Abstract
Alkaline stress has serious-negative effects on citrus production. Ziyang xiangcheng (Citrus junos Sieb. ex Tanaka) (Cj) is a rootstock that is tolerant to alkaline stress and iron deficiency. Trifoliate orange (Poncirus trifoliata (L.) Raf.) (Pt), the most widely used rootstock in China, is sensitive to alkaline stress. To investigate the molecular mechanism underlying the tolerance of Cj to alkaline stress, next-generation sequencing was employed to profile the root transcriptomes and small RNAs of Cj and Pt seedlings that were cultured in nutrient solutions along a three pH gradient. This two-level regulation data set provides a system-level view of molecular events with a precise resolution. The data suggest that the auxin pathway may play a central role in the inhibitory effect of alkaline stress on root growth and that the regulation of auxin homeostasis under alkaline stress is important for the adaptation of citrus to alkaline stress. Moreover, the jasmonate (JA) pathway exhibits the opposite response to alkaline stress in Cj and Pt and may contribute to the differences in the alkaline stress tolerance and iron acquisition between Cj and Pt. The dataset provides a wealth of genomic resources and new clues to further study the mechanisms underlying alkaline stress resistance in Cj.
Subject terms: Abiotic, Plant hormones
Molecular biology: Rooting out alkaline resistance mechanisms in citrus trees
The ability of citrus trees to tolerate alkaline soils may hinge upon plant hormones called auxins which regulate root growth. The discovery could aid the development of more resilient rootstocks. Alkaline stress, caused by e.g. industrial run-off, is a growing problem worldwide because it reduces the growth and survival of crops – including the most widely used citrus rootstock in China, Poncirus trifoliata. To better understand the mechanisms underpinning tolerance to soil alkalinity, Hualin Yi at Huazhong Agricultural University in Wuhan, China, and colleagues used next-generation sequencing to profile the transcription products of P. trifolata seedlings and those of an alkaline-tolerant rootstock, Ziyang xiangcheng, when they were grown in three different nutrient solutions. Auxin homeostasis appears to be a key element of citrus adaption to alkaline stress, they found - probably by encouraging lateral root branching.
Introduction
Saline–alkaline soils are widespread throughout the world, limiting agricultural productivity globally1–3. Of the ~831 million ha of saline–alkaline soils, more than half are alkalinized4,5. Under alkaline stress, most plants cannot survive because of the high pH. Alkaline stress has a much stronger inhibitory effect on plant growth than salt stress2,3,6. However, research examining the effect of alkaline stress on plants and the adaptation mechanism of plants to alkaline stress are much scarcer than research on salt stress2,7. Thus, elucidation of the molecular mechanisms of plant responses to alkaline stress is urgently needed.
The root is an organ for the uptake and transport of water and nutrients, and it is also the first organ that perceives soil stresses. Root branching through lateral root (LR) formation is important for the root system to cope with various abiotic stresses8,9. Many studies have revealed that auxin acts as an integrator of many endogenous and exogenous signals for lateral root development regulation9. For instance, cytokinin (CK) inhibits LR formation by reducing auxin transport mediated by PIN110 and by upregulating MIZ111, a gene involved in the hydrotropism that reduces auxin accumulation in the roots. ABA can inhibit LR formation by upregulating miR393, which can suppress the expression of the TIR1/AFB auxin receptor12. Shkolnik-Inbar and Bar-Zvi13 have shown that ABA and CK inhibit LR formation by upregulating the ABI4 transcription factor via reducing polar auxin transport, leading to a reduction in LR development. In addition, many nutrients also influence LR development by interfering with the auxin pathway, such as nitrates14, potassium15, and phosphate16.
Jasmonate (JA) is a significant signaling substance that is involved in plant responses to several abiotic and biotic stresses, such as drought stress, salt stress, wounding (mechanical stress), and pathogen infection17. JA is also involved in the regulation of developmental processes, including root growth, pollen production and senescence18. Many studies have shown that JA plays a positive role in salt stress responses in plants19–25. However, few studies have examined the role of JAs in response to alkaline stress compared with those on salt stress. Recently, Zhu et al.26 reported that TIFY10 proteins, which are JAZ proteins, positively regulate plant alkaline stress responses. A similar result was obtained by Zhu et al.27, who reported that the overexpression of GsJAZ2 in Arabidopsis results in enhanced plant tolerance to salt and alkaline stress. In addition, JAs play important roles in root growth regulation. In Arabidopsis, treatment with JA can inhibit primary root growth28, whereas JA promotes lateral root formation at submicromolar concentrations by directly inducing the auxin biosynthesis gene ASA1 and/or by modulating the PIN2 protein29,30. Raya-Gonzalez et al.31 also showed that low concentrations of JA inhibit primary root growth and promote lateral root formation in Arabidopsis seedlings. In rice, RSS3 interacts with the JA pathway to play a significant role in the regulation of root cell elongation under stressful conditions32.
Generally, alkaline stress accompanies iron deficiency. Iron is required for essential everyday processes in plants and is abundant in most soils. However, most iron exists as Fe3+ hydroxides, which are only somewhat soluble at neutral pH. The release of protons from roots increases the solubility of the Fe3+ hydroxides around the rhizosphere. The Fe3+ is reduced to Fe2+ by FRO233, which was the rate-limiting step for iron acquisition from the soil34. Iron is then transported into the root epidermal cells by IRT135,36. The FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR (FIT) plays a central role in upregulating the root-expressed genes involved in iron acquisition, which regulates FRO2 at the transcription level and IRT1 at the protein level37. Indeed, a fit Arabidopsis mutant fails to take up iron and develops a lethal Fe deficiency, leading to leaf chlorosis38.
Citrus is one of the most important economic fruit crops worldwide. Grafting is the most important reproduction mode for citrus in the production industry. Moreover, rootstocks are significant for scions in citrus and can influence the fruit quality, canopy size, and resistance, among other aspects39. Ziyang xiangcheng (Citrus junos Sieb. ex Tanaka) (abbreviated Cj) is a special citrus germplasm native to China that is widely used as an iron-deficiency tolerant and alkaline-tolerant citrus rootstock in areas of China with calcareous soil. Trifoliate orange (Poncirus trifoliata (L.) Raf.) (abbreviated Pt) is the most widely used rootstock in China. However, Pt is sensitive to alkaline stress. Scions grafted on Pt show nutritional deficiency and growth retardation phenotypes in calcareous soil. However, little is known about the molecular mechanism underlying the differences in phenotypes between Cj and Pt regarding alkaline stress. Thus, in this study, the seedlings of Cj and Pt cultured in different pH nutrient solutions were used to perform comparative analyses, including assessments of the transcriptomes and small RNAs.
Results
Difference between Cj and Pt grown hydroponically with different pH gradients
After 8 weeks of culture in nutrient solutions of different pH (6.5, 8.0, and 9.5), Cj and Pt showed different performances. As shown in Fig. 1a, b, the lengths of the stems and roots were markedly decreased in both Cj and Pt under extremely alkaline conditions (pH 9.5). At pH 6.5, the lengths of the total roots and stems did not differ between the Cj and Pt, but the number of lateral roots and total root surface area of the Cj were significantly increased compared with those of the Pt (Fig. 1b). As the pH value increased, the root architecture was altered in both Cj and Pt, and the differences gradually increased between Cj and Pt, including the total root and stem lengths, total root surface area and number of lateral roots (Fig. 1b). In particular, the number of lateral roots of Cj was much higher than that of Pt (approximately fourfold higher at pH 8.0 and pH 9.5) (Fig. 1a, b).
Fig. 1. Phenotypic characterization of seedlings of Cj and Pt cultured in solutions along a pH gradient.
a The morphological differences between Cj and Pt along a pH gradient b and the trends of several mineral element levels in the root, stem and leaf of Cj and Pt along a pH gradient c. In this figure, the samples are named “Sample_tissue.”. Pt_root indicates the root tissue of Pt. Bars represent the standard error (n = 3). A single asterisk (*) represents statistically significant differences (P < 0.05), and double asterisks (**) represent highly statistically significant differences (P < 0.01), analyzed using Student’s t-test. Lowercase represents statistically significant differences (P < 0.05), analyzed using one-way ANOVA
The mineral elements in Cj and Pt were also measured. As shown in Fig. 1c, the contents of all of the mineral elements exhibited significant differences between Cj and Pt in the root tissue and mixed tissue (stem and leaf) under all three conditions. In the root tissue, the levels of most mineral elements exhibited a decreasing trend as the pH increased, with the exception of Cu, Na, and Mg, and the trend curves of the mineral elements were similar for the Cj and Pt. The Na and Mg levels increased as the pH value increased. The Cu levels increased with increasing pH in Cj, whereas they decreased in Pt (Fig. 1c). Notably, the Fe levels in both tissues of the Cj were much higher than those in the Pt under all three conditions, and the Fe levels in both the Cj and Pt markedly decreased under alkaline stress (Fig. 1c).
Global analysis of root transcriptomes, small RNAs, and degradomes of Cj and Pt
To investigate the underlying molecular changes that accompanied the morphological and physiological changes described above, we used RNA-seq to generate transcriptome, small RNA and degradome profiles of the root tissues of Cj and Pt. All sequencing data are summarized in Supplementary Table S1. Regarding the transcriptomes, after the low-quality reads had been removed, 40–55 million clean reads per sample were mapped against the Citrus sinensis reference genome40 (Supplementary Table S1). The correlation dendrogram illustrates the global relative relationships among the 18 transcriptomes (Fig. 2a). All three biological replicates clustered together, indicating good repeatability of the RNA-seq data. The Cj and Pt samples were clustered together, respectively (Fig. 2a). A total of 28,470 genes were identified in these six samples (Supplementary Table S2). Between 19,348 (Pt3) and 20,674 (Cj1) genes were detected in each sample (Fig. 2b). In total, 18,019 genes were common among all six samples (Fig. 2b, Supplementary Table S2).
Fig. 2. Analysis of global gene expression in Cj and Pt.
a Cluster dendrogram depicting the global relationships between biological replicates and among different samples. In this figure, samples are named as “Cultivar_pH condition_replicate.” Cj1R1 indicates Cj_pH 6.5_Replicate 1. b Venn diagram depicting the number of commonly and uniquely expressed genes among the six samples. c Model profiles of genes in Cj along a pH gradient generated by the STEM clustering algorithm and top 10 enriched KEGG pathways of the Cj up and Cj down cluster profiles. d Model profiles of genes in Pt along a pH gradient generated by the STEM clustering algorithm and top 10 enriched KEGG pathways of the Pt up and Pt down cluster profiles. Each box corresponds to a model expression profile. The X-axis of each box from left to right is pH 6.5, pH 8.0, and pH 9.5. Colored model profiles contain a statistically significant number of assigned genes (p-value <0.05). Model profiles with the same color belong to the same cluster of profiles. The number in the top left corner indicates the model profile ID
For small RNAs, a total of 18 small RNA libraries were constructed for six samples. Each sample was assessed in the three biological replicates. In total, 11–26 million clean reads per replicate were obtained by RNA-seq (Supplementary Table S1). A search of the Rfam, Silva, GtRNAdb, and Repbase databases was used to remove structural RNAs (rRNAs, tRNAs, scRNAs, snRNAs, and snoRNAs). Unannotated sRNAs (4.3–16.4 million reads) were mapped to the Citrus sinensis genome and were used to identify known miRNAs and predict novel miRNAs by miRDEEP241 based on the structure and expression criteria42 (Supplementary Table S1). In total, 49 known miRNAs and 52 novel miRNAs were identified in the Cj and Pt samples (Supplementary Table S8 and Figure S5).
To identify the target genes of the miRNAs, degradome sequencing43,44 was performed to experimentally validate the target genes of the miRNAs by capturing mRNA cleavage segments. To maximize the identification of the miRNA targets, we pooled RNA from Cj1, Cj2, and Cj3 into one Cj library and pooled RNA from Pt1, Pt2, and Pt3 into one Pt library. In total, 27.04 and 26.15 million clean reads were obtained from the Cj and Pt libraries, respectively (Supplementary Table S1). After removing miRNAs, structural RNAs and others, 16.06 and 15.88 million reads of the Cj and Pt, respectively, were mapped to the Citrus sinensis genome (Supplementary Table S1). The reads that mapped to the cDNA (the reads in the cDNA_sense category in Supplementary Table S1) were analyzed to detect candidate targets of miRNAs.
Identification of temporal expression trends accompanying gradient alkaline stress
STEM45 was used to perform clustering to identify the different gene expression profiles across all three pH conditions in the gradient. After the STEM clustering algorithm was applied, 16 model profiles with 5 statistically significant model profiles (colored profiles) were identified in Cj, and 16 model profiles with 7 statistically significant model profiles were identified in Pt (Fig. 2c, d, Supplementary Table S3). Model profiles with the same color belonged to the same cluster of profiles. Therefore, two cluster profiles (an upregulated profile and downregulated profile) were identified in Cj and Pt, respectively (Fig. 2c, d). These four cluster profiles were named Cj up, Cj down, Pt up, and Pt down. A total of 506, 859, 950, and 1417 genes were clustered into the Cj up, Cj down, Pt up, and Pt down profiles, respectively (Fig. 2c, d). A Venn diagram revealed that only 157 genes exhibited common expression trends in Cj and Pt (Supplementary Figure S1a). This result revealed that large differences existed between Cj and Pt at the transcription level in response to alkaline stress.
These four gene sets (Cj up, Cj down, Pt up, and Pt down) were subject to KEGG pathway enrichment analysis (Supplementary Table S4). As shown in Fig. 2c, d, the top 10 over-represented KEGG pathways are listed. Notably, the “Phenylpropanoid biosynthesis” and “Plant hormone signal transduction” pathways were enriched in all four gene sets. Then, we used a heatmap to display the gene expression trends of the genes that were enriched in these two pathways (Supplementary Figure S2). As shown in Figure S2a, b, in the phenylpropanoid biosynthesis pathway, 22 peroxidase genes (22/50) were downregulated in Pt with increasing pH values, whereas only 4 peroxidase genes (4/18) were downregulated in Cj. In the plant hormone signal transduction pathway, many auxin signal transduction genes (14 genes in Pt and 10 genes in Cj) were enriched (Supplementary Figure S2c, d). In Pt, most auxin signal transduction genes (12/14) were downregulated. In contrast, one-half of the auxin signal transduction genes (5/10) were downregulated in Cj. These results indicate that peroxidase and auxin signal transduction genes may play significant roles in response to alkaline stress.
In addition, we identified transcription factors (TFs) in these four profiles. In total, 66 and 97 TFs were identified in Cj and Pt, respectively (Supplementary Figure S1b, c, Table S3). Among these TFs, the number of ERF family TFs was considerably greater compared with the number of TFs in the other families (Supplementary Figure S1b, c). Moreover, 24 TFs were identified both in Cj and Pt (Supplementary Table S3). As shown in Supplementary Figure S1d, the expression trends of most of these 24 TFs differed between Cj and Pt. Dynamic expression changes associated with these TFs may reveal their key functions. For example, MYC2 (orange1.1t01021), GBF4 (orange1.1t01148), WRKY46 (orange1.1t00472), and ZAT12 (Cs8g17960) were specifically downregulated in Cj and upregulated in Pt as the pH value increased (Supplementary Figure S1d, Table S3). Notably, MYC2 is a key gene in JA signal transduction. This finding indicates that JA signaling may also be involved in response to alkaline stress.
Analysis of transcriptomic changes in response to alkaline stress
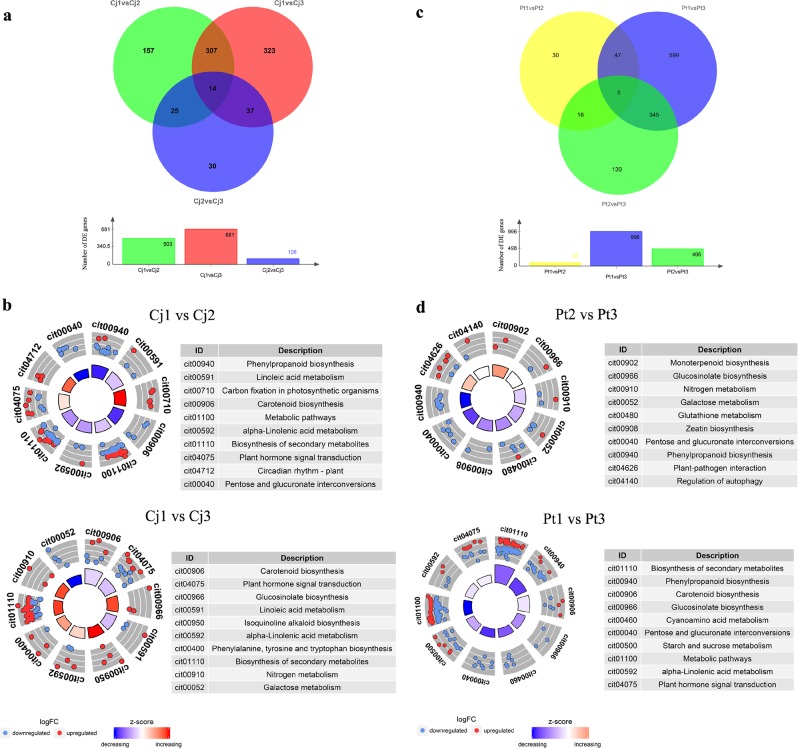
As alkaline stress increased, the phenotypes were largely altered in both Cj and Pt. Simultaneously, the transcriptome profiles of Cj and Pt were also significantly altered. In Cj, 503, 681, and 106 DEGs (differentially expressed genes) were identified in the Cj1 vs. Cj2, Cj1 vs. Cj3, and Cj2 vs. Cj3 comparison groups, respectively (Fig. 3a). In Pt, 98, 996, and 496 DEGs were identified in the Pt1 vs. Pt2, Pt1 vs. Pt3, and Pt2 vs. Pt3 comparison groups, respectively (Fig. 3c). The smallest difference was noted between the Cj2 and Cj3 transcriptomes as well as between the P1 and Pt2 transcriptomes. This result suggests that Cj responds to alkaline stress more quickly than Pt.
Fig. 3. Differential expression genes of Cj and Pt among the different pH conditions.
Venn diagrams depicting the number of common and unique DEGs among the different comparison groups in Cj a and Pt c, and the circular visualization of the KEGG pathway enrichment results of DEGs from different comparison groups in Cj b and Pt d. In Sample1 vs. Sample2, Sample1 serves as the control group. The term logFC means log2(FPKM_Sample2/FPKM_Sample1). The Z-score is an easily calculated value that provides a hint regarding the pathway that is more likely to be decreased (negative value, blue color) or increased (positive value, red color). DEG differentially expressed gene
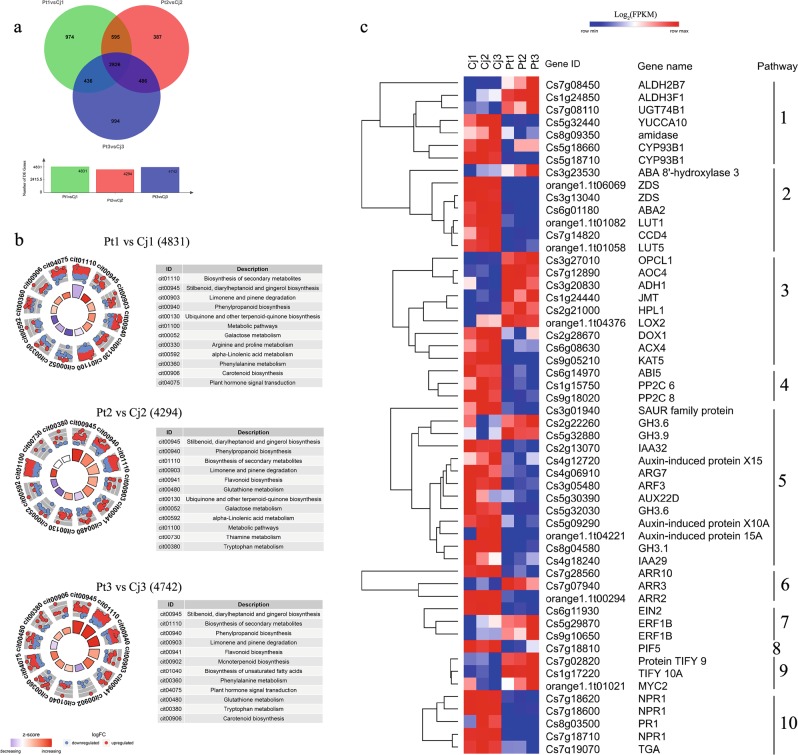
To further characterize the significant roles of Cj and Pt in response to alkaline stress, the DEGs of these six comparison groups of Cj and Pt were used to perform GO-based term and KEGG-based pathway enrichment analyses (Supplementary Tables S6 and S7). Because the number of enriched pathways in Cj2 vs. Cj3 and P1 vs. Pt2 was minimal, the pathways enriched in the other four comparison groups were used to identify important pathways. The top 10 most enriched pathways and the DEGs distributed in these pathways were visualized using the GOplot package46 (Fig. 3b, d). The number of upregulated DEGs was more than that of downregulated DEGs in the top 10 pathways in the Cj1 vs. Cj3 group, whereas the opposite pattern was observed in the Pt1 vs. Pt3 group (Fig. 3b, d). Several key pathways were screened from these four comparison groups, including “Glucosinolate biosynthesis”, “Phenylpropanoid biosynthesis”, “Carotenoid biosynthesis”, “alpha-Linolenic acid metabolism”, and “Plant hormone signal transduction” (Fig. 3b, d). In the glucosinolate biosynthesis pathway, three genes (Cs7g29760, Cs7g29770, and orange1.1t01456) encoding phenylalanine N-monooxygenase (CYP79A2) were enriched. These genes were all upregulated in Cj and downregulated in Pt as the pH increased. CYP79A2 is a key gene in the biosynthesis of glucotropaeolin47. The expression patterns of the DEGs enriched in “Phenylpropanoid biosynthesis”, “Carotenoid biosynthesis”, “alpha-Linolenic acid metabolism”, and “Plant hormone signal transduction” were visualized by heatmaps (Fig. 4). As shown in Fig. 4, in the phenylpropanoid biosynthesis pathway, many peroxidase genes were enriched, displaying V-shaped expression patterns in Cj. However, these genes displayed downregulation trends in Pt. Moreover, several key genes involved in JA, ABA, and strigolactone (SL) biosynthesis were significantly altered by alkaline stress, such as LOX3 (Cs1g17380) and JMT (Cs7g31430, Cs1g24440) for JA biosynthesis, NCED1 (Cs5g14370, Cs2g03270) and ABA 8’-hydroxylase 3 (Cs3g23530) for ABA biosynthesis, and D27 (Cs5g30540) for SL biosynthesis (Fig. 4, Table 1). According to the expression trends of these key genes, the levels of ABA, JA, and SL may be altered by alkaline stress. This result indicates that ABA, JA, and SL may play important roles in tolerance to alkaline stress. In addition, many DEGs were enriched in plant hormone signal transduction, including auxin, CK, ABA, JA, ethylene (ETH) and salicylic acid signals (Fig. 4). More DEGs were enriched in auxin signal transduction in both Cj and Pt, and these DEGs displayed mixed expression patterns (some were upregulated, and some were downregulated). In Pt, the number of downregulated auxin signal genes was greater than that of the upregulated genes, and the opposite finding was observed for Cj. This result indicates that the auxin signal in Pt is more downregulated by alkaline stress compared with that in Cj. Moreover, the JA, ABA, CK, and ETH signals were also altered by alkaline stress in both Cj and Pt (Fig. 4).
Fig. 4. Hierarchical clustering of DEGs in several key pathways across the different pH conditions in Cj.
a and Pt b Each row of the heatmap represents an individual gene, and the coloring represents the expression level. Pathways: 1, alpha-Linolenic acid metabolism; 2, Strigolactone biosynthesis; 3, ABA biosynthesis; 4, Lignin biosynthesis; 5, Auxin signal transduction; 6, Ethylene signal transduction; 7, Cytokinin signal transduction; 8, ABA signal transduction; 9, JA signal transduction; 10, Coumarin biosynthesis; 11, Salicylic acid signal transduction. DEG differentially expressed gene
Table 1.
A list of important differentially expressed genes in Cj or Pt under different pH conditions
| Gene ID | Gene name | FPKM | |||||
|---|---|---|---|---|---|---|---|
| Cj1 | Cj2 | Cj3 | Pt1 | Pt2 | Pt3 | ||
| Plant hormone biosynthesis | |||||||
| Cs1g17380 | LOX3 | 126.20 | 60.53 | 43.40 | 51.93 | 76.67 | 177.22 |
| orange1.1t03774 | LOX2 | 231.01 | 289.28 | 324.75 | 234.67 | 380.47 | 483.81 |
| Cs1g24440 | JMT | 4.87 | 8.52 | 5.89 | 79.69 | 37.49 | 77.35 |
| Cs2g03270 | NCED1 | 1.42 | 0.55 | 0.45 | 3.31 | 1.45 | 0.87 |
| Cs5g14370 | NCED1 | 59.44 | 19.47 | 7.08 | 19.29 | 12.78 | 22.50 |
| Cs3g23530 | ABA 8’-hydroxylase 3 | 0.29 | 0.87 | 0.96 | 1.75 | 3.48 | 8.42 |
| Auxin signal transduction | |||||||
| Cs2g06880 | TIR1 | 14.56 | 11.52 | 12.04 | 12.48 | 9.11 | 5.72 |
| Cs1g13960 | IAA14 | 83.72 | 57.11 | 38.22 | 88.59 | 54.95 | 37.91 |
| Cs3g01940 | SAUR family protein | 192.92 | 331.85 | 309.91 | 75.05 | 54.95 | 49.84 |
| Cs4g18240 | IAA29 | 35.45 | 11.17 | 4.40 | 0.45 | 1.04 | 2.04 |
| Cs5g30390 | AUX22D | 33.49 | 13.10 | 4.49 | 5.12 | 3.04 | 1.70 |
| JA signal transduction | |||||||
| Cs1g17220 | TIFY 10 A | 72.55 | 78.69 | 62.94 | 187.85 | 179.28 | 190.65 |
| Cs7g02820 | TIFY 9 | 104.68 | 109.34 | 71.07 | 267.47 | 279.69 | 337.96 |
| orange1.1t01021 | MYC2 | 159.59 | 83.55 | 67.92 | 123.03 | 170.53 | 227.57 |
| Cytokinin signal transduction | |||||||
| Cs5g04810 | ARR5 | 68.03 | 114.63 | 111.99 | 107.62 | 57.38 | 26.45 |
| Cs3g01890 | ARR9 | 12.95 | 20.49 | 19.15 | 86.02 | 38.91 | 25.45 |
| orange1.1t01850 | ARR9 | 24.43 | 46.81 | 44.90 | 131.95 | 84.08 | 35.45 |
| ABA signal transduction | |||||||
| Cs7g30500 | PYL4 | 10.59 | 15.90 | 31.14 | 15.66 | 15.16 | 10.68 |
| Cs7g31880 | PP2C 37 | 137.34 | 79.47 | 67.85 | 70.22 | 62.68 | 65.05 |
| Cs4g14980 | PP2C 77 | 17.38 | 13.60 | 9.11 | 2.93 | 2.20 | 6.36 |
| orange1.1t01148 | GBF4 | 25.27 | 17.10 | 11.24 | 7.30 | 11.06 | 14.95 |
| ETH signal transduction | |||||||
| Cs5g33560 | ERS1 | 53.88 | 53.74 | 62.50 | 41.84 | 42.04 | 84.30 |
| Cs9g08850 | ETR2 | 25.49 | 22.29 | 25.90 | 13.98 | 14.13 | 28.92 |
| Cs9g10650 | ERF1B | 0.59 | 2.40 | 2.28 | 11.72 | 6.31 | 35.82 |
| Cs5g29870 | ERF1B | 3.91 | 8.79 | 11.45 | 17.88 | 19.25 | 28.21 |
| Ion transport | |||||||
| Cs6g09150 | Ferritin-1 | 308.96 | 263.21 | 132.31 | 69.33 | 74.37 | 8.59 |
| Cs8g15600 | FIT1 | 10.02 | 14.90 | 25.94 | 5.30 | 5.71 | 1.88 |
| orange1.1t00399 | FRO2 | 1.32 | 1.44 | 14.88 | 2.38 | 5.40 | 1.61 |
| Cs9g19350 | FRO4 | 12.08 | 32.08 | 20.97 | 4.75 | 12.10 | 3.35 |
| Cs3g01120 | FRO6 | 45.44 | 64.64 | 77.65 | 54.26 | 36.29 | 35.95 |
| Cs2g17670 | Vacuolar iron transporter 1 | 7.96 | 8.24 | 6.91 | 3.24 | 6.01 | 1.08 |
| Cs2g17680 | Vacuolar iron transporter 1 | 1.03 | 0.87 | 0.88 | 2.59 | 4.52 | 0.39 |
| Cs4g04460 | HKT1 | 8.73 | 4.83 | 3.48 | 13.89 | 12.01 | 6.33 |
| Cs1g17440 | Potassium transporter | 1.71 | 3.46 | 6.56 | 5.03 | 5.39 | 1.97 |
| Cs8g19470 | COPT1 | 105.38 | 280.36 | 146.19 | 239.85 | 307.49 | 89.99 |
| Cs6g11670 | PIP2-2 | 1.68 | 1.79 | 2.14 | 247.81 | 211.03 | 61.79 |
| Cs6g11700 | PIP2-2 | 742.85 | 515.42 | 512.74 | 327.20 | 237.80 | 98.21 |
| Cs7g28650 | TIP1-3 | 318.73 | 206.19 | 209.54 | 865.85 | 888.13 | 374.02 |
| Cs8g17900 | TIP1-3 | 525.44 | 309.47 | 308.31 | 445.38 | 478.62 | 193.00 |
| Cs5g08710 | TIP2-2 | 249.87 | 159.79 | 208.14 | 290.55 | 318.31 | 129.69 |
| Cs6g20570 | PMA1 | 93.09 | 84.97 | 103.70 | 71.72 | 80.69 | 156.91 |
| Cs2g10720 | ZIP10 | 1.29 | 9.05 | 68.08 | 3.88 | 23.69 | 8.59 |
| Cs2g11620 | ZIP5 | 17.59 | 34.08 | 62.11 | 58.95 | 59.75 | 92.42 |
| Cs4g08930 | ZIP5 | 52.70 | 110.94 | 76.30 | 77.90 | 83.16 | 66.52 |
| Cs6g11470 | ZIP5 | 71.45 | 130.31 | 184.01 | 77.77 | 123.44 | 232.80 |
| Response to stimulus | |||||||
| orange1.1t03944 | ORG2 | 1.63 | 1.93 | 12.84 | 7.15 | 8.57 | 35.16 |
| Cs1g11190 | ZFP1 | 4.63 | 9.85 | 12.34 | 16.25 | 15.37 | 5.34 |
| Cs6g05660 | ZAT11 | 22.12 | 5.86 | 10.95 | 66.02 | 17.73 | 9.48 |
| Cs1g18580 | DOF1.7 | 44.31 | 20.81 | 11.51 | 10.18 | 11.45 | 24.69 |
| Cs3g19420 | ERF012 | 31.00 | 26.08 | 15.58 | 8.55 | 20.41 | 29.27 |
| Cs3g21660 | ERF110 | 0.61 | 1.99 | 2.25 | 11.41 | 3.06 | 2.17 |
| Cs6g15360 | Dehydration-responsive element-binding protein 1D | 4.75 | 0.79 | 1.05 | 10.52 | 5.59 | 27.18 |
| Cs6g15410 | ERF024 | 1.06 | 7.37 | 5.34 | 4.86 | 8.53 | 1.97 |
| Cs8g05910 | ERF109 | 17.31 | 4.67 | 2.56 | 7.31 | 11.04 | 25.29 |
| Cs9g13620 | ERF5 | 361.93 | 154.96 | 168.66 | 45.62 | 46.22 | 117.12 |
| Cs4g12130 | Chitin-inducible gibberellin-responsive protein 1 | 172.13 | 61.51 | 40.99 | 54.90 | 46.03 | 92.31 |
| Cs6g15680 | HRA1 | 9.24 | 2.59 | 3.18 | 1.54 | 1.76 | 4.28 |
| orange1.1t00472 | WRKY46 | 224.68 | 138.04 | 83.44 | 113.21 | 101.83 | 265.74 |
As shown in Fig. 1c, the uptake of mineral elements, especially metal elements such as Fe, Zn, and Cu, is largely influenced by alkaline stress. The GO enrichment analysis results indicated that 49 and 68 DEGs were significantly enriched in the “ion transport” term in Cj and Pt, respectively (Supplementary Figure S3, Table S7). As shown in Supplementary Figure S4a, iron-transport-related genes (FIT1, FRO2/4/6, Ferritin-1), zinc-transport-related genes (ZIP1/5/10), and copper-transport-related genes (COPT1) were upregulated in Cj under alkaline stress (Table 1). However, iron-transport-related genes (FIT1, FRO2/4, Ferritin1, and vacuolar iron transport 1) were largely downregulated at pH 9.5 in Pt (Supplementary Figure S4b, Table 1). Zinc-transport-related genes (ZIP5/10) and copper-transport-related genes (COPT1/6) were also upregulated in Pt under alkaline stress. In addition, six aquaporin genes (two TIP1-3s, two PIP2-2s, TIP2-2, and NIP2-1) were largely downregulated in Pt at pH 9.5 (Supplementary Figure S4b, Table 1). These results suggest that Pt may be more susceptible to iron and water deficiencies under alkaline stress.
Comparing Cj and Pt transcriptomes
Cj is more tolerant to alkaline stress compared with Pt in citrus production. To compare the differences of Cj and Pt at the transcriptome level, we identified some key pathways and genes involved in the response to alkaline stress. As shown in Fig. 5a, 4831, 4294 and 4742 DEGs were identified in the Pt1 vs. Cj1, Pt2 vs. Cj2 and Pt3 vs. Cj3 comparison groups, respectively. The DEGs of these three groups were subject to GO-based term and KEGG-based pathway enrichment analyses (Supplementary Tables S6 and S7). The top 12 most enriched pathways and DEGs enriched in these pathways were visualized using the GOplot package46 (Fig. 5b). The top 4 most enriched pathways were the same in the three comparison groups. As the pH increased, the number of upregulated DEGs in the top 4 pathways increased (Fig. 5b). This result indicates that there are intrinsic differences in these pathways between Cj and Pt, which are magnified under alkaline stress. In particular, the “Biosynthesis of secondary metabolites” pathway was largely altered by alkaline stress in both Cj and Pt (Figs. 3b, d and 5b). In addition, the “Phenylpropanoid biosynthesis”, “alpha-Linolenic acid metabolism”, “Carotenoid biosynthesis”, and “Plant hormone signal transduction” remained enriched in these three or two comparison groups, which was consistent with the above result (Figs. 3b, d and 5b). In addition, the auxin biosynthesis pathway (“Tryptophan metabolism” pathway) was enriched in the Pt2 vs. Cj2 and Pt3 vs. Cj3 comparison groups. Therefore, we further analyzed the expression trends of the DEGs that were distributed in these five pathways and were differentially expressed in all three comparison groups. As shown in Fig. 5c and Supplementary Figure S4c, the expression of these genes exhibited significant differences between Cj and Pt. In the plant hormone signal transduction pathway, auxin signal genes comprised the largest group, and most of these genes (11/13) were downregulated in Pt. The ABA, ETH, JA, CK, and SA signaling genes were also differentially expressed between Cj and Pt. Moreover, several DEGs were enriched in the auxin, ABA, and JA biosynthesis pathways (Fig. 5c). In the phenylpropanoid biosynthesis pathway, the expression of most of the genes (28/40) in the Pt was reduced compared with that of the genes in Cj, and most of these genes were peroxidases (Supplementary Figure S4c). These results further demonstrate that auxin, ABA, JA signal pathways, and peroxidases may play important roles in tolerance to alkaline stress.
Fig. 5. Gene expression comparisons between Cj and Pt.
a A Venn diagram depicting the number of common and unique DEGs among the three comparison groups. b Circular visualization of the results of the KEGG pathway enrichment of DEGs from the different comparison groups between Cj and Pt. c A heatmap depicting the expression patterns of DEGs in several key pathways across all six samples. Pathways: 1, Auxin biosynthesis; 2, Carotenoid biosynthesis; 3, alpha-Linolenic acid metabolism; 4, ABA signal transduction; 5, Auxin signal transduction; 6, Cytokinin signal transduction; 7, Ethylene signal transduction; 8, Gibberellin signal transduction; 9, JA signal transduction; 10, Salicylic acid signal transduction. DEG differentially expressed gene
According to the results of the GO enrichment analysis, “response to stimulus” was the most enriched term (Supplementary Figure S3, Table S7). A total of 118 DEGs were enriched in the “response to stimulus” term in both the Pt2 vs. Cj2 and Pt3 vs. Cj3 comparison groups. As shown in Supplementary Figure S4d, the genes responding to the stimulus included disease resistance proteins; plant hormone biosynthesis and signal transduction genes; ion transport genes; and cell wall metabolism genes, such as NCED1, LOX3, FRO2/4, and peroxidases. Among these DEGs, TFs with increased expression in Cj3 or Pt3 included six ERF genes (ERF012, ERF110, Dehydration-responsive element-binding protein 1D, ERF024, ERF109, and ERF5), two bHLH genes (FIT1 and ORG2), two C2H2 genes (ZFP1 and ZAT11), WRKY46, HRA1 and chitin-inducible gibberellin-responsive protein 1 (Supplementary Figure S4d, Table 1). FIT1 is an essential gene that regulates iron uptake in Arabidopsis thaliana37, and WRKY46 modulates the growth of Arabidopsis lateral roots under osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis48. Hence, these TFs may also play important roles in citrus in response to alkaline stress.
Identification and expression analysis of miRNAs in the roots of Cj and Pt
A total of 101 miRNAs (49 known miRNAs and 52 novel miRNAs) belonging to 37 families were identified from the six samples of Cj and Pt (Supplementary Table S8). Only the miRNAs identified in all three biological replicates were retained. All precursors of novel miRNAs exhibited regular stem-loop secondary structures. The miRNA sequences are shown in blue, and the sequences of the miRNAs* are shown in red (Supplementary Figure S5). Based on the criteria of significant difference (FDR < 0.05), 12, 16, 5, 11, 33, 32, and 28 DE miRNAs (differentially expressed miRNAs) were identified in the Cj1 vs. Cj2, Cj1 vs. Cj3, Pt2 vs. Pt3, Pt1 vs. Pt3, Pt1 vs. Cj1, Pt2 vs. Cj2, and Pt3 vs. Cj3 comparison groups, respectively (Fig. 6g, Supplementary Table S9). However, no DE miRNA was identified in the Cj2 vs. Cj3 and Pt1 vs. Pt2 comparison groups, indicating that the difference between Cj2 and Cj3 or Pt1 and Pt2 was minimal (Fig. 6g). This result was consistent with the transcriptome results (Figs. 3a, c and 6g). Venn diagrams and heatmaps were used to demonstrate the inclusion relations and expression patterns of DE miRNAs in these seven comparison groups (Fig. 6a–f), respectively. In total, 11 and 18 DE miRNAs were identified among the different pH conditions in Pt and Cj, respectively, and the number of downregulated miRNAs was much more than that of upregulated miRNAs in both Cj and Pt (Fig. 6a, b, d, e). A total of 44 DE miRNAs was identified between Cj and Pt, and the expression of most of the DE miRNAs in Pt was higher than that of the DE miRNAs in Cj (Fig. 6c, f).
Fig. 6. Global analysis of differentially expressed miRNAs.
a, b, and c Venn diagrams depicting the number of differentially expressed miRNAs in different comparison groups. d, e, and f Heatmaps depicting the expression patterns of differentially expressed miRNAs across Cj samples, Pt samples and all six samples, respectively. g The number of differentially expressed miRNAs in each comparison group
Identification of PHAS genes
From the eighteen deep sequencing sRNA libraries of Cj and Pt, 28 PHAS genes (the gene targeted by miRNAs to generate phasiRNAs) were identified that were capable of secondary siRNA production, with a P-value ≤0.0001 (Supplementary Table S13). In total, 24 PHAS genes that encoded resistance proteins were targeted by the MIR482 family (csi-482-3p, csi-482b/c, and csi-miR48). In addition, csi-miR393a triggered two TIR/AFB auxin receptor proteins, and csi-miR1515 triggered two dicer-like proteins (Supplementary Table S13). Moreover, the expression of csi-miR393a in Pt was higher than that in Cj under all three conditions (Supplementary Table S8). In this study, many genes in the auxin signal pathway were significantly altered by alkaline stress in both Cj and Pt, including TIR1 (Table 1). Hence, this result suggests that csi-miR393a may be involved in the regulation of auxin signals in response to alkaline stress.
Identification and functional analysis of the targets of miRNAs
From the degradome data, a total of 260 transcripts from 150 genes were predicted to be targeted by 62 miRNAs (39 known miRNAs and 23 novel miRNAs) in Cj, with 416 miRNA-target pairs (Supplementary Figure S6, Table S10). A total of 215 transcripts from 121 genes were predicted to be targeted by 56 miRNAs (38 known miRNAs and 18 novel miRNAs) in Pt, with 353 miRNA-target pairs (Supplementary Figure S7, Table S10). These miRNA-transcript pairs were classified into five categories (Category 0, 1, 2, 3, and 4) based on the confidence evaluation of the degradome data. In Categories 3 and 4, the miRNA-target pairs are not reliable; therefore, only those miRNA-target pairs that were distributed in Categories 0, 1, and 2 were retained. Finally, we identified 157 target genes (128 targets from Cj and 103 from Pt) for 58 miRNAs (52 miRNAs of Cj and 49 miRNAs of Pt) (Supplementary Figure S8a, b).
To further elucidate the roles of miRNAs in response to alkaline stress, the targets of the DE miRNAs were used to perform GO-based term and KEGG-based pathway enrichment analyses. These targets of the DE miRNAs of each comparison group are listed in Supplementary Table S11 and are shown in Supplementary Figure S8c, d. Given that the number of targets was small in each set, no enrichment pathways were identified. According to the GO enrichment analysis results, several GO terms were highly enriched, including the response to stimulus, root cap development, lignin catabolic process, and reactive oxygen species metabolic process (Supplementary Figure S9, Table S12). The expression patterns of several important miRNAs and their targets were verified in Cj and Pt (Fig. 7). The targets of csi-miRN43/45 included four ARFs (ARF10/17) that are important TFs for root cap development in Arabidopsis49 (Fig. 7a). LAC4/7/17/22 and P5CS1 are the targets of csi-miR397 (Fig. 7c). LACs are involved in lignin catabolism, and P5CS1 is a key gene for proline biosynthesis that plays important roles in scavenging reactive oxygen species (ROS)50. The csi-miR398 targeted several ROS scavenging genes, such as SOD1/2 and catalase (Fig. 7b). These results further suggest that the auxin pathway and ROS scavenging system may be important in the resistance to alkaline stress.
Fig. 7. Expression patterns of miRNAs and their target genes in Cj and Pt under different pH conditions.
a csi-miRN43/45; b csi-miR398; c csi-miR397. A single asterisk (*) represents statistically significant differences (P < 0.05), and double asterisks (**) represent highly statistically significant differences (P < 0.01), analyzed using Student’s t-test
Changes of phytohormone and antioxidants in Cj and Pt under alkaline stress
According to the comprehensive analysis of transcriptomes and small RNAs of Cj and Pt, the auxin, ABA, and JA signal pathways and antioxidants may play more important roles in tolerance to alkaline stress. To further verify the difference of phytohormone and antioxidants between Cj and Pt in response to alkaline stress, we measured the JA, ABA, IAA, POD, SOD, and CAT levels in the roots of Cj and Pt under different pH conditions. As shown in Fig. 8a, the IAA levels in Cj were significantly higher than those in Pt at pH 8.0, and no significant difference was noted between the IAA levels in Cj and Pt under the other two conditions. The ABA levels in Cj were significantly lower than those in Pt under all three pH conditions (Fig. 8b). The IAA and ABA levels decreased in both Cj and Pt as the pH increased. The JA levels in Cj were much higher than those in Pt (from approximately 12-fold to approximately 40-fold) under all three pH conditions, and the JA levels in Pt increased as the pH increased. By contrast, the JA levels in Cj were downregulated under high pH conditions (Fig. 8c). Regarding antioxidants, the levels of POD, SOD, and CAT decreased markedly in Pt as the pH increased; however, the levels of these antioxidants were minimally altered in Cj (Fig. 8d–f). These results are consistent with the results of the bioinformatics analysis.
Fig. 8. Content of several types of plant hormones.
a–c and antioxidants d–f in the roots of Cj and Pt under different pH conditions. A single asterisk (*) represents statistically significant differences (P < 0.05), and double asterisks (**) represent highly statistically significant differences (P < 0.01), analyzed using Student’s t-test
Exogenous JA and auxin analogs enhance the tolerance of citrus to alkaline stress
To further verify the roles of JA and auxin in alkaline stress tolerance in citrus, we designed a treatment experiment using JA and auxin analogs (NAA and IBA) to treat Pt seedlings at pH 8.5 (Fig. 9a). After 12 weeks of culture, the JA + NAA + IBA-treatment group exhibited the best growth, generating the largest number of LRs with healthy root caps and the lowest ROS level (Fig. 9). In addition, the status of the JA-treatment group was not good, which generated tiny short LRs with necrotic root caps; whereas, the ROS levels were lower than those of the control and NAA + IBA-treatment groups (Fig. 9). This result demonstrated that JA played a positive role in scavenging ROS and a negative role in LR growth and formation under alkaline stress. Moreover, the simultaneous application of auxin and JA to Pt can promote LR growth and scavenge ROS under alkaline stress. These results indicated that auxin and JA synergistically played important roles in the tolerance of citrus to alkaline stress.
Fig. 9. Exogenous JA, NAA, and IBA treatment of Pt seedlings under alkaline stress.
a The phenotypes. b Number of LRs of Pt seedlings cultured at pH 8.5 for 12 weeks, and three treatment groups (JA (5 μM), NAA + IBA (0.5 mg/L + 0.1 mg/L) and JA + NAA + IBA (5 μM + 0.5 mg/L + 0.1 mg/L)) were defined. c In situ accumulation of H2O2 and O2·–; examined by histochemical staining with DAB and NBT. d–f The content of H2O2, anti-O2·– and MDA. Lowercase and capital letters represent statistically significant differences (P < 0.05) and highly statistically significant differences (P < 0.01), respectively. Data were analyzed using one-way ANOVA
Discussion
Rootstock is important for the citrus industry and can significantly affect the characteristics of scions39. Cj has increasingly been used as rootstock for many scion cultivars in recent years in China given its excellent tolerance to alkaline stress and iron deficiency. However, the molecular mechanism has rarely been elucidated. Our work reported here provides crucial molecular insights into the tolerance of Cj to alkaline stress. The comprehensive transcriptomes and small RNA data sets of Cj and Pt along a pH gradient enabled genome-scale analyses at a high resolution. The genome-scale approach allowed us to examine the expression profiles of all of the members of gene/miRNA families in an unbiased manner and to simultaneously analyze multiple pathways. In addition, the integrated analysis of the transcriptomes and small RNAs provided more information from different regulation levels in response to alkaline stress.
The auxin pathway may play a central role in hormonal regulation of LR development in citrus in response to alkaline stress
In this study, alkaline stress largely inhibited the formation and growth of LR in both Cj and Pt. In Arabidopsis, auxin serves as an integrator of numerous signals that are involved in LR development, such as ABA, CK, JA, and nutrients. These signals affect the LR development by interacting with auxin homeostasis (synthesis, conjugation, and degradation), transport, or response9,16,29,51. In this study, the LR development of Cj and Pt was closely related to the content of IAA (Figs. 1a, b and 8a). Moreover, a number of genes/miRNAs related to the auxin signal transduction pathway were differentially expressed in the roots of Cj and Pt under gradient alkaline stress, such as IAA14 (Cs1g13960), csi-miR43 and others (Figs. 4 and 6, Supplementary Figure S2). A previous study reported that IAA14 is a key regulator in auxin-regulated growth and development, particularly in lateral root formation and that the slr-1/iaa14 mutant of Arabidopsis completely lacked lateral roots52. In addition, under alkaline stress, treatment with exogenous JA, IBA, and NAA significantly increased the number of LRs (Fig. 9). These results revealed that auxin played an important role in citrus lateral root development under alkaline stress. Moreover, in this study, the expression of genes involved in IAA biosynthesis was slightly altered in Cj and Pt under gradient alkaline stress. However, some GH3 genes that function in conjugating amino acids to IAA53–55 were upregulated at pH 9.5 in Cj and differentially expressed between Cj and Pt, such as GH3.6 (Cs2g22260) and GH3.9 (Cs5g32880) (Figs. 4 and 5c). This result suggests that the regulation of IAA homeostasis may contribute to alterations of the free IAA content in Cj and Pt. Overexpression GH3.6 in Arabidopsis severely reduces the number of prebranch sites and LRs56. Moreover, several IAA-amino acid conjugates function in inhibiting root elongation, such as IAA-Ala and IAA-Leu57.
Lateral root formation and development appear to be the result of the balancing effects of auxin-promoting and ABA-repressive signaling pathways. For instance, ABA inhibits LR formation by reducing the expression of the TIR1/AFB auxin receptor through miR393 upregulation58 and upregulating the expression of ABI4, which functions to repress PIN113. In this study, several key ABA biosynthesis genes were downregulated under alkaline conditions, and the ABA levels were also reduced in Cj and Pt (Figs. 2 and 8b). However, the ABA levels in Pt were higher than those in Cj for all three conditions (Fig. 8b). This result indicates that ABA may also play an important role in LR development under alkaline stress by interacting with the auxin pathway.
Jasmonate may play a significant role in tolerance to alkaline stress
Previous studies have demonstrated that JA is closely related to salt stress19,20; however, few studies have assessed the relationship between JA and alkaline stress. In this study, under alkaline stress, exogenous JA treatment could significantly reduce the level of H2O2 and MDA in Pt, and JA with auxin analogs treatment could promote LR generation and growth (Fig. 9). These results showed a positive role of JA in alkaline stress responses in citrus. However, under alkaline stress, exogenous JA treatment played a negative role in LR formation and growth in Pt (Fig. 9a, b). In addition, the expression of key genes involved in JA biosynthesis and signal transduction and the JA levels also showed opposite patterns in Cj and Pt in response to alkaline stress; these genes included LOX3 and MYC2, which were downregulated in Cj, but upregulated in Pt (Table 1). This result elucidated that JA signaling was enhanced in Pt, but weakened in Cj under alkaline stress. Jasmonate-induced primary root growth inhibition has been well studied in Arabidopsis. JA reduces both the cell number and cell length of the root through MYC2 repressing the expression of PLT1 and PLT259. On the other hand, JA inhibits primary root growth and regulates LR formation in a dose-dependent manner, with low micromolar concentrations of JA inhibiting the primary root growth and promoting LR formation in Arabidopsis30,31. Moreover, JA is involved in LR growth inhibition under Al stress and salt stress60,61. Hence, in this study, weakened JA signaling in Cj may play a positive role in the tolerance to alkaline stress. The differential response to exogenous JA may be due to the dose-dependent activity of JA.
In addition, many TFs, such as ZAT12, WRKY46, FIT1, ERF105, and others, also exhibited opposite patterns in Cj and Pt (Supplementary Figure S1d, Table 1, Supplementary Table S3). The expression patterns of these TFs were closely related with the JA levels in Cj and Pt. This result indicated that interactions may exist between these TFs and the JA pathway. Ding et al.48 reported that WRKY46 contributes to the feed-forward inhibition of osmotic/salt stress-dependent LR inhibition in Arabidopsis via regulation of ABA signaling and auxin homeostasis. ZAT12 is induced by H2O2 and functions as a negative regulator of iron acquisition by repressing FIT expression62.
ROS metabolic process is important for citrus to resist alkaline stress
When plants are exposed to stress conditions, reactive oxygen species (ROS) are generated63. A lower level of ROS following stress exposure is generally regarded as better tolerance. ROS scavenging enzymes, such as POD, SOD, and CAT, are indispensable for ROS detoxification so that plants can combat the ROS-associated cellular damage and maintain better survival under stressful conditions64. In this study, the expression of genes/miRNAs of ROS metabolic process were affected by alkaline stress in Cj and Pt, such as peroxidase genes and csi-miR398 (Figs. 4 and 7b, Supplementary Figure S2). Moreover, most ROS metabolic process genes/miRNAs were more downregulated/upregulated in Pt than those in Cj in response to alkaline stress (Figs. 4 and 7b, Supplementary Figure S2). In addition, the SOD, POD, and CAT levels were also much higher in Cj than those in Pt under alkaline stress, and these antioxidant levels changed slightly in Cj in response to alkaline stress (Fig. 8d–f). Therefore, these results showed the ROS scavenging system of Cj was stronger than that of Pt under alkaline stress. Several studies have shown that exogenous JA treatment of soybean significantly increases the activities of SOD, POD, APX, and CAT under salt or drought stress65,66. In this study, under alkaline stress, exogenous JA treatment could significantly reduce the levels of MDA and H2O2 in Pt seedlings (Fig. 9d, f). Hence, the interaction between JA and ROS metabolic process may play roles in the tolerance of citrus to alkaline stress.
Differential response of Cj and Pt to iron deficiency under alkaline stress
Cj is more tolerant to iron deficiency compared with Pt under alkaline stress. In this study, a number of genes related to iron acquisition were differentially expressed among the different pH conditions in Cj and Pt, including FIT1 and FRO2/4/6 (Table 1). Moreover, these genes were upregulated in Cj but downregulated in Pt as the pH increased (Table 1, Supplementary Figure S4a, b). This result indicates improved iron acquisition in Cj compared with Pt under alkaline stress. Previous studies demonstrated that FIT plays a central role in iron acquisition38. FRO2 and IRT1 are targets of FIT that regulate FRO2 at the level of mRNA accumulation and IRT1 at the level of protein accumulation37. FIT expression is upregulated by iron deficiency37 and is downregulated by jasmonate67,68. In this study, the JA levels were decreased in Cj but increased in Pt as the pH increased (Fig. 8c). This result suggests that JA may repress FIT1 expression in citrus under alkaline stress. The interaction between FIT1 and JA should be studied in the future.
Taken together, our results demonstrate that the inhibitory effect of alkaline stress on root growth involves the auxin, ABA and JA signaling pathways in citrus. Regulation of auxin homeostasis under alkaline stress is important for adapting to alkaline stress in citrus. Moreover, the JA pathway, which exhibits the opposite response to alkaline stress in Cj and Pt, may contribute to the differences in the alkaline stress and iron deficiency tolerance between Cj and Pt. Further studies are needed to elucidate the regulation network of JA in resistance to alkaline stress and iron deficiency in citrus.
Materials and methods
Plant materials, growth conditions, and sample collections
According to the method described by Zhou et al.69, the seeds of Cj and Pt were surface sterilized, and their germination was accelerated. The seedlings were grown in a growth chamber until they had four leaves. Then, seedlings were selected based on a uniform size and transferred into a hydroponics system with a 4 L solution in a greenhouse. Three different pH gradients (6.5, 8.0, and 9.5) were established. Given that most natural soils have weak acidity, we used pH 6.5 as the normal condition. After 8 weeks of culture, three biological replicates (three seedling plants per replicate) were collected randomly for each treatment. Cj1, Cj2, and Cj3 represented Cj seedlings cultured at pH 6.5, 8.0, and 9.5, respectively. A similar naming scheme was employed for the Pt seedlings. The roots, stems, and leaves of the seedlings were sampled separately. A portion of root samples was scanned with an Epson digital scanner (Expression 10000XL 1.0, Epson, Inc., Japan), and WinRhizo Pro (S) v. 2009c (Regent Instruments, Inc., Canada) software was used to analyze the root morphology. Another portion of samples were frozen by liquid nitrogen and immediately stored under −80 ℃ for other experiments.
Measurement of mineral element concentrations
The plant mineral element concentrations were measured as described in previous studies69,70, with some modification. Briefly, fresh samples were placed into a forced air oven at 105 °C for 30 min, and then at 75 °C until a constant weight was reached to determine the sample dry weight. All the dried samples were ground into fine powder. Then, 0.50 g of each sample was dry-ashed in a muffle furnace at 200 °C for 1 h, 300 °C for 1 h, and 500 °C for 8 h, followed by dissolution in 10 mL 0.1 N HCl. The mineral elements (except nitrogen) were determined using inductively coupled plasma atomic spectroscopy (ICP-AES; Thermo, Inc., IRIS Advan, USA). The total nitrogen content of the plant samples, the pH of the soil samples and the soil mineral element concentrations were measured using the methods described by Bao71. Each sample was assayed using three replicates.
RNA-seq, data processing, and gene annotation
Total RNA isolation was performed as described previously72. Six samples of the root tissues of Cj and Pt were collected, and three biological replicates were collected for each sample. A total of 18 transcriptomes, 18 small RNAs and two degradomes profiles were obtained by RNA-seq using Illumina HiSeq X-ten at Beijing Biomarker Technologies Limited Company (Beijing) in 2017. The sequencing raw data have been submitted to NCBI Gene Expression Omnibus (GEO), and the accession number is GSE115050. The method for data processing and gene annotation was listed in Supplementary Methods 1.
Expression data analysis
The dendrogram in Fig. 2a was made by the function cor() in R with default settings. The y-axis is computed as 1 minus cor (correlation) to reflect the degree of variance. Log2 (FPKM) was used in the computation. The expression modules in Fig. 2c, d were generated by the Short Time-series Expression Miner (v1.3.7) (STEM)45 based on FPKM, with the STEM Clustering Method, with default settings used for the Filtering, Model Profiles and Clustering Profiles. Figure 3b, d was generated by the function GOCircle () of R package GOplot (1.0.2)46. Hierarchical clustering of the gene sets was performed using Morpheus (https://software.broadinstitute.org/morpheus/) with one minus the Pearson correlation as the distance metric and average as the linkage method. Log2 (FPKM) of the differentially expressed genes from the comparison groups was used for clustering. The Z-score in here was calculated for each miRNA per sample using the formula (X–Xav)/SD, where X is the TPM value in a particular sample, and Xav and SD are the mean and standard deviation of the TPM values across all samples used for clustering, respectively.
Verification of miRNA and target gene expression by qRT-PCR
Stem-loop qRT-PCR was performed to validate the expressions of miRNAs with three biological replicates based on our previous method73. U6 was used as the endogenous reference gene74. Next, qRT-PCR was performed with three biological replicates according to our previous study75, and CsActin was used as the endogenous reference gene76. The primers are listed in Supplementary Table S14. Data are presented as the means ± standard error (SE) (n = 3).
Quantification of ABA, JA, and IAA
The samples for ABA, JA, and IAA quantification were prepared according to the method described by Pan et al.77, with slight modifications, which were described in detail in our previous study75. For this purpose, d6-ABA (Icon Isotopes, cat. no. ID1001) was used as an internal standard for ABA, d5-IAA (Aldrich, cat. no. 492817) was used as an internal standard for IAA, and H2JA (dihydrojasmonic acid, OlChemim, cat. no. 0145324) was used as an internal standard for JA. The reaction monitoring conditions (Q1/Q3) for ABA and d6-ABA, IAA and d5-IAA, and JA and H2JA were 262.8/152.6 and 269.1/158.8, 174.0/129.6 and 179.0/134.8, and 209.0/59.0 and 211.0/58.8, respectively. Each sample was characterized using four replicates.
Physiological analyses and histochemical staining of ROS
Malondialdehyde (MDA) levels were determined according to the method described by Liu et al.78. Peroxidase (POD), superoxide dismutase (SOD), catalase (CAT) and H2O2 were measured using relevant detection kits (A064-1 for H2O2, A084-3 for POD, A001-1 for SOD, and A007-1 for CAT, Nanjing Jiancheng Bioengineering Institute, Jiangsu, China) following the manufacturer’s instructions. Anti-O2·– measurement and total protein concentration measurement were performed according to Geng and Liu64. Histochemical staining with 3,3’-diaminobenzidine (DAB) and nitroblue tetrazolium (NBT) was used to examine the in situ accumulation of H2O2 and O2·–, respectively79.
JA, naphthylacetic acid (NAA), and indolebutyric acid (IBA) treatment of the seedlings of Cj and Pt
A stock solution of 1 mM JA was made in ethanol and diluted with water before application. For the Pt seedlings cultured at pH 8.5, we set up the JA (5 μM), NAA + IBA (0.5 mg/L + 0.1 mg/L) and JA + NAA + IBA (5 μM + 0.5 mg/L + 0.1 mg/L) treatment groups, and the seedlings were cultured for 12 weeks. The solution was replaced every week.
Supplementary information
Instruction of Supplementary information
Acknowledgements
This research was supported by the National Modern Citrus Industry System (CARS-26), NSFC (Natural Science Foundation of China) (31601729), NSFC (31521092) and the China Postdoctoral Science Foundation (2015M582242). We thank Yuan Zhou (a graduate of the College of Resources and Environment) for her help in the determination of the content of nutrition elements in the soil.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41438-018-0116-0).
References
- 1.Ge, Y. et al. Global transcriptome profiling of wild soybean (Glycine soja) roots under NaHCO3 treatment. BMC Plant Biol.10, 10.1186/1471-2229-10-153 (2010). [DOI] [PMC free article] [PubMed]
- 2.Xu WF, et al. PIN2 is required for the adaptation of Arabidopsis roots to alkaline stress by modulating proton secretion. J. Exp. Bot. 2012;63:6105–6114. doi: 10.1093/jxb/ers259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu WF, et al. The tomato 14-3-3 protein TFT4 modulates H+Efflux, basipetal auxin transport, and the PKS5-J3 pathway in the root growth response to alkaline stress. Plant Physiol. 2013;163:1817–1828. doi: 10.1104/pp.113.224758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin H, et al. Comparative EST profiles of leaf and root of Leymus chinensis, a xerophilous grass adapted to high pH sodic soil. Plant Sci. 2006;170:1081–1086. doi: 10.1016/j.plantsci.2006.01.002. [DOI] [Google Scholar]
- 5.Wang YC, et al. Analysis of gene expression profile of limonium bicolor under NaHCO3 stress using cDNA microarray. Plant Mol. Biol. Rep. 2008;26:241–254. doi: 10.1007/s11105-008-0037-4. [DOI] [Google Scholar]
- 6.Yang CW, Wang P, Li CY, Shi DC, Wang DL. Comparison of effects of salt and alkali stresses on the growth and photosynthesis of wheat. Photosynthetica. 2008;46:107–114. doi: 10.1007/s11099-008-0018-8. [DOI] [Google Scholar]
- 7.Zhu JK. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Den Herder G, Van Isterdael G, Beeckman T, De Smet I. The roots of a new green revolution. Trends Plant. Sci. 2010;15:600–607. doi: 10.1016/j.tplants.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Lavenus J, et al. Lateral root development in Arabidopsis: fifty shades of auxin. Trends Plant Sci. 2013;18:450–458. doi: 10.1016/j.tplants.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Laplaze L, et al. Cytokinins act directly on lateral root founder cells to inhibit root initiation. Plant Cell. 2007;19:3889–3900. doi: 10.1105/tpc.107.055863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moriwaki T, et al. Hormonal regulation of lateral root development in arabidopsis modulated by MIZ1 and requirement of GNOM activity for MIZ1 function. Plant Physiol. 2011;157:1209–1220. doi: 10.1104/pp.111.186270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen H, Li ZF, Xiong LM. A plant microRNA regulates the adaptation of roots to drought stress. FEBS Lett. 2012;586:1742–1747. doi: 10.1016/j.febslet.2012.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Shkolnik-Inbar D, Bar-Zvi D. ABI4 mediates abscisic acid and cytokinin inhibition of lateral root formation by reducing polar auxin transport in Arabidopsis. Plant Cell. 2010;22:3560–3573. doi: 10.1105/tpc.110.074641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD. Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl Acad. Sci. USA. 2008;105:803–808. doi: 10.1073/pnas.0709559105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shin R, et al. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell. 2007;19:2440–2453. doi: 10.1105/tpc.107.050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez-Torres CA, et al. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell. 2008;20:3258–3272. doi: 10.1105/tpc.108.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Creelman RA, Mullet JE. Jasmonic acid distribution and action in plants: regulation during development and response to biotic and abiotic stress. Proc. Natl Acad. Sci. USA. 1995;92:4114–4119. doi: 10.1073/pnas.92.10.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wasternack C. Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007;100:681–697. doi: 10.1093/aob/mcm079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pedranzani H, et al. Salt tolerant tomato plants show increased levels of jasmonic acid. Plant Growth Regul. 2003;41:149–158. doi: 10.1023/A:1027311319940. [DOI] [Google Scholar]
- 20.Kang DJ, et al. Jasmonic acid differentially affects growth, ion uptake and abscisic acid concentration in salt-tolerant and salt-sensitive rice cultivars. J. Agron. Crop Sci. 2005;191:273–282. doi: 10.1111/j.1439-037X.2005.00153.x. [DOI] [Google Scholar]
- 21.Yoon JY, Hamayun M, Lee SK, Lee IJ. Methyl jasmonate alleviated salinity stress in soybean. J. Crop Sci. Biotechnol. 2009;12:63–68. doi: 10.1007/s12892-009-0060-5. [DOI] [Google Scholar]
- 22.Javid MG, Sorooshzadeh A, Moradi F, Sanavy SAMM, Allahdadi I. The role of phytohormones in alleviating salt stress in crop plants. Aust. J. Crop Sci. 2011;5:726–734. [Google Scholar]
- 23.Qiu ZB, Guo JL, Zhu AJ, Zhang L, Zhang MM. Exogenous jasmonic acid can enhance tolerance of wheat seedlings to salt stress. Ecotoxicol. Environ. Saf. 2014;104:202–208. doi: 10.1016/j.ecoenv.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 24.Ismail A, Riemann M, Nick P. The jasmonate pathway mediates salt tolerance in grapevines. J. Exp. Bot. 2012;63:2127–2139. doi: 10.1093/jxb/err426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu H, Ye HY, Yao RF, Zhang T, Xiong LZ. OsJAZ9 acts as a transcriptional regulator in jasmonate signaling and modulates salt stress tolerance in rice. Plant Sci. 2015;232:1–12. doi: 10.1016/j.plantsci.2014.12.010. [DOI] [PubMed] [Google Scholar]
- 26.Zhu, D. et al. The positive regulatory roles of the TIFY10 proteins in plant responses to alkaline stress. PLoS ONE9, 10.1371/journal.pone.0111984 (2014). [DOI] [PMC free article] [PubMed]
- 27.Zhu D, et al. Over-expression of a novel JAZ family gene from Glycine soja, increases salt and alkali stress tolerance. Biochem. Biophys. Res. Commun. 2012;426:273–279. doi: 10.1016/j.bbrc.2012.08.086. [DOI] [PubMed] [Google Scholar]
- 28.Yan Y, et al. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell. 2007;19:2470–2483. doi: 10.1105/tpc.107.050708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun JQ, et al. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell. 2009;21:1495–1511. doi: 10.1105/tpc.108.064303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun JQ, et al. Jasmonate modulates endocytosis and plasma membrane accumulation of the Arabidopsis PIN2 protein. New Phytol. 2011;191:360–375. doi: 10.1111/j.1469-8137.2011.03713.x. [DOI] [PubMed] [Google Scholar]
- 31.Raya-Gonzalez J, Pelagio-Flores R, Lopez-Bucio J. The jasmonate receptor COI1 plays a role in jasmonate-induced lateral root formation and lateral root positioning in Arabidopsis thaliana. J. Plant Physiol. 2012;169:1348–1358. doi: 10.1016/j.jplph.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 32.Toda Y, et al. RICE SALT SENSITIVE3 forms a ternary complex with JAZ and Class-C bHLH factors and regulates jasmonate-induced gene expression and root cell elongation. Plant Cell. 2013;25:1709–1725. doi: 10.1105/tpc.113.112052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robinson NJ, Procter CM, Connolly EL, Guerinot ML. A ferric-chelate reductase for iron uptake from soils. Nature. 1999;397:694–697. doi: 10.1038/17800. [DOI] [PubMed] [Google Scholar]
- 34.Connolly EL, Campbell NH, Grotz N, Prichard CL, Guerinot ML. Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol. 2003;133:1102–1110. doi: 10.1104/pp.103.025122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eide D, Broderius M, Fett J, Guerinot ML. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl Acad. Sci. USA. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vert G, et al. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell. 2002;14:1223–1233. doi: 10.1105/tpc.001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colangelo EP, Guerinot ML. The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell. 2004;16:3400–3412. doi: 10.1105/tpc.104.024315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan YX, Zhang J, Wang DW, Ling HQ. AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res. 2005;15:613–621. doi: 10.1038/sj.cr.7290331. [DOI] [PubMed] [Google Scholar]
- 39.Castle WS. Rootstock as a fruit quality factor in citrus and deciduous tree crops. N.Z. J. Crop Hortic. Sci. 1995;23:383–394. doi: 10.1080/01140671.1995.9513914. [DOI] [Google Scholar]
- 40.Xu Q, et al. The draft genome of sweet orange (Citrus sinensis) Nat. Genet. 2013;45:59–U92. doi: 10.1038/ng.2472. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Z, Jiang L, Wang J, Gu P, Chen M. MTide: an integrated tool for the identification of miRNA-target interaction in plants. Bioinformatics. 2015;31:290–291. doi: 10.1093/bioinformatics/btu633. [DOI] [PubMed] [Google Scholar]
- 42.Meyers BC, et al. Criteria for annotation of plant MicroRNAs. Plant Cell. 2008;20:3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.German MA, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 44.Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ernst J, Bar-Joseph Z. STEM: a tool for the analysis of short time series gene expression data. BMC Bioinform. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walter W, Sanchez-Cabo F, Ricote M. GOplot: an R package for visually combining expression data with functional analysis. Bioinformatics. 2015;31:2912–2914. doi: 10.1093/bioinformatics/btv300. [DOI] [PubMed] [Google Scholar]
- 47.Wittstock U, Halkier BA. Cytochrome P450 CYP79A2 from Arabidopsis thaliana L. Catalyzes the conversion of L-phenylalanine to phenylacetaldoxime in the biosynthesis of benzylglucosinolate. J. Biol. Chem. 2000;275:14659–14666. doi: 10.1074/jbc.275.19.14659. [DOI] [PubMed] [Google Scholar]
- 48.Ding ZJ, et al. Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 2015;84:56–69. doi: 10.1111/tpj.12958. [DOI] [PubMed] [Google Scholar]
- 49.Wang JW, et al. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell. 2005;17:2204–2216. doi: 10.1105/tpc.105.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Verbruggen N, Hermans C. Proline accumulation in plants: a review. Amino Acids. 2008;35:753–759. doi: 10.1007/s00726-008-0061-6. [DOI] [PubMed] [Google Scholar]
- 51.Bishopp A, Benkova E, Helariutta Y. Sending mixed messages: auxin-cytokinin crosstalk in roots. Curr. Opin. Plant Biol. 2011;14:10–16. doi: 10.1016/j.pbi.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 52.Hidehiro F, Satoshi T, Haruka M, Masao T. Lateral root formation is blocked by a gain‐of‐function mutation in the SOLITARY‐ROOT/IAA14 gene of Arabidopsis. Plant J. 2002;29:153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- 53.Khan S, Stone JM. Arabidopsis thaliana GH3.9 in Auxin and Jasmonate Cross Talk. Plant Signal. Behav. 2007;2:483–485. doi: 10.4161/psb.2.6.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Staswick PE, et al. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17:616–627. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gutierrez L, et al. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell. 2012;24:2515–2527. doi: 10.1105/tpc.112.099119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xuan W, et al. Root cap-derived auxin pre-patterns the longitudinal axis of the Arabidopsis root. Curr. Biol. 2015;25:1381–1388. doi: 10.1016/j.cub.2015.03.046. [DOI] [PubMed] [Google Scholar]
- 57.LeClere S, Tellez R, Rampey RA, Matsuda SPT, Bartel B. Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J. Biol. Chem. 2002;277:20446–20452. doi: 10.1074/jbc.M111955200. [DOI] [PubMed] [Google Scholar]
- 58.Vidal EA, et al. Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2010;107:4477–4482. doi: 10.1073/pnas.0909571107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Q, et al. The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell. 2011;23:3335–3352. doi: 10.1105/tpc.111.089870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang ZB, He CM, Ma YQ, Herde M, Ding ZJ. Jasmonic acid enhances al-induced root growth inhibition. Plant Physiol. 2017;173:1420–1433. doi: 10.1104/pp.16.01756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valenzuela CE, et al. Salt stress response triggers activation of the jasmonate signaling pathway leading to inhibition of cell elongation in Arabidopsis primary root. J. Exp. Bot. 2016;67:4209–4220. doi: 10.1093/jxb/erw202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Le CTT, et al. ZINC FINGER OF ARABIDOPSIS THALIANA12 (ZAT12) interacts with FER-LIKE IRON DEFICIENCY- INDUCED TRANSCRIPTION FACTOR (FIT) linking iron deficiency and oxidative stress responses. Plant Physiol. 2016;170:540–557. doi: 10.1104/pp.15.01589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 64.Geng JJ, Liu JH. The transcription factor CsbHLH18 of sweet orange functions in modulation of cold tolerance and homeostasis of reactive oxygen species by regulating the antioxidant gene. J. Exp. Bot. 2018;69:2677–2692. doi: 10.1093/jxb/ery065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Anjum SA, Wang L, Farooq M, Khan I, Xue L. Methyl jasmonate-induced alteration in lipid peroxidation, antioxidative defence system and yield in soybean under drought. J. Agron. Crop Sci. 2011;197:296–301. doi: 10.1111/j.1439-037X.2011.00468.x. [DOI] [Google Scholar]
- 66.Noriega G, Cruz DS, Batlle A, Tomaro M, Balestrasse K. Heme oxygenase is involved in the protection exerted by jasmonic acid against cadmium stress in soybean roots. J. Plant Growth Regul. 2012;31:79–89. doi: 10.1007/s00344-011-9221-0. [DOI] [Google Scholar]
- 67.Maurer F, Müller S, Bauer P. Suppression of Fe deficiency gene expression by jasmonate. Plant Physiol. Biochem. 2011;49:530–536. doi: 10.1016/j.plaphy.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 68.Brumbarova T, Bauer P, Ivanov R. Molecular mechanisms governing Arabidopsis iron uptake. Trends Plant Sci. 2015;20:124–133. doi: 10.1016/j.tplants.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 69.Zhou, G. F. et al. Transcription profiles of boron-deficiency-responsive genes in citrus rootstock root by suppression subtractive hybridization and cDNA microarray. Front. Plant Sci.5, 10.3389/Fpls.2014.00795 (2015). [DOI] [PMC free article] [PubMed]
- 70.Storey R, Treeby MT. Seasonal changes in nutrient concentrations of navel orange fruit. Sci. Hortic. 2000;84:67–82. doi: 10.1016/S0304-4238(99)00093-X. [DOI] [Google Scholar]
- 71.Bao, S. D. Soil agrochemical analysis. third edn, (China Agriculture Press (in Chinese), 2008).
- 72.Liu Y, Liu Q, Tao N. Efficient isolation of RNA from fruit peel and pulp of ripening navel orange (Citrus sinensis Osbeck) J. Huazhong Agric. Univ. 2006;25:300–304. [Google Scholar]
- 73.Wu, J. X., Zheng, S. S., Feng, G. Z. & Yi, H. L. Comparative analysis of miRNAs and their target transcripts between a spontaneous late-ripening sweet orange mutant and its wild-type using small rna and degradome sequencing. Front. Plant Sci.7, 10.3389/fpls.2016.01416 (2016). [DOI] [PMC free article] [PubMed]
- 74.Kou SJ, et al. Selection and validation of suitable reference genes for miRNA expression normalization by quantitative RT-PCR in citrus somatic embryogenic and adult tissues. Plant Cell Rep. 2012;31:2151–2163. doi: 10.1007/s00299-012-1325-x. [DOI] [PubMed] [Google Scholar]
- 75.Wu J, et al. An integrative analysis of the transcriptome and proteome of the pulp of a spontaneous late-ripening sweet orange mutant and its wild type improves our understanding of fruit ripening in citrus. J. Exp. Bot. 2014;65:1651–1671. doi: 10.1093/jxb/eru044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu J, et al. Selection of reliable reference genes for gene expression studies using quantitative real-time PCR in navel orange fruit development and pummelo floral organs. Sci. Hortic. 2014;176:180–188. doi: 10.1016/j.scienta.2014.06.040. [DOI] [Google Scholar]
- 77.Pan X, Welti R, Wang X. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography-mass spectrometry. Nat. Protocol. 2010;5:986–992. doi: 10.1038/nprot.2010.37. [DOI] [PubMed] [Google Scholar]
- 78.Liu JH, et al. Polyamine biosynthesis of apple callus under salt stress: importance of the arginine decarboxylase pathway in stress response. J. Exp. Bot. 2006;57:2589–2599. doi: 10.1093/jxb/erl018. [DOI] [PubMed] [Google Scholar]
- 79.Wang J, et al. An arginine decarboxylase gene PtADC from Poncirus trifoliata confers abiotic stress tolerance and promotes primary root growth in Arabidopsis. J. Exp. Bot. 2011;62:2899–2914. doi: 10.1093/jxb/erq463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Instruction of Supplementary information