Abstract
Microfluidic devices have advanced significantly in recent years and are a promising technology for the field of tissue engineering. Highly sophisticated microfabrication techniques have paved the way for the development of complex ex vivo models capable of incorporating and measuring the real-time response of multiple cell types interacting together in a single system. Muscle-on-a-chip technology has drastically improved and serves as a drug screening platform for many muscular diseases such as muscular dystrophy, tendinosis, fibromyalgia, mitochondrial myopathy, and myasthenia gravis. This review seeks to communicate the gaps in knowledge of current muscular disease models and highlight the power of microfluidic devices in enabling researchers to better understand disease pathology and provide high throughput screening of therapeutics for muscular myopathies.
Keywords: muscle, microfluidics, muscle-on-a-chip, ex vivo system, myopathy
Introduction
Muscle cells are the most abundant cell type in the body. The primary function of muscle is to generate force; however, when muscle is damaged or diseased, normal functions are compromised1.There are many musculoskeletal diseases that tissue engineers are working to better understand and for which they seek to find new and improved treatments. The ultimate goal for most tissue engineers is to translate their research into a clinical setting. However, many steps must be taken before new products are tested on humans. Much of the research in muscle is conducted in 2D cell culture, but many physiological conditions are absent in these circumstances and thus do not translate well between 2D culture and in vivo work. For example, satellite cells have demonstrated high regenerative potential in vivo, but lose their regenerative capacity when cultured in vitro2. For this reason, many scientists look to animal models, often transgenic mice, to study the local and systemic physiological effects of treatments. While these tests satisfy many of the safety guidelines currently in place for preclinical testing, they do not always paint an accurate picture of how the device will perform in humans. For example, transgenic mice for muscular dystrophy (C57BL/10ScSn mdx) reveal the same genetic and biochemical expression as humans with the disease1. However, the effects of the mutation are significantly lessened in mice, resulting in almost normal mechanical function and lifespan1. Another example is seen in mouse models for myasthenia gravis, an autoimmune disease that targets neuromuscular junctions3. While mouse models can replicate the genetic markers and phenotypic responses of the disease, the conditions in which the animals are bred and housed lead to significantly different immune responses than observed in humans3. Additionally, animal models may be difficult to work with, expensive to operate, and the data that is gathered is not always clear4. An attractive alternative is 2D culture using human cells. These models allow for high throughput assays, provide a clearer picture of individual components that are being tested, and the cellular response is more translatable to in vivo human studies compared to mouse models4. On the other hand, 2D culture does not allow for the incorporation of an accurate microenvironment and subsequent cell and extracellular matrix (ECM) interactions that would be found in vivo. Muscle cells often grow in disorganized sheets in vitro rather than in highly organized structures as they do in vivo5,6. For this reason, ex vivo models have been widely explored in the last several years. Ex vivo models have the advantage of high throughput analysis as in 2D culture, but with the added advantage of mimicking the physicochemical cues found in vivo7–9.
Significant advances have been made in the field of microfluidics in the last several years. Ex vivo microfluidic devices have been used to recreate the cellular microenvironment and allow for real-time analysis of cellular behavior and responses to external stimuli5,10–12. Microfluidic devices have been implemented to induce stem cell differentiation5,13–15, study organ development and function16,17, mimic cardiac muscle15,18,19, achieve innervation20–22, study tumorigenesis10, and induce the formation of vascular-like structures23. The optically clear materials that the devices are made from allow for colorimetric scans and imaging without disturbing the samples, and the fluid flow in the systems allows for the isolation of factors excreted by the cells or markers for gene expression to be monitored in real-time10. Microfluidics also allow for rapid drug screening in disease models and provide controlled variables, such as shear stress, for cell culture5,12,16. Many in vitro disease models do not incorporate multiple cells types, factor presentation, and mechanical and electrical stimulation seen in vivo10,12,24. As shown in Figure 1, ex vivo microfluidic devices allow for all of these criteria to be incorporated, and also allow for their impact on cell behavior and response to therapeutics to be isolated and studied10,12,16,17. As a result, microfluidic devices show promise for the future of tissue engineering and epidemiology. This review aims to present an overview of current musculoskeletal models with the intention of communicating the promise of ex vivo microfluidics to create improved muscular disease models.
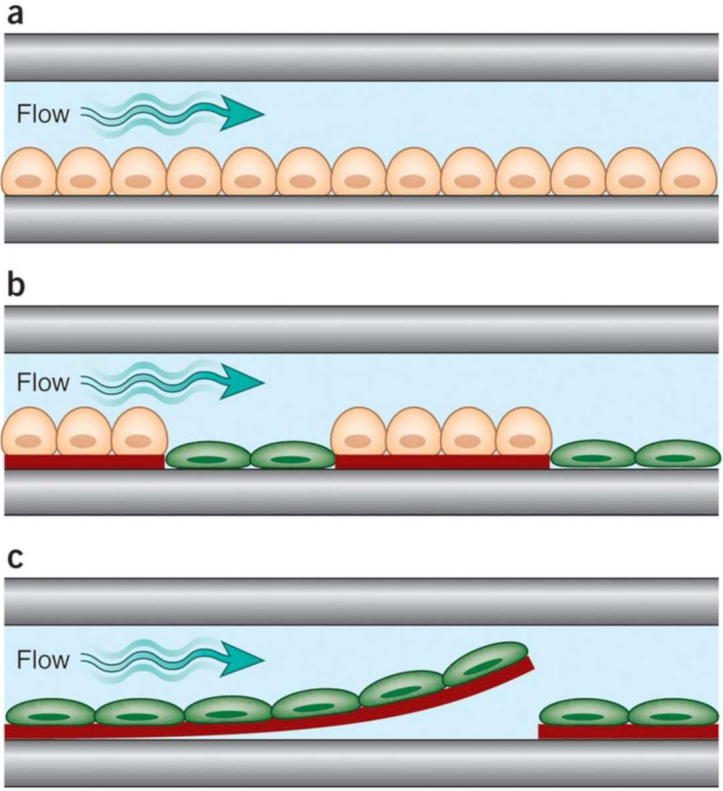
Figure 1.
Examples of increasingly complex single-channel, organ-on-chip designs. (a) Cells of a single type are cultured as a monolayer on a planar rigid (e.g., glass) or flexible [e.g., Poly(dimethyl siloxane) (PDMS)] substrate on one side of a microfluidic channel through which medium is perfused. (b) Cells of two types are cultured in direct juxtaposition by micropatterning ECM adhesive islands within the microfluidic chamber that preferentially support one cell population (e.g., hepatocyte). These cells are delivered first, and the empty spaces are then filled with the second cell population (e.g., fibroblast). (c) Cells in a tissue construct engineered with ECM are cultured in a microfluidic channel. In this example, microcontact printing of ECM in a linear pattern on a thin PDMS layer coated over the substrate is used to orient muscle cells to create an anisotropic muscle tissue layer. When parts of the PDMS film are released from the substrate, they bend up when the cells contract, allowing measurement of cell contraction forces under flow (reproduced with permission from Bhatia, et al., 201412).
Building a Musculoskeletal Model
Myocytes can be cultured in vitro with varying levels of success. Myocyte maturity is often terminated early in development before reaching an adult phenotype and eliciting a myogenic response. It has been well-documented that muscle cell maturation and differentiation rely on multiple cues, including substrate stiffness 14,25, mechanical stimulation 26,27, and transcription factors28,29 as depicted in Figure 2. These cues often originate from, or are influenced by, biomaterials. The phenotype and differentiation fate of muscle cells is heavily dependent on the mechanical properties of the substrate on which it is cultured. In 2D cultures, cells grow in disorganized sheets on substrates with very different mechanical properties than those experienced in vivo. Jacot et al. has shown that myocyte phenotype and contractile force generation reaches a maximum when cultured on substrates with stiffnesses of around 10 kPa, which is in the range of native muscle30. This group used various compositions of polyacrylamide scaffolds to grow and test primary myocytes. Many hydrogels or soft polymers used in ex vivo models and microfluidic devices are designed to be easily mechanically tuned so that the differentiation of multiple cell types can be controlled through these material mechanical cues. Modulation of mechanical properties is advantageous to provide appropriate signals for myocyte precursors and to control the fate of the cell population.
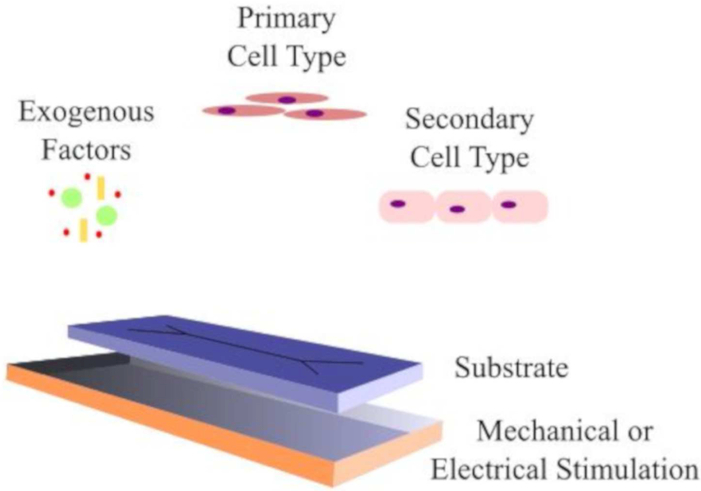
Figure 2.
Representative ex vivo muscle model containing essential components for myogenic growth and differentiation.
Another method for modulating cellular cues and promoting a myogenic phenotype is the introduction of mechanical stimulation into an ex vivo system. As shown in Figure 3, mechanical stimulation paired with the innate mechanical cues provided by the biomaterial substrate better mimics the dynamic in vivo environment in which cells simultaneously experience stretch, compression and shear flow from the ECM10,12,24. Powell et al. applied mechanical stimulation to a 3D culture system consisting of human primary skeletal muscle cells suspended in collagen/Matrigel hydrogels31. Matrigel systems are often chosen because they provide essential cell-binding motifs and inherent growth factors. Collagen was incorporated to provide mechanical integrity to the scaffold while maintaining biocompatibility. The muscle cells and collagen/Matrigel constructs were then placed in a silicon mechanical cell stimulator device and exposed to unidirectional mechanical loading to promote formation of aligned multinucleated myofibers. A similar response can also be seen by applying electrical stimulation rather than mechanical stimulation. Electrical stimulation can act in place of neuronal stimulation within the muscle mimic system32. Langelaan et al. demonstrated that electrical stimulation promoted faster myotube formation, mature phenotype and myogenic behavior, and increased culture length of mature myocytes32. This group explored the effects of electrical pulses in C2C12 murine fibroblasts and muscle progenitor cells grown in 2D and in 3D in collagen/Matrigel hydrogels. Overall, electrical stimulation improved myogenic behavior with the most success seen in muscle progenitor cells grown in 3D hydrogels. These studies demonstrate that selection of cell type, and electrical and mechanical stimulation are important when building a model for skeletal muscle. Ex vivo microfluidic models allow for the ability to incorporate and tune all of these criteria, demonstrating the platform’s promise in skeletal muscle tissue modeling10,12,24.
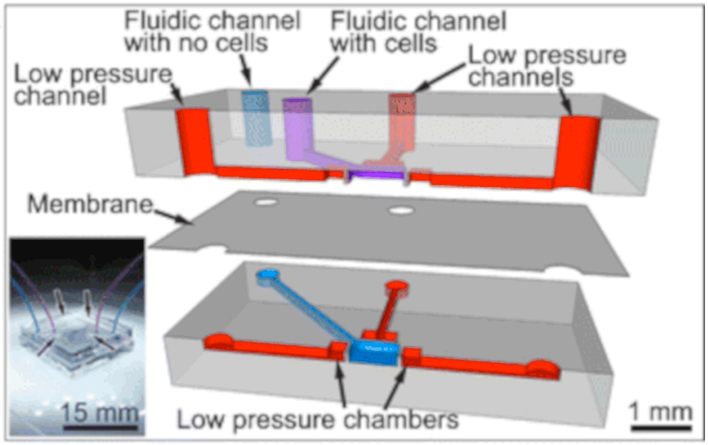
Figure 3.
Exploded cross-section of the multi-layer PDMS-based cell stretching device. Low pressure is applied to the low pressure channels (red) to induce a deformation in the walls located at each of the four sides of the cell stretching chamber (800 × 800 μm; 10 μm thick membrane). The top and bottom fluidic channels (purple and blue) are isolated from each other by a suspended membrane. The bottom fluidic channel (blue) serves to equilibrate pressures when seeding cells. Insert: Photographic image of the assembled device with the bottom and top fluidic channels (blue and purple channels) connected and the four low pressure channel inlets (reproduced with permission from Tremblay et al., 201468).
Transcription factors and chemical cues have also been shown to play an active role in myocyte culture and maturation33. There is a family of transcription factors known as the myogenic regulatory factors (MRFs) that play a role in regulating muscle differentiation. The skeletal muscle that becomes the limbs are derived from somites, which arise from the segmented paraxial mesoderm. This process involves a very finely controlled cascade of the four MRFs: MyoD, Myf5, Myogenin, and Myogenic Regulatory Factor 433. These transcription factors can be used in conjunction to control the proliferation and myogenic progression of muscle cells and precursor cells.
All of these components – substrate stiffness, external stimulation, and exogenous factors – play vital roles in muscle tissue modeling and can be finely tuned in ex vivo systems. The combination of biomaterial substrate selection, scaffold architecture, and external mechanical and electrical stimulation can be modulated to drive the appropriate cell responses34. Complex systems have been constructed to better mimic the native tissue environment, such as mimicking the native mineralization for bone tissue engineering35, presentation of vascular endothelial growth factor mimetic peptides to induce angiogenesis36, or the utilization of collagen mimetic peptides to promote endothelial cell growth37. Ex vivo microfluidic models show much promise for the future of disease modeling given their tunability; the systems can promote a more representative microenvironment by incorporating factors for mechanical, chemical, and electrical stimuli, as well as provide controlled presentation of factors.
Muscular System Diseases and Current Ex Vivo, In Vitro, and In Vivo Models
Muscular Dystrophy
Muscular dystrophy (MD) results from the disruption or deletion of the genes for dystrophin38. Along with other proteins, such as utrophin complex, dystrophin forms linkages between the cytoskeleton and the extracellular matrix in muscle tissue38–40. The loss of these connections results in separation of myoblasts from the ECM, causing muscle cell death and reduction in muscle mass for patients40. Mouse models are the most commonly used model for studying MD, but ex vivo cell models41,42 and zebrafish43,44 have also shown success. Gene therapy is most often utilized in exploring treatments because the disease involves genetic deletions or mutations39. These therapies are often developed ex vivo, to ensure proper transfection and modification of cells prior to injection back into the transgenic mouse model.
Filareto et al. used a more sophisticated MD mouse model that inhibits the expression of both dystrophin and utrophin and causes severe degeneration38. Genetically modified induced pluripotent stem cells (iPSCs) derived from fibroblasts were the target treatments. The iPSCs were modified ex vivo with gene sequence μUTRN, which has been shown to alleviate some of the symptoms of MD by targeting both dystrophin and utrophin production. The mouse model utilized presents more extreme symptoms so damage to the muscle tissue prior to injection was not necessary. Immunosuppressants were administered to alleviate the immune response associated with protein production. Treatment resulted in utrophin production and cell infiltration in the muscle treated with the genetically modified iPSCs. Due to intensive screening of the iPSCs prior to injection as well, tumor formation was not found to occur up to 3 months following the injection. Fatigue time and cross-sectional area of the muscle were not found to increase for the treated muscle, but contractility did improve38. The transfection was non-viral, and further production of dystrophin-related proteins would rely solely on the injected iPSCs and their resulting cells rather than native myoblasts.
Nelson et al. utilized CRISPR-Cas9 to genetically modify muscle cells in an MD mouse model39. The group used a viral vector for delivery and found specific deletion of the exon causing dystrophin disruption in ~2% of the alleles after 8 weeks. The mRNA demonstrated 59% deletion of the desired exon following transcription, resulting in ~8% dystrophin restoration in the muscle. The dystrophin-associated complexes were found to return in the muscle of interest. The muscles demonstrated increased twitch and spasm force, as well as improved resistance to damage under cyclic loading. Neonatal MD mice were also treated and showed improved muscle formation and behavior in the abdominal muscles, heart, and diaphragm after 7 weeks. Lastly, treatment was intravenously injected and dystrophin levels increased in the heart of the adult mice. Only 4% of normal dystrophin levels is reported to be necessary for improved muscle function and 30% of normal dystrophin levels would result in complete loss of symptoms in MD patients. Although only 2% of the alleles were modified, 8% of the dystrophin was regained, making CRISPR-Cas9 a promising tool for developing new MD therapies39. These gene editing methods can be easily implemented and tested in ex vivo microfluidic models and show promise for the future of MD disease and treatment testing.
Tendinosis
Tendinopathies, or tendinosis, are a class of diseases affecting the tendons and are characterized by inflammation of the tissue and tenocyte apoptosis45,46. Compositional changes in the tissue take place, including the upregulation of hypoxia induced factor HIF-1α and the overproduction of collagen type III. The role of apoptosis and hypoxia in tendinopathies has been debated within the field, but most scientists believe that hypoxic conditions trigger apoptosis of tenocytes, as well as compositional changes within the tendon, which lead to tissue damage and inflammation45,46. The disease is still poorly characterized and understood, however, motivating the development of ex vivo models to better understand the pathology.
Ex vivo models are important for studying tendinopathies due to the dynamic interplay between tenocytes and the ECM46. There are three main mechanisms of inducing tendinopathictype symptoms – cyclic loading, creep loading, and stress deprivation. For the cyclic loading model performed by Devkota et al., tendons were cyclically loaded for 24 hours with the ECM intact47, (Figure 4). Tenocyte death and the production of collagenase indicated tendon degradation. Similar findings were reported for the creep testing studies. Stress deprivation was also proposed as a mechanism of inducing tendinopathy; however, the mechanical properties decreased while collagen content remained the same, unlike disease presentation in the body. Scott et al. confirmed tendinosis characteristics following high-strain mechanical loading in a rat tendon ex vivo model48. After high-strain loading, tenocytes assumed a shrunken apoptotic morphology and cell density in the tendons decreased overall as the result of apoptosis. The strains applied to the tendons were supraphysiological and further work in mechanically induced tendinosis models is needed with physiologically relevant levels of strain.
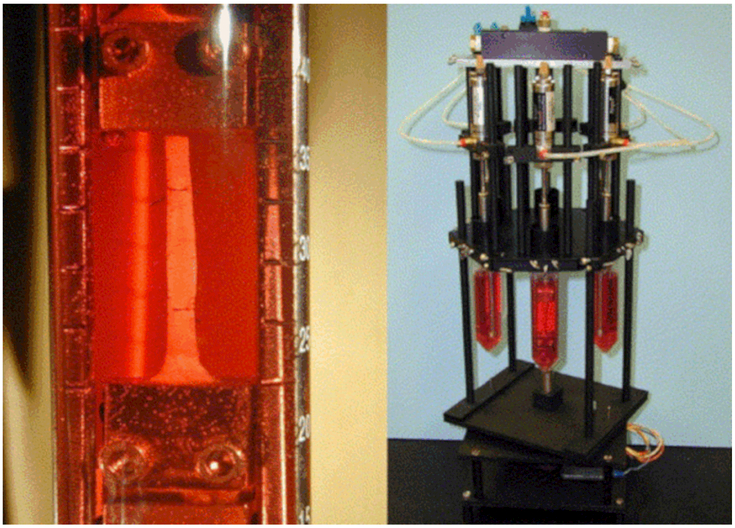
Figure 4.
Close-up of gripped tendon with midsubstance suture markers and tissue loading device on rotating stage (reproduced with permission from Devkota et al., 201847).
Millar et al. cultured primary human tenocytes in an ex vivo model to determine the effect of hypoxia on cell behavior45. Hypoxic conditions increased the number of apoptotic cells, pro-apoptotic factors HIF-1α, and anti-apoptotic proteins clusterin and Bcl-2. Collagen type III was found to be expressed in greater concentrations in the tenocytes cultured in hypoxic conditions, aligning with the higher levels of collagen III found in tendinosis patients. Inflammation is a well-characterized symptom of tendinopathies; interestingly, the group found leukocyte recruitment factors to be expressed in the hypoxic cell culture conditions, but inflammatory markers and cytokines were absent, suggesting that the expression of cytokines in vivo is due to the recruited leukocytes. Hypoxia appears to induce a shift in collagen production from type I to type III. Collagen III is more susceptible to rupture than collagen I, potentially explaining the altered mechanical properties of the tendon, which lend it susceptible to injury. Hypoxic conditions appear to be an important factor in model construction when studying tendinopathies and should continue to be accounted for in future ex vivo models.
Fibromyalgia Syndrome
Fibromyalgia is a disease that causes chronic musculoskeletal pain throughout the body. The pathology predominantly affects women and is associated with anxiety, depression, sleep disturbance, irritable bowel syndrome, temporomandibular disease, and many other comorbidities49–51. Treatment predominantly relies on pain management, antidepressants, and antianxiety medication. Models used to study the disease are scarce due to the multifactorial aspect of the disease. The most often utilized model is a mouse model49,51. However, all factors such as the chronic pain, anxiety, depression, sleep disturbance, and irritable bowel syndrome are often induced separately, limiting the verifiability of the treatments studied on these models. Despite the challenges, scientists have developed several promising models that could be utilized and built upon to better understand fibromyalgia and its treatment.
Green et al. developed a fibromyalgia rat model by exposing rats to unexpected sound stressors and water avoidance tests49. The tests resulted in hypersensitivity to physical pain and irritable bowel syndrome. The rats also exhibited long term anxiety following the application of the sound stressors. Temporomandibular disease was also present and supported by masseter muscle hyperalgesia. This rat model shows promise for future studies due to the presence of irritable bowel disorder, temporomandibular disease, anxiety, and musculoskeletal pain.
Nishiyori et al. created a fibromyalgia mouse model by exposing the mice to cold stresses repeatedly51. Temperatures were maintained around 4 °C and produced hyperalgesia and allodynia, as well as activation of pain receptors in new regions. Interestingly, under constant cold stress, the mice regained pain tolerance. If the cold stresses were applied intermittently, the pain tolerance remained low throughout the course of the study – 12 days for allodynia and 15 days for hyperalgesia. In addition, a gonadectomy in male mice resulted in decreased pain tolerance compared to the male control group. However, the pain tolerances between the males did not significantly differ from that of the female mice. Gabapentin was administered after confirming hyperalgesia and allodynia and was found to reduce the pain symptoms in the mice at lower concentrations. This model shows much promise for the future of studying fibromyalgia but does not incorporate many of the other comorbidities of the disease mentioned above, such as depression and anxiety. The complexity of fibromyalgia provides challenges for ex vivo models, but microfluidic devices may lend themselves valuable for optimization of drug selection if multiple devices and systems are combined to allow for isolation of treatments for each of the comorbidities.
Mitochondrial Myopathy
Mitochondria are responsible for producing much of the energy required by the cell in the form of adenosine triphosphate (ATP)52,53. Mitochondria are also involved in calcium homeostasis for the cell, cell death, and the regulation of the cell cycle. Mitochondrial dysfunction can occur as a result of mitochondrial DNA mutations, membrane integrity loss, or the disruption of the proton gradient involved in ATP production. Neuronal cells, skeletal muscle, and cardiac tissue are especially affected by mitochondrial dysfunction in the body due to their high energy demand. Due to the mitochondria’s crucial role in the body, developing a model to study the organelle and its associated diseases is of great interest to scientists.
Maglioni et al. highlights the roundworm Caenorhabditis elegans (C. elegans) as a valuable model in studying mitochondrial disorders52. C. elegans have been studied intensively in developmental and basic biology. As a result, its entire genome has been sequenced and many genes have been identified as analogs to human genes. The nervous system of these worms is similar to humans in that they possess motor neurons, sensory neurons, and interneurons. Their DNA is also relatively easy to manipulate, making them a versatile and valuable tool to study disease52. While the majority of studies have been performed with a focus on neural side effects of mitochondrial diseases, some tests have been designed to assess the effect of mitochondrial disease on movement, mating, and mechanical and chemical sensation. Given the promise in using C. elegans to study a variety of disorders53,54, the organism can also be incorporated into microfluidic devices for testing treatments for and studying mitochondria-related disorders54. Incorporating a complex living system into a microfluidic device allows for great potential in modeling mitochondrial myopathy due to the high-throughput, and physiologically relevant experiments available.
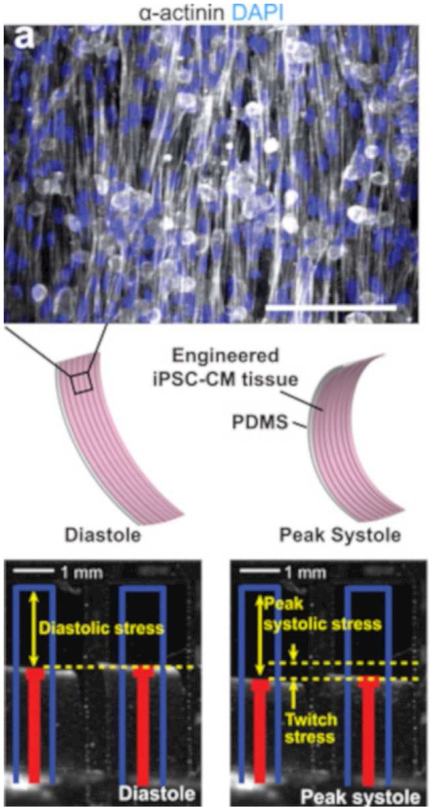
Wang et al. designed a heart-on-a-chip microfluidic device using iPSCs to study Barth syndrome, a mitochondrial cardiomyopathy53. The disease is caused by a mutation in the TAZ gene, which codes for tafazzin, an enzyme that is essential for the modification of cardiolipin, a major component of the mitochondrial inner membrane53. iPSCs were sourced from patients with Barth syndrome and differentiated into cardiomyocytes to ensure that the TAZ gene was mutated. The production of cardiolipin significantly decreased ex vivo, aligning with the clinical data of patients with the disease. Rat cardiomyocytes were utilized to verify that cardiolipin production and mitochondrial function were reliant on the TAZ gene. In an effort to better understand the cardiomyopathy, a heart-on-a-chip microfluidic device was constructed with the iPSC-derived cardiomyocytes. Myocardial tissue was formed, and contraction was permitted by peeling the cell layer from the surface of the device. The cell strips possessing the TAZ mutation demonstrated lower twitch and peak systolic stress, similar to the pathological presentation of Barth syndrome. The cells also exhibited sarcomere malformation and reactive oxidative species (ROS) production. After 5 days of incubation with a TAZ-correcting sequence, however, contractility and twitch improved. Their system setup can be seen in Figure 5. The microfluidic platform was then used to screen three different drug therapies for Barth syndrome. Linoleic acid and a therapeutic called MitoTEMPO were shown to increase twitch. This microfluidic model recapitulates key aspects of Barth syndrome and will be a valuable tool for testing drugs and gene therapies to address the disease. However, modeling the muscular and neuromuscular mitochondrial myopathies are still rare and lend themselves as promising targets for future microfluidic devices.
Figure 5.
BTHS (mitochondrial myopathy) myocardial tissue constructs exhibit depressed contractile stress generation. (a) Top, α-actinin–stained image of a muscular thin film (MTF). Middle, induced pluripotent stem cell and cardiomyocytes (iPSC-CM) seeded onto thin PDMS elastomers with patterned lines of fibronectin self-organized into anisotropic myocardial tissues. Cardiomyocyte stress generation reduces the radius of curvature of the construct as it contracts from diastole to peak systole. Scale bar, 100 μm. Bottom, still images of MTFs in diastole and systole. Red lines indicate automated MTF tracking projected onto the horizontal plane. Blue lines indicate lengths of MTFs before peeling from substrate. (Reproduced with permission from Wang et al., 201453).
Myasthenia Gravis
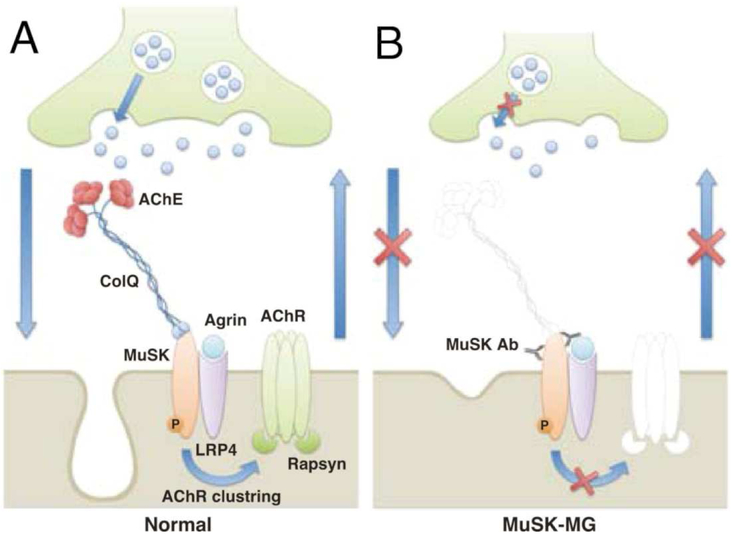
Myasthenia gravis (MG) is an autoimmune disease characterized by pain, muscular weakness, and fatigue55,56. MG results from the production of antibodies for acetylcholine receptors (AChR) or muscle specific kinase (MuSK) proteins, which hinder the transmission of signals in the neuromuscular junction (Figure 6). The pathologies of these two disease-causing malfunctions are very different. Treatment options are available for those who suffer from AChR attack, but little is known or available for those who suffer from the MuSK-related MG. The use of AChR inhibitors for patients with MuSK-related MG has been shown to cause respiratory issues, muscle twitches, and dysphagia. This inconsistency between disease pathology and treatment motivates research to better understand the pathophysiological processes of MG development and subsequent development of improved treatment options.
Figure 6.
Pathogenic mechanism of MG induced by MuSK antibodies (Abs). (A) In normal neuromuscular junctions (NMJs), agrin binds to LRP4 to activate MuSK. MuSK regulates the maintenance of both presynaptic and postsynaptic structures and functions bidirectionally. (B) MuSK Abs bind to the ectodomain of MuSK, causing MuSK degradation by antigenic modulation and/or direct inhibition of MuSK function. This inhibits MuSK, resulting in structural disruptions (e.g., dispersal of AChR clusters, loss of synaptic folds, and degeneration of nerve terminals) and functional ab-normalities (e.g., decrease in ACh release) that eventually lead to MG. Furthermore, the decreased levels of acetylcholinesterase (AchE), which is anchored to MuSK by ColQ, can induce a cholinergic crisis mediated by AChE inhibitors, exacerbating the symptoms of MG. (Reproduced with permissions from Mori et al., 201255).
Mori et al. created a mouse model that specifically possessed the form of MG resulting from MuSK antibodies55. The model was generated by injecting a recombinant MuSK protein into the mice twice over a two-week period. Following the second injection, the mice exhibited muscle weakness, tremors, and a cervicothoracic hump. They also observed a reduction in acetylcholine receptors at the neuromuscular junctions, further supporting the model as adequate to represent MG. Interestingly, the group observed no difference in the resting membrane potential between the control mice and the MG model, suggesting that the disease is not due to insufficient electrical stimulation. Because AChR inhibitors are a treatment option for patients with AChR MG, but have been shown to be detrimental in patients with MuSK MG, AChR drugs were used on the model to determine if the same deleterious effects were seen. Mice regained lost muscle action potential, but abnormal EMG patterns were seen in the mice with MuSK MG and not in the control, supporting the movement away from AChR inhibitors for patients with MuSK MG. This model provides insight into the critical role of MuSK in the function of neuromuscular junctions and is promising for the future of MuSK MG modeling ex vivo systems.
Gertel-Lapter et al. highlights the immune system’s role, and specifically the T cells’ role, in MG56. In the disease, the CD4+ T cells and B cells work together to produce AChR antibodies, which hinder signal transduction through the neuromuscular junction. Naive CD4+ T cells can differentiate into Treg cells as well, and a decrease of these cells is known to contribute to the pathology of MG. The injection of Treg cells from healthy donors into disease models with MG has been shown to suppress the disease56. A rat model of the AChR form of MG was developed by injecting the rats with modified AChR. CD4+ T cells were extracted and isolated from rats of a different experimental group and the Treg cells were further isolated from the same batch. The rats injected with the recombinant AChR possessed fewer numbers of Treg cells compared to the controls. Treg cells are known to also induce B cell death and prevent antibody production in healthy individuals. Treg cells from diseased rats possessed a diminished ability to induce B cell death ex vivo than Treg cells from healthy animals. Apoptosis and cytokine expression of the Treg cells from the diseased rats were also found to be elevated compared to Treg cell controls. Treg cells have been highlighted in other autoimmune diseases for their ability to inhibit auto-antibody production. Treg cells play an important role in MG and may play a key role in the success of future MG models. Given the complex interplay of multiple cell types in the development of MG, ex vivo microfluidic models show promise for studying the disease more.
Successful Ex Vivo Microfluidic Models for Muscle
As mentioned previously, muscle is a complex tissue that is highly vascularized and consists of myocytes aligned in elongated fibers57,58. Muscle is highly metabolically active and requires extensive energy supply for healthy function. As a result, modeling muscle tissue in ex vivo models remains a challenge due to the requirements for the models to possess multiple cell types, vasculature58, transcription factors28, adequate mechanical properties of the modeling substrate42, and constant mechanical and electrical stimulation26,27. Muscle disease modeling is particularly challenging due to the dynamic interplay of multiple organ systems in muscle diseases such as fibromyalgia49–51, muscular dystrophy38–40, mitochondrial myopathy52,53, myasthenia gravis55,56, and tendinosis45,46,48.
Ex vivo models show much promise in the future of tissue engineering. Moving testing and drug screening from animal models to ex vivo models may drastically reduce the cost of drug development4,59,60 and expedite the time to bring effective drugs to market. Advances in the fields of microfabrication and microfluidics allow complex biological systems to be shrunk to a more feasible size for benchtop testing, and for multiple “organ systems” to be combined in series or within the same microfluidic device to create a more holistic view during experimentation61,62. The systems also permit real-time testing and imaging for transcription factors, gene expression, protein secretion, and much more57. Microfluidic platforms show promise for the determination of cell pathways and mechanisms involved in the development and pathology of diseases, as well as normal biological function. Muscle disease modeling is a particularly interesting area of research as much room for advancement still remains. Some of the current ex vivo microfluidic models for muscle can be divided into basic myoblast models59, mechanically robust models for drug screening57, models designed to respond to external stimulation13,18,63, and models with multiple cell types for mimicking the neuromuscular junction64. Biomaterials serve an important role in establishing growth boundaries and providing appropriate cues for tissue modelling. An overview of the biomaterials used in the studies below are summarized in Table 1.
Table 1.
Current biomaterials utilized in microfluidic devices modeling muscle tissue.
| Material | Advantages | Disadvantages |
|---|---|---|
| Gelatin methacrylate (GelMA)40 |
• Biocompatible • Tunable crosslinking density to control material perfusion and mechanical properties • Can incorporate other acrylate tethered groups to make a composite system • Cell-binding sites |
• Molecular weight and composition can vary widely batch to batch • Cannot be combined with photosensitive, or free radical sensitive enzymes or factors |
| Collagen51 | • Biocompatible • Cell-binding sites • Can tune mechanical properties by varying weight percent • More bioactive than GelMA |
• Molecular weight and composition can vary widely batch to batch • Costly compared to gelatin |
| Fibrin52 | • Biocompatible • Cell-binding sites • Very easy to encapsulate cells inside • Crosslinker, thrombin, is very biocompatible |
• Could not be combined with blood flow due to role in clotting cascade • Complex protein so could not be easily modified |
Microfluidic Muscle Models for Drug Screening
Rodriguez-Rodriguez et al. recently created an ex vivo vascular smooth muscle model comprising a microfluidic device with five chambers for high-throughput drug screening59. For this model, the muscle cells were not cultured extensively; they were simply seeded and allowed to proliferate for 24 hours before exposure to two antiproliferative drugs, colchicine and curcumin, at varying concentrations. Both drugs were found to possess antiproliferative potential at dose-dependent concentrations, with colchicine being the most potent. The model was constructed with a grid in the cell culture region to allow rapid quantification of cell spreading and proliferation. The platform lends itself to rapid, high-throughput screening of therapeutics, and the optical transparency of the system shows promise for real-time imaging and testing.
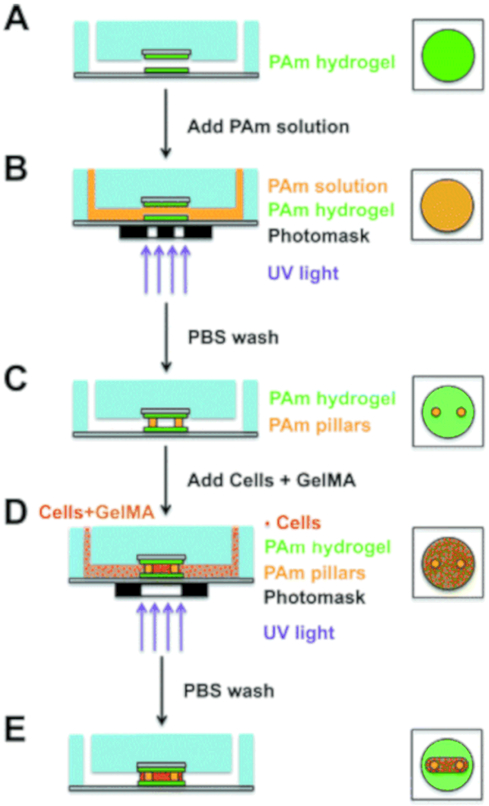
Increasing in complexity, Agrawal et al. created a muscle-on-a-chip system to study muscle formation and screen for potential therapeutics57. Mouse myoblasts were utilized inside of a microfluidic device constructed by photolithographically patterning silicon wafers with microfluidic channels. Polyacrylamide-coated glass coverslips were placed within the chambers of the device to sandwich the mice myoblasts encapsulated within gelatin methacrylate (GelMA), chosen for its biocompatibility and tunable crosslinking density and mechanical properties. Muscle cell strips were generated by photopatterning the GelMA and cell mixture and washing away the uncrosslinked portions. The cells were cultured for 12 days under continuous flow conditions with the strips in a perpendicular orientation to the flow to introduce mechanical stimulation. The cells were stained for myosin heavy chain, an indicator of a myoblast phenotype, as well as other cell markers to ensure that the cells were healthy and that muscle fibers formed. Mechanical property mapping indicated that the muscle strips were robust, with the ends wrapped around the polyacrylamide pillars being the strongest. Lastly, the system was tested for its potential for drug screening by administering cardiotoxin; the muscle strips responded in a dose-dependent manner. The ex vivo model contains a muscle strip operating under continuous fluid flow, allows real-time imaging, and shows promise for future testing with electrical, mechanical, chemical, or optical stimulation. The system’s fabrication can be found in Figure 7. The system’s response to a cardiotoxin also supports its utility as a drug-testing platform for diseases such as fibromyalgia or muscular dystrophy.
Figure 7.
3D photopatterning of support pillars and encapsulation of cells. (Left: Side view of full device; right: top view of central chamber) (A and B) To create support pillars, the bonded microfluidic chip was infused with acrylamide (Am) solution containing photoinitiator and was photopolymerized using a collimated UV light and a transparency photomask containing 100 μm diameter circle patterns. (C and D) After washing with PBS, a precursor solution composed of cells, GelMA, and photoinitiator was polymerized around the pillars using the same method as before, except with a capsule-shaped pattern. (E) PBS solution was used to wash the samples, and the device was perfused with maintenance media by using a syringe pump. (Reproduced with permission from Agrawal et al., 201757).
Stimuli-Responsive Microfluidic Muscle Models
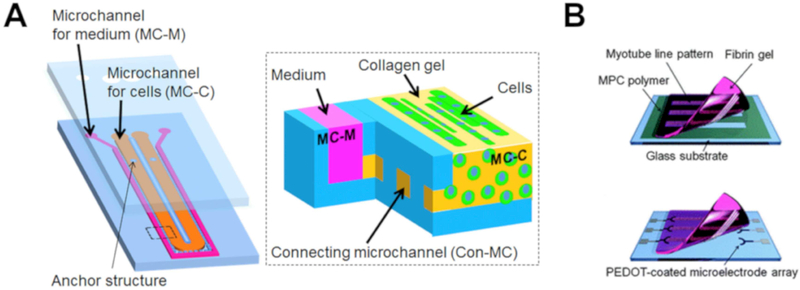
Also seeking to address the need for skeletal muscle-on-a-chip models, Shimizu et al. recently designed a system by injecting murine skeletal muscle cells and collagen I into microchannels constructed via photolithography63, shown in Figure 8A. The collagen gelled, encapsulating the cells, and media was flowed into adjacent microchannels for continuous supply of nutrients and oxygen. The channels were connected via pores to encourage nutrient and waste exchange between the cell-gel network and the media. After six days of culture, the cells began to dissociate from channel walls into bundles, which has been reported previously in myoblast culture65,66. Staining revealed that multinucleated cells were present, indicating the formation of myotubes. Immunofluorescence also revealed the formation of alpha actinin by the cells, providing further evidence of myotube formation. After introducing electrical stimulation, the muscle cells were observed to contract following both twitch and tetanus stimulation.
Figure 8.
(A) Schematic drawing of developed microfluidic device63. (B) Overview of the myotube/fibrin gel sheet combined with the PEDOT microelectrode array chip. (Top) Cell transfer from a glass substrate to a fibrin gel. (Bottom) Attachment of the myotube/fibrin gel onto the microelectrode arrays. (Reproduced with permission from (A) Shimizu et al, 2015,63 (B) Nagamine et al, 201067.)
To test the efficacy of using stem cells as a therapeutic following myocardial infarction, Ma et al. constructed a lab-on-a-chip with stem cells or cardiomyocytes seeded in gaps created within muscle fibers in the device simulating the necrotic regions of cardiac tissue following a myocardial infarction13. Conductivity of the cells was assessed, and cardiomyocytes were found to possess the highest conduction of the cells tested. The stem cells were observed to beat synchronously with the stimulation. The group also stained for connexin, a gap junction marker, and found connexin at the connecting point between the stem cells and the muscle fibers. To further assess the system, a gap junction blocker, carbenoxolone, was administered, resulting in a decrease in the conduction of the stem cells bridge. This provides evidence of gap junction formation being responsible for electrical conduction and validates the ex vivo model for drug screening.
Using muscular thin film technology, Grosberg et al. created several microfluidic devices and tested the devices using both smooth and striated muscle cells18. Vascular smooth muscle thin films were created by stamping fibronectin onto coated glass coverslips. Following the thin film formation, the strips were removed from the coverslips except for one corner to keep the cell strips anchored during testing. The muscle strips responded to electrical stimulation by contracting, and also responded to a protein kinase inhibitor, demonstrating the platform’s promise as a drug-screening tool in future studies. The group also placed cardiomyocyte muscle thin films in microfluidic channels and used the same practice of removing the cell strips from the platform in all but one corner. The cells emulated both diastolic and systolic behavior following electrical stimulation. Combining vascular smooth muscle cells and cardiomyocytes into one model, the group was able to observe the effect of the cell types on each other’s behavior. Both cell types responded to electrical stimulation by contracting. Both cell types also responded to inhibitors and exhibited reduced contractility, demonstrating the potential of the model as an ex vivo drug-screening platform capable of also incorporating multiple cell types.
Modeling the Neuromuscular Junction
Continuing the trend of models containing multiple cell types, Southam et al. created a microfluidic model for the neuromuscular junction containing glial cells, spinal motor neurons, and skeletal muscle cells64. The spinal glial cells were sourced from postnatal rats, the spinal motor neurons from embryonic rats, and the skeletal muscle cells from rat hindlimb. The motor neurons and spinal glial cells were grown in a chamber together, which was connected via channels to the chamber containing the skeletal muscle cells. The motor neurons were observed to extend axons through the channels and synapse with the muscle cells by day 10 in culture. By day 18, myotube formation occurred in the muscle cells to meet the motor neurons. Acetylcholine receptors were detected at the neuromuscular junction, indicating the platform’s promise as an ex vivo drug-screening platform for neuromuscular diseases, especially those that target the neuromuscular junctions such as MG.
Electrical stimulation has been shown as an effective alternative to neuronal innervation when modeling the neuromuscular junction. Nagamine et al. developed a microfluidic platform to electrically stimulate myotubes67, shown in Figure 8B. C2C12 myoblasts were cultured on etched glass coated in 2-methacryloyloxyethyl phosphorylcholine (MPC) to promote myotube formation. Fibrin pre-gel was then poured onto the cell system, and fibrin gels were removed, lifting the myotubes with them. The cell-gel system was then placed into a soft lithography microelectrode coated with poly(3,4-ethylenedioxythiophene) (PEDOT). PEDOT has a large electrostatic surface area and ensures a less invasive electrical stimulation of cells. The PEDOT coating was a pivotal component of the ex vivo muscle model because it allowed for stable, long-term stimulation of myotubes. The myotubes exhibited adult phenotype, myogenic behavior, and independent, synchronized contractions. In addition to modulating the cell substrate, microdevices can also be finely tuned to promote myogenic phenotype and terminal behavior.
Conclusions
In conclusion, while there are several muscle disease models currently being used for diseases such as muscular dystrophy, tendinosis, fibromyalgia, mitochondrial myopathy, and myasthenia gravis, many of these models are animal models and are unable to recapitulate all factors of the diseases. While animal models are the current gold standard for studying many diseases, they are expensive and allow limited assessment of real-time cellular responses to stimuli and monitoring of gene expression. The models are also time intensive to create, to assess their robustness, and then screen potential disease therapeutics. Current in vitro 2D and 3D cell culture models have sought to address some of the challenges of using animal models and have enabled better sensing and imaging at the cellular level, as well as monitoring gene expression and protein synthesis within cells. However, 2D and 3D culture do not always incorporate all of the complexity seen in the body such as the presence of multiple cell types, consistent mechanical and electrical stimulation, and the relationship between cells and surrounding ECM. The studies also often require the destruction of the scaffold and the cells when imaging is performed or analysis timepoints are due. Ex vivo microfluidic models seek to bridge this gap between the complexity of in vivo models and the feasibility of real-time sensing and imaging seen in in vitro 2D and 3D cell culture. Unlike 2D and 3D culture, imaging and sensing can be performed dynamically alongside the study in microfluidic devices, allowing the same study to be carried to completion with real-time analysis performed through its entirety. The technology shows much promise to address these challenges due to the ability to culture multiple cell types together, expose the cells to mechanical and electrical stimuli, and measure cellular response and genetic expression throughout the duration of testing.
Several groups have already begun modeling muscle in microfluidic devices such as the muscular thin film example, the neuromuscular junction, or the stem cell bridge myocardial infarction patch mentioned above. These groups have demonstrated the strength of microfluidic devices in being suitable to culture robust muscle, utilize the model as a drug-testing platform, explore the dynamic relationship between multiple cell types, and mimic the complexity of muscle such as the neuromuscular junction. Additional efforts in drug screening have been made to build a “human-on-a-chip” that would integrate 10 organ-on-a-chip systems connected by “vasculature”. This project has made strides in modeling the sophisticated physiological responses to drug treatments and their effects on multiple organ systems. The technology of microfluidics for muscle modeling is still young, but by building on the knowledge gained from existing muscle models and drawing from the current microfluidic muscle models developed, ex vivo microfluidic devices show much promise for the next generation of muscle disease modeling and drug screening.
Acknowledgments
We acknowledge support towards the development of tissue engineered disease models from the National Institutes of Health (R01 CA180279 and P41 EB023833). We also acknowledge support from National Science Foundation Graduate Research Fellowships (MMS, HAP) and a Ford Foundation Pre-doctoral Research Fellowship (MMS).
Footnotes
Manuscript submitted to the Special Issue of Biomaterials on “Biomaterials and Bioengineering Innovations for Ex Vivo Tissue-Chip Development” Edited by Drs. Nasim Annabi, Ankur Singh, and Mehdi Nikkah
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.C.A. C & J.E. M Duchenne’s muscular dystrophy: animal models used to investigate pathogenesis and develop therapeutic strategies. Int. J. Exp. Pathol 84, 165–172 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fishman JM et al. Skeletal muscle tissue engineering: which cell to use? Tissue Eng. PartB. Rev 19, 503–515 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Mantegazza R, Cordiglieri C, Consonni A & Baggi F Animal models of myasthenia gravis: utility and limitations. Int. J. Gen. Med 9, 53–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Skardal A, Shupe T & Atala A Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov. Today 21, 1399–1411 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uzel SGM, Pavesi A & Kamm RD Microfabrication and microfluidics for muscle tissue models. Prog. Biophys. Mol. Biol 115, 279–293 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Cooper ST et al. C2C12 co-culture on a fibroblast substratum enables sustained survival of contractile, highly differentiated myotubes with peripheral nuclei and adult fast myosin expression. Cell Motil. Cytoskeleton 58, 200–211 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Sung JH et al. Using physiologically-based pharmacokinetic-guided ‘body-on-a-chip’ systems to predict mammalian response to drug and chemical exposure. Exp. Biol. Med 239, 1225–1239 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marx U et al. ‘Human-on-a-chip’ developments: a translational cutting-edge alternative to systemic safety assessment and efficiency evaluation of substances in laboratory animals and man? Altern. Lab. Anim 40, 235–257 (2012). [DOI] [PubMed] [Google Scholar]
- 9.Kunz-Schughart LA, Freyer JP, Hofstaedter F & Ebner R The use of 3-D cultures for high-throughput screening: The multicellular spheroid model. J. Biomol. Screen 9, 273–285 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Huang CP et al. Engineering microscale cellular niches for three-dimensional multicellular co-cultures. Lab Chip 9, 1740–1748 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker M Tissue models: A living system on a chip. Nature 471, 661–665 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Bhatia SN & Ingber DE Microfluidic organs-on-chips. Nat. Biotechnol 32, 760–772 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Ma Z et al. Laser-patterned stem-cell bridges in a cardiac muscle model for on-chip electrical conductivity analyses. Lab Chip 12, 566–573 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Discher DE, Janmey P & Wang Y-L Tissue cells feel and respond to the stiffness of their substrate. Science 310, 1139–1143 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Marsano A et al. Beating heart on a chip: a novel microfluidic platform to generate functional 3D cardiac microtissues. Lab Chip 16, 599–610 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Huh D et al. Reconstituting Organ-Level Lung Functions on a Chip. Science (80-. ). 328, 1662 LP–1668 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huh D et al. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med 4, 159ra147 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grosberg A et al. Muscle on a chip: In vitro contractility assays for smooth and striated muscle. J. Pharmacol. Toxicol. Methods 65, 126–135 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boudou T et al. A microfabricated platform to measure and manipulate the mechanics of engineered cardiac microtissues. Tissue Eng. Part A 18, 910–919 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ionescu A, Zahavi EE, Gradus T, Ben-Yaakov K & Perlson E Compartmental microfluidic system for studying muscle-neuron communication and neuromuscular junction maintenance. Eur. J. Cell Biol 95, 69–88 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Zahavi EE et al. A compartmentalized microfluidic neuromuscular co-culture system reveals spatial aspects of GDNF functions. J. Cell Sci 128, 1241–1252 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morimoto Y, Kato-Negishi M, Onoe H & Takeuchi S Three-dimensional neuron–muscle constructs with neuromuscular junctions. Biomaterials 34, 9413–9419 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Choi JS, Piao Y & Seo TS Circumferential alignment of vascular smooth muscle cells in a circular microfluidic channel. Biomaterials 35, 63–70 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Mammoto T, Mammoto A & Ingber DE Mechanobiology and developmental control. Annu. Rev. Cell Dev. Biol 29, 27–61 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Romanazzo S et al. Substrate stiffness affects skeletal myoblast differentiation in vitro. Sci. Technol. Adv. Mater 13, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuda N, Hagiwara N, Shoda M, Kasanuki H & Hosoda S Enhancement of the L-type Ca2+ current by mechanical stimulation in single rabbit cardiac myocytes. Circ. Res 78, 650–659 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Sadoshima J & Izumo S The Cellular and Molecular Response of Cardiac Myocytes. Annu. Rev. Physiol 551–571 (1997). [DOI] [PubMed] [Google Scholar]
- 28.Braun T & Gautel M Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat. Rev. Mol. Cell Biol 12, 349–361 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Benam KH et al. Engineered In Vitro Disease Models. Annu. Rev. Pathol. Mech. Dis 10, 195–262 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Jacot JG, McCulloch AD & Omens JH Substrate stiffness affects the functional maturation of neonatal rat ventricular myocytes. Biophys. J 95, 3479–3487 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powell CA, Smiley BL, Mills J & Vandenburgh HH Mechanical stimulation improves tissue-engineered human skeletal muscle. AJP Cell Physiol 283, C1557–C1565 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Langelaan MLP et al. Advanced maturation by electrical stimulation: Differences in response between C2C12 and primary muscle progenitor cells. J. Tissue Eng. Regen. Med 5, 529–539 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Hernández-Hernández JM, García-González EG, Brun CE & Rudnicki MA The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin. Cell Dev. Biol 72, 10–18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qazi TH, Mooney DJ, Pumberger M, Geißler S & Duda GN Biomaterials based strategies for skeletal muscle tissue engineering: Existing technologies and future trends. Biomaterials 53, 502–521 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Pati F et al. Ornamenting 3D printed scaffolds with cell-laid extracellular matrix for bone tissue regeneration. Biomaterials 37, 230–241 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Kumar VA et al. Treatment of hind limb ischemia using angiogenic peptide nanofibers. Biomaterials 98, 113–119 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hernandez-Gordillo V & Chmielewski J Mimicking the extracellular matrix with functionalized, metal-assembled collagen peptide scaffolds. Biomaterials 35, 7363–7373 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Filareto A et al. An ex vivo gene therapy approach to treat muscular dystrophy using inducible pluripotent stem cells. Nat. Commun 4, 1549 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson CE et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 351, 403–407 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Floyd SS et al. Ex vivo gene transfer using adenovirus-mediated full-length dystrophin delivery to dystrophic muscles. Gene Ther 5, 19–30 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Bonaldo P & Sandri M Cellular and molecular mechanisms of muscle atrophy. Dis. Model. Mech 6, 25–39 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tiscornia G, Vivas EL & Belmonte JCI Diseases in a dish: Modeling human genetic disorders using induced pluripotent cells. Nat. Med 17, 1570–1576 (2011). [DOI] [PubMed] [Google Scholar]
- 43.Segalat L Invertebrate animal models of diseases as screening tools in drug discovery. ACS Chem. Biol 2, 231–236 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Guyon JR et al. Modeling human muscle disease in zebrafish. Biochim. Biophys. Acta - Mol. Basis Dis 1772, 205–215 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Millar NL et al. Hypoxia: A critical regulator of early human tendinopathy. Ann. Rheum. Dis 71, 302–310 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Dirks RC & Warden SJ Models for the study of tendinopathy. J. Musculoskelet. Neuronal Interact 11, 141–149 (2011). [PubMed] [Google Scholar]
- 47.Devkota AC & Weinhold PS A tissue explant system for assessing tendon overuse injury. Med. Eng. Phys 27, 803–808 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Scott A High strain mechanical loading rapidly induces tendon apoptosis: an ex vivo rat tibialis anterior model. Br. J. Sports Med 39, e25–e25 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Green PG, Alvarez P, Gear RW, Mendoza D & Levine JD Further validation of a model of fibromyalgia syndrome in the rat. J. Pain 12, 811–818 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skaer TL Fibromyalgia: Disease synopsis, medication cost effectiveness and economic burden. Pharmacoeconomics 32, 457–466 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Nishiyori M & Ueda H Prolonged gabapentin analgesia in an experimental mouse model of fibromyalgia. Mol. Pain 4, 1–6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maglioni S & Ventura NC elegans as a model organism for human mitochondrial associated disorders. Mitochondrion 30, 117–125 (2016). [DOI] [PubMed] [Google Scholar]
- 53.Wang G et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med 20, 616–623 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ben-Yakar A, Chronis N & Lu H Microfluidics for the analysis of behavior, nerve regeneration, and neural cell biology in C. elegans. Curr. Opin. Neurobiol 19, 561–567 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mori S et al. Antibodies against muscle-specific kinase impair both presynaptic and postsynaptic functions in a murine model of myasthenia gravis. Am. J. Pathol 180, 798–810 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Gertel-Lapter S, Mizrachi K, Berrih-Aknin S, Fuchs S & Souroujon MC Impairment of regulatory T cells in myasthenia gravis: Studies in an experimental model. Autoimmun. Rev 12, 894–903 (2013). [DOI] [PubMed] [Google Scholar]
- 57.Agrawal G, Aung A & Varghese S Skeletal muscle-on-a-chip: an in vitro model to evaluate tissue formation and injury. Lab Chip 17, 3447–3461 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Levenberg S et al. Engineering vascularized skeletal muscle tissue. Nat. Biotechnol 23, 879–884 (2005). [DOI] [PubMed] [Google Scholar]
- 59.Rodriguez-Rodriguez R et al. Cell-based microfluidic device for screening anti-proliferative activity of drugs in vascular smooth muscle cells. Biomed. Microdevices 14, 1129–1140 (2012). [DOI] [PubMed] [Google Scholar]
- 60.Ghaemmaghami AM, Hancock MJ, Harrington H, Kaji H & Khademhosseini A Biomimetic tissues on a chip for drug discovery. Drug Discov. Today 17, 173–181 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huh D, Torisawa Y, Hamilton GA, Kim HJ & Ingber DE Microengineered physiological biomimicry: organs-on-chips. Lab Chip 12, 2156–2164 (2012). [DOI] [PubMed] [Google Scholar]
- 62.van Midwoud PM, Verpoorte E & Groothuis GMM Microfluidic devices for in vitro studies on liver drug metabolism and toxicity. Integr. Biol. (Camb) 3, 509–521 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Shimizu K et al. Microfluidic devices for construction of contractile skeletal muscle microtissues. J. Biosci. Bioeng 119, 212–216 (2015). [DOI] [PubMed] [Google Scholar]
- 64.Southam KA, King AE, Blizzard CA, McCormack GH & Dickson TC Microfluidic primary culture model of the lower motor neuron-neuromuscular junction circuit. J. Neurosci. Methods 218, 164–169 (2013). [DOI] [PubMed] [Google Scholar]
- 65.Bian W & Bursac N Engineered skeletal muscle tissue networks with controllable architecture. Biomaterials 30, 1401–1412 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vandenburgh H et al. Drug-screening platform based on the contractility of tissue-engineered muscle. Muscle Nerve 37, 438–447 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Nagamine K et al. Spatiotemporally controlled contraction of micropatterned skeletal muscle cells on a hydrogel sheet. Lab Chip 11, 513–517 (2011). [DOI] [PubMed] [Google Scholar]
- 68.Tremblay D, Chagnon-Lessard S, Mirzaei M, Pelling AE & Godin M A microscale anisotropic biaxial cell stretching device for applications in mechanobiology. Biotechnol. Lett 36, 657–665 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]