Abstract
Leigh syndrome is a mitochondrial disease characterized by neurological disorders, metabolic abnormality and premature death. There is no cure for Leigh syndrome; therefore, new therapeutic targets are urgently needed. In Ndufs4-KO mice, a mouse model of Leigh syndrome, we found that Complex I deficiency led to declines in NAD+ levels and NAD+ redox imbalance. We tested the hypothesis that elevation of NAD+ levels would benefit Ndufs4-KO mice. Administration of NAD+ precursor, nicotinamide mononucleotide (NMN) extended lifespan of Ndufs4-KO mice and attenuated lactic acidosis. NMN increased lifespan by normalizing NAD+ redox imbalance and lowering HIF1a accumulation in Ndufs4-KO skeletal muscle without affecting the brain. NMN up-regulated alpha-ketoglutarate (KG) levels in Ndufs4-KO muscle, a metabolite essential for HIF1a degradation. To test whether supplementation of KG can treat Ndufs4-KO mice, a cell-permeable KG, dimethyl ketoglutarate (DMKG) was administered. DMKG extended lifespan of Ndufs4-KO mice and delayed onset of neurological phenotype. This study identified therapeutic mechanisms that can be targeted pharmacologically to treat Leigh syndrome.
Introduction
Mitochondrial diseases are caused by mutations in genes that encode mitochondrial proteins. Loss-of-function mutations occur mostly in proteins of the electron transport chain (ETC) or ATP synthase complexes, resulting in defective oxidative phosphorylation1. Leigh Syndrome (LS) is a mitochondrial disease caused by wide-spread mutations in ETC proteins, often residing in the mitochondrial Complex I (C-I)2. C-I is a large, L-shaped protein complex located in the mitochondrial inner membrane and serves as the primary site for NADH oxidation3. C-I contributes to proton gradient generation (mitochondrial membrane potential) across the inner membrane by coupling electron transfer from NADH to other components of the ETC. Mitochondrial membrane potential is utilized to generate ATP and maintain mitochondrial functions. Loss-of-function mutations in C-I in LS leads to inability to oxidize NADH, NAD(H) redox imbalance and impaired ATP production. Therefore, C-I deficiency in LS is devastating to organs with a high energy demand, such as the brain.
Apart from acting as an electron carrier in metabolic pathways, NAD+ is a co-substrate of several NAD+-dependent enzymes responsible for regulating protein post-translational modifications (PTMs) and DNA repair4,5. Mitochondrial function, through regulating NAD(H) redox balance, is critical for the availability of NAD+6–8. Loss of NAD+ induces a pseudohypoxic state and triggers accumulation of HIF1a protein under normoxic conditions9. NAD+ can be synthesized de novo, or generated by Preiss-Handler and salvage pathways. The salvage pathway is thought to be the most efficient way to replenish NAD+5,10. Recent studies showed that supplementation of NAD+ precursors in the salvage pathway is therapeutic for multiple pathologies7,11,12. Importantly, maintaining the NAD+ pool by delivering NAD+ precursors or inhibition of poly(ADP-ribose) polymerases, treats mitochondrial myopathy and aging by improving mitochondrial function9,13–15.
LS manifests an early onset neurodegeneration characterized by progressive losses of mental and motor abilities, retarded growth, lactic acidosis, followed by respiratory failure and premature death16,17. A mouse model of LS was developed by the deletion of Ndufs4 gene (Ndufs4-KO), which encodes a protein necessary for the assembly and stability of C-I. Multiple loss-of-function mutations in Ndufs4 have been found in LS patients18,19. Ndufs4-KO mice show C-I deficiency and recapitulate phenotypes observed in patients with LS20,21. We previously showed that C-I-deficiency in the heart of cardiac-specific Ndufs4-KO mice (cKO) led to elevated NADH levels, NAD(H) redox imbalance and protein hyperacetylation8. These biochemical changes contribute to the increased susceptibility of the heart to a variety of stresses, which can be rescued by increasing cellular NAD+ level.
In this study, we tested the hypothesis that NAD(H) redox imbalance is a pathogenic mechanism in LS using Ndufs4-KO mice. We found that supplementation of the NAD+ precursor, nicotinamide mononucleotide (NMN), extended the lifespan of Ndufs4-KO mice. Therapeutic benefits of NMN were mediated by elevation of alpha-ketoglutarate (KG) levels and suppression of hypoxic signaling. Our findings were further supported by the increased lifespan and the neurological benefits of KG supplementation in Ndufs4-KO mice.
Results
NMN supplementation extended lifespan of Ndufs4-KO mice
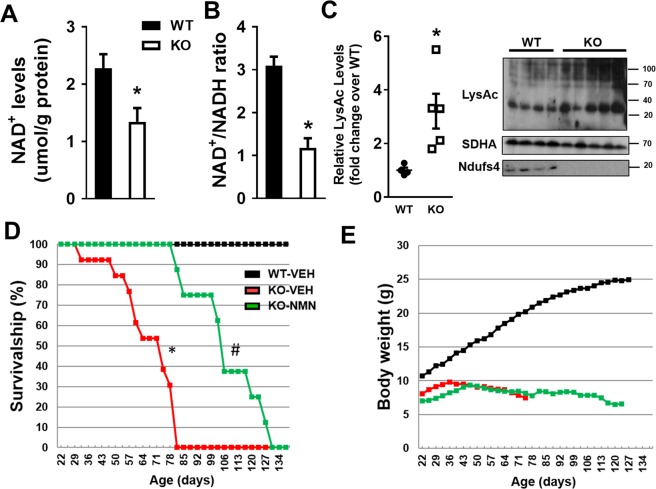
We observed a significant decline in NAD+ levels and lowered NAD+/NADH ratio in the brain of the Ndufs4-KO mice compared to that of age-matched wildtype (WT) mice (Fig. 1A,B, Supplementary Fig. 1). Similar to what was observed in the heart18,19, the NAD+ redox imbalance was associated with protein hyperacetylation in Ndufs4-KO brain (Fig. 1C). To counteract the decline of NAD+ levels in Ndufs4-KO mice, we sought to increase NAD+ pool by targeting the NAD+ salvage pathway (Supplementary Fig. 2A). We delivered NAD+ precursor, nicotinamide mononucleotide (NMN), or vehicle via intraperitoneal injection, to Ndufs4-KO mice starting from postnatal day 21 (P-21) throughout their lifetime (Supplementary Fig. 2B). NMN treatment extended the survival of Ndufs4-KO mice by approximately 2-fold (Fig. 1D; median lifespan 110 days vs 60 days). However, the benefits of NMN appeared to be partial, as the growth of body weight of Ndufs4-KO mice remained stunted after NMN treatment (Fig. 1E).
Figure 1.
Targeting altered NAD+ metabolism of Ndufs4-KO mice to extend lifespan. Brain tissues of age-matched (P-65 to -75) WT and Ndufs4-KO mice were collected. (A) NAD+ levels (B) NAD+/NADH ratio and (C) protein acetylation (LysAc) levels were measured. SDHA was used as loading control. N = 4–5. Unpaired 2-tailed t-tests were used. (D) Survival curves of WT and Ndufs4-KO mice with indicated treatments. N = 10–12. (E) Body weight plot of indicated mice. *P < 0.05 versus WT or WT-VEH. #P < 0.05 versus KO-VEH. Log-rank test was used. Full blot images are presented in Supplementary Fig. 7.
Interventions targeting the NAD+ salvage pathway failed to impact brain NAD+ level of Ndufs4-KO mice
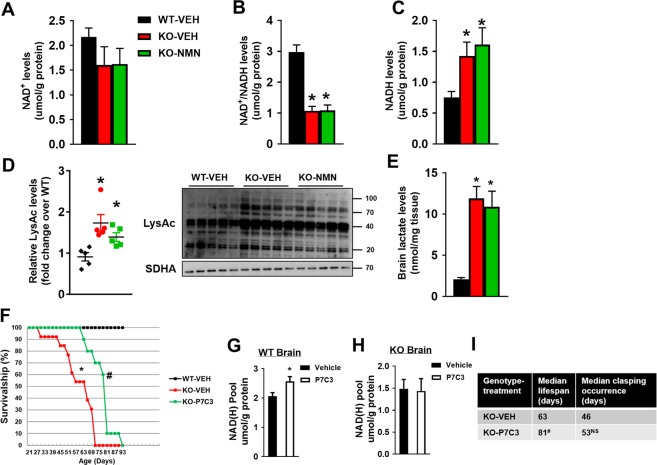
Despite prolonged survival, NMN did not elevate NAD+ levels or NAD+/NADH ratio in the brain of Ndufs4-KO mice harvested at P-50 (Fig. 2A,B, Supplementary Fig. 2C). Levels of NADH and protein acetylation in Ndufs4-KO brains remained high after NMN treatment (Fig. 2C,D). Similarly, protein acetylation levels in heart and liver of Ndufs4-KO mice remained high after NMN treatment (Supplementary Fig. 3A,B). NMN slightly elevated NAD+ levels and NAD+/NADH ratio in Ndufs4-KO hearts but had no effect in Ndufs4-KO liver (Supplementary Fig. 3C,D). Lactate levels in the brain tissue of Ndufs4-KO mice were elevated and not affected by NMN treatment (Fig. 2E). These data suggested that this regimen did not provide metabolic benefits to Ndufs4-KO brain but may have partial benefits to peripheral tissues. We speculated that this treatment regimen (Supplementary Fig. 2B) did not sufficiently deliver NMN to the brain. To test this hypothesis, we increased NMN dosing frequency to daily injections for seven consecutive days in WT mice (Supplementary Fig. 3E). Although this regimen increased NAD+ levels in the heart and liver, it still failed to increase brain NAD+ levels (Supplementary Fig. 3F). Using 1H NMR spectroscopy22 as an alternative method to measure NAD+ levels, we still did not observe elevations of NAD+ levels in the brain (Supplementary Fig. 3G).
Figure 2.
NMN or P7C3 failed to impact NAD+ metabolism in Ndufs4-KO brain. Brain tissues of indicated mice were collected at P-50. (A) NAD+ levels, (B) NAD+/NADH ratio (C) NADH levels of brain from mice as indicated were measured. (D) Protein acetylation levels and (E) lactate levels in brain from mice as indicated were quantified. N = 5. SDHA was used as loading control. *P < 0.05 versus WT-VEH. One-way ANOVA with Newman-Keuls multiple comparison test was used. Full blot images are presented in Supplementary Fig. 7. WT or Ndufs4-KO mice were treated with P7C3 daily starting from P-21. (F) Survival curves of WT and Ndufs4-KO mice treated with VEH or P7C3 were plotted. Log-rank test was used. NAD+ pools (NAD+ plus NADH levels) of brain tissues from (G) WT or (H) Ndufs4-KO mice after P7C3 treatment were measured. N = 3. P < 0.05 versus VEH; #P < 0.05 versus KO-VEH. Unpaired 2-tailed t-tests were used. (I) Table summarizing median lifespan and clasping occurrence of Ndufs4-KO mice with vehicle or P7C3 treatment. N = 10. #P < 0.05 versus KO-VEH. NS: not statistically significant versus KO-VEH.
We next sought to stimulate the NAD+ salvage pathway by activating its key enzyme, nicotinamide phosphoribosyltransferase (NAMPT), using P7C323 (Supplementary Fig. 2A). P7C3 has been shown to be a proneurogenic agent that protects mice from traumatic brain injury24. Daily delivery of P7C3 to Ndufs4-KO mice from P-21 led to a moderate lifespan extension (Fig. 2F, Supplementary Fig. 3H), which was less prominent compared to NMN treatment (median lifespan: 80 vs 110 days). Although P7C3 moderately increased NAD+ levels in the brain of WT mice, it failed to do so in the brain of Ndufs4-KO mice (Fig. 2G,H). The inability to elevate NAD+ by P7C3 was not due to the lack of NAMPT protein in Ndufs4-KO mice, as NAMPT protein levels did not change (Supplementary Fig. 3I). We also assessed the progression of neurodegeneration using the incidence of forelimb clasping as a neuro-behavior marker25. Ndufs4-KO mice showed early clasping phenotype with median occurrence around 46 days as previously observed25. P7C3 treatment did not delay the median clasping occurrence in Ndufs4-KO mice (Fig. 2I). These data collectively suggest that it is particularly challenging to boost the NAD+ levels in the brain of Ndufs4-KO mice. This prevented us from addressing the efficacy of increasing brain NAD+ level as a therapy for LS.
NMN attenuated NAD+ redox imbalance, protein hyperacetylation and suppressed lactate levels of Ndufs4-KO skeletal muscle
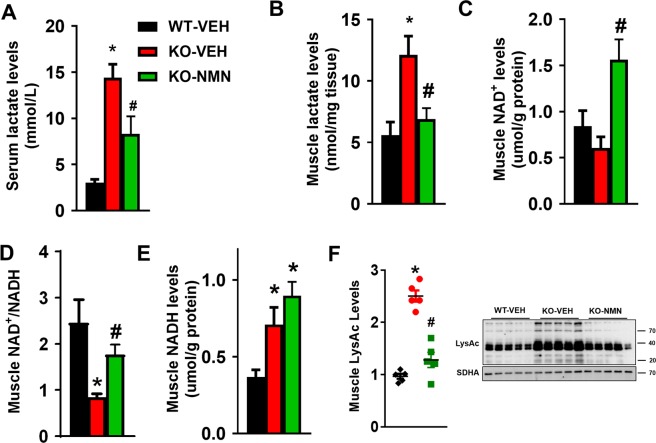
We next examined the non-neurological impacts of NMN supplementation on Ndufs4-KO mice to account for the lifespan extension (Fig. 1D). As in LS patients, serum lactate levels were elevated in Ndufs4-KO mice compared to WT mice (Fig. 3A). NMN treatment suppressed serum lactic acidosis in Ndufs4-KO mice (Fig. 3A), suggesting an improvement of systemic metabolism by the treatment. NMN suppressed lactate levels in skeletal and cardiac muscles of Ndufs4-KO mice (Fig. 3B, Supplementary Fig. 4A). In parallel to the attenuated lactic acidosis, the NAD+ pool was expanded in the skeletal muscle and the heart (Fig. 3C and Supplementary Fig. 3C). NAD+ redox balance in skeletal muscle and heart of Ndufs4-KO mice was partially restored by NMN despite a higher level of NADH (Fig. 3C–E, Supplementary Fig. 3C). Normalization of tissue NAD+/NADH ratio would reduce the conversion of pyruvate to lactate by lactate dehydrogenase (LDHA), contributing to the lowered tissue and serum lactate levels (Fig. 3B). Attenuated NAD+ redox imbalance by NMN lowered protein hyperacetylation in skeletal muscle of Ndufs4-KO mice (Fig. 3F). NMN treatment also improved cardiac function, as measured by murine echocardiography, in Ndufs4-KO mice (Supplementary Fig. 4B). These results suggest that the lifespan extension of Ndufs4-KO mice by NMN is attributable to improved metabolism and function in peripheral tissues, independent of the brain.
Figure 3.
NMN supplementation attenuated NAD+ redox imbalance and protein hyperacetylation, and suppressed lactate levels in skeletal muscle of Ndufs4-KO mice. (A,B) Serum and skeletal muscle of indicated mice were collected at P-50 and lactate levels were measured. (C) NAD+ levels, (D) NAD+/NADH ratio and (E) NADH levels of skeletal muscle from mice as indicated were measured. (F) Protein acetylation levels of skeletal muscle were measured by Western blot. N = 5. SDHA was used as loading control. Full blot images are presented in Supplementary Fig. 7. *P < 0.05 versus WT-VEH; #P < 0.05 versus KO- VEH. One-way ANOVA with Newman-Keuls multiple comparison test was used.
NMN blunted activation of hypoxic signaling in Ndufs4-KO mice
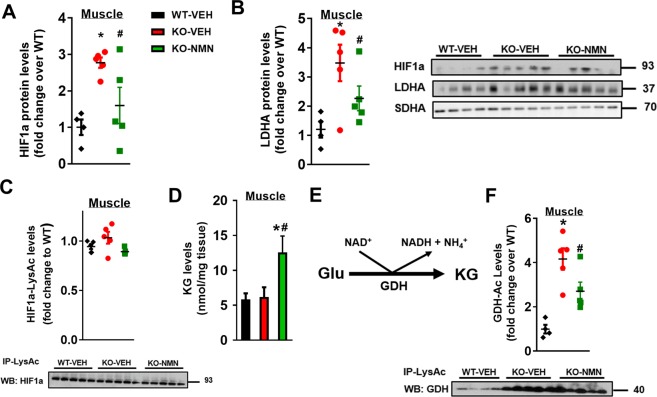
Defective oxidative metabolism in mitochondrial disease promotes anaerobic glycolysis that produces lactate from pyruvate while regenerating NAD+ from NADH. Sustained glycolysis eventually lowers NAD+/NADH ratio. The resultant lactic acidosis and abnormal NAD+ redox state are markers in LS and linked to cardiovascular risk and neurodegeneration26. We found that hypoxia inducible factor 1 alpha (HIF1a), an important transcriptional activator of glycolysis, was elevated in skeletal muscle and brain of Ndufs4-KO mice (Fig. 4A, Supplementary Fig. 5A). The HIF1a downstream protein targets, such as LDHA, were also increased in Ndufs4-KO skeletal muscle and brain (Fig. 4B, Supplementary Fig. 5A). HIF1a can be stabilized by ROS or protein acetylation27–29. Although protein acetylation levels of Ndufs4-KO skeletal muscle and brain were elevated (Figs 2D and 3F), we did not detect changes in the acetylation level of HIF1a protein (Fig. 4C, Supplementary Fig. 5B). Instead, hyperacetylation of a key mitochondrial antioxidant enzyme, superoxide dismutase 2 (SOD2), was found in the brain of Ndufs4-KO mice (Supplementary Fig. 5C). Acetylation of SOD2 has been shown to inhibit its ROS scavenging activity30. To determine whether a higher level of ROS contributed to HIF1a accumulation in Ndufs4-KO mice, protein nitrotyrosine (NT) levels, a marker of oxidative stress, were measured. Protein nitrotyrosine levels were elevated in Ndufs4-KO brain (Supplementary Fig. 5D), consistent with a prior report of increased ROS accumulation in the brain of Ndufs4-KO mice31. These results collectively suggest that ROS-induced HIF1a accumulation is a likely cause of increased glycolysis in Ndufs4-KO brain.
Figure 4.
NMN blunted the activation of hypoxia signaling in Ndufs4-KO muscle via up-regulation of α-ketoglutarate (KG) levels. Protein levels of (A) HIF1a and (B) LDHA of skeletal muscle from mice as indicated were measured by Western blot. SDHA was used as loading control. (C) Acetylation levels of HIF1a and (D) KG levels from skeletal muscle of mice as indicated were quantified. (E) Glutamate dehydrogenase (GDH) catalytic reaction. (F) Acetylation levels of GDH in skeletal muscle from mice treated as indicated were measured by Western blot analysis. N = 4–5. *P < 0.05 versus WT-VEH; #P < 0.05 versus KO-VEH. One-way ANOVA with Newman-Keuls multiple comparison test was used. Full blot images are presented in Supplementary Fig. 7.
NMN supplementation lowered levels of HIF1a and LDHA proteins in the skeletal muscle but not in the brain of Ndufs4-KO mice (Fig. 4A,B, Supplementary Fig. 5A). This finding is coincident with the expanded NAD+ pool and the lowered lactate levels in skeletal muscle of Ndufs4-KO mice by NMN treatment, but not in the brain (Figs 2–3). These results collectively suggest that hypoxic signaling and glycolysis in Ndufs4-KO mice could be manipulated via NAD+-sensitive mechanisms. We found that alpha-ketoglutarate (KG) levels were elevated in Ndufs4-KO muscle after NMN treatment (Fig. 4D). As KG is a required co-substrate for hydroxylation of HIF1a for degradation, increased KG promotes HIF1a reduction. KG can be produced from glutamate via glutamate dehydrogenase (GDH) reaction, which is coupled with conversion of NAD+ to NADH in the direction of KG production (Fig. 4E). Since NMN elevated NAD+/NADH ratio in Ndufs4-KO skeletal muscle (Fig. 3D), KG production was favored. Furthermore, hyperacetylation of GDH causes a loss of catalytic activity due to conformational change32,33. NMN treatment lowered GDH hyperacetylation in Ndufs4-KO skeletal muscle but not in brain (Fig. 4F, Supplementary Fig. 5E). These data suggest that increasing NAD+ level enhances KG production (Fig. 6), which in turn attenuates hypoxic signaling in Ndufs4-KO mice.
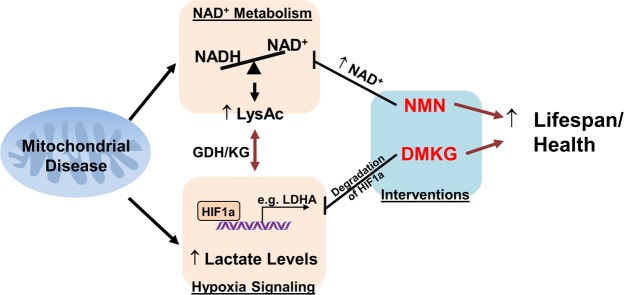
Figure 6.
Targeting NAD+ metabolism or hypoxia signaling as interventions for Leigh Syndrome, a mitochondrial disease. Complex I deficiency in Ndufs4-KO mice triggers NAD+ redox imbalance, activation of hypoxia signaling and lactate acidosis. NMN and DMKG treatment can improve lifespan and health of Ndufs4-KO mice via normalization of these pathways.
DMKG supplementation increased lifespan, improved neurological phenotype and suppressed hypoxic signaling of Ndufs4-KO mice
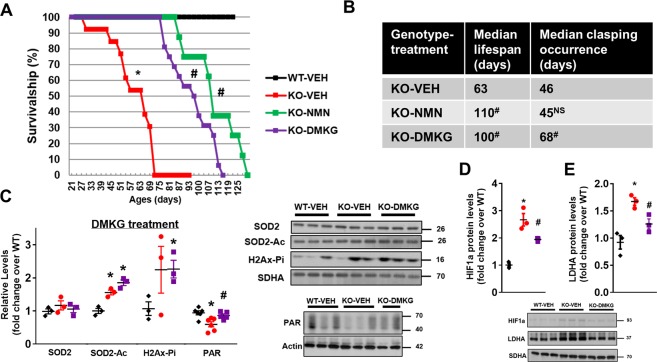
We next sought to directly increase KG level in the brain of Ndufs4-KO mice using dimethyl-ketoglutarate (DMKG), a cell permeable form of KG. DMKG was administered to Ndufs4-KO mice intraperitoneally, daily starting from P-21 (Supplementary Fig. 6A). We observed a significant lifespan extension in DMKG-treated Ndufs4-KO mice compared to vehicle-treated Ndufs4-KO mice (Fig. 5A,B). The median lifespan of DMKG-treated Ndufs4-KO mice was similar to NMN-treated mice (100 days in DMKG-treated Ndufs4-KO mice versus 110 days in NMN-treated group).
Figure 5.
Supplementation of dimethyl α-ketoglutarate (DMKG) extended lifespan and delays the onset of clasping in Ndufs4-KO mice. (A) Survival curves of WT mice, KO mice treated with vehicle (VEH), NMN or DMKG. N = 10–16. (B) Table summarizing median lifespan and clasping occurrence of Ndufs4-KO mice with vehicle, NMN or DMKG treatments. N = 10–16. Log-rank test was used. (C) Levels of SOD2 protein, SOD2 acetylation, H2Ax phosphorylation (H2Ax-Pi), and protein PAR in brain tissues in DMKG treatment cohort at P-50 were quantified. N = 3–6. Protein levels of (D) HIF1a and (E) LDHA in brain tissues were measured by Western blots. N = 3. Full blot images are presented in Supplementary Fig. 7. *P < 0.05 versus WT-VEH; #P < 0.05 versus KO-VEH. NS: not statistically significant versus KO-VEH. One-way ANOVA with Newman-Keuls multiple comparison test was used. SDHA and actin were used as loading control.
Using positive forelimb clasping as a readout25, we assessed and compared the effects of NMN and DMKG on the neurological system of Ndufs4-KO mice. It has been shown that neurological symptoms in Ndufs4-KO mice manifested around P-35, which coincided with the body weight peak of the mice (Supplementary Figs 1E and 6B) as previously shown20,25. Survivorship of Ndufs4-KO mice declined after P-35 as the incidence of clasping progressively increased (Fig. 5A, Supplementary Fig. 6B). NMN treatment did not change the incidence of clasping of Ndufs4-KO mice (median clasping occurrence of P-45 versus P-46 in vehicle, Fig. 5B, Supplementary Fig. 6B) as the brain NAD+ pool was not expanded. In contrast, DMKG treatment delayed incidence of clasping in Ndufs4-KO mice with a median clasping occurrence at P-68 (Fig. 5B, Supplementary Fig. 6B). Consistent with the unchanged NAD+ levels in brain of Ndufs4-KO mice (Fig. 2A), NMN treatment did not alter hyperacetylation of SOD2 in Ndufs4-KO brain, and DNA damage, as evidenced by elevated histone 2 A phosphorylation (H2Ax-Pi) remained high (Supplementary Figs 5 and 6C). Although NMN lowered acetylation levels of SOD2 in Ndufs4-KO muscle, protein nitrotyrosine levels (NT) were not altered in Ndufs4-KO muscle or by NMN treatment (Supplementary Fig. 6D). These data suggested that oxidative stress contributed to the impairments of Ndufs4-KO brain, but not in skeletal muscle, and the benefits of NMN treatment were not mediated by relieving oxidative stress. DMKG treatment did not alter SOD2 acetylation and histone 2A phosphorylation in Ndufs4-KO brain (Fig. 5C). However, DMKG elevated protein poly-ADP-ribosylation levels in Ndufs4-KO brain (Fig. 5C, Supplementary Fig. 6E). Importantly, DMKG treatment suppressed hypoxia signaling by lowering HIF1a and LDHA levels in Ndufs4-KO brain without affecting the NAD+ pool of Ndufs4-KO brain (Fig. 5D,E, Supplementary Fig. 6F). These results suggest that DMKG is a viable option to suppress hypoxia signaling independent of the NAD+ salvage pathway in mitochondrial disease.
Discussion
This study identifies two potential therapeutic targets to treat mitochondrial diseases, namely NAD+ metabolism and hypoxia signaling (Fig. 6). Interventions targeting these two pathways attenuate disease progression and extend lifespan in a mouse model of Leigh Syndrome. Supplementation of a NAD+ precursor, NMN, restores NAD+ redox balance and protein acetylation in muscle and attenuates lactate acidosis. These benefits are achieved by suppressing the activation of hypoxia signaling and elevations of glycolytic proteins in the muscle. A similar effect can be achieved by boosting intracellular KG levels to promote HIF1a degradation. These findings are of translational significance as compounds for increasing intracellular NAD+ or suppressing HIF1a activation are becoming available for clinical use7,34,35.
Supplementation of NMN showed the most remarkable effects on lifespan extension of Ndufs4-KO mice. However, the NAD+ pool and NAD+-dependent biochemical pathways in the brain of Ndufs4-KO mice were not responsive to NMN treatment. Consequently, we did not observe any improvement of neurological outcomes either in vivo or at cellular levels. The ineffectiveness to elevate brain NAD+ pool by NMN is likely multifactorial. One possibility is that NMN does not reach the brain. However, a recent study showed that NMN attenuated ischemia injuries and protected the blood-brain barrier11, suggesting that NMN can improve brain function. Another possibility is that supplementation of NAD+ precursor increases turnover but not the size of the NAD+ pool. Early studies indicated that the half-life of the NAD+ pool was as short as 6–10 hours36,37 and the quick turnover was likely due to robust activities of NAD+-consuming enzymes such as Sirtuins. In our study, NMN treatment did not change protein acetylation levels in the brain of Ndufs4-KO mice. Therefore, rapid NAD+ consumption by Sirtuin deacetylases is unlikely the reason for the stagnant NAD+ pool in the brain of Ndufs4-KO mice during NMN treatment. Gauging activities of Sirtuins with total acetylation levels is not without limitation, as multiple Sirtuins exist in different sub-cellular locations and Sirtuins regulate post-translational modifications other than acetylation. Therefore, we employed another pharmacologic approach that elevates NAD+ pool to test our hypothesis. Targeting the NAD+ salvage pathway by P7C3, a putative NAMPT activator, increased the brain NAD+ pool in WT mice but not in Ndufs4-KO mice at the same dose. The results suggested that impaired mitochondrial function in Ndufs4-KO mice could be responsible for the failure to expand the NAD+ pool. However, we cannot rule out any non-specific effects of P7C3 or that a higher dose is required to reverse the molecular and physiological deficits in Ndufs4-KO mice. Generation of NAD+ from NAD+ precursors requires high energy phosphate compounds such as ATP and phosphoribosyl pyrophosphate (PRPP) as substrates38. Complex I-deficiency in Ndufs4-KO results in severe energy deprivation in the brain and liver whereas muscle and heart are largely spared8,20,39. The organ-selective change in energy status in Ndufs4-KO mice matches the effectiveness of raising the intracellular NAD+ level in the present study.
Supplementation of DMKG, a KG precursor, demonstrates neurological benefit in the present study. Besides the suppression of hypoxic signaling observed here, KG was found to support mitochondrial bioenergetics in Ndufs4-KO40. Contrary to the loss of Complex I supported respiration, KG-driven respiration is unaffected in the mitochondria isolated from the brain of Ndufs4-KO mice, and it is even elevated in older Ndufs4-KO mice40. Thus, KG may be used by Ndufs4-KO mitochondria as an alternative substrate for Complex-II dependent respiration, which compensates for the deficiency of Complex-I function. It is noteworthy that elevation of KG levels has been associated with improved outcome in Ndufs4-KO mice treated by rapamycin in a recent study25 as well as by NMN in the present study. KG inhibits the TOR pathway and PARP1 is regulated by protein phosphorylation41. Thus, we speculate that DMKG supplementation increases PAR levels in Ndufs4-KO brain by suppression of PARP1 activity via inhibition of mTOR, which is consistent with the benefit of rapamycin treatment in Ndufs4-KO mice. These findings suggest that KG level is likely a therapeutic target and a readout for treatment efficacy in Leigh syndrome. Further studies are needed to define the therapeutic mechanisms of DMKG.
Mitochondrial dysfunction is known to activate hypoxia signaling under normoxic conditions. Decline in NAD+/NADH ratio, also known as pseudohypoxic state, plays an important role in mitochondria-nuclear communication in aging and diseases9. Results from the present study suggest that correction of pseudohypoxic status is therapeutic for mitochondrial disease. Interestingly, a recent study showed that the opposite strategy, i.e. subjecting Ndufs4-KO mice to chronic hypoxia environment, is also therapeutic42. These seemingly contradictory observations suggest that metabolic remodeling in organisms with mitochondrial defect is in discordant with the oxygenated environment. Blocking the aberrant remodeling, such as protein hyperacetylation and HIF1a activation, prevents the incongruity. Likewise, providing a hypoxic environment would accommodate the upregulation of hypoxia metabolism and avoid oxygen toxicity35,42.
As discussed above, several treatment strategies have improved survival of Ndufs4-KO mice, e.g. rapamycin, chronic hypoxia, NAD+ precursor (NMN) and DMKG. Although the effect of lifespan extension by supplementation of NAD+ precursor is not as dramatic as hypoxic chamber and rapamycin treatments, this strategy has a high potential for translation. Several NAD+ precursors are available as nutraceuticals. They are safe and well tolerated in humans even at high doses43. In a previous study, rapamycin treatment started on P-21 at a dose comparable to clinical use resulted in a similar lifespan extension in Ndufs4-KO as the NMN treatment described here. However, increasing the rapamycin dose several folds higher and starting the treatment from P-10 drastically improve the efficacy25. In the present study, all treatments started from P-21. It remains to be determined whether a higher dose and/or earlier treatment will yield greater improvement. As restoration of the brain NAD+ pool is likely energy-dependent, early intervention before severe energy depletion is desirable. Thus, the optimal time to initiate interventions is worth further investigation for maximum benefit.
Materials and Methods
Animals care and experiments
All animal care and procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Washington and were performed in accordance with IACUC guidelines and regulations. Ndufs4 heterozygous deletion (Ndufs4+/−) mice were generated from breeding of Ndufs4flox/flox mice (Palmiter laboratory) with Meox2-Cre expressing mice (Jackson laboratory). Breeding of Ndufs4+/− male and female mice generated Ndufs4−/− mice (KO). Mice were weaned at 21 days of age (P-21). KO mice were housed with control littermates (Ndufs4+/− or Ndufs4+/+) in the same cage. These mice were genotyped at P-21. Both male and female mice were used in the study. The survival curves were analyzed by pooling the data from both sexes (male-to-female ratio is ~1:1), as there was no sex-specific difference in survivalship. Survival of KO mice was recorded. KO mice were euthanized after veterinarian consultation if 20% loss in maximum body weight, severe dehydration and immobility were observed. Body weight and incidence of clasping25 were monitored starting from P-21 until death. Forelimb clasping is a marker of the progression of neurodegeneration25. WT or KO mice were grasped with their tails and lifted clear of surrounding objects. Forelimb position was observed. If forelimb retraction to the midline of the body is observed, it will be scored positive for clasping25. Tissue harvest was performed in the afternoon during light cycle without fasting.
Nicotinamide mononucleotide (NMN, N3501-Sigma) was dissolved in sterile saline at 50 mg/ml and delivered intraperitoneally to mice at a dose of 500 mg/kg/day twice a week. P7C3 (4076-Tocris) was dissolved in 10% Cremopho EL at 5 mg/ml and delivered intraperitoneally to mice at dose of 50 mg/kg/day daily. Dimethyl-a-ketoglutarate (DMKG, 349631-Sigma) was dissolved at 30 mg/ml in sterile saline and delivered intraperitoneally to mice at dose of 300 mg/kg/day daily.
Cardiac function was assessed by echocardiography using VEVO 2100 system (VisualSonics) on lightly-anesthetized mice. Measurements were made when heart rate was within 500–600 beats per minute. Cardiac function was measured in parasternal long axis B- and M-mode images and calculated by average of three cardiac cycles. Fractional shortening (FS) was calculated using ultrasound analysis software for image data associated with Vevo 2100 (VisualSonics).
Western blotting
Blood inside organs were removed by rinsing tissues with PBS. Tissues were snap-frozen under liquid nitrogen. Tissues were homogenized in RIPA buffer (Sigma) with protease (Roche), deacetylase (nicotinamide, Tricostatin A) and phosphatase inhibitors (Roche)7. Protein concentrations of samples were determined by Lowry assay and each amount of protein (30–50 ug per sample) were loaded for SDS-PAGE. Antibodies from the following companies were used for Western blot analysis: Phospho-H2Ax (1:500, NBP1-19255-Novus Biological), PAR (1:500, AM80100UG-Millipore), actin (1:5000, sc-8432-Santa Cruz), nitrotyrosine (1:500, 06-284-Millipore) acetyl-lysine (1:1000, 9441-Cell signaling), HIF1a (1:400, 14179-Cell Signaling), GLUT1 (1:500, ab652-Abcam), PDK4 (1:1000, 3820-Cell Signaling), LDHA (1:1000, 3582-Cell Signaling), SDHA (1:10000, ab14715-Abcam), Ndufs4 (1:1000, ab87399-Abcam,), SOD2 (1:1000, ab16956-Abcam) SOD2-Ac (1:1000, ab137037-Abcam), GDH (1:1000, 12793-Cell Signaling). Blots were blocked in 5% BSA-TBST. Antibodies were diluted in 5% BSA-TBST. Protein bands were visualized with chemiluminescence assay (Pierce) with secondary antibodies coupled with HRP. The protein abundance was analyzed by densitometry with ImageJ. SDHA or actin were used as a loading control for quantification.
Detections of metabolites
Assay kits were used to measure levels of NADH and NAD+ (BioAssay, CA, USA), alpha-ketoglutarate (Sigma, MO, USA), serum and tissue lactate (Trinity Biotech, Ireland) by following manufacturers’ instructions. Tissues for metabolite measurements were collected by snap-freezing.
For 1H NMR spectroscopy, methanol, chloroform, monosodium phosphate (NaH2PO4), disodium phosphate (Na2HPO4), sodium salt of 3-(trimethylsilyl)propionic acid-2,2,3,3-d4 (TSP) were obtained (Sigma, MO, USA). Deuterium oxide (D2O) was obtained from Cambridge Isotope laboratories, Inc. (Andover, MA).
To extract mouse heart tissues by methanol and chloroform extractions, weighed tissue specimens were mixed with a 1 mL mixture of cold methanol and chloroform (1:2 v/v; 4 °C) in 2 mL Eppendorf vials, homogenized and sonicated for 20 s. A further 800 µL (cold chloroform/distilled water mixture (1:1 v/v) was added, the sample was then vortexed and set aside for 30 min on ice to separate the solvent layers. Next, after centrifugation at 2039 rcf, the aqueous (top) layer was separated and filtered using 1.5 mL 0.2 mm syringe filters and freeze dried. The dried extracts were mixed with 220 µL of a cold phosphate buffer (0.1 M; pH = 7.45; 4 °C) in D2O containing 50 µM TSP and the solutions were transferred to 3 mm sample tubes for measurements. The phosphate buffer was degaussed by bubbling helium gas for 30 min prior to preparation of the samples for NMR analysis.
All NMR experiments were performed at 298 K. A Bruker Avance III 800 MHz spectrometer equipped with a cryogenically cooled probe and z-gradients suitable for inverse detection was used. One dimensional NOESY pulse sequence with residual water suppression using pre-saturation, 10204 Hz spectral width, 6.6 s total recycle delay, 64 transients and 32 K time domain points were used for 1H 1D NMR experiments. Chemical shifts were referenced to the internal TSP signal. Bruker Topspin versions 3.0 or 3.1 software packages were used for NMR data acquisition, processing, and analyses.
Identification of the coenzyme peaks in the NMR spectra was based on the database and peak assignments established recently22. The Chenomx NMR Suite Professional Software package (version 5.1; Chenomx Inc., Edmonton, Alberta, Canada) was used to quantify the coenzyme peaks. Peak fitting with reference to the internal TSP signal enabled the determination of absolute concentrations of the coenzymes. Concentrations of the coenzymes were expressed both relative to the internal reference TSP as well as the internal taurine.
Statistical analysis
Comparisons among the multiple groups were performed by 1-way ANOVA, followed by Newman-Keuls multiple comparison test. For comparisons involving only two groups, unpaired 2-tailed t-tests were used. Statistical comparisons of survival curves and clasping were performed using log-rank test. All analyses were performed using GraphPad Prism 7.0. All data are expressed as mean ± SEM and a p < 0.05 was considered significant.
Supplementary information
Acknowledgements
This work is supported in part by research funds from the National Institute of Health, NS098329, HL110349, HL142628 (to R.T.) and NIH/NIBIB-T32EB1650 (to L.A.), from the American Heart Association, 13POST16200007 and 17SDG33330003 (both to C.F.L.), from a recruitment grant of the Presbyterian Health Foundation (to C.F.L.) and from the Royalty Research Fund, University of Washington (to N.G.).
Author Contributions
R.T. and C.F.L. designed the experiments, interpreted the data and wrote the manuscript. R.T., C.F.L., A.C., G.A.N.G. and L.A. edited the manuscript. C.F.L., A.C., G.A.N.G. and L.A. executed the experiments and analyzed the data. All authors reviewed the manuscript.
Data Availability
The data that support the findings of this study are included in this manuscript and available from corresponding author upon request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-39419-4.
References
- 1.Lightowlers RN, Taylor RW, Turnbull DM. Mutations causing mitochondrial disease: What is new and what challenges remain? Science. 2015;349:1494–1499. doi: 10.1126/science.aac7516. [DOI] [PubMed] [Google Scholar]
- 2.Ruhoy IS, Saneto RP. The genetics of Leigh syndrome and its implications for clinical practice and risk management. Appl Clin Genet. 2014;7:221–234. doi: 10.2147/TACG.S46176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baradaran R, Berrisford JM, Minhas GS, Sazanov LA. Crystal structure of the entire respiratory complex I. Nature. 2013;494:443–448. doi: 10.1038/nature11871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nature reviews. Molecular cell biology. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- 5.Houtkooper RH, Canto C, Wanders RJ, Auwerx J. The secret life of NAD+: an old metabolite controlling new metabolic signaling pathways. Endocr Rev. 2010;31:194–223. doi: 10.1210/er.2009-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu XH, Lu M, Lee BY, Ugurbil K, Chen W. In vivo NAD assay reveals the intracellular NAD contents and redox state in healthy human brain and their age dependences. Proc Natl Acad Sci USA. 2015;112:2876–2881. doi: 10.1073/pnas.1417921112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee CF, et al. Normalization of NAD+ Redox Balance as a Therapy for Heart Failure. Circulation. 2016;134:883–894. doi: 10.1161/CIRCULATIONAHA.116.022495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karamanlidis G, et al. Mitochondrial complex I deficiency increases protein acetylation and accelerates heart failure. Cell metabolism. 2013;18:239–250. doi: 10.1016/j.cmet.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gomes AP, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verdin E. NAD(+) in aging, metabolism, and neurodegeneration. Science. 2015;350:1208–1213. doi: 10.1126/science.aac4854. [DOI] [PubMed] [Google Scholar]
- 11.Wei CC, et al. NAD replenishment with nicotinamide mononucleotide protects blood-brain barrier integrity and attenuates delayed tissue plasminogen activator-induced haemorrhagic transformation after cerebral ischaemia. Br J Pharmacol. 2017;174:3823–3836. doi: 10.1111/bph.13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Picciotto NE, et al. Nicotinamide mononucleotide supplementation reverses vascular dysfunction and oxidative stress with aging in mice. Aging Cell. 2016;15:522–530. doi: 10.1111/acel.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan NA, et al. Effective treatment of mitochondrial myopathy by nicotinamide riboside, a vitamin B3. EMBO Mol Med. 2014;6:721–731. doi: 10.1002/emmm.201403943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang H, et al. NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science. 2016;352:1436–1443. doi: 10.1126/science.aaf2693. [DOI] [PubMed] [Google Scholar]
- 15.Pirinen E, et al. Pharmacological Inhibition of poly(ADP-ribose) polymerases improves fitness and mitochondrial function in skeletal muscle. Cell metabolism. 2014;19:1034–1041. doi: 10.1016/j.cmet.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schaefer AM, Taylor RW, Turnbull DM, Chinnery PF. The epidemiology of mitochondrial disorders–past, present and future. Biochim Biophys Acta. 2004;1659:115–120. doi: 10.1016/j.bbabio.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 17.Pastores GM, et al. Leigh syndrome and hypertrophic cardiomyopathy in an infant with a mitochondrial DNA point mutation (T8993G) Am J Med Genet. 1994;50:265–271. doi: 10.1002/ajmg.1320500310. [DOI] [PubMed] [Google Scholar]
- 18.Leshinsky-Silver E, et al. NDUFS4 mutations cause Leigh syndrome with predominant brainstem involvement. Mol Genet Metab. 2009;97:185–189. doi: 10.1016/j.ymgme.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Petruzzella V, et al. A nonsense mutation in the NDUFS4 gene encoding the 18 kDa (AQDQ) subunit of complex I abolishes assembly and activity of the complex in a patient with Leigh-like syndrome. Hum Mol Genet. 2001;10:529–535. doi: 10.1093/hmg/10.5.529. [DOI] [PubMed] [Google Scholar]
- 20.Kruse SE, et al. Mice with mitochondrial complex I deficiency develop a fatal encephalomyopathy. Cell metabolism. 2008;7:312–320. doi: 10.1016/j.cmet.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quintana A, Kruse SE, Kapur RP, Sanz E, Palmiter RD. Complex I deficiency due to loss of Ndufs4 in the brain results in progressive encephalopathy resembling Leigh syndrome. Proc Natl Acad Sci USA. 2010;107:10996–11001. doi: 10.1073/pnas.1006214107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagana Gowda GA, Abell L, Lee CF, Tian R, Raftery D. Simultaneous Analysis of Major Coenzymes of Cellular Redox Reactions and Energy Using ex Vivo (1)H NMR Spectroscopy. Anal Chem. 2016;88:4817–4824. doi: 10.1021/acs.analchem.6b00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang G, et al. P7C3 neuroprotective chemicals function by activating the rate-limiting enzyme in NAD salvage. Cell. 2014;158:1324–1334. doi: 10.1016/j.cell.2014.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yin TC, et al. P7C3 neuroprotective chemicals block axonal degeneration and preserve function after traumatic brain injury. Cell reports. 2014;8:1731–1740. doi: 10.1016/j.celrep.2014.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson SC, et al. mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science. 2013;342:1524–1528. doi: 10.1126/science.1244360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson Legault J, et al. A Metabolic Signature of Mitochondrial Dysfunction Revealed through a Monogenic Form of Leigh Syndrome. Cell reports. 2015;13:981–989. doi: 10.1016/j.celrep.2015.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chandel NS, et al. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci USA. 1998;95:11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandel NS, et al. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. The Journal of biological chemistry. 2000;275:25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 29.Geng H, et al. HIF1alpha protein stability is increased by acetylation at lysine 709. The Journal of biological chemistry. 2012;287:35496–35505. doi: 10.1074/jbc.M112.400697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tao R, et al. Sirt3-mediated deacetylation of evolutionarily conserved lysine 122 regulates MnSOD activity in response to stress. Molecular cell. 2010;40:893–904. doi: 10.1016/j.molcel.2010.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi WS, Palmiter RD, Xia Z. Loss of mitochondrial complex I activity potentiates dopamine neuron death induced by microtubule dysfunction in a Parkinson’s disease model. The Journal of cell biology. 2011;192:873–882. doi: 10.1083/jcb.201009132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colman RF, Frieden C. On the role of amino groups in the structure and function of glutamate dehydrogenase. I. Effect of acetylation on catalytic and regulatory properties. The Journal of biological chemistry. 1966;241:3652–3660. [PubMed] [Google Scholar]
- 33.Pacella-Ince L, Zander-Fox DL, Lan M. Mitochondrial SIRT3 and its target glutamate dehydrogenase are altered in follicular cells of women with reduced ovarian reserve or advanced maternal age. Hum Reprod. 2014;29:1490–1499. doi: 10.1093/humrep/deu071. [DOI] [PubMed] [Google Scholar]
- 34.Masoud GN, Li W. HIF-1alpha pathway: role, regulation and intervention for cancer therapy. Acta Pharm Sin B. 2015;5:378–389. doi: 10.1016/j.apsb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Airhart SE, et al. An open-label, non-randomized study of the pharmacokinetics of the nutritional supplement nicotinamide riboside (NR) and its effects on blood NAD+ levels in healthy volunteers. PloS one. 2017;12:e0186459. doi: 10.1371/journal.pone.0186459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rechsteiner M, Hillyard D, Olivera BM. Magnitude and significance of NAD turnover in human cell line D98/AH2. Nature. 1976;259:695–696. doi: 10.1038/259695a0. [DOI] [PubMed] [Google Scholar]
- 37.Rechsteiner M, Catanzarite V. The biosynthesis and turnover of nicotinamide adenine dinucleotide in enucleated culture cells. J Cell Physiol. 1974;84:409–422. doi: 10.1002/jcp.1040840309. [DOI] [PubMed] [Google Scholar]
- 38.Canto C, Menzies KJ, Auwerx J. NAD(+) Metabolism and the Control of Energy Homeostasis: A Balancing Act between Mitochondria and the Nucleus. Cell metabolism. 2015;22:31–53. doi: 10.1016/j.cmet.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manjeri GR, et al. Increased mitochondrial ATP production capacity in brain of healthy mice and a mouse model of isolated complex I deficiency after isoflurane anesthesia. J Inherit Metab Dis. 2016;39:59–65. doi: 10.1007/s10545-015-9885-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kayser EB, Sedensky MM, Morgan PG. Region-Specific Defects of Respiratory Capacities in the Ndufs4(KO) Mouse Brain. PloS one. 2016;11:e0148219. doi: 10.1371/journal.pone.0148219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chin RM, et al. The metabolite alpha-ketoglutarate extends lifespan by inhibiting ATP synthase and TOR. Nature. 2014;510:397–401. doi: 10.1038/nature13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jain IH, et al. Hypoxia as a therapy for mitochondrial disease. Science. 2016;352:54–61. doi: 10.1126/science.aad9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Trammell SA, et al. Nicotinamide riboside is uniquely and orally bioavailable in mice and humans. Nat Commun. 2016;7:12948. doi: 10.1038/ncomms12948. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are included in this manuscript and available from corresponding author upon request.