sariyer et al., 2006). The website, https://wolfpsort.hgc.jp/ was used to predict the functional motif or domains. (b) For prediction of mitochondrial targeting sequences, the following website was used: http://mitf.cbrc.jp/MitoFates/cgi-bin/top.cgi. JCV: JC virus, ER: Endoplasmic reticulum [Color figure can be viewed at wileyonlinelibrary.com]
4.2 |. Stable dimer and oligomer formation by polyomavirus agnoproteins
Dimer and oligomer formation by proteins are common and some are essential for homeostasis of biological systems. Weak subunit interactions are, for example, necessary for regulation of the biological activity of many proteins including ion channels, enzymes, transcription factors, and receptors (Marianayagam, Sunde, & Matthews, 2004). In some cases, dimers and oligomers made by some proteins are found to be highly stable and resistant to different denaturing agents. A number of viral and eukaryotic proteins were reported to have such properties. The nitric oxide synthetase, for example, was found to form highly stable dimers in cells that are resistant to denaturing reagents and heat (Kolodziejski, Rashid, & Eissa, 2003). Small regulatory proteins from HIV‐1, including rev, vpr, vif, nef, and tat were also known to form dimers and oligomers (Cullen, 1998a; Frankel, Bredt, & Pabo, 1988; Frankel, Chen, Cotter, & Pabo, 1988; Kwak et al., 2010; S. Yang, Sun, & Zhang, 2001; Zhao, Wang, Mukherjee, & Narayan, 1994a). It is implicated that dimerization of vpr is critical for HIV gag protein recognition and the accumulation of gag at the plasma membrane (Fritz et al., 2010). Another HIV‐1 regulatory protein, Rev functions in oligomeric forms. Rev is known to function in transport HIV RNA from nucleus to cytoplasm by binding to the ~350 nt Rev response element RNA found in the intron regions of partially spliced and unspliced HIV RNA (Cullen, 1998a, 1998b, 2003a, 2003b; Cullen, 1998b; Pollard & Malim, 1998). To export its bound RNA, oligomeric form of Rev is required to bind to the human nuclear export receptor, CRM1, (Cullen, 2003b; Fornerod, Ohno, Yoshida, & Mattaj, 1997). The homodimerization of Ebola virus (EBOV) transcription factor, VP30, was also shown to be critical for its function during the viral transcription (Hartlieb, Modrof, Muhlberger, Klenk, & Becker, 2003). Some viral proteins undergo nucleic acid‐dependent oligomerization in a sequence‐specific manner, an example of which is the Borna disease virus (BDV) nucleoprotein which requires the presence of 5′‐specific BDV RNA for its oligomerization (Hock et al., 2010). Most famously, the main regulatory protein of all polyomaviruses, LT‐Ag, binds to the origin of the viral DNA replication sequences in a double hexameric forms to initiate viral DNA replication (Auborn, Markowitz, Wang, Yu, & Prives, 1988; Lynch & Frisque, 1990; Simmons, Loeber, & Tegtmeyer, 1990; Simmons, et al. 2004).
Formation of stable dimers and oligomers by the JCV, BKV, and SV40 agnoproteins has also been previously reported by Saribas, Arachea, White, Viola, and Safak (2011). Stable dimer–oligomer formation was initially observed through the analysis of the slower migration patterns of the fusions of agnoprotein, including the glutathione S‐transferase agnoprotein (GST‐Agno) and maltosebinding protein on the denaturing sodium dodecyl sulfate (SDS)–PAGE gels (Safak et al., 2001; Sariyer et al., 2006). It turned out that agnoprotein dimers and oligomers are highly resistant to, SDS and 8M urea treatment and heat denaturation (Saribas et al., 2011). These complexes were further analyzed by mass spectrometry (Saribas et al., 2011) and data revealed that the bands that migrated slower than expected sizes turned out to mainly contain the fusion proteins (GST and agnoprotein) suggesting that these slower migrating bands arise from the dimer–oligomer formation features of agnoprotein. The dimer and oligomer formation by JCV agnoprotein was also reported in infected cells in vivo (Saribas et al., 2016). Further investigation of the nature of these oligomeric structures suggested that they do not result from a covalent bond formation between the monomers, but most likely from the hydrophobic interactions and numerous strong ionic attractions (Saribas et al., 2011). Moreover, our findings regarding the dimeric–oligomeric forms of agnoprotein were also consistent with the observations by Merabova et al. (2008) and Del Valle et al. (2002) who also detected slow migrating agnoprotein bands under denaturing conditions but during that time, it was not known that these are due to dimer–oligomer formation property of agnoprotein. Cross‐linking and fluorescence resonance energy transfer studies by Suzuki et al. (2010) also hinted the formation of the dimeric–oligomeric forms in the transfected cells. Further mapping studies revealed that the Leu‐/ Ile‐/Phe‐rich domain of the protein is essential for dimer–oligomer formation and the stability of the protein (Saribas et al., 2011).
4.3 |. Agnoprotein structure
Agnoprotein is one of the important regulatory proteins of human (JCV and BKV) and monkey (SV40) polyomaviruses. It plays a critical role in the biology of these three viruses, because they are unable to sustain a productive life cycle without its expression during the viral life cycle (Carswell & Alwine, 1986; Carswell, Resnick, & Alwine, 1986; Dang, Wuthrich, Gordon, Sawa, & Koralnik, 2012; Gerits & Moens, 2012; Hay, Kessler, & Aloni, 1984; Hou‐Jong, Larsen, & Roman, 1987; Merabova et al., 2008; Myhre, Olsen, Gosert, Hirsch, & Rinaldo, 2010; Ng, Mertz, Sanden-Will, & Bina, 1985; Okada et al., 2002; Resnick & Shenk, 1986; Saribas et al., 2013; Sariyer, Saribas, White, & Safak, 2011; Suzuki et al., 2010, 2012; Unterstab et al., 2010). It has a highly basic amino acid composition. Most of the positively charged residues including Arg (R) and Lys (K) are located at the amino‐terminus of the protein and they are highly conserved among agnoprotein of JCV, BKV, and SV40 (Saribas et al., 2016; Figure 3). However, a short region toward the middle portion of the protein, aa 28 to 39 is mostly made up of hydrophobic residues and this area is designated as the Leu‐/Ile‐/Phe‐rich domain of the protein and predicted to adopt α‐helix. It is noteworthy that all three agnoproteins contain only three Phe residues and all are confined within the Leu‐/Ile‐/Phe‐rich domain. More interestingly, all also contain only one cysteine residue (Cys40), which is located right after the Leu‐/Ile‐/Phe‐rich region (Figure 2). Carboxy terminus of the agnoproteins is less conserved and predicted to have intrinsically disordered conformation.
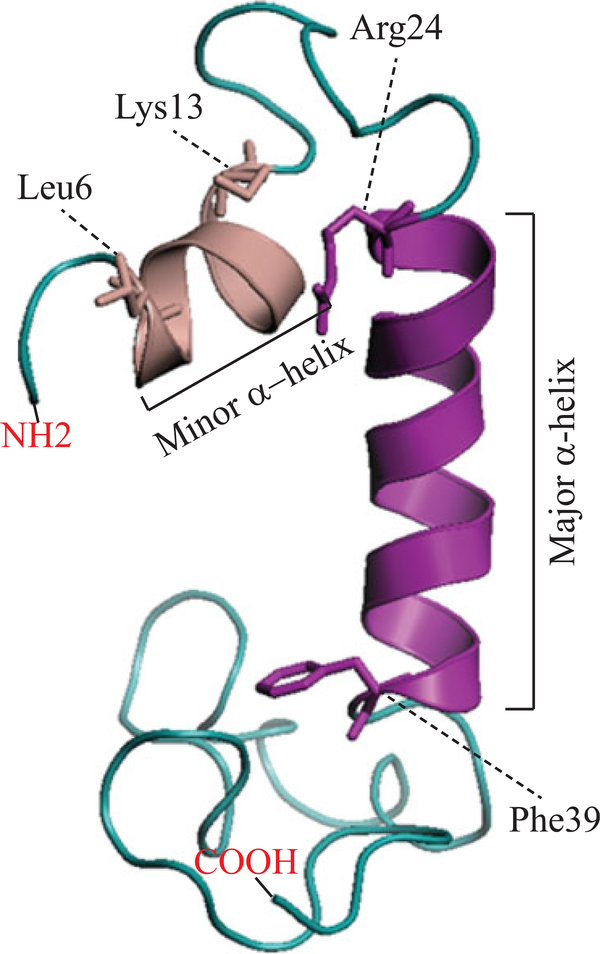
FIGURE 3.
The 3D structure of the full‐length JC virus agnoprotein was recently resolved by NMR (Coric et al., 2017). According to this 3D structure, agnoprotein contains a minor and a major α‐helix located at amino acid position Leu6‐Lys13 and Arg24-Phe39, respectively. The rest of the protein adopts an intrinsically unstructured conformation (Met1‐Gln5, Val14‐Lys23, and Cyt40-Thr71). 3D: three dimensional; NMR: nuclear magnetic resonance [Color figure can be viewed at wileyonlinelibrary.com]
4.4 |. Functional domains of agnoprotein
Over the years, the structure–function studies with agnoprotein clearly demonstrated multifunctionality of agnoprotein targeting multiviral and cellular proteins in infected cells (Gerits & Moens, 2012; Johannessen, Myhre, Dragset, Tummler, & Moens, 2008; Saribas et al., 2016; Sariyer et al., 2006). It has a relatively short amino acid sequence and is mainly localized to the cytoplasmic compartment of the cells, accumulating largely at the perinuclear area (Rinaldo, Traavik, & Hey, 1998; Saribas et al., 2012). However, a small portion of the protein is also found in the nucleus (Saribas et al., 2012). It was previously demonstrated that this protein is posttranslationally modified by protein kinases including protein kinase C (PKC) and cAMP‐dependent kinase at the predicted phosphorylation sites at the “SRK, aa 7–9,” “SVK, aa 11–13,” “RKAS, aa 8–11,” and “TKK, aa 21–23” (Johannessen et al., 2008; Sariyer et al., 2006). Agnoprotein also contains another predicted phosphorylation site by casein kinase II (CKII) at amino acids encompassing 41–44 (TGED). Prediction studies also showed that this viral protein has additional functional motifs and signatures for various predicted functions (Figure 2). For example, the far ends of the N‐terminus and C‐terminus of the protein contain an “ER membrane retention” motifs, including a “VLRQ, aa 2–5” and a “EPKA, aa 67–70.” This prediction corroborates with the data previously reported by Suzuki et al. (2010), who demonstrated the targeting of ER by agnoprotein. Another interesting predicted site is the presence of a “a weak bipartite nuclear localization signal, PNLS” located toward the N‐terminus of the protein encompassing aa 8–24 (RKASVKVSKTWSGTKKR). The functionality of this site is also supported by a previously reported data, where agnoprotein was consistently detected, in small amounts, in the nucleus of the infected cells (Saribas et al., 2012). Recent findings also revealed that certain mutants of agnoprotein can exclusively localize to the nucleus (unpublished data) further supporting the functionality and validity of this weak BNLS of agnoprotein. Additionally, the major α‐helix domain of agnoprotein (Leu‐/Ile‐/Phe‐rich region) is predicted to contain a strong nuclear export signal and exhibits high similarity to those of the HIV‐1 Rev (Daugherty, Liu, & Frankel, 2010) and bovine papillomavirus E1 proteins (Figure 2b and Table 3). In support of this prediction, it was previously reported that agnoprotein shuttles back and forth between the nucleus and cytoplasm in a phosphorylation-dependent manner (Okada et al., 2001). Another predicted the signature sequence of agnoprotein is the amidation site, located toward the middle portion of the protein (aa 47–50, DGKK). Such a chemical modification is, in general, essential for the biological activity of the hormones, neurotransmitters and growth factor peptides. Those factors are produced in inactive form and upon cleavage at their certain regions, the C‐terminus of the peptide is modified by amidation and secreted into the blood stream (Walsh & Jefferis, 2006) for the fulfillment of their activity. In addition to ER membrane retention motif described above, agnoprotein contains another organelle targeting sequence in its far N‐terminus namely “Mitochondrial Targeting Sequence” (MTS), amino acids encompassing 1–14 (MVLRQLSRKASVKV). This sequence (a) adopts a short α‐helical structure and (b) contains several positively charged residues. Both features are known to be characteristics of the mitochondrial targeting sequences. Our initial validation studies suggest that, agnoprotein not only targets the “mitochondrial protein import complexes” but also other components of mitochondria (unpublished data; Figure 2 and Table 3).
TABLE 3.
Predicted and/or validated functional motifs of agnoprotein
| Motif sequences | Amino acid position | Modification/organelle targeting | Website/reference |
|---|---|---|---|
| MVLRQLSRKASVKV | 1–14 | Mitochondria targeting signal | http://mitf.cbrc.jp/MitoFates/cgi-bin/top.cgi |
| VLRQ | 2–5 | Endoplasmic reticulum membrane retention signal | https://wolfpsort.hgc.jp/ |
| SRK | 7–9 | Phosphorylation by protein kinase C (PKC) | Sariyer et al. (2006) |
| RKAS | 8–11 | Phosphorylation by cAMP-dependent protein kinase A (cAMP-PKA) | https://wolfpsort.hgc.jp/ |
| SVK | 11–13 | Phosphorylation by PKC | Sariyer et al. (2006), Johannessen et al. (2008) |
| TKK | 21–23 | Phosphorylation by PKC | Sariyer et al. (2006), Johannessen et al. (2008) |
| RKASVKVSKTWSGTKKR | 8–24 | Bipartite nuclear localization signal | https://wolfpsort.hgc.jp/ |
| IFLLEFLLD | 30–38 | Nuclear export signal (NES) | https://wolfpsort.hgc.jp/ |
| TGED | 41–44 | Phosphorylation by Casein Kinase II (CKII) | https://wolfpsort.hgc.jp/ |
| DGKK | 48–51 | Amidation | https://wolfpsort.hgc.jp/ |
| EPKA | 66–69 | Endoplasmic reticulum membrane retention signal | https://wolfpsort.hgc.jp/ |
4.5 |. Three‐dimensional (3D) structure of agnoprotein determined by nuclear magnetic resonance (NMR)
Initial bioinformatics modeling studies predicted that agnoprotein contains two α‐helices, one of which is located at the in the N‐terminus and the other at the central portion of the protein, from Val2 to Ala10 and Gly20 to Cys40, respectively (Combet, Blanchet, Geourjon, & Deleage, 2000; Coric et al., 2014; McGuffin, Bryson, & Jones, 2000; Roy, Kucukural, & Zhang, 2010; Saribas et al., 2013; Unterstab et al., 2010). The rest of the protein is predicted to adopt an intrinsically disordered conformation from Ser11 to Ser19 and Thr41 to Thr71. Several investigators including our group failed to resolve the X‐ray crystal structure of JCV agnoprotein using purified proteins either from baculovirus or bacterial systems due to technical difficulties associated with purification of the protein and and a process called the dynamic interconversion of monomers to dimers or oligomers (Saribas et al., 2011). Finally, our group in collaboration with the NMR experts from the Université Paris Descartes, France, determined the first partial 3D structure of agnoprotein by NMR using a highly pure synthetic agnoprotein peptide encompassing amino acids Thr17 to Gln52 (Coric et al., 2014). The NMR structure of partial agnoprotein revealed an α‐helix, encompassing amino acids Lys23 to 39. The surrounding regions of the α‐helix turned out to adopt an intrinsically unstructured conformation. Helical wheel representation of the α‐helical region revealed four distinct faces: A hydrophilic face containing residues Lys23, Arg27, Glu34, and Asp38; an aromatic face containing Phe31, Phe35, Ile28, and Phe39; a hydrophobic face 1 containing Ala25, Leu29, Leu32, and Leu36; and hydrophobic face II containing Ile30, Leu33, and Leu37 (Coric et al., 2014). Very recently, our group again in collaboration with the same French group resolved the full‐length 3D structure agnoprotein using a highly pure synthetic agnoprotein peptide (Coric et al., 2017). Both NMR structures of agnoprotein revealed the formation of a relatively long α‐helix (major α‐helix) at the same position encompassing amino acids Arg24‐Phe39 (Coric et al., 2017; Figure 3). The new NMR structure also revealed the formation of another short α‐helix (minor α‐helix) at the N‐terminus of the protein, encompassing amino acids Leu6‐Lys13. The rest of the structure exhibited intrinsically disordered conformation. In other words, amino acids 1–5, 14–23, and 40–71 do not have a defined tertiary structure. This type of disordered conformation results from a combined low overall hydrophobicity and high net charge which is thought to provide considerable flexibility to agnoprotein to interact with its various partners in different conformations through which it diversifies and perhaps amplifies its functions in cells (Dyson & Wright, 2005; Fink, 2005). Existence of similar intrinsically disordered structures for other viral proteins was also reported. For example, adenovirus small protein, E4‐ORF3 forms multivalent functional matrices in infected cells by using its intrinsically disordered structured regions (Ou et al., 2012). This then leads to the inactivation of not only one but also multiple tumor suppressor proteins including, TIM24, MRE11/ Rad50/NBS1 (MRN), promyelocytic leukemia factor and p53 (Ou et al., 2012). Similarly, the amino acid sequences located at the intrinsically unstructured regions of agnoprotein may behave in a similar fashion with respect to the targeting its partners in infected cells and thereby significantly contribute to the overall viral replication cycle.
4.6 |. NMR structure‐based dimeric models of agnoprotein
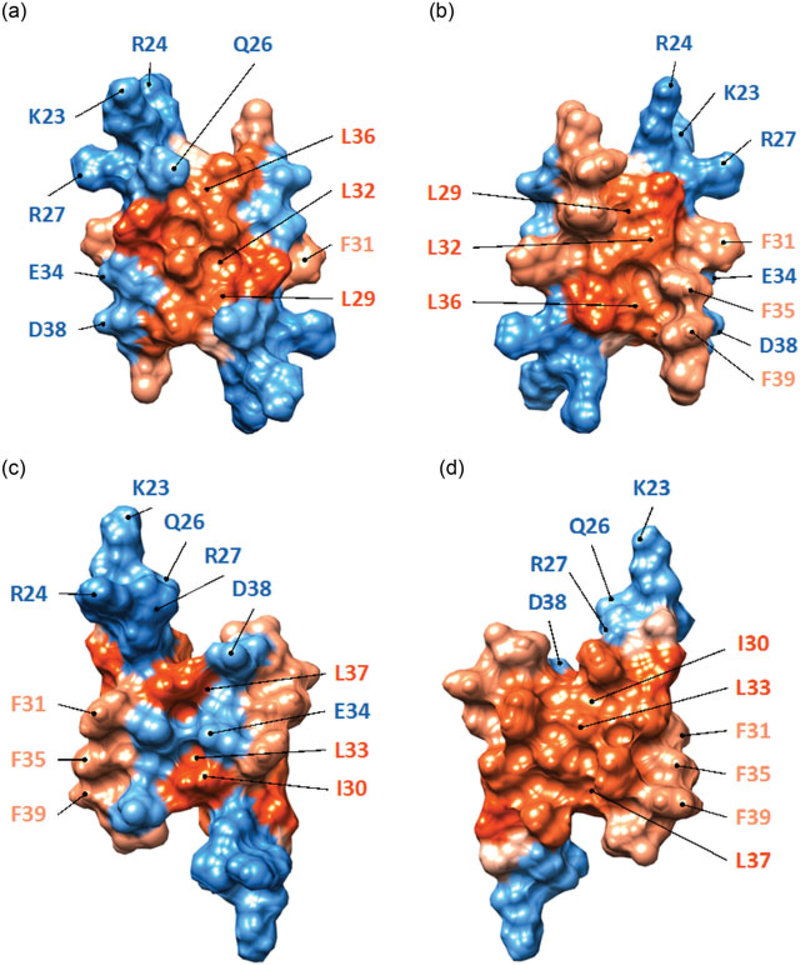
It became apparent that agnoprotein forms dimers and oligomers through the specific aa residues located within the Leu‐/Ile‐/Phe‐rich α‐helix dimerization domain (Coric et al., 2017, 2014; Saribas et al., 2013, 2011). In addition, the NMR structure of agnoprotein provided important clues regarding how the dimer formation would be possible between agnoprotein monomers. Based on the previously reported “helical wheel model” and site‐directed mutagenesis studies (Coric et al., 2014), it was predicted that agnoprotein may form homodimers through the hydrophobic face I and hydrophobic face II of the major α‐helix region. The hydrophobic surface 1 contains residues Ala25, Leu29, Leu32, and Leu36; and the hydrophobic surface II contains residues Ile30, Leu33, and Leu37 (Coric et al., 2014). Indeed, when Leu29, Leu32, and Leu36 were simultaneously mutated to Ala, agnoprotein levels were significantly reduced during the viral replication cycle (Coric et al., 2014), suggesting that hydrophobic face I is most likely involved in dimer–oligomer formation and protein stability. In addition, similar mutations were also made on the hydrophobic surface II, where Ile30 and Leu33 residues were mutated to Ala individually or combinatorially but agnoprotein expression levels did not change as dramatically as observed for hydrophobic surface I. Nonetheless, theoretical dimeric models can be built for hydrophobic face I and II by employing the program X‐PLOR (Brunger, 1992; Brunger et al., 1998; Figure 4a,b). According to these models, agnoprotein forms dimers either through the interaction of Leu29 and Leu36 residues (Figure 4a,b) or that of Ile30, Leu33, and Leu37 (Figure 4c,d) in an antiparallel manner (Coric et al., 2014; Figure 4a–d). The surface representation of these two models provides more information about the dimeric structures of agnoprotein (5a–d). Within the dimeric structure formed by the hydrophobic surface I, where the interface formed through the amino acids L29, L32, and L36, one can distinguish two opposite faces with equivalent properties. The first one is constituted of the hydrophobic residues L29, I30, L33, and L37 and the other face is composed of amino acids residues A25, I28, F31, L32, F35, L36, and F39 (5a,b) and forms a relatively extended hydrophobic surface (5a,b). The second possible dimeric structure whose interface is composed of amino acids I30, L33, and L37, exhibits two distinguishable opposite faces with different properties. The first surface displays hydrophilic properties and is constituted of the hydrophilic residues, including K23, R24, Q26, R27, E34, and D38 (5c,d) while the opposite surface is composed of the hydrophobic residues, including L29, I30, F31, L32, L33, F35, L36, L37, and F39, which forms a relatively large hydrophobic surface, area wise (5c,d).
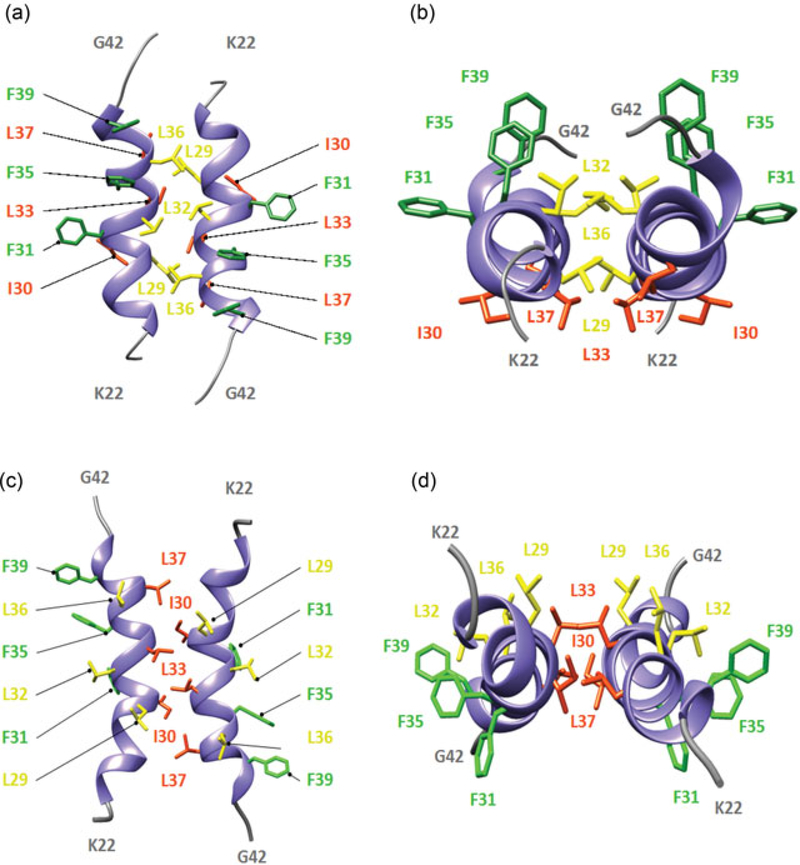
FIGURE 4.
Theoretical dimer formation by agnoprotein through the hydrophobic face I and II, both of which are located at the major α‐helix domain. (a, b) The first homodimer is formed through the interface of Leu29, Leu32 and Leu36 residues located at the hydrophobic face I and is illustrated by a radial (a) and axial (b) view. The side chains of the residues are represented in a stick conformation. The backbone of each α‐helix is represented in a ribbon representation and colored in purple. Amino acids Leu29, Leu32, and Leu36 located at the interface are colored in yellow and aromatic residues Phe31, Phe35, and Phe39 forming an aromatic cluster are colored in green. (c, d) The second homodimer structure is formed through the interface of Ile30, Leu33, and Leu37, which are located on the hydrophobic face II and is also represented in a radial (c) and axial (d) view. The backbone of the antiparallel α‐helices is illustrated in a ribbon representation and colored in purple. The hydrophobic residues Ile30, Leu33, and Leu37 forming the interface are illustrated in a stick representation and colored in orange. Phenylalanine residues, Phe31, Phe35, and Phe39, forming an aromatic cluster are colored in green. The data used to build each homodimer structure of agnoprotein are the same as the one used to build the homodimer structure for the short fragment of agnoprotein (aa 17–52; Coric et al., 2014). The structure of each theoretical dimer was calculated using simulated annealing with restrained molecular dynamics and energy minimization with the X‐PLOR program (Arkin, Sukharev, Blount, Kung, & Brunger, 1998; Fleming et al., 1998) implemented with a total of 453 distance restraints for dimer A (a) and 447 distance restraints for dimer C (c). To build each dimer, 232 intermolecular nuclear overhauser effect (NOE)‐derived distances were unambiguously defined to build dimer A with interface L29, L32, and L36 (a) and 68 for dimer C with interface I30, L33, and L37 (c) respectively [Color figure can be viewed at wileyonlinelibrary.com]
It is possible that the hydrophobic surfaces of the major α‐helix region form additional multiple potential conformational variants during the infection cycle of JCV. Since agnoprotein localizes to nucleus, cytoplasm, and perhaps into the cellular membranes, it is then most likely that agnoprotein exists in different conformations and oligomeric forms in different compartments of the infected cells, which then allow agnoprotein a considerable flexibility to fulfill its functions in a more diverse manner. In addition, the Cys40 residue was also originally thought to be involved in dimerization by forming a disulfide bridge between the homodimers of agnoprotein. In fact, Hidaka, Hojo, Fujioka, Nukuzuma, and Tsuda (2015) reported such a disulfide bridge formation through the Cys40 residue in vitro when two short synthetic peptides of agnoprotein was used (aa 22–40). However, similar studies by our group using the full‐length agnoprotein failed to demonstrate such a contribution by Cys40 to dimer–oligomer formation of agnoprotein (unpublished data), suggesting that the work performed by Hidaka et al., represents an inherent limitation in the experimental design of the work due to use of a short peptide in dimerization studies. In short, these modeling studies further suggest that particularly the Leu29, Leu32, and Leu36 face of the α‐helix can be targeted for drug development purposes to prevent the dimer formation in order to develop effective therapeutics against JCV infections.
4.7 |. Agnoprotein is released from agnoprotein-positive cells
Viruses have evolved various strategies to modify not only the host but also the neighboring cell environment to have a successful life cycle and thereby efficiently propagate their progeny. One of the mechanisms by which they achieve this task is to secrete some of their own proteins from the infected cells to modify the neighboring cellular environment so that incoming virus finds the new environment more conducive for initiation of the next round of the viral replication cycle. When released, some of those viral proteins can act as cytokine mimickers (Liu et al., 2004; Suzuki et al., 1995), cytokine inhibitors (Alcami, Symons, Collins, Williams, & Smith, 1998; Liu et al., 2000), inflammatory cell inhibitors (Lucas et al., 1996), and complement inhibitors (Al‐Mohanna, Parhar, & Kotwal, 2001; Anderson, Smith, & Kotwal, 2002) to evade the host immune system and thereby perhaps to establish a long‐term chronic infection cycle. Not only some DNA but also some RNA viruses exhibit such a behavior. In an RNA virus group, for example, a dimeric and oligomeric regulatory protein of the West Nile virus, NS1 was reported be released from the infected cells (Muller & Young, 2013). Another example to the RNA viral protein that is released from cells is the HIV‐1 Vpr protein. Like agnoprotein, vpr forms dimers and oligomers (Zhao, Wang, Mukherjee, & Narayan, 1994b) and is released from the infected cells (Levy, Refaeli, MacGregor, & Weiner, 1994). Nearly two decades ago, Levy et al. (1994) first time reported the detection of Vpr in the serum of the infected patients. It was demonstrated that vpr can modulate the activity of the treated cells. In this regard, it was found that recombinant Vpr is readily taken up by neurons and inhibits their axonal growth (Kitayama et al., 2008). It was also demonstrated that Vpr crosses the mitochondrial outer membrane via voltage‐dependent anion channel and interacts with mitochondrial protein adenine nucleotide translocator (ANT) on the inner mitochondrial membrane. This leads to the dissipation of the mitochondrial membrane potential (Δѱm), and plays a disruptive role in Ca++ metabolism and apoptosis (Brenner & Kroemer, 2003; Mukerjee et al., 2011). As stated above, some of the DNA viruses are not exception from releasing some of their own proteins from the infected cells. For example, poxvirus releases its interleukin‐18 binding proteins (IL‐18BPS) from the infected cells to modulate the host immune response (Smith, Bryant, & Alcami, 2000). Similarly, the human cytomegalovirus is also known to release a number of viral proteins from the infected cells, including UL21, UL128, UL146, and UL147. Among them, UL21 was shown to interact with CC15/ RANTES and blocks the interaction of CC15/RANTES with cellular receptors (Luganini, Terlizzi, & Gribaudo, 2016). Another DNA viral protein, adenovirus E4/49 K was also shown to be released and is bound to leukocyte common antigen (CD45) to modulate the leukocyte function (Windheim et al., 2013).
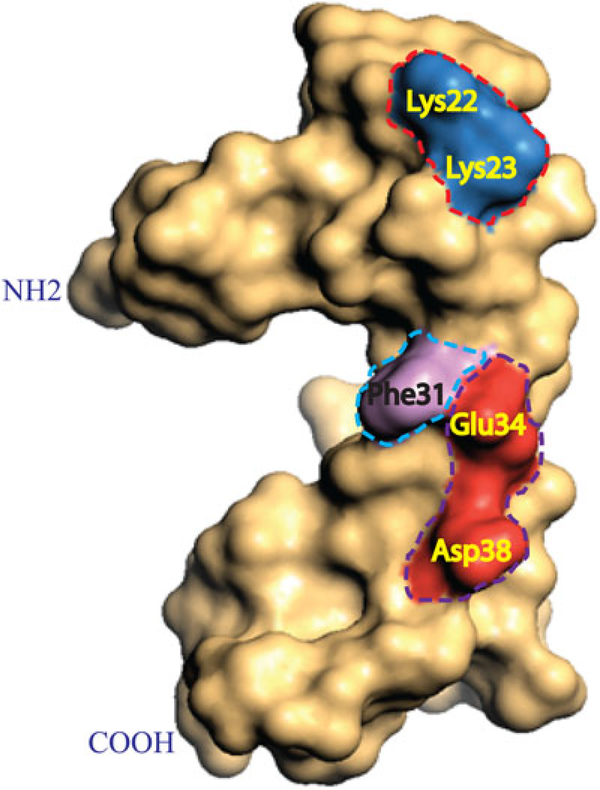
The release of JCV agnoprotein from the agnoprotein‐positive cells was previously reported by Otlu, De Simone, Otalora, Khalili, and Sariyer (2014). Following this report, Saribas, White, and Safak (2018) investigated the mechanism of its release by mapping studies. The recently resolved 3D NMR structure of agnoprotein (Coric et al., 2014) was taken as the base for mutagenesis to determine the mechanism of its release and deletion analysis indicated that the major α‐helix region of agnoprotein plays a critical role in the release process (Saribas, White, et al., 2018). As previously reported, the major α‐helix region is composed of four distinct surfaces: an aromatic, a hydrophilic and two hydrophobic surfaces (Coric et al., 2014). The specific amino acid residues on each surface were analyzed for their contribution to agnoprotein release by site‐directed mutagenesis. Results demonstrated that two positively (Lys22 and Lys23) and two negatively (Glu34 and Asp38) charged residues located on the hydrophilic surface play critical roles in agnoprotein release (Saribas, White, et al., 2018). In addition, an aromatic residue, Phe31, located adjacent to the hydrophilic surface was also identified to be a significant contributor to this process (Saribas, White, et al., 2018; Figure 6). Interestingly, amino acid residues that are involved in agnoprotein release are all but Asp38 conserved among JCV, BKV, and SV40 agnoproteins. Asp38 is substituted with Gln in SV40 and Glu in BKV agnoprotein. This high level of conservation suggests that they also play similar roles in release of BKV and SV40 agnoproteins. Note that, in addition to JCV agnoprotein, the release of BKV and SV40 agnoprotein was also observed (unpublished data; Saribas, White, et al., 2018). The exact mechanism of agnoprotein release is currently unknown, however, the involvement of the ER to Golgi pathway was previously suspected using a specific inhibitor of this pathway, Brefeldin A (Otlu et al., 2014).
FIGURE 6.
Hydrophilic residues located on the major α‐helix domain play important roles in release of agnoprotein. Three-dimensional structure of the full‐length agnoprotein is illustrated in a “surface fill‐in” representation. The hydrophilic residues located on the hydrophilic face of the major α‐helix domain of agnoprotein, including Lys22, Lys23, Glu34, and Asp38; and an aromatic residue (Phe31), which is adjacent to this hydrophilic surface were determined to be critical for the release of agnoprotein from agnoprotein‐positive cells (Saribas, White, et al., 2018). Amino acid residues involved in agnoprotein release are encased in boxes and named. Note that Lys23, Lys23, Phe31, and Glu34 are all conversed between JCV, BKV, and SV40 agnoproteins. JCV Asp38 is substituted for Glu and Gln for BKV and SV40 agnoproteins, respectively, at the same position (Saribas, White, et al., 2018). BKV: BK virus; JCV: JC virus; SV40: simian virus 40 [Color figure can be viewed at wileyonlinelibrary.com]
Questions still remain unanswered as to what the functional consequences of the released agnoprotein is. It is conceivable that released agnoprotein exerts its functions through either interacting with the cell surface molecules or entering cells; and thereby modulates the neighboring cell environment for the benefit of incoming virus for next round of infection cycle. Saribas et al., partially addressed this question by treating cells with a synthetic agnoprotein peptide and by following the behavior of agnoprotein. Results consistently revealed that agnoprotein primarily interacts with and remains bound to the cell surface rather than visibly entering cells in significant quantities (Saribas, White, et al., 2018). However, these investigators also suggested a possibility in this regard that some of the monomeric, dimeric and oligomeric forms of agnoprotein (Saribas et al., 2013) may enter cells at low levels which is hard to detect with immunocytochemistry. In either case, this does not still address the question regarding the function of the released agnoprotein. The following possibilities are plausible: (a) The released agnoprotein can modulate signaling pathways of the neighboring cells by interacting with some of the cell surface components or by entering cells, where then, directly or indirectly, modulates the host‐cell gene expression so that incoming virus replicates more efficiently in those cells. (b) The cell surface‐bound agnoprotein and/or agnoprotein that enters cells can modulate the expression levels of various chemokines and cytokines, thereby contributes to the virus spread. In similar cases, released agnoprotein can negatively affect the survival of the neighboring cells in the absence of viral infection. In fact, in support of this scenario, Merabova et al. (2012, 2008) demonstrated a negative effect of agnoprotein on chemokine secretion by rat cells where it was found that the secreted levels of CXCL5/LIX by CG4 rat oligodendroglial cells is significantly reduced. This chemokine is related to the human CXCL5/ENA78 and CXCL6/GCP‐2 chemokines and required for neuronal survival, strongly suggesting that the released agnoprotein may have long‐term effects on neuronal survival in the CNS and may contribute to neuronal–axonal injury in the pathogenesis of PML lesions as well as to some of the neurological complications including Alzheimer’s disease. Nonetheless, the functional consequences of the released extracellular agnoprotein with respect to its effect on neighboring cells need to be further explored considering the above‐stated possibilities.
4.8 |. Cellular and viral partners of agnoprotein
Interaction of agnoprotein with a number of viral and cellular proteins, including JCV Sm t‐Ag (Sariyer, Khalili, & Safak, 2008), JCV LT‐Ag (Safak & Khalili, 2001), and Tubulin (Endo et al., 2003), capsid protein, VP1 (Suzuki et al., 2012), YB‐1 (Safak, Sadowska, Barrucco, & Khalili, 2002), p53 (Darbinyan et al., 2002), FEZ1, and HP1‐α (Okada et al., 2005; Suzuki et al., 2005), adaptor protein complex 3 (AP‐3; Suzuki et al., 2013) was previously reported and has been implicated in various aspects of the JCV life cycle, including viral replication (Safak et al., 2001), viral transcription, functioning as a viroporin (Suzuki et al., 2013, 2010), virion formation (Sariyer et al., 2006; Suzuki et al., 2012). BKV Agno has also been implicated in interfering with exocytosis (Johannessen et al., 2011), inhibition of the viral DNA replication (Gerits et al., 2015) and egress of virions from the infected cells (Panou et al., 2018). In addition, JCV Agno was shown to deregulate cell cycle progression (Darbinyan et al., 2002). These cellular and viral targets by agnoprotein are summarized in Table 4.
TABLE 4.
Viral and cellular binding partners of agnoprotein and associated functions
| Polyomavirus names | Binding partners | Binding domain of agnoprotein | Associated functions | References |
|---|---|---|---|---|
| JCVPyV | JCV LT-Ag | 1–36 | Inhibition of JCV transcription and replication | Safak et al. (2001) |
| JCVPyV | JCV Sm t-Ag | ND | Inhibition of PP2A-mediated dephosphorylation of agnoprotein | Sariyer et al. (2008) |
| JCVPyV | JCV VP1 | ND | Involved in virion assembly | Suzuki et al. (2012) |
| JCVPyV | HIV-1 Tat | 18–54 | Inhibition of Tat-mediated HIV promoter activation | Kaniowska et al. (2006) |
| JCVPyV | p53 | 1–36 | Suppression of cell cycle progression | Darbinyan et al. (2002) |
| JCVPyV | Ku70 | 1–36 | Inhibition of double-strand break repair | Darbinyan et al. (2004) |
| JCVPyV | PP2A | 18–36 | Dephosphorylation of agnoprotein | Sariyer et al. (2008) |
| JCVPyV | YB-1 | 1–36) | Inhibition of YB-1-mediated transcription of JCV promoters | Safak et al. (2002) |
| JCVPyV | FEZ1 | ND | Inhibition of FEZ1-mediated neurite outgrowth | Suzuki et al. (2005) |
| JCVPyV | HP1α | 1–18 | Facilitation of nuclear export of JCV virions | Okada et al. (2005) |
| JCVPyV | AP-3 | ND | Inhibition of PB-3-mediated vesicular trafficking | Suzuki et al. (2013) |
| BKPyV | α-SNAP | 1–15 | Inhibition of vesicle transport | Johannessen et al. (2011) |
| BKPyV | PCNA | 1–15 | Inhibition of DNA replication | Gerits et al. (2015) |
In addition to these individual targets, recent proteomic studies by our lab aimed at identification of pathways and cellular proteins and complexes targeted by JCV agnoprotein revealed more interesting results. According to this unpublished data, agnoprotein appears to target a broad range of pathways involved in various fundamental processes of the cells including but not limited to eukaryotic initiation factor complex, ubiquitin degradation pathway, 26S proteasome complex subunits, mitochondrial protein import complexes, nuclear pore complexes, nuclear import and export system, and tRNA synthetase complexes (unpublished data).
5 |. CONCLUDING REMARKS
In this comprehensive review, we have summarized the recent advances in human polyomavirus genome expression with respect to the novel cis‐ and trans‐splice products of the early and late genomes and the regulatory miRNAs. Polyomavirus family contains the highly significant group of viruses, some of which significantly contributed to the modern advances in biological sciences with respect to gene expression, DNA replication, alternative splicing of RNA, RNA editing and so forth. Recent detailed studies on polyomavirus gene expression clearly demonstrated that their genomes express novel, sometimes unpredictable, alternatively spliced, and virus‐specific RNA products, some of which have been proven to encode novel proteins and play regulatory roles not only in the viral life cycle but also in cell transformation. Detailed functional analysis of these proteins in the viral replication cycle and cell transformation awaits further investigation. It is now clear that some family members are the causative agent of some of the afflicting human diseases, including PML induced by JCV, PNV caused by BKV, MCC (a benign hyperproliferative skin cancer associated with MCC) and TS characterized by reddish colored papules occurring in the central region of the face and sometimes anywhere on the body. Such affected areas sometimes produce spicules or spines made up of keratin. Therefore, investigating the molecular mechanisms governing the alternatively spliced gene products of polyomaviruses and their functions is pivotal to our understanding of the progression of the diseases caused by these viruses as well as for the development of effective therapeutics against these diseases.
A special emphasis has also been devoted to summarize the recent advances in the structure–function relationship of agnoprotein in this review, due to its importance in polyomavirus life cycle. The agnoprotein is expressed only JCV, BKV, and SV40 and these viruses require agnoprotein for the successful completion of their life cycle. This protein is a small phosphoprotein and forms highly stable dimers and oligomers in vivo and in vitro. Recent NMR structural studies provided a fascinating 3D structure for the JCV full‐length agnoprotein. According to this 3D structure, JCV agnoprotein only forms a major and a minor α‐helix within the entire structure of the protein and the rest of the residues adopts an intrinsically unstructured conformation. The major α‐helix domain is located at the central portion of the protein, Arg24‐Phe39, whereas the minor one encompasses amino acids Leu6‐Lys13. The major α‐helix region is rich in hydrophobic residues, including Leu, Ile, and Phe. Structure‐based mutational analysis of the region demonstrated that major α‐helix region plays a critical role in protein stability, dimer–oligomer formation, and protein release. Particularly, residues Leu29 and Leu36 plays a major in protein stability and dimer–oligomer formation. Theoretical modeling studies support these findings as well because these two residues appear to interact with one another when a homodimer forms in an antiparallel manner. These structural studies also provided us with ample opportunities to target the major α‐helix domain of agnoprotein with small molecules to disrupt its dimer–oligomer formation in order to develop effective therapeutics against JCV infection in PML patients.
Agnoprotein was recently found to be released from the agnoprotein‐positive cells in relatively significant amounts (Otlu et al., 2014; Saribas, White, et al., 2018). The structure‐based release analysis studies clearly showed that the hydrophilic surface of the major α‐helix domain of the protein plays critical roles in this process (Saribas, White, et al., 2018). These studies further demonstrated that agnoprotein strongly interacts with unidentified cell surface component(s), the functional consequence of which has yet to be determined. This obviously leads to several unanswered questions as follows: The main interesting question in this regard is that “what is the function of this released protein in the extracellular matrix?” Does it enter the uninfected neighboring cells or act through the cell surface molecules/receptors? How does “the entered or cell‐surface acting” agnoprotein affect the health of the neighboring cells including neurons, astrocytes, oligodendrocytes, and microglia in the brain or other cells or tissues or organs in the body through the blood stream? What kind of signaling pathways does this protein trigger through either or both pathways? Since agnoprotein is constitutively produced in the body of the infected individuals in low amounts, does this then have any long‐term impact on the wellbeing of any particular organ system in the affected individuals, such as the brain, which is more sensitive to such challenges as aging is facilitated. It is possible that the toxic and released agnoprotein may trigger the onset of age‐related neurological diseases including Alzheimer’s in the long‐term exposure cases (Jackson, 2018). Exploring answers to these relevant questions will open up new avenues for us to better understand the progression of the JCV‐ and BKV‐caused diseases.
FIGURE 5.
Surface fill‐in representation of the theoretical homodimeric structure of agnoprotein formed through the interface Leu29, Leu32, and Leu36. Two opposite faces with equivalent properties are shown colored according their hydrophobic properties. (a) The first hydrophobic face is constituted of the hydrophobic residues Leu29, Ile30, Leu33 and Leu37. (b) The opposite face of the dimer is also constituted of the hydrophobic residues Ala25, Ile28, Phe31, Leu32, Phe35, Leu36, and Phe39. (c) The surface fill‐in representation of the theoretical dimeric structure of agnoprotein formed through the interface of Ile30, Leu33, and Leu37. The surface is colored according to the hydrophobic properties of the amino acids. Two opposite faces with different properties can be distinguished. The first one is constituted of the hydrophilic residues Lys23, Arg24, Gln26, Arg27, Glu34, and Asp38 (c) while the opposite one is also composed of more hydrophobic residues Leu29, Ile30, Phe31, Leu32, Leu33, Phe35, Leu36, Leu37, and Phe39; and forms a relatively larger hydrophobic face (d) [Color figure can be viewed at wileyonlinelibrary.com]
ACKNOWLEDGMENTS
This study was made possible by grants awarded to M. S. (RO1NS090949) by NIH.
Funding information
National Institute of Health, USA, Grant/Award Number: RO1NS090949
Footnotes
CONFLICTS OF INTEREST
The authors declare that there are no conflicts of interest.
REFERENCES
- Abend JR, Joseph AE, Das D, Campbell‐Cecen DB, & Imperiale MJ (2009). A truncated T antigen expressed from an alternatively spliced BK virus early mRNA. Journal of General Virology, 90(Pt 5), 1238–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhbari P, Tobin D, Poterlowicz K, Roberts W, & Boyne JR (2018). MCV‐miR‐M1 targets the host‐cell immune response resulting in the attenuation of neutrophil chemotaxis. Journal of Investigative Dermatology, 138, 2343–2354. [DOI] [PubMed] [Google Scholar]
- Alcamí A, Symons JA, Collins PD, Williams TJ, & Smith GL (1998). Blockade of chemokine activity by a soluble chemokine binding protein from vaccinia virus. Journal of Immunology, 160(2), 624–633. [PubMed] [Google Scholar]
- Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MAA, … Andersson B (2007). Identification of a third human polyomavirus. Journal of Virology, 81(8), 4130–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Mohanna F, Parhar R, & Kotwal GJ (2001). Vaccinia virus complement control protein is capable of protecting xenoendothelial cells from antibody binding and killing by human complement and cytotoxic cells. Transplantation, 71(6), 796–801. [DOI] [PubMed] [Google Scholar]
- Anderson JB, Smith SA, & Kotwal GJ (2002). Vaccinia virus complement control protein inhibits hyperacute xenorejection. Transplantation Proceedings, 34(4), 1083–1085. [DOI] [PubMed] [Google Scholar]
- Arkin IT, Sukharev SI, Blount P, Kung C, & Brünger AT (1998). Helicity, membrane incorporation, orientation and thermal stability of the large conductance mechanosensitive ion channel from E. coli. Biochimica et Biophysica Acta/General Subjects, 1369(1), 131–140. [DOI] [PubMed] [Google Scholar]
- Van Assche G, Van Ranst M, Sciot R, Dubois B, Vermeire S, Noman M, … Rutgeerts P (2005). Progressive multifocal leukoencephalopathy after natalizumab therapy for Crohn’s disease. New England Journal of Medicine, 353(4), 362–368. [DOI] [PubMed] [Google Scholar]
- Auborn KJ, Markowitz RB, Wang E, Yu YT, & Prives C (1988). Simian virus 40 (SV40) T antigen binds specifically to double‐stranded DNA but not to single‐stranded DNA or DNA/RNA hybrids containing the SV40 regulatory sequences. Journal of Virology, 62(6), 2204–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcena‐Panero A, Echevarria JE, Van Ghelue M, Fedele G, Royuela E, Gerits N, & Moens U (2012). BK polyomavirus with archetypal and rearranged non‐coding control regions is present in cerebrospinal fluids from patients with neurological complications. Journal of General Virology, 93(Pt 8), 1780–1794. [DOI] [PubMed] [Google Scholar]
- Bauman Y, Nachmani D, Vitenshtein A, Tsukerman P, Drayman N, Stern-Ginossar N, … Mandelboim O (2011). An identical miRNA of the human JC and BK polyoma viruses targets the stress‐induced ligand ULBP3 to escape immune elimination. Cell Host & Microbe, 9(2), 93–102. [DOI] [PubMed] [Google Scholar]
- Berger JR (1992). PML in AIDS. Neurology, 42(9), 1845–1846. [DOI] [PubMed] [Google Scholar]
- Berger JR (2000). Progressive multifocal leukoencephalopathy. Current Treatment Options in Neurology, 2(4), 361–368. [DOI] [PubMed] [Google Scholar]
- Berger JR, Kaszovitz B, Post MJ, & Dickinson G (1987). Progressive multifocal leukoencephalopathy associated with immunodeficiency virus infection. A review of the literature with a report of sixteen cases. Annals of Internal Medicine, 107, 78–87. [DOI] [PubMed] [Google Scholar]
- Berger JR, Scott G, Albrecht J, Belman AL, Tornatore C, & Major EO (1992). Progressive multifocal leukoencephalopathy in HIV‐1-infected children. AIDS (London, England), 6(8), 837–841. [DOI] [PubMed] [Google Scholar]
- Black DL (2003). Mechanisms of alternative pre‐messenger RNA splicing. Annual Review of Biochemistry, 72, 291–336. [DOI] [PubMed] [Google Scholar]
- Bollag B, Kilpatrick LH, Tyagarajan SK, Tevethia MJ, & Frisque RJ (2006). JC virus T’135, T’136 and T’165 proteins interact with cellular p107 and p130 in vivo and influence viral transformation potential. Journal of Neurovirology, 12(6), 428–442. [DOI] [PubMed] [Google Scholar]
- Boyapati A, Wilson M, Yu J, & Rundell K (2003). SV40 17KT antigen complements dnaj mutations in large T antigen to restore transformation of primary human fibroblasts. Virology, 315(1), 148–158. [DOI] [PubMed] [Google Scholar]
- Brenner C, & Kroemer G (2003). The mitochondriotoxic domain of Vpr determines HIV‐1 virulence. Journal of Clinical Investigation, 111(10), 1455–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT (1992). A system for X‐ray crystallography and NMR. New Haven, CT: Yale University Press. [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, … Warren GL (1998). Crystallography & NMR system: A new software suite for macromolecular structure deter mination. Acta Crystallographica. Section D, Biological Crystallography, 54(Pt 5), 905–921. [DOI] [PubMed] [Google Scholar]
- Buck CB, Phan GQ, Raiji MT, Murphy PM, McDermott DH, & McBride AA (2012). Complete genome sequence of a tenth human polyomavirus. Journal of Virology, 86(19), 10887–10887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carswell S, & Alwine JC (1986). Simian virus 40 agnoprotein facilitates perinuclear‐nuclear localization of VP1, the major capsid protein. Journal of Virology, 60(3), 1055–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carswell S, Resnick J, & Alwine JC (1986). Construction and characterization of CV‐1P cell lines which constitutively express the simian virus 40 agnoprotein: Alteration of plaquing phenotype of viral agnogene mutants. Journal of Virology, 60(2), 415–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter JJ, Daugherty MD, Qi X, Bheda‐Malge A, Wipf GC, Robinson K, … Galloway DA (2013). Identification of an overprinting gene in Merkel cell polyomavirus provides evolutionary insight into the birth of viral genes. Proceedings of the National Academy of Sciences of the United States of America, 110(31), 12744–12749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combet C, Blanchet C, Geourjon C, & Deléage G (2000). NPS@: Network protein sequence analysis. Trends in Biochemical Sciences, 25 (3), 147–150. [DOI] [PubMed] [Google Scholar]
- Coric P, Saribas AS, Abou‐Gharbia M, Childers W, Condra JH, White MK, … Bouaziz S (2017). Nuclear magnetic resonance structure of the human polyoma JC virus agnoprotein. Journal of Cellular Biochemistry, 118(10), 3268–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coric P, Saribas AS, Abou‐Gharbia M, Childers W, White MK, Bouaziz S, & Safak M (2014). Nuclear magnetic resonance structure revealed that the human polyomavirus JC virus agnoprotein contains an alpha‐helix encompassing the Leu/Ile/Phe‐rich domain. Journal of Virology, 88(12), 6556–6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR (1998a). HIV‐1 auxiliary proteins: Making connections in dying cell. Cell, 93, 685–692. [DOI] [PubMed] [Google Scholar]
- Cullen BR (1998b). Retroviruses as model systems for the study of nuclear RNA export pathways. Virology, 249(2), 203–210. [DOI] [PubMed] [Google Scholar]
- Cullen BR (2003a). Nuclear mRNA export: Insights from virology. Trends in Biochemical Sciences, 28(8), 419–424. [DOI] [PubMed] [Google Scholar]
- Cullen BR (2003b). Nuclear RNA export. Journal of Cell Science, 116(Pt 4), 587–597. [DOI] [PubMed] [Google Scholar]
- Dang X, Vidal JE, Penalva de oliveira AC, Simpson DM, Morgello S, Hecht JH, … Koralnik IJ (2012). JC virus granule cell neuronopathy is associated with VP1 C terminus mutants. Journal of General Virology, 93(Pt 1), 175–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang X, Wüthrich C, Gordon J, Sawa H, & Koralnik IJ (2012). JC virus encephalopathy is associated with a novel agnoprotein‐deletion JCV variant. PLOS One, 7(4), e35793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R, Sadowicz D, & Hebert DN (2007). A very late viral protein triggers the lytic release fo SV40. PLOS Pathogens, 3(7), 928938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darbinyan A, Darbinian N, Safak M, Radhakrishnan S, Giordano A, & Khalili K (2002). Evidence for dysregulation of cell cycle by human polyomavirus, JCV, late auxiliary protein. Oncogene, 21(36), 5574–5581. [DOI] [PubMed] [Google Scholar]
- Darbinyan A, Siddiqui KM, Slonina D, Darbinian N, Amini S, White MK, & Khalili K (2004). Role of JC virus agnoprotein in DNA repair. J Virol, 78(16), 8593–8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty MD, Liu B, & Frankel AD (2010). Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nature Structural & Molecular Biology, 17(11), 1337–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaprio JA (2017). Merkel cell polyomavirus and Merkel cell carcinoma. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 372(1732), 20160276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C, & Laskey RA (1986). Protein import into the cell nucleus. Annual Review of Cell Biology, 2, 367–390. [DOI] [PubMed] [Google Scholar]
- Dingwall C, & Laskey RA (1991). Nuclear targeting sequences—A consensus? Trends in Biochemical Sciences, 16(12), 478–481. [DOI] [PubMed] [Google Scholar]
- Van Doorslaer K, Kraberger S, Austin C, Farkas K, Bergeman M, Paunil E, … Varsani A (2018). Fish polyomaviruses belong to two distinct evolutionary lineages. Journal of General Virology, 99(4), 567–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson HJ, & Wright PE (2005). Intrinsically unstructured proteins and their functions. Nature Reviews Molecular Cell Biology, 6(3), 197–208. [DOI] [PubMed] [Google Scholar]
- Ellis LC, & Koralnik IJ (2015). JC virus nucleotides 376‐396 are critical for VP1 capsid protein expression. Journal of Neurovirology, 21(6), 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endo S, Okada Y, Orba Y, Nishihara H, Tanaka S, Nagashima K, & Sawa H (2003). JC virus agnoprotein colocalizes with tubulin. Journal of Neurovirology, 9(Suppl. 1), 10–14. [DOI] [PubMed] [Google Scholar]
- Eul J, Graessmann M, & Graessmann A (1996a). In vitro synthesized SV40 cRNA is trans‐spliced after microinjection into the nuclei of mammalian cells. FEBS Letters, 394(2), 227–232. [DOI] [PubMed] [Google Scholar]
- Eul J, Graessmann M, & Graessmann A (1996b). Trans‐splicing and alternative‐tandem‐cis‐splicing: Two ways by which mammalian cells generate a truncated SV40 T‐antigen. Nucleic Acids Research, 24(9), 1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eul J, & Patzel V (2013). Homologous SV40 RNA trans‐splicing A new mechanism for diversification of viral sequences and phenotypes. RNA Biology, 10(11), 1689–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng H, Shuda M, Chang Y, & Moore PS (2008). Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science, 319(5866), 1096–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenczy MW, Marshall LJ, Nelson CDS, Atwood WJ, Nath A, Khalili K, & Major EO (2012). Molecular biology, epidemiology, and pathogenesis of progressive multifocal leukoencephalopathy, the JC virus‐induced demyelinating disease of the human brain. Clinical Microbiology Reviews, 25(3), 471–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante P, Delbue S, Pagani E, Mancuso R, Marzocchetti A, Borghi E, … Cinque P (2003). Analysis of JC virus genotype distribution and transcriptional control region rearrangements in human immunodeficiency virus‐positive progressive multifocal leukoencephalopathy patients with and without highly active antiretroviral treatment. Journal of Neurovirology, 9(Suppl. 1), 42–46. [DOI] [PubMed] [Google Scholar]
- Fink AL (2005). Natively unfolded proteins. Current Opinion in Structural Biology, 15(1), 35–41. [DOI] [PubMed] [Google Scholar]
- Fleming KG, Hohl TM, Yu RC, Müller SA, Wolpensinger B, Engel A, … Hanson PI (1998). A revised model for the oligomeric state of the N‐ethylmaleimide‐sensitive fusion protein, NSF. Journal of Biological Chemistry, 273(25), 15675–15681. [DOI] [PubMed] [Google Scholar]
- Fluck MM, & Schaffhausen BS (2009). Lessons in signaling and tumorigenesis from polyomavirus middle T antigen. Microbiology and Molecular Biology Reviews, 73(3), 542–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, & Mattaj IW (1997). CRM1 is an export receptor for leucine‐rich nuclear export signals. Cell, 90(6), 1051–1060. [DOI] [PubMed] [Google Scholar]
- Frankel AD, Bredt DS, & Pabo CO (1988). Tat protein from human immunodeficiency virus forms a metal‐linked dimer. Science, 240(4848), 70–73. [DOI] [PubMed] [Google Scholar]
- Frankel AD, Chen L, Cotter RJ, & Pabo CO (1988). Dimerization of the tat protein from human immunodeficiency virus: A cysteine‐rich peptide mimics the normal metal‐linked dimer interface. Proceedings of the National Academy of Sciences of the United States of America, 85(17), 6297–6300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisque RJ, Bream GL, & Cannella MT (1984). Human polyomavirus JC virus genome. Journal of Virology, 51(2), 458–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisque RJ, & White FA (1992). The molecular biology of JC virus, causative agent of progressive multifocal leukoencophalopathy In Roose RP (Ed.), Molecular neurovirology, pathogenesis of viral CNS infections (pp. 25–158). Totowa, NJ: Humana Press Inc. [Google Scholar]
- Fritz JV, Dujardin D, Godet J, Didier P, De Mey J, Darlix JL, … de Rocquigny H (2010). HIV‐1 Vpr oligomerization but not that of Gag directs the interaction between Vpr and Gag. Journal of Virology, 84(3), 1585–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner SD, Feild AM, Colleman DV, & Hulme B (1971). New human papovavirus (BK) isolated from urine after renal transplantation. Lancet, 297, 1253–1257. [DOI] [PubMed] [Google Scholar]
- Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, … Wang D (2007). Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLOS Pathogens, 3(5), e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerits N, Johannessen M, Tümmler C, Walquist M, Kostenko S, Snapkov I, … Moens U (2015). Agnoprotein of polyomavirus BK interacts with proliferating cell nuclear antigen and inhibits DNA replication. Virology Journal, 12, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerits N, & Moens U (2012). Agnoprotein of mammalian polyomaviruses. Virology, 432(2), 316–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert J, Nomura S, Anderson CW, & George K (1981). Identification of the SV40 agnoproduct: a DNA binding protein. Nature, 291, 346–349. [DOI] [PubMed] [Google Scholar]
- Giorda KM, Raghava S, & Hebert DN (2012). The simian virus 40 late viral protein VP4 disrupts the nuclear envelope for viral release. Journal of Virology, 86(6), 3180–3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik L, Goelz S, & Sandrock AW (2009). Asymptomatic reactivation of JC virus in patients treated with natalizumab. The New England Journal of Medicine, 361, 2487–2488 author reply 2489‐2490 [DOI] [PubMed] [Google Scholar]
- Gorelik L, Reid C, Testa M, Brickelmaier M, Bossolasco S, Pazzi A, … Cinque P (2011). Progressive multifocal leukoencephalopathy (PML) development is associated with mutations in JC virus capsid protein VP1 that change its receptor specificity. Journal of Infectious Diseases, 204(1), 103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb AB, Krueger JG, Wittkowski K, Dedrick R, Walicke PA, & Garovoy M (2002). Psoriasis as a model for T‐cell‐mediated disease: Immunobiologic and clinical effects of treatment with multiple doses of efalizumab, an anti‐CD11a antibody. Archives of Dermatology, 138(5), 591–600. [DOI] [PubMed] [Google Scholar]
- Hannafon B, & Ding W‐Q (2013). Intercellular communication by exosome‐derived microRNAs in cancer. International Journal of Molecular Sciences, 14(7), 14240–14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartlieb B, Modrof J, Mühlberger E, Klenk HD, & Becker S (2003). Oligomerization of Ebola virus VP30 is essential for viral transcription and can be inhibited by a synthetic peptide. Journal of Biological Chemistry, 278(43), 41830–41836. [DOI] [PubMed] [Google Scholar]
- Hay N, Kessler M, & Aloni Y (1984). SV40 deletion mutant (d1861) with agnoprotein shortened by four amino acids. Virology, 137(1), 160–170. [DOI] [PubMed] [Google Scholar]
- He L, & Hannon GJ (2004). MicroRNAs: Small RNAs with a big role in gene regulation. Nature Reviews Genetics, 5(7), 522–531. [DOI] [PubMed] [Google Scholar]
- Henriksen S, Hansen T, Bruun J‐A, & Rinaldo CH (2016). The presumed polyomavirus viroporin VP4 of simian virus 40 or human BK polyomavirus is not required for viral progeny release. Journal of Virology, 90(22), 10398–10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidaka K, Hojo K, Fujioka S, Nukuzuma S, & Tsuda Y (2015). Oligomerization of neutral peptides derived from the JC virus agnoprotein through a cysteine residue. Amino Acids, 47(10), 2205–2213. [DOI] [PubMed] [Google Scholar]
- Hock M, Kraus I, Schoehn G, Jamin M, Andrei‐Selmer C, Garten W, & Weissenhorn W (2010). RNA induced polymerization of the Borna disease virus nucleoprotein. Virology, 397(1), 64–72. [DOI] [PubMed] [Google Scholar]
- Hou‐Jong MH, Larsen SH, & Roman A (1987). Role of the agnoprotein in regulation of simian virus 40 replication and maturation pathways. Journal of Virology, 61(3), 937–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichaso N, & Dilworth SM (2001). Cell transformation by the middle T‐antigen of polyoma virus. Oncogene, 20(54), 7908–7916. [DOI] [PubMed] [Google Scholar]
- Imperiale MJ, & Major EO (2007). Polyomaviruses In Knipe DM & Howley PM (Eds.), Fields virology (5th ed., pp. 2263–2298). Philadelphia, PA: Lippincott Williams and Wilkins. [Google Scholar]
- Iwakawa HO, & Tomari Y (2015). The functions of microRNAs: mRNA decay and translational repression. Trends in Cell Biology, 25(11), 651–665. [DOI] [PubMed] [Google Scholar]
- Jackson AC (2018). JC virus infection: An expanding spectrum of neurological disorders. Canadian Journal of Neurological Sciences, 45(4), 365–366. [DOI] [PubMed] [Google Scholar]
- Johannessen M, Myhre MR, Dragset M, Tümmler C, & Moens U (2008). Phosphorylation of human polyomavirus BK agnoprotein at Ser‐11 is mediated by PKC and has an important regulative function. Virology, 379(1), 97–109. [DOI] [PubMed] [Google Scholar]
- Johannessen M, Walquist M, Gerits N, Dragset M, Spang A, & Moens U (2011). BKV agnoprotein interacts with alpha‐soluble N‐ethylmaleimide‐sensitive fusion attachment protein, and negatively influences transport of VSVG‐EGFP. PLOS One, 6(9), e24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaniowska D, Kaminski R, Amini S, Radhakrishnan S, Rappaport J, Johnson E, … Darbinyan A (2006). Cross-interaction between JC virus agnoprotein and human immunodeficiency virus type 1 (HIV-1) Tat modulates transcription of the HIV-1 long terminal repeat in glial cells. J Virol, 80(18), 9288–9299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili K, Sariyer IK, & Safak M (2008). Small tumor antigen of polyomaviruses: Role in viral life cycle and cell transformation. Journal of Cellular Physiology, 215(2), 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili K, White MK, Sawa H, Nagashima K, & Safak M (2005). The agnoprotein of polyomaviruses: A multifunctional auxiliary protein. Journal of Cellular Physiology, 204(1), 1–7. [DOI] [PubMed] [Google Scholar]
- Kitayama H, Miura Y, Ando Y, Hoshino S, Ishizaka Y, & Koyanagi Y (2008). Human immunodeficiency virus type 1 Vpr inhibits axonal outgrowth through induction of mitochondrial dysfunction. Journal of Virology, 82(5), 2528–2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmidt‐DeMasters BK, & Tyler KL (2005). Progressive multifocal leukoencephalopathy complicating treatment with natalizumab and interferon beta‐1a for multiple sclerosis. New England Journal of Medicine, 353(4), 369–374. [DOI] [PubMed] [Google Scholar]
- Kolodziejski PJ, Rashid MB, & Eissa NT (2003). Intracellular formation of “undisruptable” dimers of inducible nitric oxide synthase. Proceedings of the National Academy of Sciences of the United States of America, 100(24), 14263–14268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korup S, Rietscher J, Calvignac‐Spencer S, Trusch F, Hofmann J, Moens U, … Ehlers B (2013). Identification of a novel human polyomavirus in organs of the gastrointestinal tract. PLOS One, 8(3), e58021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M, & Hirsh D (1987). A trans‐spliced leader sequence on actin mRNA in C. elegans. Cell, 49(6), 753–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak YT, Raney A, Kuo LS, Denial SJ, Temple BR, Garcia JV, & Foster JL (2010). Self‐association of the Lentivirus protein, Nef. Retrovirology, 7, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer‐Gould A, Atlas SW, Green AJ, Bollen AW, & Pelletier D (2005). Progressive multifocal leukoencephalopathy in a patient treated with natalizumab. New England Journal of Medicine, 353(4), 375–381. [DOI] [PubMed] [Google Scholar]
- Lee S, Paulson KG, Murchison EP, Afanasiev OK, Alkan C, Leonard JH, … Nghiem P (2011). Identification and validation of a novel mature microRNA encoded by the Merkel cell polyomavirus in human Merkel cell carcinomas. Journal of Clinical Virology, 52(3), 272–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DN, Refaeli Y, MacGregor RR, & Weiner DB (1994). Serum Vpr regulates productive infection and latency of human immunodeficiency virus type 1. Proceedings of the National Academy of Sciences of the United States of America, 91(23), 10873–10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim ES, Reyes A, Antonio M, Saha D, Ikumapayi UN, Adeyemi M, … Wang D (2013). Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology, 436(2), 295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Dai E, Miller L, Seet B, Lalani A, Macauley C, … Lucas A (2004). Viral chemokine‐binding proteins inhibit inflammatory responses and aortic allograft transplant vasculopathy in rat models. Transplantation, 77(11), 1652–1660. [DOI] [PubMed] [Google Scholar]
- Liu L, Lalani A, Dai E, Seet B, Macauley C, Singh R, … Lucas A (2000). The viral anti‐inflammatory chemokine‐binding protein M‐T7 reduces intimal hyperplasia after vascular injury. Journal of Clinical Investigation, 105(11), 1613–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas A, Liu L, Macen J, Nash P, Dai E, Stewart M, … McFadden G (1996). Virus‐encoded serine proteinase inhibitor SERP‐1 inhibits atherosclerotic plaque development after balloon angioplasty. Circulation, 94(11), 2890–2900. [DOI] [PubMed] [Google Scholar]
- Luganini A, Terlizzi ME, & Gribaudo G (2016). Bioactive molecules released from cells infected with the human cytomegalovirus. Frontiers in Microbiology, 7, 715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch KJ, & Frisque RJ (1990). Identification of critical elements within the JC virus DNA replication origin. Journal of Virology, 64(12), 5812–5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova JA, Shkurnikov MU, Wicklein D, Lange T, Samatov TR, Turchinovich AA, & Tonevitsky AG (2016). Intracellular and extracellular microRNA: An update on localization and biological role. Progress in Histochemistry and Cytochemistry, 51(3‐4), 33–49. [DOI] [PubMed] [Google Scholar]
- Marianayagam NJ, Sunde M, & Matthews JM (2004). The power of two: Protein dimerization in biology. Trends in Biochemical Sciences, 29 (11), 618–625. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, & Jones DT (2000). The PSIPRED protein structure prediction server. Bioinformatics, 16(4), 404–405. [DOI] [PubMed] [Google Scholar]
- van der Meijden E, Janssens RWA, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, & Feltkamp MCW (2010). Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromized patient. PLOS Pathogens, 6(7), e1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meijden E, Kazem S, Dargel CA, van Vuren N, Hensbergen PJ, & Feltkamp MCW (2015). Characterization of T Antigens, Including Middle T and Alternative T, Expressed by the Human Polyomavirus Associated with Trichodysplasia Spinulosa. J Virol, 89(18), 9427–9439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabova N, Kaminski R, Krynska B, Amini S, Khalili K, & Darbinyan A (2012). JCV agnoprotein‐induced reduction in CXCL5/LIX secretion by oligodendrocytes is associated with activation of apoptotic signaling in neurons. Journal of Cellular Physiology, 227(8), 3119–3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merabova N, Kaniowska D, Kaminski R, Deshmane SL, White MK, Amini S, … Khalili K (2008). JC virus agnoprotein inhibits in vitro differentiation of oligodendrocytes and promotes apoptosis. Journal of Virology, 82(3), 1558–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N, Pereira M, Rhodes RH, An P, Pipas JM, Jain K, … Lipkin WI (2014). Identification of a novel polyomavirus in a pancreatic transplant recipient with retinal blindness and vasculitic myopathy. Journal of Infectious Diseases, 210(10), 1595–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco MC, Jensen PN, Hou J, Durham LC, & Major EO (1998). Detection of JC virus DNA in human tonsil tissue: Evidence for site of initial viral infection. Journal of Virology, 72(12), 9918–9923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukerjee R, Chang JR, Del Valle L, Bagashev A, Gayed MM, Lyde RB, … Sawaya BE (2011). Deregulation of microRNAs by HIV‐1 Vpr protein leads to the development of neurocognitive disorders. Journal of Biological Chemistry, 286(40), 34976–34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller DA, & Young PR (2013). The flavivirus NS1 protein: Molecular and structural biology, immunology, role in pathogenesis and application as a diagnostic biomarker. Antiviral Research, 98(2), 192–208. [DOI] [PubMed] [Google Scholar]
- Myhre MR, Olsen GH, Gosert R, Hirsch HH, & Rinaldo CH (2010). Clinical polyomavirus BK variants with agnogene deletion are nonfunctional but rescued by trans‐complementation. Virology, 398(1), 12–20. [DOI] [PubMed] [Google Scholar]
- Ng SC, Mertz JE, Sanden‐Will S, & Bina M (1985). Simian virus 40 maturation in cells harboring mutants deleted in the agnogene. Journal of Biological Chemistry, 260(2), 1127–1132. [PubMed] [Google Scholar]
- Nomura S, Khoury G, & Jay G (1983). Subcellular localization of the simian virus 40 agnoprotein. Journal of Virology, 45(1), 428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Endo S, Takahashi H, Sawa H, Umemura T, & Nagashima K (2001). Distribution and function of JCV agnoprotein. Journal of Neurovirology, 7(4), 302–306. [DOI] [PubMed] [Google Scholar]
- Okada Y, Sawa H, Endo S, Orba Y, Umemura T, Nishihara H, … Nagashima K (2002). Expression of JC virus agnoprotein in progressive multifocal leukoencephalopathy brain. Acta Neuropathologica, 104(2), 130–136. [DOI] [PubMed] [Google Scholar]
- Okada Y, Suzuki T, Sunden Y, Orba Y, Kose S, Imamoto N, … Sawa H (2005). Dissociation of heterochromatin protein 1 from lamin B receptor induced by human polyomavirus agnoprotein: Role in nuclear egress of viral particles. EMBO Reports, 6(5), 452–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otlu O, De Simone FI, Otalora YL, Khalili K, & Sariyer IK (2014). The agnoprotein of polyomavirus JC is released by infected cells: Evidence for Its cellular uptake by uninfected neighboring cells. Virology, 468‐470, 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou HD, Kwiatkowski W, Deerinck TJ, Noske A, Blain KY, Land HS, … O’Shea CC (2012). A structural basis for the assembly and functions of a viral polymer that inactivates multiple tumor suppressors. Cell, 151(2), 304–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett BL, Zu Rhein GM, Walker DL, Echroade R, & Dessel B (1971). Cultivation of papova‐like virus from human brain with progressive multifocal leukoencephalopathy. Lancet, 297, 1257–1260. [DOI] [PubMed] [Google Scholar]
- Panou M‐M, Prescott EL, Hurdiss DL, Swinscoe G, Hollinshead M, Caller LG, … Macdonald A (2018). Agnoprotein is an essential egress factor during BK polyomavirus infection. International Journal of Molecular Sciences, 19(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Pasquier RA, Corey S, Margolin DH, Williams K, Pfister LA, De Girolami U, … Koralnik IJ (2003). Productive infection of cerebellar granule cell neurons by JC virus in an HIV+ individual. Neurology, 61(6), 775–782. [DOI] [PubMed] [Google Scholar]
- Pastrana DV, Fitzgerald PC, Phan GQ, Raiji MT, Murphy PM, McDermott DH, … Buck CB. (2013). A divergent variant of the eleventh human polyomavirus species, saint louis polyomavirus. Genome Announcements, 1(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard VW, & Malim MH (1998). The HIV‐1 rev protein. Annual Review of Microbiology, 52, 491–532. [DOI] [PubMed] [Google Scholar]
- Prins C, & Frisque RJ (2001). JC virus T’ proteins encoded by alternatively spliced early mRNAs enhance T antigen‐mediated viral DNA replication in human cells. Journal of Neurovirology, 7(3), 250–264. [DOI] [PubMed] [Google Scholar]
- Raghava S, Giorda KM, Romano FB, Heuck AP, & Hebert DN (2011). The SV40 late protein VP4 is a viroporin that forms pores to disrupt membranes for viral release. PLOS Pathogens, 7(6), e1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghava S, Giorda KM, Romano FB, Heuck AP, & Hebert DN (2013). SV40 late protein VP4 forms toroidal pores to disrupt membranes for viral release. Biochemistry, 52(22), 3939–3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randhawa P, Zygmunt D, Shapiro R, Vats A, Weck K, Swalsky P, & Finkelstein S (2003). Viral regulatory region sequence variations in kidney tissue obtained from patients with BK virus nephropathy. Kidney International, 64(2), 743–747. [DOI] [PubMed] [Google Scholar]
- Reid CE, Li H, Sur G, Carmillo P, Bushnell S, Tizard R, … Carulli JP (2011). Sequencing and analysis of JC virus DNA From Natalizumabtreated PML patients. Journal of Infectious Diseases, 204(2), 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick J, & Shenk T (1986). Simian virus 40 agnoprotein facilitates normal nuclear location of the major capsid polypeptide and cell‐tocell spread of virus. Journal of Virology, 60(3), 1098–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley MI, Yoo W, Mda NY, & Folk WR (1997). Tiny T antigen: An autonomous polyomavirus T antigen amino‐terminal domain. Journal of Virology, 71(8), 6068–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldo CH, & Hirsch HH (2007). Antivirals for the treatment of polyomavirus BK replication. Expert Review of Anti‐Infective Therapy, 5(1), 105–115. [DOI] [PubMed] [Google Scholar]
- Rinaldo CH, Traavik T, & Hey A (1998). The agnogene of the human polyomavirus BK is expressed. Journal of Virology, 72(7), 6233–6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, & Zhang Y (2010). I‐TASSER: A unified platform for automated protein structure and function prediction. Nature Protocols, 5(4), 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundell K, Hearing P, & Yang YC (1980). SV40 17K protein is associated with two cellular proteins. Cold Spring Harbor Symposia on Quantitative Biology, 44(Pt 1), 211–214. [DOI] [PubMed] [Google Scholar]
- Safak M, Barrucco R, Darbinyan A, Okada Y, Nagashima K, & Khalili K (2001). Interaction of JC virus agno protein with T antigen modulates transcription and replication of the viral genome in glial cells. Journal of Virology, 75(3), 1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safak M, Gallia GL, Ansari SA, & Khalili K (1999). Physical and functional interaction between the Y‐box binding protein YB‐1 and human polyomavirus JC virus large T antigen. Journal of Virology, 73 (12), 10146–10157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safak M, Gallia GL, & Khalili K (1999). A 23‐bp sequence element from human neurotropic JC virus is responsive to NF‐kappa B subunits. Virology, 262(1), 178–189. [DOI] [PubMed] [Google Scholar]
- Safak M, & Khalili K (2001). Physical and functional interaction between viral and cellular proteins modulate JCV gene transcription. Journal of Neurovirology, 7(4), 288–292. [DOI] [PubMed] [Google Scholar]
- Safak M, Sadowska B, Barrucco R, & Khalili K (2002). Functional interaction between JC virus late regulatory agnoprotein and cellular Y‐box binding transcription factor, YB‐1. Journal of Virology, 76(8), 3828–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Abou‐Gharbia M, Childers W, Sariyer IK, White MK, & Safak M (2013). Essential roles of Leu/Ile/Phe‐rich domain of JC virus agnoprotein in dimer/oligomer formation, protein stability and splicing of viral transcripts. Virology, 443(1), 161–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Arachea BT, White MK, Viola RE, & Safak M (2011). Human polyomavirus JC small regulatory agnoprotein forms highly stable dimers and oligomers: Implications for their roles in agnoprotein function. Virology, 420(1), 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, Coric P, Hamazaspyan A, Davis W, Axman R, White MK, … Safak M (2016). Emerging from the unknown: Structural and functional features of agnoprotein of polyomaviruses. Journal of Cellular Physiology, 231(10), 2115–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, DeVoto J, Golla A, Wollebo HS, White MK, & Safak M (2018). Discovery and characterization of novel trans‐spliced products of human polyoma JC virus late transcripts from PML patients. Journal of Cellular Physiology, 233(5), 4137–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, White MK, & Safak M (2012). JC virus agnoprotein enhances large T antigen binding to the origin of viral DNA replication: Evidence for its involvement in viral DNA replication. Virology, 433(1), 12–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribas AS, White MK, & Safak M (2018). Structure‐based release analysis of the JC virus agnoprotein regions: A role for the hydrophilic surface of the major alpha helix domain in release. Journal of Cellular Physiology, 233(3), 2343–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saribaş AS, Özdemir A, Lam C, & Safak M (2010). JC virus‐induced progressive multifocal leukoencephalopathy. Future Virology, 5(3), 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyer IK, Akan I, Palermo V, Gordon J, Khalili K, & Safak M (2006). Phosphorylation mutants of JC virus agnoprotein are unable to sustain the viral infection cycle. Journal of Virology, 80(8), 3893–3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyer IK, Khalili K, & Safak M (2008). Dephosphorylation of JC virus agnoprotein by protein phosphatase 2A: Inhibition by small t antigen. Virology, 375(2), 464–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariyer IK, Saribas AS, White MK, & Safak M (2011). Infection by agnoprotein‐negative mutants of polyomavirus JC and SV40 results in the release of virions that are mostly deficient in DNA content. Virology Journal, 8, 255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, & Buck CB (2010). Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host & Microbe, 7(6), 509–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scuda N, Hofmann J, Calvignac‐Spencer S, Ruprecht K, Liman P, Kuhn J, … Ehlers B (2011). A novel human polyomavirus closely related to the African green monkey‐derived lymphotropic polyomavirus. Journal of Virology, 85(9), 4586–4590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo GJ, Chen CJ, & Sullivan CS (2009). Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology, 383(2), 183–187. [DOI] [PubMed] [Google Scholar]
- Seo GJ, Fink LHL, O’Hara B, Atwood WJ, & Sullivan CS (2008). Evolutionarily conserved function of a viral microRNA. Journal of Virology, 82(20), 9823–9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shishido‐Hara Y, Hara Y, Larson T, Yasui K, Nagashima K, & Stoner GL (2000). Analysis of capsid formation of human polyomavirus JC (Tokyo‐1 strain) by a eukaryotic expression system: Splicing of late RNAs, translation and nuclear transport of major capsid protein VP1, and capsid assembly. Journal of Virology, 74(4), 1840–1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebrasse EA, Reyes A, Lim ES, Zhao G, Mkakosya RS, Manary MJ, … Wang D (2012). Identification of MW polyomavirus, a novel polyomavirus in human stool. Journal of Virology, 86(19), 10321–10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DT, Loeber G, & Tegtmeyer P (1990). Four major sequence elements of simian virus 40 large T antigen coordinate its specific and nonspecific DNA binding. Journal of Virology, 64(5), 1973–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith VP, Bryant NA, & Alcami A (2000). Ectromelia, vaccinia and cowpox viruses encode secreted interleukin‐18‐binding proteins. Journal of General Virology, 81(Pt 5), 1223–1230. [DOI] [PubMed] [Google Scholar]
- Soeda E, Arrand JR, Smolar N, & Griffin BE (1979). Sequence from early region of polyoma virus DNA containing viral replication origin and encoding small, middle and (part of) large T antigens. Cell, 17(2), 357–370. [DOI] [PubMed] [Google Scholar]
- Soleimani‐Meigooni DN, Schwetye KE, Angeles MR, Ryschkewitsch CF, Major EO, Dang X, … Bucelli RC (2017). JC virus granule cell neuronopathy in the setting of chronic lymphopenia treated with recombinant interleukin‐7. Journal of Neurovirology, 23(1), 141–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart SE, Eddy BE, & Borgese N (1958). Neoplasms in mice inoculated with a tumor agent carried in tissue culture. Jo urnal of the National Cancer Institute, 20(6), 1223–1243. [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, & Ganem D (2005). SV40‐encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature, 435(7042), 682–686. [DOI] [PubMed] [Google Scholar]
- Sullivan CS, Sung CK, Pack CD, Grundhoff A, Lukacher AE, Benjamin TL, & Ganem D (2009). Murine Polyomavirus encodes a microRNA that cleaves early RNA transcripts but is not essential for experimental infection. Virology, 387(1), 157–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RE, & Boothroyd JC (1986). Evidence for trans splicing in trypanosomes. Cell, 47(4), 527–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Okada Y, Semba S, Orba Y, Yamanouchi S, Endo S, … Sawa H (2005). Identification of FEZ1 as a protein that interacts with JC virus agnoprotein and microtubules: Role of agnoprotein‐induced dissociation of FEZ1 from microtubules in viral propagation. Journal of Biological Chemistry, 280(26), 24948–24956. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Orba Y, Makino Y, Okada Y, Sunden Y, Hasegawa H, … Sawa H (2013). Viroporin activity of the JC polyomavirus is regulated by interactions with the adaptor protein complex 3. Proceedings of the National Academy of Sciences of the United States of America, 110(46), 18668–18673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Orba Y, Okada Y, Sunden Y, Kimura T, Tanaka S, … Sawa H (2010). The human polyoma JC virus agnoprotein acts as a viroporin. PLOS Pathogens, 6(3), e1000801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Semba S, Sunden Y, Orba Y, Kobayashi S, Nagashima K, … Sawa H (2012). Role of JC virus agnoprotein in virion formation. Microbiology and Immunology, 56(9), 639–646. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Tahara H, Narula S, Moore KW, Robbins PD, & Lotze MT (1995). Viral interleukin 10 (IL‐10), the human herpes virus 4 cellular IL‐10 homologue, induces local energy to allogeneic and syngeneic tumors. Journal of Experimental Medicine, 182(2), 477–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet BH, & Hilleman MR (1960). The vacuolating virus, S.V. 40. Proceedings of the Society for Experimental Biology and Medicine Society for Experimental Biology and Medicine (New York, N Y), 105, 420–427. [DOI] [PubMed] [Google Scholar]
- Tange S, Imai T, & Nakanishi A (2011). An SV40 mutant defective in VP4 expression exhibits a temperature‐sensitive growth defect. Virus Research, 157(1), 116–120. [DOI] [PubMed] [Google Scholar]
- Tian Y‐C, Li Y‐J, Chen H‐C, Wu H‐H, Weng C‐H, Chen Y‐C, … Yang C‐W (2014). Polyomavirus BK‐encoded microRNA suppresses autoregulation of viral replication. Biochemical and Biophysical Research Communications, 447(3), 543–549. [DOI] [PubMed] [Google Scholar]
- Tobinai K, Kobayashi Y, Narabayashi M, Ogura M, Kagami Y, Morishima Y, … Murate T (1998). Feasibility and pharmacokinetic study of a chimeric anti‐CD20 monoclonal antibody (IDEC‐C2B8, rituximab) in relapsed B‐cell lymphoma. The IDEC‐C2B8 Study Group. Annals of Oncology, 9(5), 527–534. [DOI] [PubMed] [Google Scholar]
- Trowbridge PW, & Frisque RJ (1995). Identification of three new JC virus proteins generated by alternative splicing of the early viral mRNA. Journal of Neurovirology, 1(2), 195–206. [DOI] [PubMed] [Google Scholar]
- Turchinovich A, & Cho WC (2014). The origin, function and diagnostic potential of extracellular microRNA in human body fluids. Frontiers in Genetics, 5, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich A, Weiz L, Langheinz A, & Burwinkel B (2011). Characterization of extracellular circulating microRNA. Nucleic Acids Research, 39(16), 7223–7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterstab G, Gosert R, Leuenberger D, Lorentz P, Rinaldo CH, & Hirsch HH (2010). The polyomavirus BK agnoprotein co‐localizes with lipid droplets. Virology, 399(2), 322–331. [DOI] [PubMed] [Google Scholar]
- Del Valle L, Gordon J, Enam S, Delbue S, Croul S, Abraham S, … Khalili K. (2002). Expression of human neurotropic polyomavirus JCV late gene product agnoprotein in human medulloblastoma. Journal of the National Cancer Institute, 94(4), 267–273. [DOI] [PubMed] [Google Scholar]
- Walsh G, & Jefferis R (2006). Post‐translational modifications in the context of therapeutic proteins. Nature Biotechnology, 24(10), 1241–1252. [DOI] [PubMed] [Google Scholar]
- Windheim M, Southcombe JH, Kremmer E, Chaplin L, Urlaub D, Falk CS, … Burgert H‐G (2013). A unique secreted adenovirus E3 protein binds to the leukocyte common antigen CD45 and modulates leukocyte functions. Proceedings of the National Academy of Sciences of the United States of America, 110(50), E4884–E4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich C, Batson S, Anderson MP, White LR, & Koralnik IJ (2016). JC virus infects neurons and glial cells in the hippocampus. Journal of Neuropathology and Experimental Neurology, 75, 712–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wüthrich C, Cheng YM, Joseph JT, Kesari S, Beckwith C, Stopa E, … Koralnik IJ (2009). Frequent infection of cerebellar granule cell neurons by polyomavirus JC in progressive multifocal leukoencephalopathy. Journal of Neuropathology and Experimental Neurology, 68(1), 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Sun Y, & Zhang H (2001). The multimerization of human immunodeficiency virus type I Vif protein: A requirement for Vif function in the viral life cycle. Journal of Biological Chemistry, 276(7), 4889–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YC, Hearing P, & Rundell K (1979). Cellular proteins associated with simian virus 40 early gene products in newly infected cells. Journal of Virology, 32(1), 147–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Greninger AL, Isa P, Phan TG, Martínez MA, de la Luz Sanchez M, Chiu CY (2012). Discovery of a novel polyomavirus in acute diarrheal samples from children. PLOS One, 7(11), e49449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerrahn J, Knippschild U, Winkler T, & Deppert W (1993). Independent expression of the transforming amino‐terminal domain of SV40 large I antigen from an alternatively spliced third SV40 early mRNA. EMBO Journal, 12(12), 4739–4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li S, Li L, Li M, Guo C, Yao J, & Mi S (2015). Exosome and exosomal microRNA: Trafficking, sorting, and function. Genomics, Proteomics & Bioinformatics, 13(1), 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao LJ, Wang L, Mukherjee S, & Narayan O (1994a). Biochemical mechanism of HIV‐1 Vpr function. Oligomerization mediated by the N‐terminal domain. Journal of Biological Chemistry, 269(51), 32131–32137. [PubMed] [Google Scholar]
- Zhao LJ, Wang L, Mukherjee S, & Narayan O (1994b). Biochemical mechanism of HIV‐1 Vpr function. Oligomerization mediated by the Nterminal domain. Journal of Biological Chemistry, 269(51), 32131–32137. [PubMed] [Google Scholar]
- Zhou AY, Ichaso N, Adamarek A, Zila V, Forstova J, Dibb NJ, & Dilworth SM (2011). Polyomavirus middle T‐antigen is a transmembrane protein that binds signaling proteins in discrete subcellular membrane sites. Journal of Virology, 85(7), 3046–3054. [DOI] [PMC free article] [PubMed] [Google Scholar]