Abstract
Autism spectrum disorder (ASD) is a neurodevelopmental disorder inexplicably biased towards males. Although prenatal exposure to bisphenol A (BPA) has recently been associated with the ASD risk, whether BPA dysregulates ASD-related genes in the developing brain remains unclear. In this study, transcriptome profiling by RNA-seq analysis of hippocampi isolated from neonatal pups prenatally exposed to BPA was conducted and revealed a list of differentially expressed genes (DEGs) associated with ASD. Among the DEGs, several ASD candidate genes, including Auts2 and Foxp2, were dysregulated and showed sex differences in response to BPA exposure. The interactome and pathway analyses of DEGs using Ingenuity Pathway Analysis software revealed significant associations between the DEGs in males and neurological functions/disorders associated with ASD. Moreover, the reanalysis of transcriptome profiling data from previously published BPA studies consistently showed that BPA-responsive genes were significantly associated with ASD-related genes. The findings from this study indicate that prenatal BPA exposure alters the expression of ASD-linked genes in the hippocampus and suggest that maternal BPA exposure may increase ASD susceptibility by dysregulating genes associated with neurological functions known to be negatively impacted in ASD, which deserves further investigations.
Introduction
Autism spectrum disorder (ASD) is an early-onset neurodevelopmental disorder characterized by 2 main symptoms: i) social interaction and communication impairments, and ii) restricted interests and stereotyped behaviors. The Centers for Disease Control and Prevention (CDC) recently reported that the prevalence of ASD is as high as 1 in 59 children in the United States1. ASD is inexplicably biased towards males, with a prevalence in males approximately four times higher than that in females1. Although there is accumulating evidence that genetic factors are associated with ASD etiology or susceptibility, the majority (80–90%) of ASD cases remain idiopathic. Moreover, recent studies have reported that epigenetic regulatory mechanisms, including DNA methylation2–5, histone modifications6,7, and RNA-associated mechanisms8, are associated with ASD. Epigenetic mechanisms play an important role in gene-environment interactions and susceptibility to environmental stresses, and environmental factors are also thought to be associated with ASD etiology and/or susceptibility. In fact, several recent studies have reported that exposure to certain environmental pollutants and industrial chemicals is associated with increased risk of ASD5,9,10. Examples of environmental chemicals that have been associated with ASD include endocrine-disrupting compounds (EDCs)11, lead12, mercury13, pesticides14, and cigarette smoke15.
EDCs are a group of chemicals that can be found in various products widely used in daily life. With chemical structures similar to sex hormones, particularly estrogen, EDCs are thought to disrupt hormone regulatory systems in the body by interfering in several processes, including hormone synthesis, secretion, transport, metabolism, binding process, and elimination of natural hormones that are present in the body16. Given that sex hormones are known to play critical roles in homeostasis, reproduction, and developmental processes, EDCs are thought to disrupt hormone-related biological functions and pose a risk for many diseases/disorders, including ASD5,17. EDCs that have been associated with ASD include bisphenol A (BPA), phthalates, polybrominated diphenyl ethers (PBDEs), and polychlorinated biphenyls (PCBs).
BPA ((CH3)2C(C6H4OH)2) is an organic compound consisting of two hydroxyphenyl groups. It is widely used in polycarbonate plastic and epoxy resin products, including linings inside beverage and food cans, plastic bottles, and dental sealants. Under high heat and alkaline conditions, BPA can be hydrolyzed and leach from products, posing a risk of exposure to consumers18–20. After ingestion, BPA is metabolized by UDP glucuronosyltransferases in the liver to BPA-glucuronide which is then excreted in urine21. The U.S. Food and Drug Administration (FDA) and European Food Safety Authority (EFSA) have determined the No-Observed-Adverse-Effect Level (NOAEL) for BPA to be 5,000 µg/kg body weight/day and established the Tolerable Daily Intake (TDI) of 50 µg/kg body weight/day which is derived by applying a 100-fold uncertainty factor to the NOAEL22.
BPA can circulate throughout the body and pass through the placenta and blood-brain barrier23,24. Although the safety of exposure to BPA remains unclear, accumulating evidence indicates that BPA alters synaptic plasticity25, neonatal brain development26, neurogenesis27,28, learning29, memory30, anxiety31,32, and social interaction33, all of which have been implicated in ASD34–36. Moreover, recent studies have reported that BPA exposure is associated with the risk of ASD37–40. Stein et al. (2015) determined the concentrations of free and total BPA in urine obtained from 46 children with ASD and 52 typically developing children using liquid chromatography-mass spectrometry (LC-MS/MS) analysis. They found that approximately 20% of the ASD children had BPA levels beyond the 90th percentile (>50 ng/ml) of the frequency distribution for the total sample of 98 children39. Moreover, Kardas et al. (2016) measured the levels of BPA, mono-(2-ethylhexyl)-phthalate (MEHP), and di-(2-ethylhexyl)-phthalate (DEHP) in the sera of 48 children with ASD and 41 typically developing children using high-performance liquid chromatography (HPLC). Interestingly, the levels of serum BPA, MEHP, and DEHP were significantly increased in children with ASD compared with controls37. Similar to Kardas et al. (2016), Kondolot et al. (2016) measured the plasma concentrations of BPA and phthalates in 51 ASD children and 50 age-/sex-matched typically developing children using HPLC analysis38. They found that the average plasma BPA level of children with a specific disorder in the autism spectrum called PDD-NOS (pervasive developmental disorder not otherwise specified) was significantly higher than that of the control, and the average level of BPA detected in the plasma was as high as 6.91 ng/ml38. Furthermore, it was reported that in vitro BPA exposure induced oxidative stress and mitochondrial dysfunctions in lymphoblastoid cell lines derived from individuals with ASD and unaffected siblings, suggesting that BPA may act as an environmental risk factor for ASD40. Nevertheless, whether BPA exposure can cause changes at the molecular level reminiscent of those observed in individuals with ASD has not been investigated.
In this study, we therefore sought to determine the effects of prenatal BPA exposure on transcriptome profiles in the context of ASD. First, we conducted a transcriptome profiling analysis of hippocampal tissues isolated from neonatal rats prenatally exposed to BPA or control vehicle to investigate the effects of BPA exposure on gene expression profiles in the hippocampus. The lists of differentially expressed genes (DEGs) in males and females were compared with autism candidate genes to determine the associations between the DEGs and ASD and the sex differences in the effects of BPA on the expression of ASD candidate genes. Biological pathways and interactome analysis were predicted using Ingenuity Pathway Analysis (IPA) software. DEGs that were associated with ASD were selected for further confirmation by qRT-PCR analysis. Moreover, we conducted a data-mining analysis of transcriptome profiles published in the NCBI GEO DataSets database to identify genes that were differentially expressed in response to BPA exposure. The list of significant genes was then overlapped with lists of ASD candidate genes obtained from two different ASD databases to predict whether BPA-responsive genes were also significantly associated with ASD candidate genes.
Results
Prenatal BPA exposure alters hippocampal transcriptome profiles in a sex-dependent manner
To examine whether prenatal BPA exposure could lead to dysregulation of ASD candidate genes in the developing brain in vivo, we conducted an RNA-seq analysis of hippocampal tissues isolated from male and female neonatal rats exposed to 5,000 µg/kg·maternal BW of BPA in utero or vehicle control. Notably, the dose of BPA used to treat rats in this study is equal to the No-Observed-Adverse-Effect Level (NOAEL) in humans as determined by the FDA and ESFA. We found that when all male and female rat pups under the same treatment condition were combined into one group, as many as 5,624 transcripts corresponding to 4,525 genes were significantly differentially expressed in the hippocampi of BPA-treated rats compared with the controls. In addition, to determine whether prenatal BPA exposure alters hippocampal transcriptome profiles in a sex-dependent manner, DEGs in each sex were identified. We found that 2,496 transcripts (corresponding to 2,078 genes) and 4,021 transcripts (corresponding to 3,522 genes) were significantly differentially expressed in the hippocampi of BPA-treated male and female pups, respectively, compared to controls (P-value < 0.05 and FDR < 0.05). This finding indicates that the brain transcriptome profiles of males and females were unequally disturbed by prenatal BPA exposure. The lists of DEGs are shown in Supplementary Table S1.
BPA-responsive DEGs in the hippocampus exhibit sex differences in ASD-associated genes
To determine whether DEGs in response to prenatal BPA exposure are associated with ASD, the lists of BPA-responsive genes were overlapped with the lists of ASD candidate genes from two ASD databases, including the SFARI and AutismKB databases. When all male and female pups were combined, a total of 298 and 700 genes among the DEGs were found to be ASD candidate genes in the SFARI and AutismKB databases, respectively. We next performed hypergeometric distribution analyses to assess the over-representation of ASD candidate genes among DEGs responsive to BPA. Hypergeometric distribution analysis of the list of DEGs in the combined male and female pups with respect to autism candidate genes showed no significant association. However, when each sex was analyzed separately, DEGs from male and female hippocampal tissues exhibited significant enrichment in ASD-related genes from the SFARI database (Table 1, Supplementary Table S2). Notably, DEGs in male hippocampal tissues tended to exhibit stronger associations with ASD genes than those in female tissues. This male bias was also observed when the list of DEGs was analyzed for enrichment of syndromic ASD genes in the AutismKB database. These results indicated that DEGs due to BPA exposure showed sex differences in their associations with ASD genes. In addition, to determine whether enrichment of ASD-related genes exists on the X chromosome in these DEG lists, we conducted hypergeometric distribution analyses between ASD-related genes on the X chromosome and each of these lists. Interestingly, we found significant enrichment of ASD-related genes on the X chromosome in the list of ASD-related DEGs in both sexes (9 from 298 genes; P-value = 6.45E-06), DEGs in males only (11 from 183 genes; P-value = 8.46E-10), and DEGs in females only (15 from 266 genes; P-value = 1.20E-12), suggesting that the X chromosome may be involved in the underlying mechanism of BPA-associated risk for ASD.
Table 1.
Association analysis between differentially expressed genes in hippocampi of offspring prenatally exposed to BPA and ASD-related genes.
| Overlap with (number of genes) | Gene list category | P-value from hypergeometric analysis (number of overlapping genes from both sexes) | P-value from hypergeometric analysis (number of overlapping genes from males) | P-value from hypergeometric analysis (number of overlapping genes from females) |
|---|---|---|---|---|
| SFARI database (1,007 genes) | All | 0.50 (298) | 1.32E-05 (183) | 3.83E-03 (266) |
| Syndromic | 0.57 (39) | 1.82E-11 (51) | 6.34E-02 (41) | |
| Score 1 | 0.12 (10) | 5.40E-06 (13) | 1.57E-02 (11) | |
| Score 2 | 0.58 (16) | 5.73E-05 (20) | 1.84E-02 (21) | |
| Score 3 | 1.07E-02 (63) | 1.22E-02 (35) | 4.49E-04 (60) | |
| Score 4 | 0.48 (113) | 0.13 (63) | 8.57E-02 (105) | |
| Score 5 | 0.75 (40) | 0.95 (15) | 0.95 (28) | |
| AutismKB database (3,055 genes) | All | 0.99 (700) | 0.99 (322) | 0.99 (562) |
| Syndromic | 0.80 (24) | 1.23E-02 (22) | 8.38E-02 (29) | |
| Non-syndromic | 0.99 (694) | 0.99 (317) | 0.99 (558) |
We overlapped the lists of significantly differentially expressed BPA-responsive genes in the neonatal hippocampus and ASD-related genes (SFARI and AutismKB databases). The lists of significantly differentially expressed genes in both sexes were analyzed using MeV software with a standard Bonferroni test (P-value < 0.05), and the lists of sex-specific significantly differentially expressed genes from the RNA-seq process were analyzed using Poisson distribution (FDR < 0.05, P-value < 0.05). P-values of association were calculated using hypergeometric distribution analysis and are shown in the table. SFARI scores represent the level of confidence. Score 1 = High confidence; Score 2 = Strong candidates; Score 3 = Suggestive evidence; Score 4 = Minimal evidence; Score 5 = Hypothesized; Syndromic: all syndromic genes associated with ASD.
BPA-responsive genes in the hippocampus are involved in biological functions, canonical pathways, and networks associated with ASD
To predict biological functions, pathways, and interactome networks associated with BPA-responsive genes in the hippocampus, the lists of DEGs were analyzed using IPA software. DEGs in the hippocampus were associated with several functions impacted in ASD, including “nervous system development and function”, “inflammatory response”, and “digestive system development and function”. Interestingly, the top canonical pathways significantly associated with DEGs in the male hippocampus included “glutamate receptor signaling”, “axonal guidance signaling”, and “circadian rhythm signaling”, all of which have been associated with ASD. Similarly, “glutamate receptor signaling” and “axonal guidance signaling” were also present among the top canonical pathways significantly associated with DEGs in the female hippocampus (P-value < 0.05; Supplementary Table S3). Neurological diseases/disorders associated with DEGs included “autism or intellectual disability”, “mental retardation”, and “developmental delay”. It was interesting to note that several neurological functions, including “morphogenesis of neurons”, “neuritogenesis”, and “formation of brain”, were significantly associated with DEGs in males only (P-value < 0.05; Table 2). Additionally, the IPA comparison analysis between canonical pathways associated with DEGs in males and in females revealed several pathways that exhibited significant associations in a sex-dependent manner. Such canonical pathways included “DNA methylation and transcriptional repression signaling”, “IGF-1 signaling”, “synaptic long-term potentiation”, and “androgen signaling”, all of which have been associated with ASD (Supplementary Table S4).
Table 2.
Comparison of neurological diseases/disorders and nervous system development functions between both sexes and males and females separately. NS = Not significant.
| Disease or Function Annotation | P-values (number of genes) | ||
|---|---|---|---|
| Both Sexes | Males | Females | |
| Autism or intellectual disability | 5.19E-04 (144) | 4.71E-16 (97) | 9.30E-10 (120) |
| Mental retardation | 9.66E-04 (133) | 1.49E-14 (89) | 1.11E-09 (113) |
| Familial syndromic intellectual disability | 6.51E-03 (84) | 1.40E-10 (58) | 3.09E-07 (73) |
| Disorder of stature | 5.78E-03 (55) | 5.05E-03 (25) | 8.70E-06 (47) |
| Autism | NS | 5.77E-03 (15) | NS |
| Global developmental delay | NS | 4.40E-04 (4) | NS |
| Developmental delay | 9.55E-04 (20) | NS | NS |
| Development of central nervous system | NS | 4.06E-03 (21) | NS |
| Development of neurons | NS | 5.14E-03 (21) | NS |
| Morphogenesis of neurons | NS | 4.18E-04 (19) | NS |
| Neuritogenesis | NS | 9.01E-04 (18) | NS |
| Formation of brain | NS | 1.60E-03 (14) | NS |
| Migration of neurons | NS | NS | 1.83E-05 (11) |
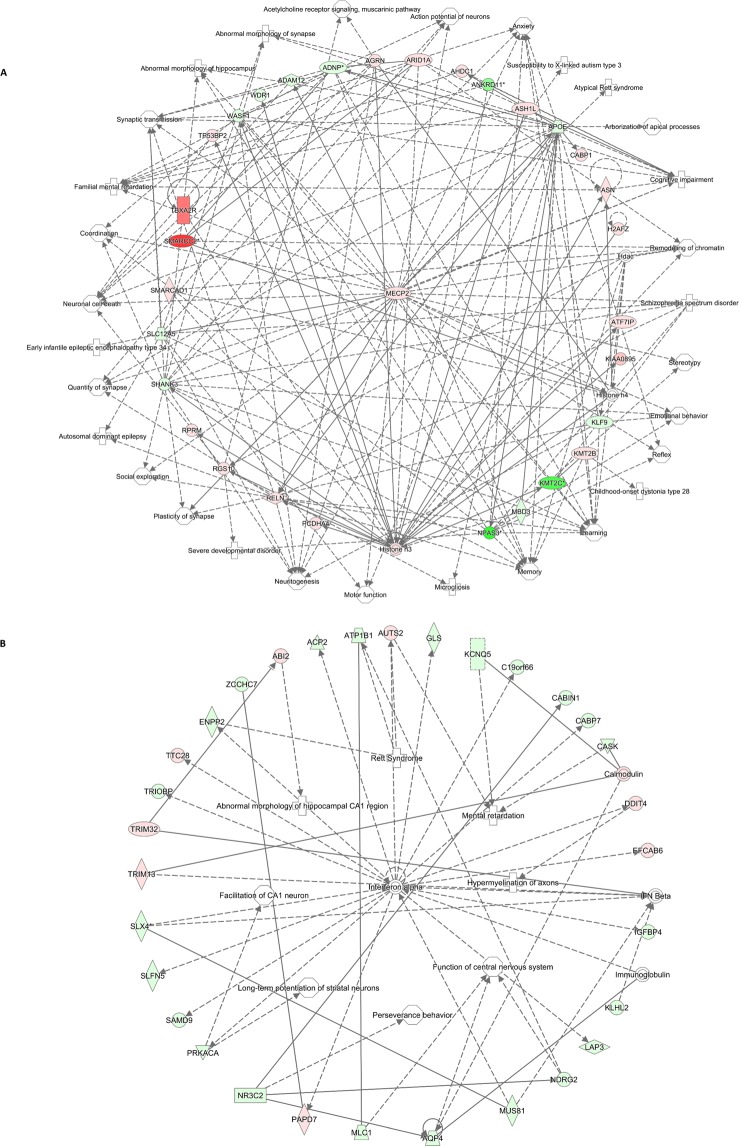
Interactome networks, which are collections of genes that interact with each other or with specific biological functions, were created using the lists of significant DEGs in males and females (Fig. 1). A representative interactome network of DEGs in the male hippocampus revealed gene interactions among DEGs and associations with disorders/diseases, neurological functions, and behaviors, including mental retardation, neuritogenesis, social exploration, learning, and motor functions (Fig. 1). Similarly, the interactome network of DEGs in the female hippocampus showed associations with Rett syndrome, perseverance behavior, and mental retardation (Fig. 1). Interestingly, the hub gene in the interactome generated using DEGs from the male hippocampus is MeCP2, which is the key gene responsible for Rett syndrome. These findings suggest that prenatal BPA exposure alters the expression of genes in the brain, which may in turn disrupt gene regulatory networks/pathways and neurological functions underlying the pathobiology of ASD.
Figure 1.
The regulatory network of DEGs in hippocampal tissues is related to neurological diseases/disorders and functions that are impacted in ASD. The gene regulatory network was predicted by IPA software using the list of DEGs from RNA-seq (colored; red = up-regulation; green = down-regulation), and the IPA showed that these genes are associated with functions that are impacted in individuals with ASD. (A) male (B) female.
In addition, to investigate whether BPA-responsive genes have divergent effects on biological pathways and networks in males and females, the separate lists of DEGs in males and females were used to predict disorders/diseases associated with ASD using IPA. Interestingly, we found that the DEGs in male but not female hippocampal tissues exclusively associated with autism (P-value = 1.18E-02, 10 genes) (Table 3). However, DEGs in both males and females were significantly associated with pervasive developmental disorder (P-value = 2.20E-02, 17 genes, and P-value = 1.44E-04, 41 genes, respectively), which is currently considered a component of ASD.
Table 3.
Comparison of neurological diseases/disorders of DEGs uniquely found in males or females.
| Diseases or Functions Annotation | P-values (number of genes) | Diseases or Functions Annotation | P-values (number of genes) |
|---|---|---|---|
| Males | Females | ||
| Huntington’s Disease | 6.67E-03 (40) | Disorder of basal ganglia | 2.72E-04 (122) |
| Pervasive developmental disorder | 2.20E-02 (17) | Dementia | 1.90E-04 (118) |
| Autism | 1.18E-02 (10) | Tauopathy | 3.82E-05 (117) |
| Alcohol withdrawal syndrome | 3.15E-02 (4) | Alzheimer disease | 1.66E-04 (110) |
| Susceptibility to Alzheimer disease | 7.03E-03 (3) | Pervasive developmental disorder | 1.44E-04 (41) |
The lists of genes that were dysregulated only in males or females were used to predict the neurological diseases/disorders associated with ASD using IPA. Significance was determined by the Fisher’ exact test, with a P-value = 0.05 as the cutoff.
To determine whether the gene expression profiles in the hippocampi of rats prenatally exposed to BPA reflect those in the brains of ASD individuals, we obtained the lists of genes that are differentially expressed in post-mortem brain tissues of ASD individuals from two previously published ASD brain transcriptome profiling studies41,42, and overlapped them with our list of BPA-responsive genes. Interestingly, we found that as many as 206, 159, and 80 genes differentially expressed due to BPA exposure in both sexes, in females, and in males, respectively, were also dysregulated in ASD post-mortem brain tissues identified by Voineague, I., et al.41. In addition, as many as 1,045, 690, and 393 genes differentially expressed by BPA exposure in both sexes, in females, and in males, respectively, were also dysregulated in ASD post-mortem brain tissues identified by Parikshak, N. N., et al.42. The lists of DEGs identified by both ASD brain transcriptome studies and genes overlapping with BPA-responsive genes are shown in Supplementary Table S5. This finding suggests that prenatal BPA exposure may result in dysregulation of at least some genes reminiscent of those altered in the brains of ASD individuals.
Quantitative RT-PCR analysis of BPA-responsive genes
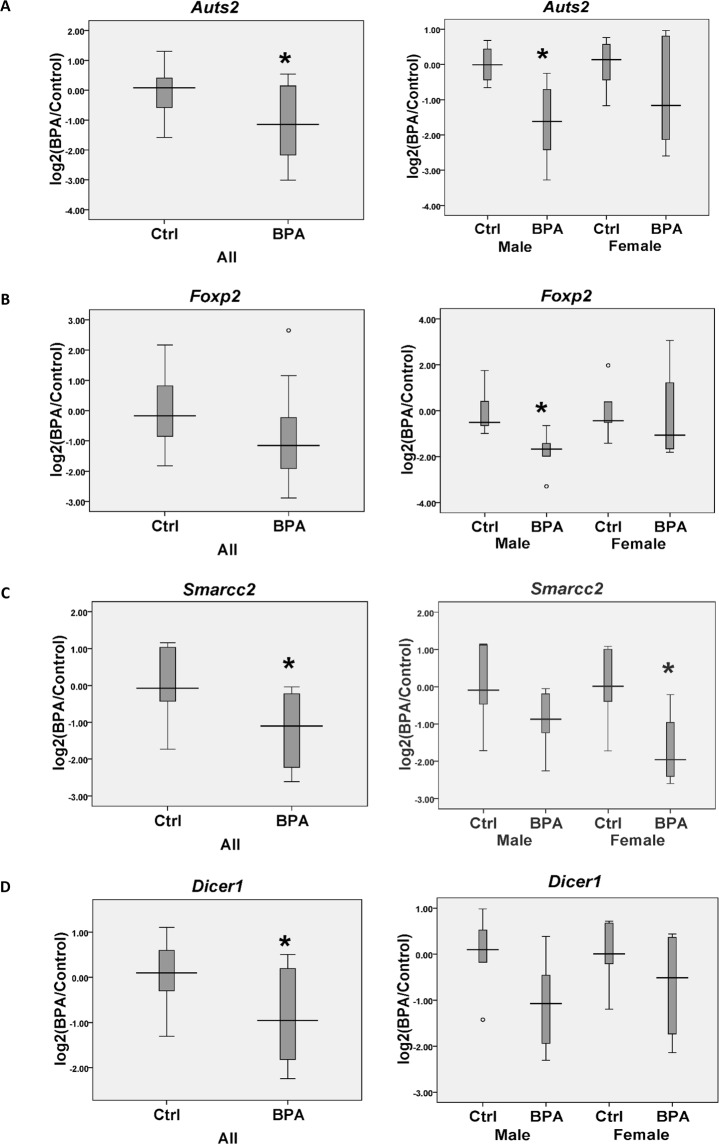
To further examine whether prenatal BPA exposure causes the dysregulation of genes in the hippocampus, four DEGs (i.e., Auts2, Foxp2, Smarcc2, and Dicer1) identified by RNA-seq analysis were selected for further confirmation by qRT-PCR analysis in another set of hippocampal tissue samples (Fig. 2). Auts2 (Autism Susceptibility Gene 2), Foxp2 (Forkhead Box P2), and Smarcc2 (SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin subfamily C member 2) have been identified as ASD candidate genes, whereas Dicer1 (Dicer 1, Ribonuclease III) is involved in a post-transcriptional gene silencing mechanism that has been associated with ASD. We found that when both males and females were combined, the expression levels of the Auts2, Smarcc2, and Dicer1 genes were significantly reduced in the hippocampi of rats prenatally exposed to BPA (Fig. 2). Foxp2 expression tended to decrease in the BPA group, although the difference was not statistically significant. Interestingly, sex-specific dysregulation of genes was observed when qRT-PCR data from each sex were analyzed separately. The expression levels of Auts2 and Foxp2 were significantly decreased in males but not in females (Fig. 2), whereas Smarcc2 expression was significantly decreased in females but not in males (Fig. 2). These results indicate that prenatal BPA exposure causes the dysregulation of genes associated with ASD in the hippocampus in a sex-dependent manner.
Figure 2.
Box plot of ASD-related gene expression in hippocampal tissues. The expression levels of Auts2 (A), Foxp2 (B), Smarcc2 (C), and Dicer1 (D) were determined in both sexes and separately in males and females. The qRT-PCR analyses revealed that Auts2 and Foxp2 were significantly down-regulated in the hippocampi of both sexes and males that were prenatally exposed to BPA. In contrast, Smarcc2 was significantly reduced in both sexes and in females, and Dicer1 was significantly reduced in both sexes. * P-value < 0.05.
DEGs in response to BPA exposure based on the integration of data from multiple transcriptomic studies revealed an association with ASD candidate genes
To determine whether BPA-responsive genes identified by other independent investigators were also associated with ASD, transcriptome profiling data from cell lines, primary cells, or tissues from animal models treated with BPA were obtained from six independent transcriptomic studies previously deposited in the NCBI GEO DataSets database (https://www.ncbi.nlm.nih.gov/gds/). The details of each study, including the title, sample size, and sample type, are shown in Supplementary Table S6. Significantly differentially expressed genes in the BPA treatment group compared with the corresponding control group from each transcriptomic study were then identified using a common statistical program for large-scale expression analyses. The lists of BPA-responsive genes from the transcriptomic studies are shown in Supplementary Table S7. We next overlapped the list of DEGs from each study with ASD candidate genes previously deposited in two different ASD databases: SFARI (https://gene.sfari.org/) and AutismKB (http://autismkb.cbi.pku.edu.cn/). Furthermore, hypergeometric distribution analyses were performed to determine whether ASD candidate genes were associated with the BPA-responsive genes from each study. Interestingly, several to hundreds of ASD candidate genes were found to be differentially expressed in response to BPA, and the hypergeometric distribution analyses revealed that the ASD candidate genes obtained from each ASD database were significantly enriched (P-value < 0.05) in the lists of BPA-responsive genes identified from four of six transcriptomic studies (Table 4).
Table 4.
Hypergeometric distribution analyses between significantly differentially expressed genes from BPA studies and autism candidate genes.
| Overlap with | GSE44387 | GSE63852 | GSE58642 | GSE50527 | GSE58516 | GSE86923 |
|---|---|---|---|---|---|---|
| SFARI database (1,007 genes) | 44 | 26 | 2 | 75 | 1 | 88 |
| *P-value from hypergeometric distribution analysis | 5.60E-10 | 1.15E-07 | 0.26 | 1.42E-15 | 0.73 | 4.35E-11 |
| AutismKB database (3,055 genes) | 142 | 76 | 3 | 168 | 7 | 289 |
| *P-value from hypergeometric distribution analysis | 4.60E-24 | 1.23E-17 | 0.59 | 1.36E-19 | 0.09 | 5.23E-41 |
Hypergeometric distribution analyses were used to analyze associations between differentially expressed genes from six previously published BPA transcriptome studies and autism candidate genes. Statistically significant associations were determined by hypergeometric distribution analysis (P-value < 0.05).
To determine whether the BPA-responsive genes identified by our study were also dysregulated in the independent studies, the lists of BPA-responsive genes in the hippocampi of rats prenatally exposed to BPA were overlapped with the BPA-responsive genes from the previously published transcriptome studies. The numbers of overlapping genes are shown in Table 5. When the DEGs from the published studies were combined, as many as 914 DEGs identified by our study were also found to be dysregulated in at least one of the independent studies (Supplementary Table S8). IPA revealed that this set of genes was significantly associated with several canonical pathways, including “Aldosterone Signaling in Epithelial Cells” (P-value = 2.14E-04), “PTEN Signaling” (P-value = 1.62E-03), “PPARα/RXRα Activation” (P-value = 5.75E-03), “Dendritic Cell Maturation” (P-value = 1.02E-02), and “Circadian Rhythm Signaling” (P-value = 1.78E-02) (Table 6). Taken together, the results of these bioinformatic analyses suggest that BPA exposure may cause dysregulation of genes associated with ASD-related biological functions in the brain as well as other tissues.
Table 5.
Numbers of overlapping genes between our list of DEGs in the hippocampus and the lists of BPA-responsive genes identified by other transcriptome profiling studies.
Table 6.
Significant canonical pathways associated with our DEGs that were also identified as BPA-responsive genes in other independent studies.
| Canonical Pathways | P-values | Genes |
|---|---|---|
| Aldosterone Signaling in Epithelial Cells | 2.14E-04 | RAF1, HSPH1, SLC12A2, TRAP1, DNAJC13, HSPD1, HSPA2, HSPA8, PIK3R3, HSP90B1, PLCB4, PIK3C3, PRKCD, PIK3CD, DNAJB6, PRKD3, HSPB6, HSPA4L, HSPB1 |
| PTEN Signalingop | 1.62E-03 | RAF1, YWHAH, BAD, ITGA5, NFKB2, CCND1, SYNJ2, PIK3R3, GHR, PIK3CD, INSR, FGFRL1, FASLG, PDGFRB |
| PPARα/RXRα Activation | 5.75E-03 | RAF1, IL1RL1, MED1, NFKB2, TGS1, PRKAG1, HSP90B1, PLCB4, GHR, GPD2, LPL, SMAD4, NCOR1, INSR, NFKBIB, MED24 |
| Dendritic Cell Maturation | 1.02E-02 | LEP, FCGR2A, HLA-A, TYROBP, HLA-DQA1, NFKB2, MAPK11, PIK3R3, PLCB4, PIK3C3, CD86, ATF4, PIK3CD, TLR3, IL23A, NFKBIB |
| Circadian Rhythm Signaling | 1.78E-02 | PER1, GRIN2A, ATF4, VIP, PER2 |
The list of DEGs in the hippocampus was overlapped with the list of BPA-responsive genes identified by other studies. Canonical pathways associated with the overlapping genes were analyzed by IPA software. P-values were calculated using Fisher’s exact test (P < 0.05).
Discussion
Accumulating evidence from both in vitro and in vivo studies indicates that exposure to BPA, even at low doses, disrupts the expression of multiple genes in the brain and alters the behaviors of offspring from exposed females43,44. Increased BPA levels have been reported in the blood and urine of ASD children compared with typically developing children37–39, prompting the hypothesis that BPA may be an environmental risk factor for ASD and that exposure to BPA, especially during pregnancy, may cause and/or increase the risk of ASD. However, whether prenatal BPA exposure causes the dysregulation of genes associated with ASD in the brain that could lead to the pathobiological conditions associated with ASD has never been investigated.
This is the first study to demonstrate that BPA exposure can cause sex-dependent changes in the transcriptome profiles of many genes involved in biological functions known to be negatively impacted in ASD, and that significant associations exists between BPA-responsive genes and dysregulated genes observed in individuals with ASD. Using rats as an experimental model, we demonstrated that prenatal BPA exposure in pregnant dams dysregulated the transcriptome profiles of ASD candidate genes in the brains of the offspring. Specifically, RNA-seq analysis of hippocampal tissues isolated from prenatally exposed neonatal rats showed sex differences in the response to BPA exposure, with 2,078 and 3,522 DEGs in the hippocampi of males and females, respectively, indicating that prenatal BPA exposure affects brain transcriptome profiles in a sex-dependent manner. Sex differences in the effects of prenatal BPA exposure on brain transcriptome profiles have also been reported in recent studies43,45. Arambula et al. (2016) conducted a transcriptome profiling analysis of hypothalami and hippocampi isolated from neonatal rats prenatally exposed to BPA43 and found that BPA induced sex-specific effects on hypothalamic ERα and ERβ (Esr1 and Esr2) expression and hippocampal and hypothalamic oxytocin (Oxt) expression. Moreover, prenatal BPA exposure was reported to disrupt the transcriptome of the neonate amygdala in a sex-specific manner45.
Interestingly, when overlapped with the lists of ASD candidate genes, the list of DEGs in males identified in this study exhibited stronger associations with ASD genes than the DEGs in females. Moreover, we found significant enrichment of ASD genes on the X chromosome in the lists of ASD-related DEGs in both males and females, suggesting that BPA exerts its effect on the brain partly through X-linked genes, which provides a plausible explanation for the sex difference in BPA effects on the brain transcriptome. Notably, the X chromosome theory of ASD46–48 posits that the male bias of ASD partly involves genes on the X chromosome, the dysregulation of which increases susceptibility to ASD. This result suggests that prenatal BPA exposure may elevate the risk of ASD in males and may help explain the higher male prevalence of ASD, which deserves further study. Additionally, IPA showed that DEGs in the hippocampus were significantly associated with ASD and mental retardation. Canonical pathways associated with DEGs in both males and females included glutamate receptor signaling, axonal guidance signaling, and circadian rhythm signaling, all of which have been associated with ASD49–51. Interestingly, several neuro/biological functions and disorders, including “autism”, “global developmental delay”, “formation of brain”, “neuritogenesis”, and “inflammatory response”, were associated with DEGs in the male hippocampus only. The canonical pathway analysis also revealed significant associations of DEGs with “DNA methylation and transcriptional repression signaling” and “4-aminobutyrate degradation” in male only, both of which have been associated with ASD3,52–54. We then overlapped the DEGs in males together with those in females, and the lists of genes that were found to be dysregulated in only males or females were separately analyzed to demonstrate diseases/disorders specific to male and female DEGs. The results revealed that genes that were dysregulated in males were significantly associated with “Autism” (P-value = 1.18E-02) while the dysregulated genes in females were associated with “Pervasive developmental disorder” (P- value = 1.44E-04). Pervasive development disorder is a group of disorders characterized by developmental delays of socialization and communication skills, consisting of autism, Asperger syndrome, Rett syndrome, childhood disintegrative disorder, and pervasive developmental disorder-not other wised specified (PDD-NOS). In the DSM-5, all of these neurodevelopmental conditions, except for Rett syndrome, were grouped into the new classification for autism spectrum disorder (ASD) which has an overall prevalence of approximately 1 in 59 children and is 4 times higher in males than females1. This result suggests that exposure to BPA during pregnancy can cause divergent effects on the expression of genes associated with ASD in both sexes, but may be more directly associated with classic autism (typically considered the most severe subtype) in males. Interactome analysis showed that Mecp2, a gene located on the X chromosome encoding the methyl-CpG binding protein 2, served as the hub gene in a biological network of DEGs in the hippocampus. This protein mediates transcriptional repression through interaction with histone deacetylase55,56 and plays a role in the maintenance of synapses and normal brain function57,58. Loss-of-function mutations of MeCP2 in humans are known to cause Rett syndrome, a childhood neurodevelopmental disorder with some ASD-related symptoms that affects females almost exclusively. An increased MeCP2 gene copy number was reported in males with neurodevelopmental delay who exhibited autistic-like features, absent speech, stereotypic movements, and infantile hypotonia59. Moreover, increased binding of MeCP2 to the promoters of GAD1 and RELN which are candidate genes for ASD was also found in the ASD cerebellum60. This evidence suggests that the up-regulation of Mecp2 due to prenatal exposure to BPA may lead to ASD-like symptoms, which should be further studied.
We then conducted quantitative RT-PCR analyses to further investigate the expression levels of four DEGs (Auts2, Foxp2, Smarcc2, and Dicer1) in the hippocampi of neonatal rats prenatally exposed to BPA compared with vehicle control. Auts2, Smarcc2, and Dicer1 were significantly reduced in the hippocampi of the BPA group compared with the control, whereas Foxp2 tended to decrease but did not show a statistically significant difference. Although the expression levels of these four genes seemed to be reduced in rats of both sexes exposed to BPA, there were some sex differences in the effects of BPA exposure on the expression levels of these genes. Auts2 and Foxp2 were significantly decreased in the hippocampi of male rats exposed to BPA compared with sex-matched controls, but these differences were not observed in females. Smarcc2, in contrast, was significantly decreased in females prenatally exposed to BPA, but not in males. These findings suggest that prenatal BPA exposure may pose an increased risk of ASD in males and females by disrupting the expression profiles of ASD-related genes, providing a plausible explanation for how an environmental factor can contribute to ASD susceptibility. The molecular mechanisms underlying how BPA affects differential gene expression between males and females should be studied further, but evidence indicates that exposure to BPA can alter genes related to global DNA methylation and histone modification processes44,61.
Auts2 (Autism Susceptibility Candidate 2) is an ASD candidate gene that has been associated with ASD and other neurodevelopmental disorders that are comorbid with ASD, including intellectual disability62 and developmental delay62. Auts2 is abundantly expressed in the developing brain and is mostly expressed in the hippocampus, prefrontal cortex, and cerebellum63, which are brain regions known to be impacted in individuals with ASD64. Recent studies have revealed that Auts2 is important for neuronal development. Knockout of both coding and noncoding sequences of the Auts2 gene in zebrafish caused microcephaly and a decreased number of neuronal cells65, both of which are consistently found in ASD patients66.
Foxp2 (Forkhead Box P2) encodes a member of the forkhead/winged-helix (FOX) family of transcription factors that is widely reported as a candidate gene associated with language development67. Foxp2 is expressed in the fetal and adult brain and is required for the development of speech and language regions of the brain during embryogenesis. Mutation of this gene has been reported in speech-language disorder 1 (SPCH1), also known as autosomal dominant speech and language disorder with orofacial dyspraxia. A single-nucleotide polymorphism (SNP) in the FOXP2 gene has been associated with social deficits in ASD patients68,69. Moreover, the disruption of Foxp2 in mice caused altered ultrasonic vocalization70.
Smarcc2 (SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin subfamily C member 2) encodes a member of the SWI/SNF family of proteins. The functions of this gene include transcriptional activation and repression by chromatin remodeling process71. Smarcc2 is highly expressed in the brain and is required for the differentiation of stem/progenitor cells into mature neural cells during neural development. Recent studies reported that mutation of Smarcc2 resulted in alteration of chromatin remodeling complexes in ASD6. A de novo splice-site variant in this gene was also observed in ASD cases72.
The Dicer1 (Dicer 1, Ribonuclease III) gene encodes a protein involved in the repression of gene expression. The protein acts as a ribonuclease that is required for RNA interference and small temporal RNA (stRNA) in the small RNA component production pathway. There is evidence that post-transcriptional mechanisms are associated with ASD. Recent studies revealed dysregulated miRNAs in the ASD brain73 and in lymphoblastoid cell lines derived from individuals with ASD8,74.
To further understand the systemic effects of BPA, we identified BPA-responsive genes using the transcriptome profiles of cells/tissues isolated from animals exposed to BPA because of the limitation of brain transcriptome data in the GEO DataSets database. In addition, we attempted to use several statistical tests, such as t-test with standard Bonferroni correction, to identify the DEGs, but we were unable to identify any DEGs from the studies under these stringent conditions for multiple testing correction. We then used student’s t-test to re-analyze the significant DEGs from other studies with the goal of identifying some genes that are dysregulated due to BPA exposure in other cells/tissues. Hypergeometric distribution analyses were then performed using BPA-responsive genes from each transcriptomic study and lists of ASD candidate genes obtained from two different ASD bioinformatic databases. We found that ASD candidate genes were significantly enriched in BPA-responsive genes in four transcriptomic studies. Interestingly, one of these four transcriptomic studies investigated the effects BPA exposure on the transcriptome profiles of mouse placenta75. That study found that in utero exposure to BPA disrupted blood vessel development and morphology in the placenta. BPA exposure caused narrowing of blood vessels and disrupted the embryonic head and forelimb structures76. A recent study revealed that altered maternal vascular malperfusion was significantly associated with the pathobiology of ASD and increased the risk of ASD77.
Moreover, we overlapped the DEGs from our study with DEGs from other BPA studies in different cell types or tissues. Interestingly, we found some overlapping genes among these sets of genes, suggesting that genes that are found to be differentially expressed in the brain also show differential expression in response to BPA in other tissues. The set of overlapping genes was significantly associated with pathways impacted in ASD. There is some evidence showing that “Aldosterone Signaling”78, “PTEN signaling”79 and “Circadian Rhythm”51 are implicated in ASD patients. These findings suggest that BPA exposure may cause changes in the transcriptome profiles of genes involved in biological functions known to be impacted in ASD.
In addition to changes in transcriptome profiles, recent studies have shown that prenatal BPA exposure altered neurological functions, including neurogenesis in the hippocampus and hypothalamus and synaptic density in mouse models31,80,81. Moreover, prenatal BPA exposure induced behavioral impairments in offspring, such as in learning and memory82 and in social interaction82, along with anxiety-like behavior31. Whether the changes in the transcriptome profiles observed in this study could lead to altered neurological functions and behaviors should be investigated further. Moreover, in this study we used oral administration of BPA at 5,000 µg/kg of maternal BW/day which is equal to the NOAEL in humans determined by the FDA and ESFA. The TDI in humans is 50 µg/kg BW/day, and the estimated BPA exposure levels from use in food-contacting materials in infants and adults are 2.42 µg/kg BW/day and 0.185 µg/kg BW/day, respectively22. The effects of prenatal BPA exposure at the TDI and these estimated daily doses in humans on the brain transcriptome and functions warrant further investigation. Moreover, the molecular mechanisms through which BPA disrupts the expression of genes associated with ASD deserve further study.
Conclusions
In this study, transcriptomic profiling analysis of hippocampi isolated from rats prenatally exposed to BPA revealed sex-dependent dysregulation of gene expression, with a greater number of differentially expressed genes in females. However, the genes that were disrupted in the male hippocampus showed more significant association with ASD than those in females. Interestingly, the expression of ASD candidate genes selected for validation by quantitative RT-PCR, including Auts2, Foxp2, and Smarcc2, was also sex-dependent in response to prenatal BPA exposure. Finally, re-analyses of transcriptomic data obtained from multiple published studies on the effects of BPA in various cellular, tissue, and animal models support our current findings that BPA-responsive genes are significantly associated with ASD candidate genes as well as ASD-related neurological functions and disorders. Taken together, this study shows that prenatal BPA exposure causes changes in the hippocampal expression of genes associated with ASD in a sex-specific fashion, supporting the hypothesis that BPA is an environmental risk factor for ASD, and thus providing a plausible explanation for how BPA exposure may contribute to the sex bias of ASD.
Methods
Animal husbandry and treatment
Eight-week-old female and male Wistar rats were purchased from the National Laboratory Animal Center (NLAC), Thailand. All animals were housed at the Chulalongkorn University Laboratory Animal Center (CULAC) under standard temperature (21 ± 1 °C) and humidity (30–70%) conditions in a 12-h light/dark cycle with food and RO-UV water available ad libitum. Female rats (gestational day 1 (GD1); n = 8) were divided into 2 groups (control group and BPA treatment group) with a total of 4 rats per group. The weight of each rat was measured daily and used to calculate the amount of BPA or vehicle control needed to treat each rat. For BPA treatment, BPA (Sigma-Aldrich, USA) was dissolved in absolute ethanol (Merck Millipore, USA) to a final concentration of 250 mg/ml to make a stock BPA solution. Then, the stock solution was further diluted with corn oil to a final concentration of 5,000 µg/kg·maternal BW of BPA to treat each rat. The vehicle control treatment was prepared by mixing absolute ethanol with corn oil in amounts equivalent to those used for preparing BPA. After mating, each rat was intragastrically administered either BPA or the vehicle control from GD1 until parturition. To prevent cross-contamination of the treatment conditions, rats in the BPA and control groups were raised separately in individual ventilated cages in a biohazard containment housing system. Separate sets of stainless steel needles and all consumable products were used for oral gavage. All reusable materials were cleaned with ethanol and rinsed with copious amounts of Milli-Q deionized water before use. All experimental procedures were approved by the Chulalongkorn University Animal Care and Use Committee (Animal Use Protocol No. 1673007 and No. 1773011), Chulalongkorn University. We confirm that all experiments were performed in accordance with the relevant guidelines and regulations.
RNA isolation and transcriptome profiling analysis
Male and female neonatal pups were euthanized (BPA n = 6; control n = 6), and the hippocampi were isolated as previously described with slight modifications83. Briefly, neonatal pups were euthanized by decapitation on ice following intraperitoneal injection of 100 mg/kg·BW sodium pentobarbital. The brain was quickly removed from the head and placed in a pre-chilled tube containing ice-cold, freshly prepared 1X HBSS (Invitrogen, USA) containing 30 mM glucose (Sigma-Aldrich, USA), 2 mM HEPES (GE Healthcare Bio-Sciences, USA), and 26 mM NaHCO3 (Sigma-Aldrich, USA). The brain was then dissected, and the hippocampus was isolated under a Nikon SMZ18 Stereo Microscope (Nikon, Japan). Meninges were removed completely, and the hippocampal tissues were immediately placed in a tube with RNAlater (Ambion, USA) and stored at −80 °C, according to the manufacturer’s protocol, until use.
Total RNA from the hippocampus was isolated and purified using the mirVana miRNA Isolation Kit (Ambion, USA) according to the manufacturer’s protocol. The RNA integrity was assessed using an Agilent Bioanalyzer (BGI, Hong Kong). To identify DEGs in the hippocampus in response to prenatal BPA exposure, a transcriptome profiling analysis of total RNA isolated from the hippocampi of neonatal rats from six independent litters prenatally exposed to BPA or vehicle control was performed by BGI Genomics Co., Ltd using the Illumina HiSeq 4000 next-generation sequencing platform with 4 G reads (Illumina, Inc.) according to the manufacturer’s protocol. Briefly, total RNA was treated with DNase I, and oligo(dT) treatment was used for mRNA isolation. Next, the RNA was mixed with fragmentation buffer to fragment the mRNA. Then, cDNA was synthesized using the mRNA fragments as templates. Subsequently, sequencing reads were filtered and subjected to quality control. Clean reads in a FASTQ file were mapped to the rat reference genome (RefSeq ID: 1174938) using Bowtie 284 and gene expression levels were then calculated using RSEM85. We then compared the transcriptome profiles between the BPA and the control groups with Poisson distribution. Comparisons were performed with all male and female pups with the same treatment condition combined into one group and separately for each sex. P-values were calculated using a Poisson distribution method. DEGs with a P-value < 0.05 and FDR < 0.05 were considered statistically significant.
Quantitative RT-PCR analysis
Four DEGs in the hippocampus identified by RNA-seq transcriptomic analysis were selected for further confirmation by quantitative RT-PCR analysis. These four DEGs were selected for further validation based on differential expression between males and females as well as known association with ASD. Total RNA was used for cDNA synthesis with the AccuPower® RT PreMix (Bioneer, Korea) according to the manufacturer’s protocol. Briefly, 0.5 µg total RNA was mixed with 0.5 µg (100 pmol) oligo dT18 primer, and DEPC-treated water was added to 15 µl. Then, the reaction was incubated at 70 °C for 5 min and placed on ice. To perform the cDNA synthesis, the mixture (15 µl) was then transferred to an AccuPower® RT PreMix tube, and DEPC-treated water was added to 20 µl. The cDNA synthesis reaction was performed by incubating the reaction at 42 °C for 60 min, followed by 94 °C for 5 min. The cDNA reaction mixture was further diluted to a volume of 50 μl with nuclease-free water and was used as a template for subsequent qPCR analyses. Quantitative PCR analysis was conducted in triplicate using AccuPower® 2X GreenStar™ qPCR MasterMix (Bioneer, Korea) according to the manufacturer’s instructions. Briefly, 1 μl of the cDNA was mixed with 2X Greenstar Master Mix, forward primer, reverse primer, and nuclease-free water. The reaction was then incubated in a Bio-Rad CFX Connect Real-Time System (Bio-Rad, USA). The PCR amplification conditions were set as follows: an initial denaturing step at 95 °C for 15 min, followed by 40 cycles of 10 s at 95 °C for denaturing and 30 s at 55 °C for annealing/extension. Product formation was confirmed by melting curve analysis (65 to 95 °C). The expression levels were calculated by the 2−ΔΔCt method using the 18 S ribosomal RNA (Rn18s) gene as an endogenous control. The specific primers in the qPCR analyses were designed using the UCSC Genome Browser (https://genome.ucsc.edu/), Ensembl (https://asia.ensembl.org/index.html), and Primer3 software (http://bioinfo.ut.ee/primer3-0.4.0/). Forward and reverse primers were designed for rat Auts2, Foxp2, Smarcc2, and Dicer1, and Rn18s. The sequences of the qPCR primers are shown in Supplementary Table S9.
Prediction of biological functions and interactome analysis
Biological functions, disorders, canonical pathways, and interactome networks associated with DEGs were predicted using IPA software (Qiagen Inc., USA, https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis/). The list of DEGs was overlapped with the list of genes experimentally validated to be associated with each function/disorder/canonical pathway in the Ingenuity’s Knowledge Base database. Fisher’s exact test was then performed to calculate P-values, and a P-value < 0.05 was considered statistically significant.
Transcriptome data collection
Transcriptome profiling data of cells/tissues dissected from animals exposed to BPA or vehicle controls were obtained from the NCBI Gene Expression Omnibus database (GEO DataSets: http://www.ncbi.nlm.nih.gov/gds) in a search performed on May 13, 2017, using the keyword “bisphenol A” and the following criteria: i) the experimental models were animals, primary cells, or cell lines; and ii) each treatment group consisted of more than three samples. Transcriptome profiling data of cells exposed to chemicals other than BPA, when present in any selected study, were excluded prior to subsequent differential expression analyses.
Identification of BPA-responsive genes and association with ASD candidate genes
To identify significant BPA-responsive genes in cells/tissues exposed to BPA, the transcriptome profile from each BPA study was analyzed separately using Multiple Experiment Viewer (MeV) (http://mev.tm4.org/)86. All transcriptome profiling data were filtered using a 70% cutoff, which removed transcripts for which intensity values were missing in >30% of the samples. The available transcripts were then used for identifying DEGs in the BPA group with two-tailed t-tests. Lists of ASD-related genes were obtained from two different ASD databases: the SFARI database (updated on April 17, 2018) (https://gene.sfari.org/) and the AutismKB database (from May 25, 2012) (http://autismkb.cbi.pku.edu.cn/). To determine whether the BPA-responsive genes identified in each transcriptomic study were significantly associated with ASD candidate genes, the list of BPA-responsive genes was overlapped with the list of ASD candidate genes from each ASD database, and a hypergeometric distribution analysis was conducted using the Hypergeometric Distribution Calculator program in the Keisan Online Calculator package (http://keisan.casio.com/exec/system/1180573201). There are four variables in the Hypergeometric Distribution Calculator: number of overlapping genes, total number of DEGs in the experiment, total number of ASD-candidate genes, and total number of genes from RNA-seq analysis.
Statistical analyses
Statistical analyses were conducted using SPSS version 16.0. The criterion for statistical significance was a P-value < 0.05. A two-tailed Student’s t-test was used to determine the statistical significance of differences between the mean values of two groups. A hypergeometric distribution analysis was performed to determine the association of DEGs with ASD candidate genes obtained from the SFARI (https://gene.sfari.org/) and AutismKB (http://autismkb.cbi.pku.edu.cn/) databases using the Hypergeometric Distribution Calculator in the Keisan Online Calculator program (http://keisan.casio.com/exec/system/1180573201). A P-value < 0.05 was considered statistically significant.
Ethics approval and informed consent
All animal experimental procedures were approved by the Chulalongkorn University Animal Care and Use Committee (Animal Use Protocol No. 1673007 and No. 1773011), Chulalongkorn University.
Supplementary information
Acknowledgements
S.T. and S.K. are graduate students in the Ph.D. Program in Clinical Biochemistry and Molecular Medicine, Department of Clinical Chemistry, Faculty of Allied Health Sciences, Chulalongkorn University. We wish to thank Dr. Noriko Osumi and Dr. Takako Kikkawa, Department of Developmental Neuroscience, Tohoku University, Sendai, Japan, for their suggestions regarding the animal experiments. We also wish to acknowledge Asst. Prof. Dr. Anusak Kijtawornrat, Dr. Nitira Anakkul and Mr. Choopet Nitsakulthong, Chulalongkorn University Laboratory Animal Center, for their help with the ethics approval process and for training S.T., S.K., and T.S. in the proper care and use of laboratory animals. This work was supported by a Research Grant for New Scholars by The Thailand Research Fund and Office of the Higher Education Commission (MRG6080118) and partly supported by the Asahi Glass Foundation (RES_60_341_37) to TS. The conclusions in this article were made by the authors. The Thailand Research Fund and Office of the Higher Education Commission may not necessarily agree. ST was financially supported by The 90th Anniversary Chulalongkorn University Fund (Ratchadaphiseksomphot Endowment Fund: GCUGR1125601056M 53-2), “The Scholarship from the Graduate School, Chulalongkorn University to commemorate the 72nd anniversary of His Majesty King Bhumibhol Adulyadej”, and “The Overseas Research Experience Scholarship for Graduate Students from the Graduate School and Faculty of Allied Health Sciences, Chulalongkorn University”. SK was supported by The 100th Anniversary Chulalongkorn University Fund for Doctoral Scholarship. VWH is supported by NIEHS grants R21 ES023061 and R21 ES028124.
Author Contributions
S.T. performed all the experiments, analyzed the data, and drafted the manuscript under the supervision of T.S. and D.J. S.K. isolated RNA, conducted the quantitative RT-PCR analysis, and assisted S.T. in animal treatment and tissue collection. T.T. served as the mentor of T.S. for the Research Grant for New Scholars and supervised to S.T. and S.K. V.W.H. provided the Ingenuity Pathway Analysis program and participated in the editing of this manuscript. T.S. conceived of the study, designed the experiments, analyzed the data, interpreted the results, determined the conclusion, and participated in the writing and editing of this manuscript. All the authors read and approved the final manuscript.
Data Availability
The transcriptome profiling data used in this study have been published in the NCBI GEO DataSets database (GSE44387, GSE63852, GSE58642, GSE50527, GSE58516, and GSE86923). The RNA-seq data will be made publicly available in the GEO upon acceptance of this manuscript for publication.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-39386-w.
References
- 1.Baio J, et al. Prevalence of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities MonitoringNetwork, 11 Sites, United States, 2014. Morbidity and mortality weekly report. Surveillance summaries (Washington, D.C.: 2002) 2018;67:1–23. doi: 10.15585/mmwr.ss6706a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tangsuwansri C, et al. Investigation of epigenetic regulatory networks associated with autism spectrum disorder (ASD) by integrated global LINE-1 methylation and gene expression profiling analyses. PloS one. 2018;13:e0201071. doi: 10.1371/journal.pone.0201071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saeliw T, et al. Integrated genome-wide Alu methylation and transcriptome profiling analyses reveal novel epigenetic regulatory networks associated with autism spectrum disorder. Molecular autism. 2018;9:27. doi: 10.1186/s13229-018-0213-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nguyen A, Rauch TA, Pfeifer GP, Hu VW. Global methylation profiling of lymphoblastoid cell lines reveals epigenetic contributions to autism spectrum disorders and a novel autism candidate gene, RORA, whose protein product is reduced in autistic brain. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2010;24:3036–3051. doi: 10.1096/fj.10-154484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moosa A, Shu H, Sarachana T, Hu VW. Are endocrine disrupting compounds environmental risk factors for autism spectrum disorder? Hormones and behavior. 2018;101:13–21. doi: 10.1016/j.yhbeh.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.LaSalle JM. Autism genes keep turning up chromatin. OA autism. 2013;1:14-. doi: 10.13172/2052-7810-1-2-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun W, et al. Histone Acetylome-wide Association Study of Autism Spectrum Disorder. Cell. 2016;167:1385–1397.e1311. doi: 10.1016/j.cell.2016.10.031. [DOI] [PubMed] [Google Scholar]
- 8.Sarachana T, Zhou R, Chen G, Manji HK, Hu VW. Investigation of post-transcriptional gene regulatory networks associated with autism spectrum disorders by microRNA expression profiling of lymphoblastoid cell lines. Genome medicine. 2010;2:23. doi: 10.1186/gm144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalkbrenner AE, Schmidt RJ, Penlesky AC. Environmental chemical exposures and autism spectrum disorders: a review of the epidemiological evidence. Current problems in pediatric and adolescent health care. 2014;44:277–318. doi: 10.1016/j.cppeds.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujiwara T, Morisaki N, Honda Y, Sampei M, Tani Y. Chemicals, Nutrition, and Autism Spectrum Disorder: A Mini-Review. Frontiers in Neuroscience. 2016;10:174. doi: 10.3389/fnins.2016.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miodovnik A, et al. Endocrine disruptors and childhood social impairment. Neurotoxicology. 2011;32:261–267. doi: 10.1016/j.neuro.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams JB, et al. Toxicological status of children with autism vs. neurotypical children and the association with autism severity. Biological trace element research. 2013;151:171–180. doi: 10.1007/s12011-012-9551-1. [DOI] [PubMed] [Google Scholar]
- 13.Geier DA, Audhya T, Kern JK, Geier MR. Blood mercury levels in autism spectrum disorder: Is there a threshold level? Acta neurobiologiae experimentalis. 2010;70:177–186. doi: 10.55782/ane-2010-1789. [DOI] [PubMed] [Google Scholar]
- 14.Roberts EM, et al. Maternal residence near agricultural pesticide applications and autism spectrum disorders among children in the California Central Valley. Environmental Health Perspectives. 2007;115:1482–1489. doi: 10.1289/ehp.10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalkbrenner AE, et al. Maternal smoking during pregnancy and the prevalence of autism spectrum disorders, using data from the autism and developmental disabilities monitoring network. Environmental Health Perspectives. 2012;120:1042–1048. doi: 10.1289/ehp.1104556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diamanti-Kandarakis E, et al. Endocrine-Disrupting Chemicals: An Endocrine Society Scientific Statement. Endocrine Reviews. 2009;30:293–342. doi: 10.1210/er.2009-0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schug TT, Janesick A, Blumberg B, Heindel JJ. Endocrine disrupting chemicals and disease susceptibility. The Journal of steroid biochemistry and molecular biology. 2011;127:204–215. doi: 10.1016/j.jsbmb.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brotons JA, Olea-Serrano MF, Villalobos M, Pedraza V, Olea N. Xenoestrogens released from lacquer coatings in food cans. Environmental Health Perspectives. 1995;103:608–612. doi: 10.1289/ehp.95103608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Olea N, et al. Estrogenicity of resin-based composites and sealants used in dentistry. Environmental Health Perspectives. 1996;104:298–305. doi: 10.1289/ehp.96104298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnan AV, Stathis P, Permuth SF, Tokes L, Feldman D. Bisphenol-A: an estrogenic substance is released from polycarbonate flasks during autoclaving. Endocrinology. 1993;132:2279–2286. doi: 10.1210/endo.132.6.8504731. [DOI] [PubMed] [Google Scholar]
- 21.Konieczna A, Rutkowska A, Rachon D. Health risk of exposure to Bisphenol A (BPA) Roczniki Panstwowego Zakladu Higieny. 2015;66:5–11. [PubMed] [Google Scholar]
- 22.World Health Organization. Food and Agriculture Organization of United Nations: Bisphenol A (BPA) Current state of knowledge and future actions by WHO and FAO. International Food Safety Authorities Network (INFOSAN) (2009).
- 23.Nishikawa M, et al. Placental transfer of conjugated bisphenol A and subsequent reactivation in the rat fetus. Environmental health perspectives. 2010;118:1196–1203. doi: 10.1289/ehp.0901575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y, Nakashima MN, Takahashi M, Kuroda N, Nakashima K. Determination of bisphenol A in rat brain by microdialysis and column switching high-performance liquid chromatography with fluorescence detection. Biomedical chromatography: BMC. 2002;16:319–326. doi: 10.1002/bmc.161. [DOI] [PubMed] [Google Scholar]
- 25.Hu F, et al. Bisphenol A Impairs Synaptic Plasticity by Both Pre- and Postsynaptic Mechanisms. Advanced Science. 2017;4:1600493. doi: 10.1002/advs.201600493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tando S, et al. Effects of pre- and neonatal exposure to bisphenol A on murine brain development. Brain & development. 2007;29:352–356. doi: 10.1016/j.braindev.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Kim K, et al. Potencies of bisphenol A on the neuronal differentiation and hippocampal neurogenesis. Journal of toxicology and environmental health. Part A. 2009;72:1343–1351. doi: 10.1080/15287390903212501. [DOI] [PubMed] [Google Scholar]
- 28.Komada M, et al. Maternal bisphenol A oral dosing relates to the acceleration of neurogenesis in the developing neocortex of mouse fetuses. Toxicology. 2012;295:31–38. doi: 10.1016/j.tox.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 29.Xu XH, Zhang J, Wang YM, Ye YP, Luo QQ. Perinatal exposure to bisphenol-A impairs learning-memory by concomitant down-regulation of N-methyl-D-aspartate receptors of hippocampus in male offspring mice. Hormones and behavior. 2010;58:326–333. doi: 10.1016/j.yhbeh.2010.02.012. [DOI] [PubMed] [Google Scholar]
- 30.Jardim NS, Sartori G, Sari MHM, Muller SG, Nogueira CW. Bisphenol A impairs the memory function and glutamatergic homeostasis in a sex-dependent manner in mice: Beneficial effects of diphenyl diselenide. Toxicology and applied pharmacology. 2017;329:75–84. doi: 10.1016/j.taap.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 31.Kumar D, Thakur MK. Anxiety like behavior due to perinatal exposure to Bisphenol-A is associated with decrease in excitatory to inhibitory synaptic density of male mouse brain. Toxicology. 2017;378:107–113. doi: 10.1016/j.tox.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 32.Fan, Y. & Tian, C. Preconception paternal bisphenol A exposure induces sex-specific anxiety and depression behaviors in adult rats. 13, e0192434, 10.1371/journal.pone.0192434 (2018). [DOI] [PMC free article] [PubMed]
- 33.Wolstenholme JT, et al. Gestational exposure to bisphenol a produces transgenerational changes in behaviors and gene expression. Endocrinology. 2012;153:3828–3838. doi: 10.1210/en.2012-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jamain S, et al. Reduced social interaction and ultrasonic communication in a mouse model of monogenic heritable autism. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1710–1715. doi: 10.1073/pnas.0711555105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams DL, Goldstein G, Minshew NJ. The Profile of Memory Function in Children With Autism. Neuropsychology. 2006;20:21–29. doi: 10.1037/0894-4105.20.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.White SW, Oswald D, Ollendick T, Scahill L. Anxiety in children and adolescents with autism spectrum disorders. Clinical psychology review. 2009;29:216–229. doi: 10.1016/j.cpr.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kardas F, et al. Increased Serum Phthalates (MEHP, DEHP) and Bisphenol A Concentrations in Children With Autism Spectrum Disorder: The Role of Endocrine Disruptors in Autism Etiopathogenesis. Journal of child neurology. 2016;31:629–635. doi: 10.1177/0883073815609150. [DOI] [PubMed] [Google Scholar]
- 38.Kondolot M, et al. Plasma phthalate and bisphenol a levels and oxidant-antioxidant status in autistic children. Environmental toxicology and pharmacology. 2016;43:149–158. doi: 10.1016/j.etap.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 39.Stein TP, Schluter MD, Steer RA, Guo L, Ming X. Bisphenol A Exposure in Children With Autism Spectrum Disorders. Autism research: official journal of the International Society for Autism Research. 2015;8:272–283. doi: 10.1002/aur.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaur K, Chauhan V, Gu F, Chauhan A. Bisphenol A induces oxidative stress and mitochondrial dysfunction in lymphoblasts from children with autism and unaffected siblings. Free radical biology & medicine. 2014;76:25–33. doi: 10.1016/j.freeradbiomed.2014.07.030. [DOI] [PubMed] [Google Scholar]
- 41.Voineagu I, et al. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474:380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parikshak NN, et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 2016;540:423–427. doi: 10.1038/nature20612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arambula SE, Belcher SM, Planchart A, Turner SD, Patisaul HB. Impact of Low Dose Oral Exposure to Bisphenol A (BPA) on the Neonatal Rat Hypothalamic and Hippocampal Transcriptome: A CLARITY-BPA Consortium Study. Endocrinology. 2016;157:3856–3872. doi: 10.1210/en.2016-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kundakovic M, et al. Sex-specific epigenetic disruption and behavioral changes following low-dose in utero bisphenol A exposure. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:9956–9961. doi: 10.1073/pnas.1214056110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arambula SE, Jima D, Patisaul HB. Prenatal bisphenol A (BPA) exposure alters the transcriptome of the neonate rat amygdala in a sex-specific manner: a CLARITY-BPA consortium study. Neurotoxicology. 2018;65:207–220. doi: 10.1016/j.neuro.2017.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baron-Cohen S, et al. Why are autism spectrum conditions more prevalent in males? PLoS biology. 2011;9:e1001081. doi: 10.1371/journal.pbio.1001081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gecz J, Shoubridge C, Corbett M. The genetic landscape of intellectual disability arising from chromosome X. Trends in genetics: TIG. 2009;25:308–316. doi: 10.1016/j.tig.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 48.Ropers HH, Hamel BC. X-linked mental retardation. Nature reviews. Genetics. 2005;6:46–57. doi: 10.1038/nrg1501. [DOI] [PubMed] [Google Scholar]
- 49.Mejias R, et al. Gain-of-function glutamate receptor interacting protein 1 variants alter GluA2 recycling and surface distribution in patients with autism. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4920–4925. doi: 10.1073/pnas.1102233108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Suda S, et al. Decreased expression of axon-guidance receptors in the anterior cingulate cortex in autism. Molecular autism. 2011;2:14. doi: 10.1186/2040-2392-2-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu VW, et al. Gene expression profiling differentiates autism case-controls and phenotypic variants of autism spectrum disorders: evidence for circadian rhythm dysfunction in severe autism. Autism research: official journal of the International Society for Autism Research. 2009;2:78–97. doi: 10.1002/aur.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fatemi SH, Reutiman TJ, Folsom TD, Thuras PD. GABA(A) receptor downregulation in brains of subjects with autism. Journal of autism and developmental disorders. 2009;39:223–230. doi: 10.1007/s10803-008-0646-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ladd-Acosta C, et al. Common DNA methylation alterations in multiple brain regions in autism. Molecular psychiatry. 2014;19:862–871. doi: 10.1038/mp.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nardone S, Sams DS, Zito A, Reuveni E, Elliott E. Dysregulation of Cortical Neuron DNA Methylation Profile in Autism Spectrum Disorder. Cerebral cortex (New York, N.Y.: 1991) 2017;27:5739–5754. doi: 10.1093/cercor/bhx250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nan X, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 56.Fuks F, et al. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. Journal of Biological Chemistry. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- 57.Chao H-T, Zoghbi HY, Rosenmund C. MeCP2 controls excitatory synaptic strength by regulating glutamatergic synapse number. Neuron. 2007;56:58–65. doi: 10.1016/j.neuron.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen MVC, et al. MeCP2 is critical for maintaining mature neuronal networks and global brain anatomy during late stages of postnatal brain development and in the mature adult brain. Journal of Neuroscience. 2012;32:10021–10034. doi: 10.1523/JNEUROSCI.1316-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.del Gaudio, D. et al. Increased MECP2 gene copy number as the result of genomic duplication in neurodevelopmentally delayed males. Genetics in medicine: official journal of the American College of Medical Genetics8, 784–792, 10.109701.gim.0000250502.28516.3c (2006). [DOI] [PubMed]
- 60.Zhubi A, et al. Increased binding of MeCP2 to the GAD1 and RELN promoters may be mediated by an enrichment of 5-hmC in autism spectrum disorder (ASD)cerebellum. Translational psychiatry. 2014;4:e349. doi: 10.1038/tp.2013.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Senyildiz M, Karaman EF, Bas SS, Pirincci PA, Ozden S. Effects of BPA on global DNA methylation and global histone 3 lysine modifications in SH-SY5Y cells: An epigenetic mechanism linking the regulation of chromatin modifiying genes. Toxicology in vitro: an international journal published in association with BIBRA. 2017;44:313–321. doi: 10.1016/j.tiv.2017.07.028. [DOI] [PubMed] [Google Scholar]
- 62.Fan Y, Qiu W, Wang L, Gu X, Yu Y. Exonic deletions of AUTS2 in Chinese patients with developmental delay and intellectual disability. American journal of medical genetics. Part A. 2016;170a:515–522. doi: 10.1002/ajmg.a.37454. [DOI] [PubMed] [Google Scholar]
- 63.Bedogni F, et al. Autism susceptibility candidate 2 (Auts2) encodes a nuclear protein expressed in developing brain regions implicated in autism neuropathology. Gene expression patterns: GEP. 2010;10:9–15. doi: 10.1016/j.gep.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Courchesne E, et al. Neuron number and size in prefrontal cortex of children with autism. JAMA. 2011;306:2001–2010. doi: 10.1001/jama.2011.1638. [DOI] [PubMed] [Google Scholar]
- 65.Oksenberg N, Stevison L, Wall JD, Ahituv N. Function and regulation of AUTS2, a gene implicated in autism and human evolution. PLoS genetics. 2013;9:e1003221. doi: 10.1371/journal.pgen.1003221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y, et al. De novo exon 1 deletion of AUTS2 gene in a patient with autism spectrum disorder and developmental delay: a case report and a brief literature review. American journal of medical genetics. Part A. 2015;167:1381–1385. doi: 10.1002/ajmg.a.37050. [DOI] [PubMed] [Google Scholar]
- 67.Enard W, et al. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 68.Chien YL, et al. The central nervous system patterning gene variants associated with clinical symptom severity of autism spectrum disorders. Journal of the Formosan Medical Association=Taiwan yi zhi. 2017;116:755–764. doi: 10.1016/j.jfma.2016.11.015. [DOI] [PubMed] [Google Scholar]
- 69.Gong X, et al. Association between the FOXP2 gene and autistic disorder in Chinese population. American journal of medical genetics. Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2004;127b:113–116. doi: 10.1002/ajmg.b.20162. [DOI] [PubMed] [Google Scholar]
- 70.Shu W, et al. Altered ultrasonic vocalization in mice with a disruption in the Foxp2 gene. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:9643–9648. doi: 10.1073/pnas.0503739102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kadam S, et al. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes & development. 2000;14:2441–2451. doi: 10.1101/gad.828000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu YE, Parikshak NN, Belgard TG. Genome-wide, integrative analysis implicates microRNA dysregulation in autism spectrum disorder. 2016;19:1463–1476. doi: 10.1038/nn.4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Talebizadeh Z, Butler MG, Theodoro MF. Feasibility and relevance of examining lymphoblastoid cell lines to study role of microRNAs in autism. Autism research: official journal of the International Society for Autism Research. 2008;1:240–250. doi: 10.1002/aur.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tait S, Tassinari R, Maranghi F, Mantovani A. Toxicogenomic analysis of placenta samples from mice exposed to different doses of BPA. Genomics data. 2015;4:109–111. doi: 10.1016/j.gdata.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tait S, Tassinari R, Maranghi F, Mantovani A. Bisphenol A affects placental layers morphology and angiogenesis during early pregnancy phase in mice. Journal of applied toxicology: JAT. 2015;35:1278–1291. doi: 10.1002/jat.3176. [DOI] [PubMed] [Google Scholar]
- 77.Straughen JK, et al. The association between placental histopathology and autism spectrum disorder. Placenta. 2017;57:183–188. doi: 10.1016/j.placenta.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 78.Pinggera A, et al. CACNA1D de novo mutations in autism spectrum disorders activate Cav1.3 L-type calcium channels. Biological psychiatry. 2015;77:816–822. doi: 10.1016/j.biopsych.2014.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Butler MG, et al. Subset of individuals with autism spectrum disorders and extreme macrocephaly associated with germline PTEN tumour suppressor gene mutations. Journal of medical genetics. 2005;42:318–321. doi: 10.1136/jmg.2004.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Desai M, Ferrini MG, Han G, Jellyman JK, Ross MG. In vivo maternal and in vitro BPA exposure effects on hypothalamic neurogenesis and appetite regulators. Environmental research. 2018;164:45–52. doi: 10.1016/j.envres.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jang YJ, et al. High dose bisphenol A impairs hippocampal neurogenesis in female mice across generations. Toxicology. 2012;296:73–82. doi: 10.1016/j.tox.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 82.Kundakovic M, et al. DNA methylation of BDNF as a biomarker of early-life adversity. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:6807–6813. doi: 10.1073/pnas.1408355111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Guo W, Patzlaff NE, Jobe EM, Zhao X. Isolation of multipotent neural stem/progenitor cells from both the dentate gyrus and subventricular zone of a single adult mouse. Nature protocols. 2012;7:2005–2012. doi: 10.1038/nprot.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Saeed AI, et al. TM4: a free, open-source system for microarray data management and analysis. BioTechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The transcriptome profiling data used in this study have been published in the NCBI GEO DataSets database (GSE44387, GSE63852, GSE58642, GSE50527, GSE58516, and GSE86923). The RNA-seq data will be made publicly available in the GEO upon acceptance of this manuscript for publication.