Abstract
Heart failure affects 2–3% of adult Western population. Prevalence of heart failure with preserved left ventricular (LV) ejection fraction (HFpEF) increases. Studies suggest HFpEF patients to have altered myocardial structure and functional changes such as incomplete relaxation and increased cardiac stiffness. We hypothesised that patients undergoing elective coronary bypass surgery (CABG) with HFpEF characteristics would show distinctive gene expression compared to patients with normal LV physiology. Myocardial biopsies for mRNA expression analysis were obtained from sixteen patients with LV ejection fraction ≥45%. Five out of 16 patients (31%) had echocardiographic characteristics and increased NTproBNP levels indicative of HFpEF and this group was used as HFpEF proxy, while 11 patients had Normal LV physiology. Utilising principal component analysis, the gene expression data clustered into two groups, corresponding to HFpEF proxy and Normal physiology, and 743 differentially expressed genes were identified. The associated top biological functions were cardiac muscle contraction, oxidative phosphorylation, cellular remodelling and matrix organisation. Our results also indicate that upstream regulatory events, including inhibition of transcription factors STAT4, SRF and TP53, and activation of transcription repressors HEY2 and KDM5A, could provide explanatory mechanisms to observed gene expression differences and ultimately cardiac dysfunction in the HFpEF proxy group.
Introduction
Coronary artery disease and hypertension are the most common etiologic factors for heart failure (HF). HF is characterised according to left ventricular ejection fraction (LVEF) as preserved (HFpEF, LVEF ≥ 45%) or reduced (HFrEF < 45%), as defined in our PREFERS design paper1. Heart failure affects 2–3% of the adult Western population2,3 and prevalence increases with age4,5. In particular, the proportion of HFpEF is increasing6,7, with poor prognosis8 comparable to HFrEF. In contrast to HFrEF, there is currently no available evidence based therapy for HFpEF9, which may be explained by different pathophysiology between these HF conditions. It was proposed that co-morbidities such as ischemia, hypertension and diabetes may be the drivers of disease progression in HFpEF10 through diverse mechanisms more specific for HFpEF than HFrEF involving coronary microvascular inflammation and endothelial dysfunction leading to intra- and extracellular rearrangements. This has been demonstrated in incomplete relaxation of myocardial strips11 and increased passive cardiac stiffness by titin changes and increased interstitial diffuse fibrosis10,12. Patients undergoing coronary bypass grafting (CABG) commonly have disturbances in LV function and thus may serve as models to study HF particularly since myocardial biopsies obtained during surgery provide a unique opportunity to study the gene expression in the different types of heart failure.
At the molecular level, gene expression programs are systematically regulated by transcription factors, chromatin regulators and other factors that are important for the establishment and maintenance of the cell state. Dysregulation of these programs can result in different diseases13. Studies of gene expression in heart tissue has a great potential to uncover the underlying molecular mechanisms leading to HF. In this regard, previous few studies have primarily attempted to identify gene expression differences between failing hearts (HFrEF) and normal hearts using RNA-seq analyses with limited number of samples14–16. There are also reports on RNA-seq expression in a mouse model for cardiac hypertrophy17.
In this study, we hypothesised that patients with HFpEF characteristics would show distinctive gene expression compared to patients with Normal physiology. Myocardial biopsies were obtained from patients undergoing elective CABG who all had LVEF ≥ 45%1. We performed RNA-seq based transcriptomics analysis in order to identify genes that are dysregulated in HFpEF compared to Normal.
Results
Patients
From a total of 16 patients, 5 were prospectively classified as HFpEF proxy and 11 as Normal physiology. The clinical patients’ characteristics are summarised in Table 1. The majority were male with a median age of 75 years in the HFpEF proxy group and 65 years in the Normal physiology group (p-value = 0.089). Three patients had a clinical diagnosis of heart failure, all in the HFpEF proxy group (patients 9, 11 and 15, Table 2). A history of hypertension (100% and 82%) and diabetes (60% and 45%) was present in HFpEF proxy and Normal physiology groups, respectively. One patient with Normal physiology had a history of myocardial infarction. Echocardiographic variables revealing HFpEF proxy or Normal physiology are shown in Table 2.
Table 1.
Patients' characteristics. Data are expressed as median and quartiles (Q1;Q3) or number (%).
| All patients (n = 16) | HFpEF pathophysiology (n = 5) | Normal (n = 11) | p-value | |
|---|---|---|---|---|
| Age (years) (median (Q1;Q3)) | 68 (63;73) | 75 (72;77) | 65 (61;68) | 0.089 |
| Sex Men/Women (n (%)) | 15/1 (94%/6%) | 4/1 (80%/20%) | 11/0 (100%/0%) | 0.312 |
| Smoking Current (n (%)) | 1 (7%) | 0 | 1 (10%) | 0.592 |
| Smoking Previous (n (%)) | 10 (67%) | 3 (60%) | 7 (70%) | |
| Previous | ||||
| Heart failure | 3 (19%) | 3 (60%) | 0 | 0.018 |
| Atrial fibrillation | 4 (25%) | 2 (40%) | 2 (18%) | 0.547 |
| Myocardial infarction | 1 (7%) | 0 | 1 (10%) | 1.000 |
| Percutaneous coronary intervention | 3 (19%) | 2 (40%) | 1 (10%) | 0.524 |
| CABG | 0 (0%) | 0 | 0 | 1.000 |
| Stroke/TIA | 1 (7%) | 0 | 1 (10%) | 1.000 |
| Peripheral artery disease | 2 (13%) | 2 (40%) | 0 | 0.083 |
| Hypertension | 14 (88%) | 5 (100%) | 9 (82%) | 1.000 |
| Diabetes type 1 type 2 |
8 (50%) 1 (7%) 7 (44%) |
3 (60%) 0 3 (60%) |
5 (45%) 1 (10%) 4 (36%) |
1.000 |
| COPD | 0 (0%) | 0 | 0 | |
| Anemia | 0 (0%) | 0 | 0 | |
| Cancer | 1 (7%) | 0 | 1 (10%) | 1.000 |
| At enrolment (median (Q1;Q3)) | ||||
| BMI | 27 (24;30) | 27 (26;28) | 28 (23;32) | 0.739 |
| Systolic blood pressure (mmHg) | 135 (130;145) | 148 (138;156) | 130 (125;140) | 0.112 |
| Diastolic blood pressure (mmHg) | 80 (75;80) | 83 (73;86) | 80 (75;80) | 0.537 |
| Heart rate (beats/minute) | 66 (57;77) | 66 (51;81) | 66 (63;72) | 0.847 |
| Creatinine (μmol/L) | 82 (75;102) | 84 (76;104) | 79 (74;100) | 0.867 |
| Hb (g/L) | 144 (137;153) | 151 (136;159) | 144 (137;149) | 0.609 |
| Sodium (mmol/L) | 140 (138;141) | 140 (140;141) | 140 (136;141) | 0.733 |
| Potassium (mmol/L) | 4.0 (3.8;4.3) | 3.9 (3.3;4.3) | 4.0 (3.8;4.3) | 0.592 |
| Troponin T (ng/L) | 10 (7;15) | 16 (10;17) | 9 (5;11) | 0.052 |
| NT-proBNP (pmol/L) | 205 (127;367) | 298 (190;697) | 181 (84;338) | 0.090 |
| LDL (mmol/L) | 2.0 (1.5;2.8) | 1.5 (1.4;2.0) | 2.3 (1.5;3.0) | 0.408 |
| Triglycerides (mmol/L) | 1.3 (1.0;2.0) | 1.2 (1.0;1.3) | 1.4 (1.1;2.0) | 0.413 |
| HbA1c (mmol/mol) | 41 (37;46) | 41 (40;46) | 43 (35;46) | 0.780 |
| Urate (μmol/L) | 359 (261;423) | 405 (249;496) | 355 (273;419) | 0.579 |
| Treament (n (%)) | ||||
| Nitrates (long standing) | 8 (50%) | 3 (60%) | 5 (45%) | 1.000 |
| Antiplatelets | 13 (81%) | 5 (100%) | 10 (91%) | 1.000 |
| Anticoagulants | 2 (13%) | 2 (40%) | 0 | 0.083 |
| Betablockers | 15 (94%) | 4 (80%) | 11 (100%) | 0.313 |
| ACE inhibitors | 8 (50%) | 3 (60%) | 5 (45%) | 1.000 |
| ARBs | 10 (63%) | 4 (80%) | 6 (55%) | 0.588 |
| Statins | 16 (100%) | 5 (100%) | 11 (100%) | 1.000 |
| Loop diuretics | 0 (0%) | 0 | 0 | |
| Tiazide diuretics | 4 (25%) | 1 (20%) | 3 (27%) | 1.000 |
ACE = Angiotensin converting enzyme; ARB = Angiotensin II receptor blocker; BMI = Body Mass Index; COPD = Chronic Obstructive Pulmonary Disease; CABG = Coronary Artery Bypass Surgery; NT-proBNP = N-Terminal pro-Brain Natriuretic Peptide; TIA = Transient Ischemic Attack.
Table 2.
List of patients included in this study with notations of disease status (HFpEF/Normal), age, gender, comorbidities, and echocardiographic parameters.
| Patient nr | Group | Age | Gender | Previous medical history | LVEF (%) | LAVI (mL/m2) | E/e′ | E′sept (m/s) | E′lat (m/s) | TR_Vmax(m/s) | NTproBNP (ng/L) | LVGLS % | LVMI (g/m′) | RWT | LVDed (mm) | IVSTed (mm) | PWTed (mm) | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hyper-tension | Diabetes type I | Diabetes type II | Atrial fibri-llation* | Stroke/TIA | Peri-pheral vascular disease | |||||||||||||||||
| 1 | HFpEF | 81 | M | x | x | 68 | 43.5 | 10.6 | 0.060 | 0.070 | 2.4 | 697 | −21.1 | 102 | 0.38 | 48 | 11 | 9 | ||||
| 2 | HFpEF | 58 | F | x | x | 63 | 52.1 | 13.1 | 0.062 | 0.075 | na | 161 | −20.4 | 94 | 0.45 | 44 | 13 | 10 | ||||
| 9 | HFpEF | 71 | M | x | 54 | 40.3 | 8.8 | 0.070 | 0.080 | 2.5 | 190 | −12.9 | 107 | 0.42 | 52 | 11 | 11 | |||||
| 11 | HFpEF | 77 | M | x | x | x | x | 49 | 44.1 | 11.4 | 0.064 | 0.098 | 2.9 | 2160 | −15.2 | 89 | 0.49 | 45 | 11 | 11 | ||
| 15 | HFpEF | 75 | M | x | x | x | 48 | 37.1 | 12.0 | 0.048 | 0.060 | 2.8 | 298 | −15.8 | 121 | 0.32 | 56 | 13 | 9 | |||
| 3 | Normal | 67 | M | x | x | 57 | 31.5 | 5.5 | 0.050 | 0.070 | na | 338 | −16.6 | 83 | 0.44 | 45 | 11 | 10 | ||||
| 4 | Normal | 68 | M | x | 60 | 25.0 | 5.4 | 0.070 | 0.140 | na | na | −16.8 | 101 | 0.39 | 46 | 14 | 9 | |||||
| 5 | Normal | 73 | M | x | 57 | 33.1 | 5.8 | 0.065 | 0.120 | na | 396 | −18.4 | 76 | 0.33 | 48 | 9 | 8 | |||||
| 6 | Normal | 49 | M | x | x | 48 | 32.0 | 7.6 | 0.098 | 0.100 | 2.0 | 181 | −11.9 | 84 | 0.32 | 50 | 12 | 8 | ||||
| 7 | Normal | 64 | M | x | 56 | 27.9 | 9.1 | 0.082 | 0.104 | na | 147 | −17.6 | 80 | 0.42 | 43 | 14 | 9 | |||||
| 8 | Normal | 65 | M | 60 | 27.3 | 7.8 | 0.080 | 0.100 | na | 262 | −17.0 | 58 | 0.33 | 43 | 10 | 7 | ||||||
| 10 | Normal | 60 | M | x | x | 66 | 33.5 | 8.0 | 0.072 | 0.124 | na | 219 | −19.5 | 77 | 0.36 | 45 | 10 | 8 | ||||
| 12 | Normal | 67 | M | x | x | x | 53 | 25.5 | 8.7 | 0.056 | 0.047 | na | 77 | −18.5 | 103 | 0.36 | 50 | 12 | 9 | |||
| 13 | Normal | 61 | M | x | x | 65 | 30.0 | 6.8 | 0.092 | 0.116 | na | 107 | −20.0 | 100 | 0.38 | 47 | 12 | 9 | ||||
| 14 | Normal | 63 | M | x | x | 59 | 40.6 | 6.8 | 0.097 | 0.099 | na | 66 | −19.9 | 84 | 0.37 | 49 | 11 | 9 | ||||
| 16 | Normal | 68 | M | x | 61 | 35.8 | 7.6 | 0.094 | 0.113 | na | 84 | −22.0 | 117 | 0.44 | 50 | 11 | 11 | |||||
LVEF – left ventricular ejection fraction, LAVI – left atrial volume index, E – early mitral inflow velocity, e – early diastolic tissue velocity, sept – septal, lat – lateral, TR_Vmax - tricuspid regurgitation maximal velocity, LVGLS –left ventricular global longitudinal strain, LVMI - left ventricular mass index, RWT – relative wall thickness, LVDed – left ventricular diameter, enddiastolic, IVSTed – intraventricular septum thickness, enddiastolic, PWTed – posterior wall thickness, venddiastolic. *Only patient 11 had atrial fibrillation at echocardiography examination. Patients 9, 11 and 15 had a clinical diagnosis of heart failure.
Gene expression profiles of LV tissue in HFpEF proxy and Normal physiology groups
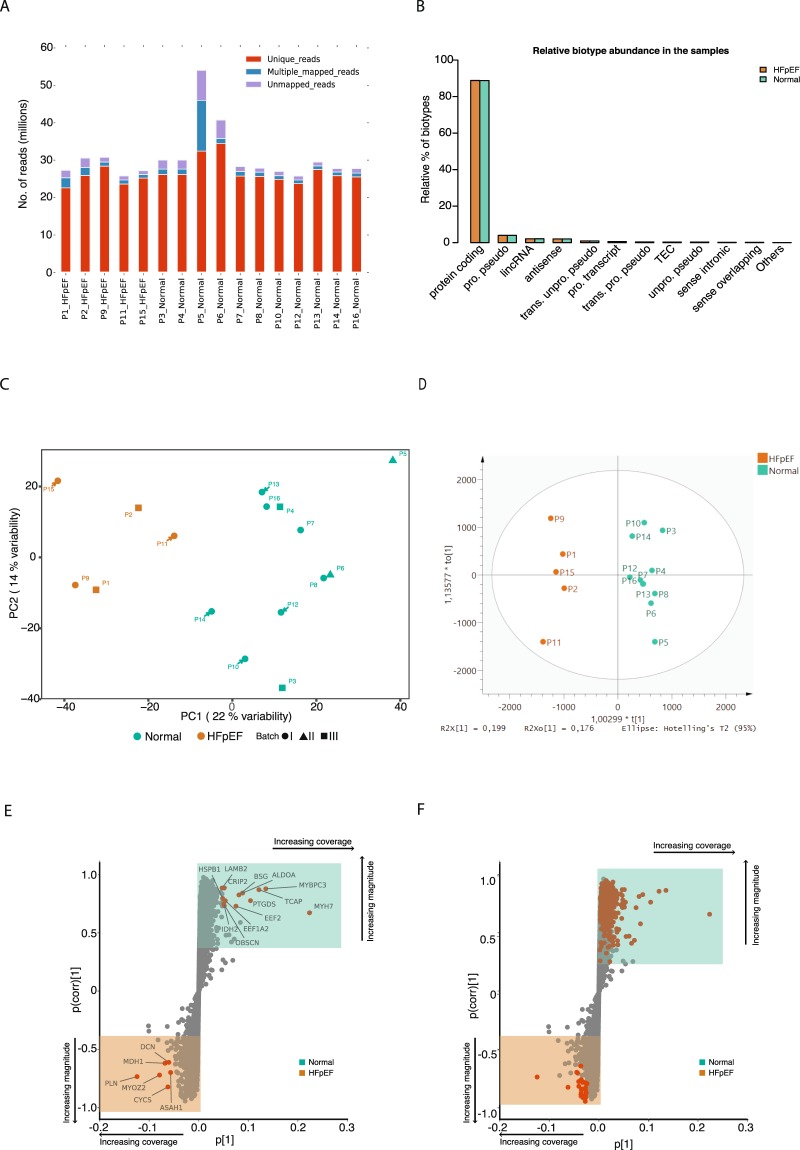
The transcriptome sequencing resulted in an average of 26 million paired-end reads per biopsy sample. Number of mapped reads ranged from 17 to 30 million with an average of 24 million (Fig. 1A) which covered more than 85% of the sequenced reads. Normalisation and batch correction was performed using Trim mean of the M-values (TMM) and ARSyNseq, respectively, after filtering out lowly expressed genes (Supplementary Fig. S1). The majority (~90%) of the expressed genes were protein coding while less than 5% were detected as long noncoding RNA (lncRNA) or antisense RNA (Fig. 1B, Supplementary Fig. S2).
Figure 1.
Gene expression profiles of left ventricle tissues discriminate HFpEF proxy from Normal physiology. (A) Stack barplot showing uniquely mapped, multiple mapped and unmapped reads. The x axis shows the samples and the y axis the number of reads. (B) Bar plot showing the relative abundance of each biotype (y axis) in the samples (x axis). (C) Principal component analysis (PCA) score plot with the two principal components (PC1 and PC2) plotted on the x- and y-axis, respectively. Each data point represents one sample, which is colour-coded according to the condition and shaped according to the sequencing batch. Green and orange colours correspond to Normal physiology and HFpEF proxy, respectively. (D) Orthogonal projections to latent structures discriminant analysis (OPLS-DA) score plot for the groups HFpEF proxy (orange) and Normal physiology (green). (E) S-plot of the OPLS-DA data showing the magnitude of each gene’s contribution to the separation, p[1], in relationship to its significance, p(corr)[1]. Genes contributing to the highest magnitude of the separation for the respective groups are highlighted in red/orange. Shaded boxes indicate up-regulated genes in the HFpEF proxy group (bottom orange box) and in the Normal physiology group (top green box). (F) The same plot as in “E” but genes overlapping with differentially expressed genes are highlighted with in red/orange.
We first characterised differences between the gene expression profiles of the 16 samples, utilising an unsupervised classification method – Principal Component Analysis (PCA). PCA using batch corrected normalised gene expression revealed two clusters, corresponding to HFpEF proxy and Normal physiology along the first principal component 1 (PC1). The PCA model revealed that the largest variation of the PC1 could explain 22% of the variation while principal component 2 explained 14%. Additionally, any bias between these groups and sequencing batches was investigated and no batch effect contributing to this clustering was found (Fig. 1C).
The Orthogonal Projections to Latent Structures Discriminant Analysis (OPLS-DA) model with 7-fold cross-validation depicted in Fig. 1B distinguished HFpEF proxy from Normal physiology using the first predictive component. An S-plot was generated to identify signature genes in both groups (Fig. 1D). In the S-plot, the magnitude of the contribution of each gene to the OPLS-DA model (p[1]) was plotted against significance of the corresponding genes (p(corr)[1]). As can be seen in Fig. 1E, genes (marked in red) contributing most to the discrimination of HFpEF proxy were MYH7, MYBPC3, TCAP, PTGDS, BSG, ALDOA, EEF2, IDH2, CRIP2, EEF1A2, PLN, CYCS, MYOZ2, MDH1 and DCN. Several of these genes were also found among the significantly differentially expressed genes (Fig. 1F; see below).
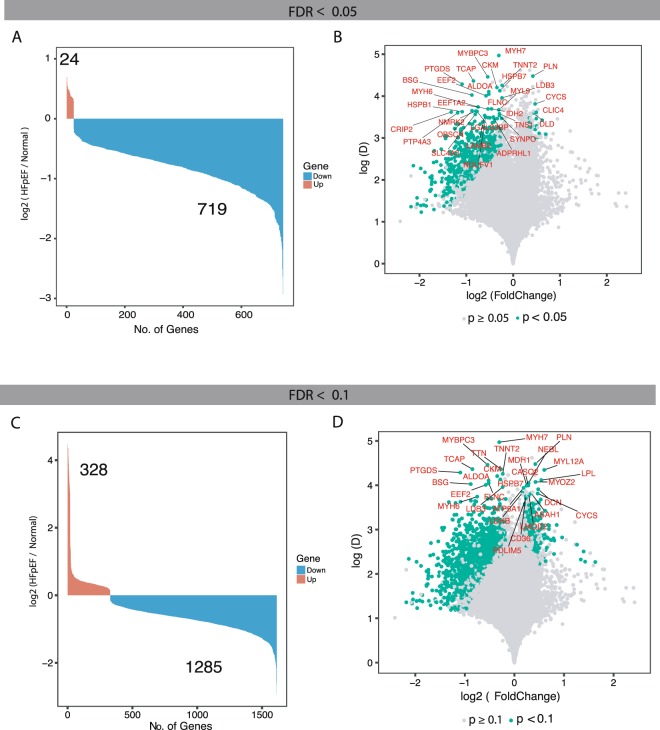
Genes dysregulated in HFpEF proxy patients
Differentially expressed genes (DEGs) were identified using NOIseqbio with a false discovery rate (FDR) < 0.05. Our analysis identified 743 DEGs discriminating between HFpEF proxy and Normal physiology whereof the majority were down-regulated in HFpEF proxy samples compared with Normal physiology samples (Fig. 2A). The distribution of fold change as a function of mean expression difference in all the cases revealed that more than 90% of significant DEGs had fold changes ranging from 1.30 to –2.60 (Fig. 2A,B). For comparison, we also calculated DEGs using FDR < 0.1 (Fig. 2C,D), resulting in 328 up-regulated and 1285 down-regulated genes in HFpEF proxy samples. The functional analysis (cf. below) of these DEGs showed similar results to those using FDR < 0.05, and we therefore kept the more stringent FDR < 0.05 DEGs in the current analysis.
Figure 2.
Identification and annotation of the dysregulated genes in HFpEF proxy versus Normal physiology. (A) Bar plot showing number of differentially expressed genes between HFpEF proxy and Normal physiology predicted using NOISeq with false discovery rate (FDR) adjusted p-value < 0.05. The x axis represents significantly dysregulated genes and the y axis showing the fold change in log2 scale of the corresponding genes. Down-regulated genes in blue and up-regulated genes in orange. (B) Volcano plot of the differentially expressed genes. The x axis represents fold change in log2 scale of HFpEF proxy versus Normal physiology while the y axis indicates the differences of mean expression between HFpEF proxy and Normal physiology. Each point represents a gene, and significantly expressed genes are highlighted in green. Genes that are significantly expressed (FDR-adjusted p-value < 0.05) and with a difference of mean expression above 3.5 are labelled with gene symbols. (C) Similar to plot “A”, but FDR-adjusted p-value < 0.1. (D) Similar to plot “B” but FDR-adjusted p-value < 0.1. Genes that are significantly expressed and with a difference of mean expression above 3.7 are labelled with gene symbols.
Among the 743 DEGs, 69 genes were transcriptional regulators, of which 67 were down- and 2 up-regulated. A comparison between the predicted DEGs with the genes in the S-plot derived from OPLS-DA showed that most DEGs with few exceptions are located at largest distances from the axes centre. Thus, the dysregulated genes were predicted by two independent methods (Fig. 1F). The complete list of up- and down-regulated genes is given in the Supplementary Table S1.
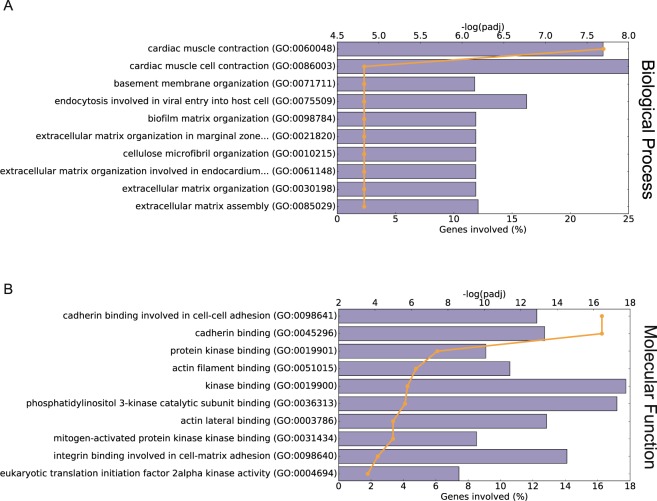
The top biological processes associated with down-regulated genes in the HFpEF proxy group were cardiac muscle contraction and extracellular matrix assembly/organization (Fig. 3A, Supplementary Table S2). Enriched Gene Ontology (GO) terms reflecting molecular function revealed genes for cadherin binding, kinase binding, actin filament and integrin binding (Fig. 3B, Supplementary Table S2).
Figure 3.
Functional classification of the differentially expressed genes. (A,B) GO annotations of biological process and molecular function, respectively, of the down-regulated genes in HFpEF proxy group. The horizontal bars show percentage of the down-regulated genes with the corresponding GO annotations (scale at bottom x axis), The orange lines represent significance of the corresponding GO annotations (scale at top x axis) as calculated by Enrichr53.
We found down-regulation of genes involved in cardiac muscle contraction: myosin heavy chain 6 and 7 (MYH6,MYH7), cardiac myosin-binding protein C (MYBPC3), cardiac troponin T2 (TNNT2), titin cap (TCAP) and two potassium voltage-gated channels (KCNH2,KCNQ1).
Matrix-related genes that were down-regulated in the HFpEF proxy group LAMA5, LAMB2, coding for two laminin subunits (α5 and β2), SPARC coding for secreted protein acidic and rich in cysteine, and a number of collagens – Iα1, IIIα1, IVα, IVβ1, VIα1, VIβ1 and XVIIIα1.
Among the up-regulated genes we identified lumican (LUM), phospholamban (PLN), myozenin 2 (MYOZ2), and cytochrome c (CYCS).
Upstream regulators and regulatory effect networks
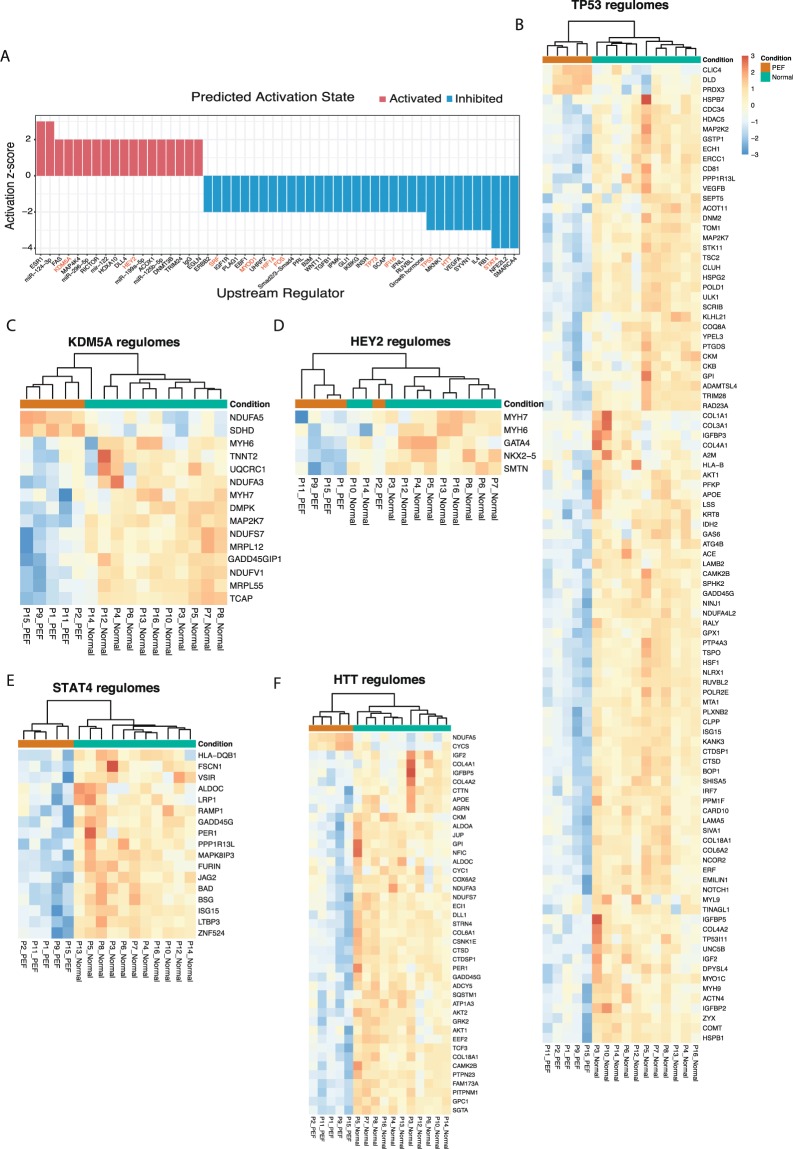
The Ingenuity Pathways Analysis (IPA) analysis suggested that out of the total set of 736 DEGs (7 Ensembl gene IDs were not recognised by IPA and hence excluded from this analysis), 381 DEGs (52%) belong to known upstream regulators predicted to be activated or inhibited (activation Z-score ≥ ±2), at a Fisher’s Exact test p-value ≤ 0.0518. A comprehensive list of 52 upstream regulators along with predicted activation states is found in Supplementary Table S3. Of these regulators, 34 were predicted to be inhibited and 18 were to be activated (Fig. 4A).
Figure 4.
Predicted upstream regulators and gene expression of their regulomes. (A) Bar plot showing activation or inhibition scores of the upstream regulators. Transcription factors are highlighted in red. (B–F) Heat maps showing the expression profiles of the genes regulated by predicted transcription factors from A. Additional heat maps are shown in Supplementary Fig. S3. Samples in the HFpEF proxy group are coloured orange in Condition, and samples in the Normal physiology group are coloured green. The regulators genes were identified using IPA. Patient numbers and conditions are shown at the bottom of each heatmap.
Among the upstream regulators predicted in our analysis to be inhibited in the HFpEF proxy group were transcription factor STAT4, HTT and tumour suppressor TP53 (Fig. 4), the latter regulating a total of 96 DE genes (13% of the DE gene set). Additional predicted inhibited upstream regulators included specific transcription factors (SRF, MYOD1, FOS and HIF1A; Supplementary Fig. S3). Furthermore, histone demethylase KDM5A and the basic helix–loop–helix (bHLH) type transcription factor HEY2 (Fig. 4), both transcriptional repressors involved in Notch signalling, were among the identified activated upstream regulators.
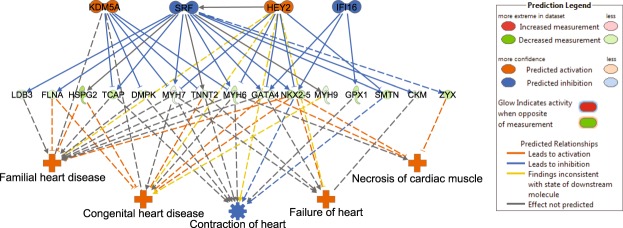
Based on the predicted upstream regulator, a total of 8 regulatory effect networks were identified (Supplementary Table S4). These networks depict potential paths by which activation or inhibition of specific transcription factors lead to impaired cardiac function, heart failure and other heart diseases (networks 1–4,7), as well as impaired vessel formation/endothelial cell function (networks 3,5,6,8). The most causally consistent and densely connected network is shown in Fig. 5.
Figure 5.
Transcription factor regulatory effect network identified using Ingenuity Pathway Analysis (IPA). In the network nodes, the upper panel shows transcription factors, the middle panel shows differentially expressed genes, and the lower panel shows biological functions and diseases. For the network edges, a solid line indicates direct interaction, while a dashed line indicate indirect interaction. Node colours in upper and lower panels: predicted activation in orange; predicted inhibition in blue. Node colours in middle panel: downregulated in data set coloured green; upregulated in data set coloured red (not represented in this network). Edge colours; predicted activation in orange; predicted inhibition in blue, findings inconsistent with state of downstream node in yellow; effect not predicted in grey.
Discussion
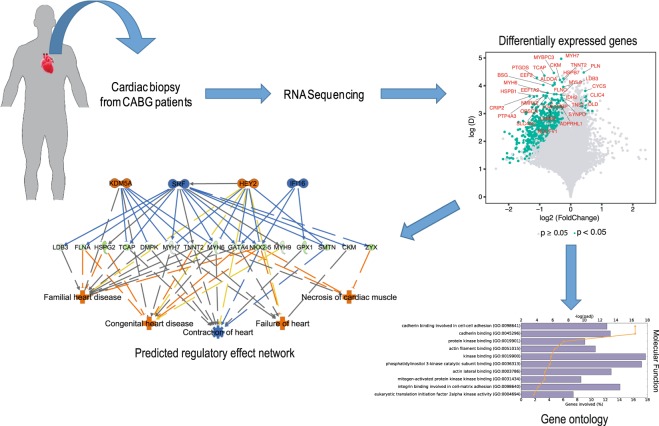
To our knowledge, this is the first study using RNA-seq to identify dysregulated genes in patients with HFpEF characteristics, as schematically summarised in Fig. 6. In this exploratory translational study of elective CABG patients undergoing perioperative myocardial biopsies, we found that patients in the HFpEF proxy group displayed distinctive gene expression compared to patients with Normal physiology. The top biological functions associated with down-regulated genes in HFpEF proxy patients were cardiac muscle contraction, oxidative phosphorylation, endocytosis/cell remodelling, matrix organization and fibrosis. Further, genes regulated by transcription factor STAT4 and tumour suppressor TP53 were found to be down-regulated.
Figure 6.
Schematic summary of the current study. Cardiac biopsies from CABG patients were submitted to RNA sequencing to detect differentially expressed genes between HFpEF and Normal. These differentially expressed genes were characterised using gene ontology and predicted transcription factor regulatory effect network.
Patients
The patients investigated in this study were the initial group in whom the myocardial biopsies were obtained within the ongoing CABG-PREFERS study1. They represent patients with a clinical indication for elective CABG. Hence, very few had a previous myocardial infarction or coronary intervention and few had a previous HF diagnosis. The three patients who had a HF diagnosis were all in the HFpEF proxy group.
HFpEF
HFpEF is more frequent today, which may be due to increasing life span of the population, improved survival after myocardial infarction and increasing rates of HF risk factors like hypertension, overweight, and diabetes. However, the pathophysiology of this disease in not well understood at the transcriptome level. Already in the 1980s, it was recognised that ischemia might lead to diastolic dysfunction. We identified HFpEF characteristics in 31% of the group of patients planned for elective CABG, implying that other prevalent comorbidities except coronary artery disease, such as hypertension and diabetes may also play a role for development of HFpEF suggesting a link to microvascular dysfunction19.
Imaging
HFpEF constitutes a diagnostic challenge and in an individual patient, there may be problematic measure overlaps and grey zones. Poor echocardiographic windows, tachyarrhythmias and atrial fibrillation makes measurements more difficult. The present guidelines advocate the use of at least 4 up to 8 parameters of structural LV dysfunction and diastolic dysfunction for diagnosis and risk prediction, some of these parameters may be used interchangeably5,20. In summary, the number of altered variables may increase the precision of the HFpEF diagnosis. In the current study we therefore used state-of-the-art guideline criteria for HFpEF and a majority of the 4–8 criteria achieved in an individual patient should be positive for rendering a HFpEF proxy diagnosis. Our definition is further strengthened by the fact that we used a consensus method in ambiguous patients. Although the five HFpEF proxy patients fulfilled the defined criteria they only had mild dysfunction and low NT-proBNP levels. However, we believe that such early stage HFpEF is highly relevant since these patients seem more sensitive to treatment with RAAS blockade21. Thus, RNA sequencing may be of particular value in early or mild HFpEF patients for finding new pathophysiologic translational mechanisms.
Genes dysregulated in HFpEF proxy patients
To get an overview of the gene expression profiles, we performed PCA and OPLS-DA to distinguish HFpEF proxy patients from those with Normal physiology. Our results (Fig. 1C,D) suggest that the two groups have different gene expression profiles. The downregulated genes (Supplementary Table S1) were analysed with respect to enrichment of GO terms (Fig. 3A,B), revealing that cardiac muscle contraction, oxidative phosphorylation, endocytosis and extracellular matrix organization were associated with the dysregulated genes (Fig. 3A, Supplementary Table S2). These first two GO terms were also found to be enriched in a mouse model of pathological hypertrophy17.
Downregulated cardiac contraction genes may correlate with impaired systolic pump function in HFpEF patients as has been described in previous studies22. The gene TNNT2 encodes for the tropomyosin binding subunit in the troponin complex which is located in the thin filament of the striated muscle and regulates muscle contraction in response to alteration in the intracellular calcium ion concentration23. However, we found no differences between the two groups for measures of systolic function at rest. EF and global longitudinal strain (GLS) were similar in the two groups. The described downregulated genes for myocardial contraction may be related to more long-term changes caused by passive cardiac stiffness by fibrosis or oxidative titin changes12. Short-time adaptations of systolic function, cardiac relaxation and diastolic function may counteract long-term adaptations and could be caused by other regulatory physiological mechanisms rather than transcriptome proliferative changes.
Our study shows that RASD1, coding for ras-related dexamethasone-induced protein 1, is downregulated in HFpEF proxy patients. RASD1 is a modulator of cardiac endocrine function in response to volume overload underlying anti-natriuretic peptide excretion such as atrial natriuretic peptide (ANP) and brain natriuretic peptide (BNP)24. RASD1 has also been shown to be downregulated in volume overloaded rat hearts24.
Furthermore, PLN, coding for phospholamban was upregulated in HFpEF proxy patients. PLN is a pentamer and a major substrate for the c-AMP dependent kinase in cardiac muscle. Phospholamban modulates the activity of the sarcoplasmatic reticulum ATPase type 2 (SERCA 2a), which in turn modulates Ca2+ handling by the sarcoplasmatic reticulum and increases both contractility and relaxation, at least in studies of electric cardiac contractility modulation (CCM) therapy25. In contrast, these genes have been seen to be downregulated in HFpEF26.
During the cardiac cycle there is an active coupling between contractility during systole and relaxation during early diastole. Cardiac contraction and relaxation are both closely linked active energy dependent processes, i.e. in the ischemic cascade. The finding of downregulated genes in the HFpEF proxy group for oxidative phosphorylation and energy supply may thus be considered crucial for development for HFpEF as regards both systolic and diastolic cardiac function and in relation to ischemia in these patients. Further, transcriptional changes may develop more slowly and be counteracted by short-term alterations of contractility and relaxation by changes in neurohormonal activation and sympathetic tone, calcium fluxes, and ischemia27. Sympathetic tone is increased in both aging and heart failure, by increasing circulating catecholamines but also decreased β-adrenoreceptor sensitivity28. Calcium handling in diastole is essential to remove Ca2+ from the cytosol to ensure cardiomyocyte relaxation by SERCA back into the Sarcoplasmatic reticulum29. Traditional RAAS- and betablockade does not show benefits in HFpEF, but new knowledge of role of β-arrestins and G-protein coupled receptor kinases (GRKs) may open new targets for treatment of HFpEF30.
Our results show several downregulated genes involved in extracellular matrix assembly, which may influence remodelling and dilatation of the heart rather than increasing passive myocardial stiffness as shown for HFpEF12 due to increased synthesis of collagens typically with predominance for collagen I and III in the myocardium. This finding is therefore surprising and counterintuitive. However, the process of myocardial fibrosis is complex and involves dynamics of fibrous tissue turnover including sevveral steps; active synthesis, crosslinking and active degrading of collagen31. The genes LAMA5 and LAMB2, coding for two laminin subunits (α5 and β2), were also downregulated in HFpEF proxy patients. The secreted protein acidic and rich in cysteine (SPARC) is a matrix-cellular collagen binding protein serving a key role in collagen assembly into the extracellular matrix. Recent studies demonstrated that disruption of the SPARC gene is associated with decreased capacity to generate organised, mature collagen fibres32.
ANKRD1 is a transcription factor known to interact with sarcomeric proteins in the myofibrillar stretch sensor system33. It has been observed that the expression of ANKRD1 in both transcription and protein levels were increased in failing heart15,34. In our data we also see a trend of upregulated mRNA expression in the HFpEF proxy patients.
We find that the gene LUM coding for lumican is among the most up-regulated genes in the HFpEF proxy group compared to Normal physiology. Lumican is an extracellular matrix localised proteoglycan associated with inflammatory conditions known to bind collagen. In a recent study, cardiac lumican was increased in experimental and clinical HF35. This study also indicated that inflammatory and mechanical stimuli induce lumican production by cardiac fibroblasts indicating a role in HF development. However, we are unaware of whether lumican has previously been shown to be up-regulated in CABG and HFpEF patients.
Upstream regulators and regulatory effect networks
The tumour suppressor TP53 has recently been described as an important regulatory factor in the heart36. In our study, 96 DE genes (13%) belong to those regulated by TP53 (Fig. 3B). It has been described to be increased in human dilated cardiomyopathy (DCM)37, suggesting that elevation of TP53 also plays a key role in the common path toward heart failure. There are data showing that TP53 inhibits angiogenesis by suppressing HIF-1 resulting in myocardial hypoxia and cardiac dysfunction which may be a novel molecular mechanism underlying transition of cardiac hypertrophy to HF38.
STAT4 is important in both innate and adaptive immune responses39,40. However, STAT4 did not show transcriptional repression in the HFpEF proxy group, indicating that additional mechanisms, e.g. post-translational modifications, protein–protein interactions, might be involved in providing the inhibitory effect.
We further hypothesised a regulatory effect network model for mechanistic understanding of the disease and dysregulated genes using the IPA regulatory effect tool. The network illustrates potential mechanism(s) by which transcription regulator activation (KDM5A, HEY) and inhibition (SRF, IFI16) may lead to impaired cardiac function (Fig. 5).
KDM5A (retinoblastoma-binding protein 2/RBP2) encodes a histone demethylase that is part of the core Notch–RBP-J repressor complex41 and has implicated in transcriptional regulation of Hox genes and cytokines42. It also plays a role in tumour progression and selective inhibition blocks cancer cell growth43. Little is known regarding the role of this gene in heart. In a recent study, likely disease-causal KDM5A variants were uncovered in whole exome sequencing in patients with congenital heart disease (CHD)44.
HEY2 encodes a basic helix-loop-helix (bHLH)-type transcription factor that is preferentially expressed in the developing and adult cardiovascular system45. It acts as a transcriptional repressor downstream of Notch signalling pathway46 and likely plays a central role in the cardiac transcriptional machinery47. For example, HEY2 expression levels influence cardiac hypertrophy and the progression to heart failure in response to pressure overload through modulation of apoptosis and GATA4 activity48. Our network analysis results suggest that activation of HEY2 may have contributed to cardiac dysfunction in HFpEF proxy via transcriptional repression of key cardiac transcription activators GATA4 and NKX2.5, among others.
Serum response factor (SRF) is a central cardiac transcription factor required for appearance of beating sarcomeres in the heart49. Based on our analysis, inhibition of SRF with accompanied downregulation of its target genes (GATA4, NKX2.5, MYH6, MYH7, TNNT2, TCAP and others) could be one additional explanatory mechanism underlying cardiac dysfunction in the HFpEF proxy group.
Although the exact function of p53 and IFN-inducible gene IFI16 is not currently known, it has been proposed to act as transcriptional repressor and tumour suppressor via activation of p53 signals and inflammasome50,51.
In addition to IPA, we also explored network-based approaches for transcription factors enrichment; ChEA-db which is transcription factor targets database inferred from integrating literature curated Chip-X data52 and transcription factor protein-protein interaction networks53. The enrichment analysis revealed TP53, SMAD2, SMAD3 and ESR1 among the enriched transcription factors which were also predicted by IPA as upstream regulators.
Limitations and strengths
The strength of this study consists of the revealed gene expression differences between HFpEF and Normal groups found in myocardial biopsies. However, there are some limitations in the study, such as relatively small number of patients used and unequal distribution between HFpEF and Normal. In order to obtain myocardial biopsies, we have chosen patients undergoing elective CABG enabling us to safely obtain tissue samples. Based on data from the SWEDEHEART (Swedish Web-system for Enhancement and Development of Evidence-based care in Heart disease Evaluated According to Recommended Therapies) registry we knew that around 20–30% of elelective CABG patients had LVEF ≥ 45%. To identify the HFpEF group, we used echocardiographic evaluation according to current international Guidelines. We could confirm that a proportion of patients indeed had HFpEF characterstics. Due to limited amounts of biopsy material, we had to use the entire biopsy to get deeper RNA sequencing for better sensitivity. Consequently, we were unable to perform any validation experiments which may be considered a limitation. A larger cohort of patients, which we aim at in the future, will add more insight into differences in gene expression between these two groups.
Conclusions
This exploratory study could confirm our hypothesis that patients undergoing CABG with HFpEF characteristics compared to patients with Normal physiology had distinctive gene expression in cardiac biopsies with downregulated genes for myocardial contraction, energy supply, remodelling and fibrosis. We consider differences within these functional areas relevant for possible pathophysiological mechanisms in HFpEF. However, down-regulation of these functions in patients with HFpEF characteristics is complex to describe and understand. Our findings lend to support our further studies in a larger patient cohort to find pathophysiological mechanisms that can explain and ultimately lead to treatment for HFpEF.
Methods
Patients
Patients enrolled were scheduled for elective CABG without concomitant valve surgery and with preserved LVEF. They all had angina pectoris with or without a previous myocardial infarction. Cardiac biopsies were obtained during CABG for analysis of mRNA expression in the myocardial tissue. All patients were assessed at a baseline visit 4–8 weeks prior to CABG by clinical characteristics, echocardiography and blood sampling including natriuretic peptides. From the ongoing study CABG-PREFERS1 we now report data from the initial patients.
Descriptive data are presented as median and quartiles (Q1;Q3) or number (%), and comparisons between groups were performed by Wilcoxon rank sum test and Fisher’s exact test as appropriate.
Definitions
Preserved LVEF was defined at the time of study design as LVEF ≥ 45%1. The patients were divided into two groups according to echocardiography, NTproBNP levels and HF guidelines definitions5. The group with echocardiographic characteristics and increased NTproBNP levels indicative of HFpEF1,5 was called HFpEF proxy for the purpose of this study and was used as a representative for HFpEF even when not showing signs or symptoms of heart failure. The Normal physiology group had LVEF ≥ 45% and no echocardiographic signs of HFpEF54.
The HFpEF proxy definition was based on LVEF ≥ 45% and the combination of the following five echocardiographic criteria; 1. left atrial volume indexed for body surface area (LAVI) > 34 ml/m2, 2. LV mass index ≥115 g/m2 for males or ≥95 g/m2 for females, 3. ratio of early mitral inflow wave velocity (E) to myocardial tissue early diastolic wave velocity (e′) defined as E/e′ ≥ 8, 4. e′ septal <0.07 m/s or e′ mean septal/lateral <0.09 m/s, 5. tricuspid regurgitation velocity >2.8 m/s, and additionally NTproBNP > 125 ng/L. Three abnormal criteria of these five were required to fulfil the definition (majority rule). In equivocal cases, classification was performed by consensus of two experts (M.J.E. and H.P., blinded for clinical characteristics) in line with previous experiences from the CHARM substudy20.
Echocardiography
Transthoracic Doppler echocardiography was performed according to guidelines as previously reported1. A Vivid 9 ultrasound system (Vingmed-General Electric, Horten, Norway) was used in all studies. Images were digitally stored on a dedicated server, and data analysis was performed offline on the EchoPAC workstation (GE EchoPAC sw only, Norway) by one experienced sonographer. The mean value of 3 cardiac cycles was calculated for each variable.
Tissue collection
From patients undergoing CABG, core needle biopsies were taken from the lateral wall of the left ventricle before initiation of cardiac arrest and stored in −70 °C as previously described1 and used for mRNA analysis. Patients were prepared for surgery according to standard clinical routines with placement of a central venous line in the internal jugular vein, an arterial line in the distal radial artery and a peripheral venous line in the brachial vein. A midline sternotomy was performed and one or two mammary arteries were procured for usage as conduits55.
RNA extraction and sequencing
Total RNA was extracted using the RNeasy Fibrous Tissue Mini Kit (#74704, Qiagen). RNA libraries for sequencing were prepared using poly-A selection and the Illumina RNA strand-specific TruSeq Stranded mRNA Sample prep kit with 96 dual indexes (Illumina, CA, USA) according to the manufacturer’s instructions with the following changes. The protocols were automated using an Agilent NGS workstation (Agilent, CA, USA) using purification steps as described56,57.
Clustering was done by ‘cBot’ and samples were sequenced on HiSeq. 2500 (HiSeq Control Software 2.2.58/RTA 1.18.64) with a 2 × 125/2 × 150 setup using ‘HiSeq SBS Kit v4’ chemistry. The Bcl to FastQ conversion was performed using bcl2fastq-v2.17.1.14 from the CASAVA software suite. The quality scale used was Sanger/phred33/Illumina 1.8 + .
Analysis of transcriptome (RNA-Seq) data
Whole transcriptome sequencing was performed for each biopsy. Initial quality checking of the sequencing raw reads was performed to identify potential outliers before doing further analysis using FastQC. Sequencing paired-end reads were mapped towards the human reference genome (version GR38) using Star Aligner58 with default options, and Ensembl genome annotation (version 37) was used for subsequent analysis. Reads that mapped to the exons of the coding genes were counted using HTSeq.59. Genes with count values of zero (i.e. no read detected) in all samples were filtered out before further analysis. Genes were then categorised into different biotypes and distribution over the reference genomes was calculated. Count data was also investigated for several potential biases such as RNA-degradation, GC content etc. using NOISeq.60 package in R (http://www.R-project.org). Before normalisation, lowly expressed genes were filtered out using proportion test per condition and multiple testing correction as per NOIseq manual60. Normalisation of the count data was performed using the Trim mean of M-values (TMM) approach61.
Batch correction of the data was performed using ARSyNseq function of the NOISeq package60. Differential gene expression analysis was done using the function noiseqbio with parameters k = 0.5, norm = ‘n’, lc = 0, r = 20, adj = 1.5, a0per = 0.9, which is recommended for clinical RNAseq samples60. For NOISeq, we set the parameter ‘q’ to 0.95, corresponding to a false discovery rate (FDR) < 0.05. Analysis and plots were generated in R environment using ggplot2.
Principal component analysis (PCA) was performed on the log2 transformed values of normalised batch corrected expression values using “princomp” function in R environment.
Orthogonal projections to latent structures discriminant analysis (OPLS-DA) was performed using SIMCA v14.1 (Umetrics, Umeå, Sweden) on the already normalised and batch corrected data to identify genes showing high variation in a pairwise manner. The Pareto scaling method was used in this case, since it reduces the relative importance of large values, but keeps data structure partially intact, and we wanted to detect small to medium feature differences.
Clustering analysis of the differentially expressed genes (DEGs) was performed using unsupervised hierarchical clustering. Normalised expression data were standardised before plotting as heatmap using pheatmap package for R environment.
Functional analysis of the dysregulated genes
Gene set enrichment analysis for Gene Ontology (GO) terms with focus on biological process (BP) and cellular component (CC) was performed for the DEGs (using gene symbols as input) using Enrichr53 with the probability density function as p-value model. The enrichment was tested using Fisher’s exact test with corrected p-value < 0.05. The DEGs were further analysed through the use of IPA18 (Ingenuity Pathways Analysis; QIAGEN Inc., https://www.qiagenbioinformatics.com/products/ingenuity-pathway-analysis). This tool uses the information in the Ingenuity® Knowledge Base to assess signalling and metabolic pathways, upstream regulators, regulatory effect networks, and disease and biological functions that are likely to be perturbed based on a data set of interest (in our case, the DEGs). The IPA upstream analysis18 was performed to predict which regulators (i.e. any gene, protein or miRNA) that are activated or inhibited based on a calculated Activation Z-score, to explain the observed DEG changes in HFpEF proxy group vs. Normal physiology group. The IPA regulatory effect network analysis generated hypotheses for how a phenotype, function or disease is regulated by activated or inhibited upstream regulators. In our study, regulatory effect network analysis was used to specifically study the impact of the identified upstream transcription factor regulators on downstream heart disease functions, given the observed gene expression changes in HFpEF proxy group vs. Normal physiology group.
Ethics statement
This study was conducted according to the Declaration of Helsinki and approved by the regional Ethics Committee Stockholm. All patients were included following oral and written informed consent.
Supplementary information
Acknowledgements
This work was supported by funding from the Science for Life Laboratory–Astra Zeneca; Mölndal, Sweden collaborative grant No. 1377. The authors acknowledge assistance with massively parallel sequencing from the National Genomics Infrastructure in Stockholm funded by Science for Life Laboratory, the Knut and Alice Wallenberg Foundation and the Swedish Research Council, and from SNIC/UPPMAX for access to computational infrastructure.
Author Contributions
S.D., C.F., A.W., C.K. and B.P. have performed the bioinformatics analyses; M.J.E., E.M. and H.P. have done the echocardiographic diagnostics; M.C. has performed the CABG surgery and taken the biopsies; C.H. has been responsible for patient recruitment; M.A. and J.L. have done RNA sequencing; B.P., C.L. and H.P. have conceived the study; B.P., C.L., H.P., C.H., M.J.E., C.F. and S.D. have written the manuscript. All authors have approved the final version of the manuscript.
Data Availability
RNA-seq data have been deposited in the EMBL-EBI ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-7454.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Sarbashis Das and Christoffer Frisk contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-39445-2.
References
- 1.Linde C, et al. Rationale and design of the PREFERS (Preserved and Reduced Ejection Fraction Epidemiological Regional Study) Stockholm heart failure study: an epidemiological regional study in Stockholm county of 2.1 million inhabitants. Eur J Heart Fail. 2016;18:1287–1297. doi: 10.1002/ejhf.599. [DOI] [PubMed] [Google Scholar]
- 2.Paren P, et al. Trends in prevalence from 1990 to 2007 of patients hospitalized with heart failure in Sweden. Eur J Heart Fail. 2014;16:737–742. doi: 10.1002/ejhf.109. [DOI] [PubMed] [Google Scholar]
- 3.Benjamin EJ, et al. Heart Disease and Stroke Statistics-2017 Update: A Report From the American Heart Association. Circulation. 2017;135:e146–e603. doi: 10.1161/CIR.0000000000000485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heidenreich PA, et al. Forecasting the impact of heart failure in the United States: a policy statement from the American Heart Association. Circ Heart Fail. 2013;6:606–619. doi: 10.1161/HHF.0b013e318291329a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ponikowski P, et al. 2016 ESC Guidelines for the diagnosis and treatment of acute and chronic heart failure: The Task Force for the diagnosis and treatment of acute and chronic heart failure of the European Society of Cardiology (ESC). Developed with the special contribution of the Heart Failure Association (HFA) of the ESC. Eur J Heart Fail. 2016;18:891–975. doi: 10.1002/ejhf.592. [DOI] [PubMed] [Google Scholar]
- 6.Gerber Y, et al. A contemporary appraisal of the heart failure epidemic in Olmsted County, Minnesota, 2000–2010. JAMA Intern Med. 2015;175:996–1004. doi: 10.1001/jamainternmed.2015.0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsao CW, et al. Temporal Trends in the Incidence of and Mortality Associated With Heart Failure With Preserved and Reduced Ejection Fraction. JACC Heart Fail. 2018;6:678–685. doi: 10.1016/j.jchf.2018.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Owan TE, et al. Trends in prevalence and outcome of heart failure with preserved ejection fraction. N Engl J Med. 2006;355:251–259. doi: 10.1056/NEJMoa052256. [DOI] [PubMed] [Google Scholar]
- 9.Yang J, Xu WW, Hu SJ. Heart failure: advanced development in genetics and epigenetics. Biomed Res Int. 2015;2015:352734. doi: 10.1155/2015/352734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulus WJ, Tschope C. A novel paradigm for heart failure with preserved ejection fraction: comorbidities drive myocardial dysfunction and remodeling through coronary microvascular endothelial inflammation. J Am Coll Cardiol. 2013;62:263–271. doi: 10.1016/j.jacc.2013.02.092. [DOI] [PubMed] [Google Scholar]
- 11.van Heerebeek L, et al. Myocardial structure and function differ in systolic and diastolic heart failure. Circulation. 2006;113:1966–1973. doi: 10.1161/CIRCULATIONAHA.105.587519. [DOI] [PubMed] [Google Scholar]
- 12.Zile MR, et al. Myocardial stiffness in patients with heart failure and a preserved ejection fraction: contributions of collagen and titin. Circulation. 2015;131:1247–1259. doi: 10.1161/CIRCULATIONAHA.114.013215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, et al. RNA-Seq identifies novel myocardial gene expression signatures of heart failure. Genomics. 2015;105:83–89. doi: 10.1016/j.ygeno.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrer I, et al. RNA-sequencing analysis reveals new alterations in cardiomyocyte cytoskeletal genes in patients with heart failure. Lab Invest. 2014;94:645–653. doi: 10.1038/labinvest.2014.54. [DOI] [PubMed] [Google Scholar]
- 16.Dhar K, et al. Targeted myocardial gene expression in failing hearts by RNA sequencing. J Transl Med. 2016;14:327. doi: 10.1186/s12967-016-1083-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song HK, Hong SE, Kim T, Kim DH. Deep RNA sequencing reveals novel cardiac transcriptomic signatures for physiological and pathological hypertrophy. PLoS One. 2012;7:e35552. doi: 10.1371/journal.pone.0035552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kramer A, Green J, Pollard J, Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics. 2014;30:523–530. doi: 10.1093/bioinformatics/btt703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandesara PB, et al. The Prognostic Significance of Diabetes and Microvascular Complications in Patients With Heart Failure With Preserved Ejection Fraction. Diabetes Care. 2018;41:150–155. doi: 10.2337/dc17-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Persson H, et al. Diastolic dysfunction in heart failure with preserved systolic function: need for objective evidence:results from the CHARM Echocardiographic Substudy-CHARMES. J Am Coll Cardiol. 2007;49:687–694. doi: 10.1016/j.jacc.2006.08.062. [DOI] [PubMed] [Google Scholar]
- 21.Anand IS, et al. Prognostic value of baseline plasma amino-terminal pro-brain natriuretic peptide and its interactions with irbesartan treatment effects in patients with heart failure and preserved ejection fraction: findings from the I-PRESERVE trial. Circ Heart Fail. 2011;4:569–577. doi: 10.1161/CIRCHEARTFAILURE.111.962654. [DOI] [PubMed] [Google Scholar]
- 22.Toma M, et al. Differentiating heart failure phenotypes using sex-specific transcriptomic and proteomic biomarker panels. ESC Heart Fail. 2017;4:301–311. doi: 10.1002/ehf2.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liew CC, Dzau VJ. Molecular genetics and genomics of heart failure. Nat Rev Genet. 2004;5:811–825. doi: 10.1038/nrg1470. [DOI] [PubMed] [Google Scholar]
- 24.McGrath MF, Ogawa T, de Bold AJ. Ras dexamethasone-induced protein 1 is a modulator of hormone secretion in the volume overloaded heart. Am J Physiol Heart Circ Physiol. 2012;302:H1826–1837. doi: 10.1152/ajpheart.01085.2011. [DOI] [PubMed] [Google Scholar]
- 25.Imai M, et al. Therapy with cardiac contractility modulation electrical signals improves left ventricular function and remodeling in dogs with chronic heart failure. J Am Coll Cardiol. 2007;49:2120–2128. doi: 10.1016/j.jacc.2006.10.082. [DOI] [PubMed] [Google Scholar]
- 26.Tschope C, et al. Cardiac contractility modulation signals improve exercise intolerance and maladaptive regulation of cardiac key proteins for systolic and diastolic function in HFpEF. Int J Cardiol. 2016;203:1061–1066. doi: 10.1016/j.ijcard.2015.10.208. [DOI] [PubMed] [Google Scholar]
- 27.Katz AM, Lorell BH. Regulation of cardiac contraction and relaxation. Circulation. 2000;102:IV69–74. doi: 10.1161/01.CIR.102.suppl_4.IV-69. [DOI] [PubMed] [Google Scholar]
- 28.Santulli G, Iaccarino G. Adrenergic signaling in heart failure and cardiovascular aging. Maturitas. 2016;93:65–72. doi: 10.1016/j.maturitas.2016.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gambardella J, et al. Functional Role of Mitochondria in Arrhythmogenesis. Adv Exp Med Biol. 2017;982:191–202. doi: 10.1007/978-3-319-55330-6_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangmool S, Parichatikanond W, Kurose H. Therapeutic Targets for Treatment of Heart Failure: Focus on GRKs and beta-Arrestins Affecting betaAR Signaling. Front Pharmacol. 2018;9:1336. doi: 10.3389/fphar.2018.01336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez A, Schelbert EB, Diez J, Butler J. Myocardial Interstitial Fibrosis in Heart Failure: Biological and Translational Perspectives. J Am Coll Cardiol. 2018;71:1696–1706. doi: 10.1016/j.jacc.2018.02.021. [DOI] [PubMed] [Google Scholar]
- 32.Bradshaw AD. The role of secreted protein acidic and rich in cysteine (SPARC) in cardiac repair and fibrosis: Does expression of SPARC by macrophages influence outcomes? J Mol Cell Cardiol. 2016;93:156–161. doi: 10.1016/j.yjmcc.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 33.Mikhailov AT, Torrado M. The enigmatic role of the ankyrin repeat domain 1 gene in heart development and disease. Int J Dev Biol. 2008;52:811–821. doi: 10.1387/ijdb.082655am. [DOI] [PubMed] [Google Scholar]
- 34.Zolk O, et al. Cardiac ankyrin repeat protein, a negative regulator of cardiac gene expression, is augmented in human heart failure. Biochem Biophys Res Commun. 2002;293:1377–1382. doi: 10.1016/S0006-291X(02)00387-X. [DOI] [PubMed] [Google Scholar]
- 35.Engebretsen KV, et al. Lumican is increased in experimental and clinical heart failure, and its production by cardiac fibroblasts is induced by mechanical and proinflammatory stimuli. FEBS J. 2013;280:2382–2398. doi: 10.1111/febs.12235. [DOI] [PubMed] [Google Scholar]
- 36.Mak TW, Hauck L, Grothe D, Billia F. p53 regulates the cardiac transcriptome. Proc Natl Acad Sci USA. 2017;114:2331–2336. doi: 10.1073/pnas.1621436114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsipis A, et al. Apoptosis-related factorsp53, bcl-2 and the defects of force transmission in dilated cardiomyopathy. Pathol Res Pract. 2010;206:625–630. doi: 10.1016/j.prp.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 38.Oka T, Morita H, Komuro I. Novel molecular mechanisms and regeneration therapy for heart failure. J Mol Cell Cardiol. 2016;92:46–51. doi: 10.1016/j.yjmcc.2016.01.028. [DOI] [PubMed] [Google Scholar]
- 39.Liang Y, Pan HF, Ye DQ. Therapeutic potential of STAT4 in autoimmunity. Expert Opin Ther Targets. 2014;18:945–960. doi: 10.1517/14728222.2014.920325. [DOI] [PubMed] [Google Scholar]
- 40.Rocha Rodrigues DB, et al. In situ expression of regulatory cytokines by heart inflammatory cells in Chagas’ disease patients with heart failure. Clin Dev Immunol. 2012;2012:361730. doi: 10.1155/2012/361730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liefke R, et al. Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev. 2010;24:590–601. doi: 10.1101/gad.563210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang GG, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gale M, et al. Screen-identified selective inhibitor of lysine demethylase 5A blocks cancer cell growth and drug resistance. Oncotarget. 2016;7:39931–39944. doi: 10.18632/oncotarget.9539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szot JO, et al. A Screening Approach to Identify Clinically Actionable Variants Causing Congenital Heart Disease in Exome Data. Circ Genom Precis Med. 2018;11:e001978. doi: 10.1161/CIRCGEN.117.001978. [DOI] [PubMed] [Google Scholar]
- 45.Chin MT, et al. Cardiovascular basic helix loop helix factor 1, a novel transcriptional repressor expressed preferentially in the developing and adult cardiovascular system. J Biol Chem. 2000;275:6381–6387. doi: 10.1074/jbc.275.9.6381. [DOI] [PubMed] [Google Scholar]
- 46.Nakagawa O, et al. Members of the HRT family of basic helix-loop-helix proteins act as transcriptional repressors downstream of Notch signaling. Proc Natl Acad Sci USA. 2000;97:13655–13660. doi: 10.1073/pnas.250485597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson DJ, et al. NKX2-5 regulates human cardiomyogenesis via a HEY2 dependent transcriptional network. Nat Commun. 2018;9:1373. doi: 10.1038/s41467-018-03714-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Yu M, Wu L, Chin MT. The bHLH transcription factor CHF1/Hey2 regulates susceptibility to apoptosis and heart failure after pressure overload. Am J Physiol Heart Circ Physiol. 2010;298:H2082–2092. doi: 10.1152/ajpheart.00747.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niu Z, Li A, Zhang SX, Schwartz RJ. Serum response factor micromanaging cardiogenesis. Curr Opin Cell Biol. 2007;19:618–627. doi: 10.1016/j.ceb.2007.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choubey D, Panchanathan R. IFI16, an amplifier of DNA-damage response: Role in cellular senescence and aging-associated inflammatory diseases. Ageing Res Rev. 2016;28:27–36. doi: 10.1016/j.arr.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 51.Lin, W. et al. IFI16 restoration in hepatocellular carcinoma induces tumour inhibition via activation of p53 signals and inflammasome. Cell Prolif50, 10.1111/cpr.12392 (2017). [DOI] [PMC free article] [PubMed]
- 52.Lachmann A, et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuleshov MV, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagueh SF, et al. Recommendations for the Evaluation of Left Ventricular Diastolic Function by Echocardiography: An Update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. J Am Soc Echocardiogr. 2016;29:277–314. doi: 10.1016/j.echo.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 55.Popov S, et al. Lack of salt-inducible kinase 2 (SIK2) prevents the development of cardiac hypertrophy in response to chronic high-salt intake. PLoS One. 2014;9:e95771. doi: 10.1371/journal.pone.0095771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lundin S, Stranneheim H, Pettersson E, Klevebring D, Lundeberg J. Increased throughput by parallelization of library preparation for massive sequencing. PLoS One. 2010;5:e10029. doi: 10.1371/journal.pone.0010029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Borgstrom E, Lundin S, Lundeberg J. Large scale library generation for high throughput sequencing. PLoS One. 2011;6:e19119. doi: 10.1371/journal.pone.0019119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tarazona S, et al. Data quality aware analysis of differential expression in RNA-seq with NOISeq R/Bioc package. Nucleic Acids Res. 2015;43:e140. doi: 10.1093/nar/gkv711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in the EMBL-EBI ArrayExpress database (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-7454.