Abstract
In addition to the standard set of translation factors common in eukaryotic organisms, protein synthesis in the yeast Saccharomyces cerevisiae requires an ABCF ATPase factor eEF3, eukaryotic Elongation Factor 3. eEF3 is an E-site binder that was originally identified as an essential factor involved in the elongation stage of protein synthesis. Recent biochemical experiments suggest an additional function of eEF3 in ribosome recycling. We have characterised the global effects of eEF3 depletion on translation using ribosome profiling. Depletion of eEF3 results in decreased ribosome density at the stop codon, indicating that ribosome recycling does not become rate limiting when eEF3 levels are low. Consistent with a defect in translation elongation, eEF3 depletion causes a moderate redistribution of ribosomes towards the 5′ part of the open reading frames. We observed no E-site codon- or amino acid-specific ribosome stalling upon eEF3 depletion, supporting its role as a general elongation factor. Surprisingly, depletion of eEF3 leads to a relative decrease in P-site proline stalling, which we hypothesise is a secondary effect of generally decreased translation and/or decreased competition for the E-site with eIF5A.
Introduction
Protein synthesis – translation – is universally performed by the ribosome, which is assisted by specialised proteins referred to as translation factors. Some translation factors are universally conserved, e.g. the elongation factor eEF2/EF-G1 – and some are lineage-specific, such as elongation factor 3, eEF3, a member of the ABCF ATPase family2,3. While initial analysis of eEF3 distribution suggested it a fungi-specific translational factor4, its distribution is broader, with eEF3-like homologues found in non-fungal species, such as oomycete Phytophthora infestans5, choanoflagellates, and various distantly related algae3.
The protein is essential both for the viability of Saccharomyces cerevisiae6 and for peptide elongation in a reconstituted yeast translational system7,8. Despite decades of research it is not clear why eEF3 is essential for translation elongation in yeast, since it is not a part of the translational apparatus in the vast majority of eukaryotes, including animals and land plants3. In the test tube eEF3 stimulates aminoacyl-tRNA delivery by elongation factor eEF1A8,9, with the C-terminal region of eEF3 directly interacting with eEF1A10,11. In addition to elongation, biochemical experiments suggest a secondary function of eEF3 in ribosome recycling12, inviting an analogy with the multifunctional bacterial GTPase EF-G that participates both in elongation13 and ribosome recycling14. In a reconstituted biochemical system, eEF3-mediated ribosome recycling does not lead to an accumulation of ribosome ‘halfmers’ – that is 40 S subunits associated with mRNA after the 60 S release12,15. Therefore, Kurata and colleagues proposed that eEF3 mediates a recycling pathway via ejection of P-site tRNA and mRNA from the 80 S termination complex. Cryoelectron microscopy reconstruction of ribosome-associated eEF3 localises the factor in the vicinity of the ribosomal E-site16, providing a structural explanation for the biochemical observation that eEF3 competes with the E-site tRNA on the ribosome in the presence of ATP17.
We have analysed the global effects of eEF3 depletion on translation in S. cerevisiae using ribosome profiling (Ribo-Seq), a functional genomics approach that provides a bird’s-eye view of mRNA translation in the cell by means of isolation and sequencing of mRNA fragments protected by translating ribosomes18–21. We took advantage of our high-coverage dataset to ask the following questions: which stage of the ribosomal functional cycle is more sensitive to eEF3 depletion – elongation or ribosome recycling? And is eEF3’s function in elongation codon- or amino acid-specific?
Results
Construction and characterisation of the PMET25-YEF3 strain for tunable eEF3 expression
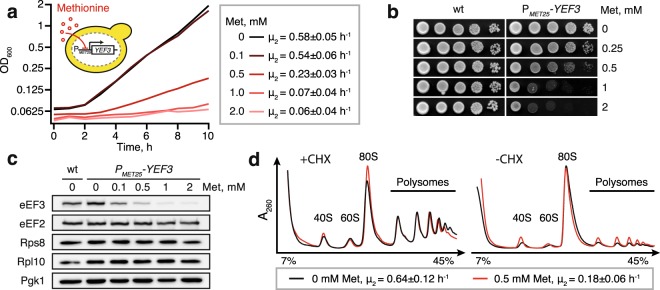
To investigate the role of eEF3 in translation, we set out to develop a system that would allow quick, specific and efficient depletion of eEF3. As our first approach, we constructed a set of strains in which the synthesis of different destabilised forms of eEF3 is post-transcriptionally inhibited by addition of tetracycline to the medium22 (see Supplementary information). However, even in the case of the most responsive strain, eEF3 depletion was inefficient and did not cause growth inhibition until after 7–8 hours. Therefore, rather than rely on rapid eEF3 depletion, we opted for controlling the steady-state level of the protein. We constructed a strain in which the sequence upstream of the endogenous YEF3 ORF, encoding eEF3, was replaced with the sequence of the promoter and 5′-UTR of the methionine-repressible MET25 gene23. The resulting PMET25-YEF3 strain shows concentration-dependent growth inhibition upon addition of methionine to liquid (Fig. 1a) and solid (Fig. 1b) medium. In good agreement with the growth assays, western blotting revealed that the abundance of eEF3 decreases with increasing concentration of methionine (Fig. 1c and Supplementary Fig. S1). Moreover, the eEF3 levels in the PMET25-YEF3 strain grown in the absence of methionine are comparable to those in wild-type cells. Importantly, the levels of eukaryotic elongation factor 2 (eEF2), ribosomal proteins Rps8 and Rps10 as well as phosphoglycerate kinase 1 (Pgk1), are largely unaffected by the methionine concentration in the medium, demonstrating the specificity of eEF3 depletion.
Figure 1.
Tunable repression of eEF3 expression leads to a gradual decrease in growth rate. (a) The PMET25-YEF3 (VKY8) strain was grown at 30 °C in liquid synthetic complete medium lacking methionine and cysteine (SC-met-cys) supplemented with methionine at different concentrations (see insert). The growth rate (µ2) was calculated as the slope of the linear regression of log2-transformed OD600 measurements. (b) Wild type (VKY9) and PMET25-YEF3 (VKY8) strains were grown overnight in SC-met-cys medium, 10-fold serially diluted, spotted on SC-met-cys plates supplemented with the indicated concentration of methionine, and incubated at 30 °C for two days. (c) Western blot analysis of the PMET25-YEF3 strain grown in SC-met-cys medium supplemented with indicated methionine concentrations. In addition to eEF3, the blot was probed for eEF2, Pgk1, Rps8 and Rpl10. Full-length western blots are presented in Supplementary Fig. S1. (d) Polysome profile analyses of the PMET25-YEF3 strain grown in the presence and absence of 0.5 mM methionine. Before harvesting, the cells were either treated with 100 µg/ml cycloheximide for 10 minutes (+CHX) or left untreated (–CHX, ‘run-off conditions’). Whole cell extracts were resolved on sucrose gradients and the absorbance at 260 nm was measured during fractionation. The profiles are normalised to total area under the curve excluding non-ribosomal top fraction and are representatives of two biological replicates.
Depletion of eEF3 decreases growth without dramatically perturbing translation
To assess the overall effect of eEF3 depletion on translation, we used velocity sedimentation of whole cell lysates in sucrose gradients to analyze the polysome profiles of the PMET25-YEF3 strain grown in the absence or presence of 0.5 mM methionine. In these experiments, the presence of 0.5 mM methionine in the medium increased the generation time from 1.6 to 5.6 hours and decreased the eEF3 levels at least five times (Supplementary Fig. S2). Prior to the preparation of the lysates, the cultures were either pre-treated for 10 minutes with 100 µg/ml cycloheximide, CHX, in order to stabilise polysomes, or left untreated (‘run-off conditions’) (Fig. 1d). Depletion of eEF3 results in a slight decrease of the polysomal fraction in the presence of CHX. However, in run-off conditions the polysomal fraction is slightly larger in eEF3 depleted cells. In both CHX-treated and CHX-untreated conditions, eEF3 depletion is associated with a slight increase of the 40 S peak in relation to the 60 S, indicative of effects on either synthesis or degradation of ribosomal subunits.
The effect of eEF3 depletion on polysome run-off is consistent with the previous finding that polysomes are stabilised in cells with a mutant form of eEF311. While reduced polysome run-off is likely a consequence of a defect in translation elongation, it can, in principle, also reflect a ribosome recycling defect, i.e., the queueing of elongating ribosomes behind ribosomes stalled at stop codons.
Generation and technical analysis of Ribo-Seq and RNA-Seq datasets
Ribo-Seq analysis allows sensitive identification of specific rate-limiting steps in translation manifested in an increased read density of ribosome protected fragments (RPFs). To uncover the specific effects of eEF3 depletion, we applied ribosome profiling to the PMET25-YEF3 strain growing exponentially in the presence or absence of 0.5 mM methionine. The yeast cultures used for Ribo-Seq were not treated with CHX prior to harvesting to avoid the specific codon enrichment caused by drug treatment24,25. However, CHX was added in the lysis buffer to inhibit translation elongation and avoid ‘run-off’ during purification of the ribosome protected RNA fragments. Ribo-Seq and RNA-Seq libraries were prepared and sequenced from two biological replicates. Ribo-Seq (25–35 nt) and RNA-Seq (50 nt) reads were mapped to the genome allowing only unique alignments. The detailed description of the NGS data analysis pipeline is described in the Methods section as well as at the GitHub depository page (https://github.com/GCA-VH-lab/RiboSeqPy).
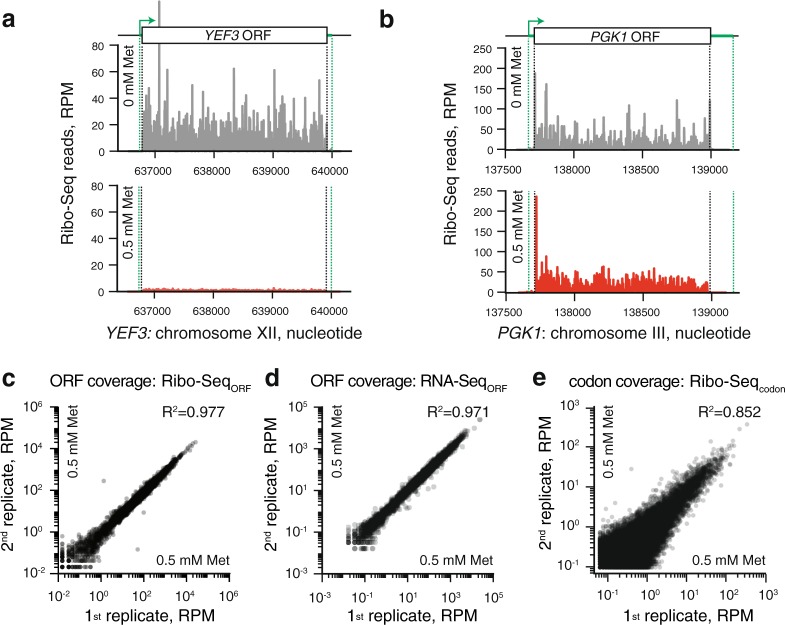
Prior to the analysis of the specific effects of eEF3 depletion on translation, we scrutinised the specificity and reproducibility of our Ribo-Seq and RNA-Seq datasets. In good agreement with the western blotting results, growth in medium containing 0.5 mM methionine dramatically reduced the number of ribosome footprints mapped to the YEF3 ORF (Fig. 2a). Moreover, the ribosome occupancy in the PGK1 ORF is comparable between the two growth conditions (Fig. 2b). The two biological Ribo-Seq and RNA-Seq replicates demonstrate good reproducibility. The R2 for the Ribo-Seq data, quantified in reads per million (RPM) for individual ORFs (Ribo-SeqORF), is 0.977 in the presence, and 0.993 in the absence of methionine (Fig. 2c and Supplementary Fig. S3a). The RNA-SeqORF data also show high reproducibility between the two replicates (Fig. 2d and Supplementary Fig. S3b, R20.5Met = 0.971 and R20Met = 0.988). To uncover codon-specific effects in the ribosome profiling datasets, we used individual 5′ offsets (12–15 nucleotides) to assign the position of the ribosomal P-site in the 28–35 nucleotide (nt) reads, and the quality of codon periodicity for individual read lengths was assessed using Rp-Bp26 (Supplementary Figs S4 and S5). The reproducibility of the Ribo-Seq libraries quantified per individual codon (Ribo-Seqcodon) is lower than that quantified per ORF (Fig. 2e and Supplementary Fig. S3c, R20.5Met = 0.852 and R20Met = 0.803). To examine effects on translation efficiency, we also calculated the ribosomal load, defined as the ratio between the Ribo-Seq and RNA-Seq coverage. The ribosomal load was calculated either per individual ORF (ribosomal loadORF, used for differential gene expression analyses, Fig. 3c, see below) or per individual nucleotide position (ribosomal loadposition, used for metagene and polarity score plots, Fig. 4, see below). The ribosomal load is highly reproducible when calculated per ORF (R2ribosomal load ORF 0 mM Met = 0.932, Supplementary Fig. S3d and R2ribosomal load ORF 0.5 mM Met = 0.889, Supplementary Fig. S3e).
Figure 2.
Reproducibility and specificity of Ribo-Seq and RNA-Seq datasets. Ribosome footprint density along the YEF3 (a) and PGK1 (b) genes in the absence of methionine (upper panel, grey trace, no repression of eEF3 expression) and presence of 0.5 mM methionine (lower panel, red trace, repression of eEF3 expression). Green dotted lines indicate mRNA 3′ and 5′ ends. Reproducibility of ribosome footprint (c and e) and RNA-Seq (d) densities between the two biological replicates. Read density is normalised in reads per million (RPM) of total reads, and is quantified either for individual ORFs (c, d) or individual codons (e).
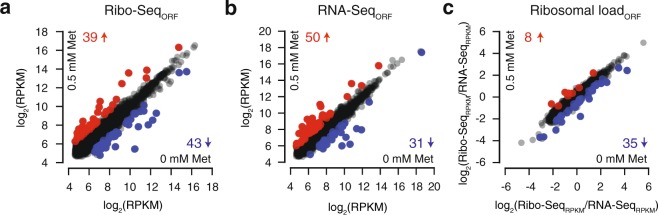
Figure 3.
Genome-wide effects of eEF3 depletion on gene expression. The effects of eEF3 repression to ribosomal density (a), on the levels of mRNA (b) and ribosomal load (c) calculated for individual genes (2581 in total). Changes in gene coverage is considered significant when the Z-score of the differences is above 2 or below -2 in both replicates. Up-regulated genes in eEF3 depleted conditions are marked with red dots and down-regulated genes with blue dots, and the counts of genes presented as numbers in respective colors. Gene IDs of up-regulated and down-regulated genes, as well as associated GO IDs and p-values are provided in Supplementary Dataset 2. Read density is normalised as reads per kilobase per million (RPKM) of total reads for individual ORFs (a,b).
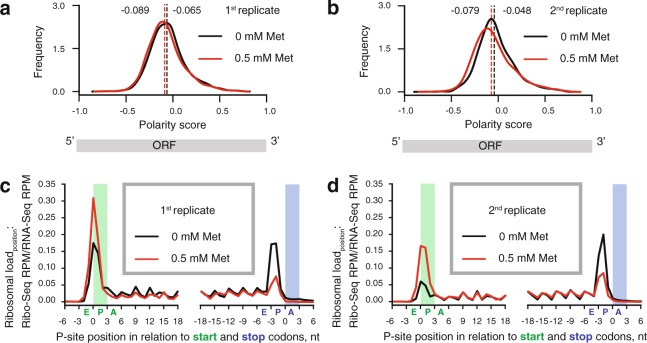
Figure 4.
eEF3 depletion causes moderate redistribution of the ribosome density along mRNAs. The graphs show distributions of polarity scores for individual ORFs (a,b, two experimental replicates), and average ribosome density in the vicinity of start and stop codons (c,d, two experimental replicates). The expression of eEF3 was either repressed by the addition of 0.5 mM methionine (red trace) or unrepressed (black trace, no methionine). The locations of start and stop codons are highlighted with green and blue backdrops, and the corresponding A-, P- and E-site positions of initiating and terminating ribosomes are indicated with letters under the x-axis.
Changes in gene expression upon eEF3 depletion are driven by metabolic adjustments and reduced growth rate
To uncover the effects of reduced abundance of eEF3 on gene expression we compared our RNA-SeqORF, Ribo-SeqORF and ribosomal loadORF datasets between the eEF3-deficient and eEF3-proficient cells. A considerable fraction of S. cerevisiae proteins is encoded by paralogous gene pairs, including highly expressed genes encoding ribosomal proteins and translation factors27,28. Therefore, if only single-mapping is allowed, a significant fraction of reads corresponding to these genes is lost (Supplementary Fig. S6). In order to avoid this systematic bias, for gene expression analysis we re-mapped the raw Ribo-Seq and RNA-Seq data retaining the reads that map twice. Differentially expressed (DE) genes were defined by Z-score < −2 or > 2 of gene coverage difference between eEF3-deficient and eEF3-proficient conditions in both biological replicates (Supplementary Dataset 1).
Comparison of the Ribo-Seq datasets identifies 82 ORFs with altered ribosome density in the eEF3-deficient condition compared to eEF3-proficient: 39 with increased and 43 with decreased ribosome density (Fig. 3a). The majority of the effect is driven by the mRNA copy number since 35 of the genes with increased ribosome density are also up-regulated as per RNA-Seq analysis (90% of the genes identified by Ribo-Seq) and 27 of the genes with decreased ribosome density are also picked up by RNA-Seq (63%) (Fig. 3b). The differential expression of at least a subset of these transcripts is likely caused by the absence/presence of methionine in the medium and not the altered eEF3 levels, although the latter is also possible, especially given the ability of eEF3 to interact directly with mRNA, which could potentially affect transcript stability29.
Computing the ribosomal load subtracts the transcriptional effects to identify cases of specific translational regulation (Fig. 3c). Only eight ORFs display increased ribosomal load, consistent with a predominantly transcriptional origin of the effect observed on the Ribo-Seq level (see above). Out of 35 ORFs with reduced ribosomal load only 4 are common with the Ribo-Seq dataset (43 ORFs, Fig. 3a), indicating that the genes with decreased ribosomal load are predominantly down-regulated on the translational level.
We applied the YeastMine Gene Ontology (GO) enrichment tool30 to identify the molecular functions, biological processes, and cellular components enriched in the differentially affected genes (Supplementary Dataset 2). GO annotations of genes with increased read density in Ribo-Seq and RNA-Seq point to biological processes related to the cell response to heat GO:0034605 (HSP104, DDR2, HSP12, HSP26, SSA4, TPS2) and stress GO:0070413 (HSP104, TPS2, TSL1). Induction of the heat shock response and elevated expression of chaperones upon eEF3 depletion could be a consequence of an accumulation of misfolded proteins due to defects in protein synthesis31. Genes with decreased RNA-Seq read density are dominated by GO categories related to metabolic processes, especially amino acid synthesis (GO:0017144, GO:0000096, GO:1901605, GO:0044281, GO:0006555 and GO:0006520; p-values from 4.33e−6 to 9.0e−4). This is an expected consequence of repressing eEF3 by the addition of 0.5 mM methionine to cell cultures. The gene set that displays a specific decrease in the ribosomal load shows a different pattern. The GO annotations are dominated by the cellular component categories: extracellular region, cell wall and cell surface (GO:0005576, GO:0009277, GO:0005618, GO:0030312 and GO:0009986; p-values from 5.3e−5 to 6.0e−3), most likely reflecting the significant growth rate decrease upon eEF3 depletion. No specific GO enrichments were identified for the eight genes that display specifically increased ribosomal load.
eEF3 depletion causes a decrease in the efficiency of translation elongation
Defects in translation elongation lead to an accumulation of ribosomes on the 5′ part of ORFs19,25,32. This effect can be detected in ribosome profiling data by computing the so-called polarity score metric that ranges from −1 (corresponding to all of the ribosome density localised at the 5′ half of the ORF) to +1 (corresponding to all of the ribosome density localised at the 3′ half of the ORF)32,33. To account for potential differences in mRNA integrity, e.g. differential effects on co-translational mRNA degradation34,35, we normalised the Ribo-Seq reads to the RNA-Seq coverage for each individual nucleotide position (Ribosomal loadposition). Ribosome density corresponding to 15 nucleotides from both the 5′ and 3′ ends of ORFs were excluded from the analysis in order to avoid effects acting on initiation and termination, respectively. Depletion of eEF3 correlates with a slight but reproducible shift in ribosome distribution towards the 5′ end of ORFs (Fig. 4a,b). To test the statistical significance of the effect, we applied the nonparametric Wilcoxon signed-rank test of the null hypothesis i.e. that the two polarity score distributions originate from the same underlying distribution. In order to quantify biological variance we compared the two biological replicates. The p-values for eEF3-proficient (5.3e−2) and eEF3-deficient (5.5e−4) conditions provide an estimate of the biological variability. The difference between eEF3-proficient and eEF3-deficient conditions is significantly higher for both biological replicates (p-values 7.5e−21 and 2.1e−21). This suggests that the shift in ribosome distribution towards the 5′ end of ORFs is, indeed, specific to eEF3 depletion. While the effect is consistent with a defect of translation elongation, we cannot exclude alternative explanations such as an increase in initiation and elongation at the ramp region or ribosome dissociation from mRNAs.
To determine the ribosomal load around the stop and start codons, we performed a metagene analysis of ribosome density around the stop and start codons for all genes with sufficient coverage (Fig. 4c,d). Depletion of the ribosome recycling factor Rli1/ABCE1 leads to an increased ribosomal occupancy at stop codons and the appearance of aperiodic ribosome density in 3′-UTR regions36; if subunit dissociation during ribosome recycling is the primary function of eEF312, we would expect a similar pattern. However, eEF3 depletion results in decreased ribosome occupancy at stop codons and does not lead to increased ribosome occupancy in the 3′-UTR regions, suggesting that eEF3 depletion compromises ribosome recycling less than it does elongation.
eEF3 depletion leads to a moderate decrease in ribosomal stalling on P-site proline residues
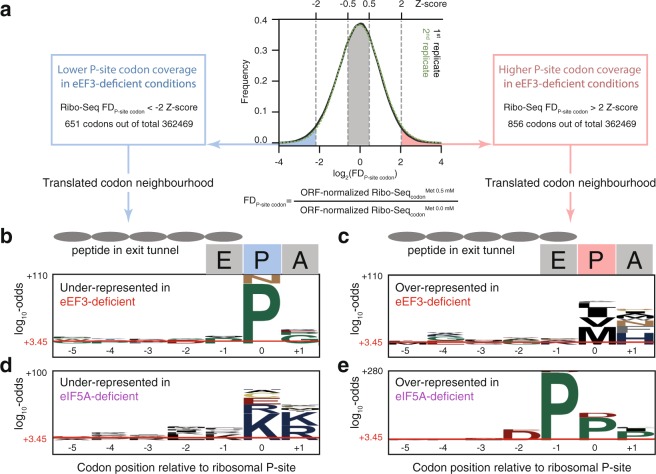
To uncover possible sequence-specific effects of eEF3 depletion we followed the approach originally developed by the Vázquez-Laslop and Mankin labs37,38. We computed changes in ribosome density at individual P-site codons between eEF3-deficient and eEF3-proficient cells – the relative fold difference, Ribo-Seq FDP-site codon or just FD for simplicity (Fig. 5a). Positive log2(FD) values signify a relatively higher coverage of the individual codon by ribosomes under eEF3-deficient conditions, commonly interpreted as codon-specific ribosomal stalling – or at least slowing down or pausing of translating ribosomes37,38. Conversely, negative log2(FD) values indicate relatively lower ribosomal coverage under eEF3-deficient conditions. To discount the contribution of ribosome occupancy at initiation codons, the first 10 codons in all ORFs were excluded from the analysis. Assuming a near-normal distribution of the log2(FD), we identified codons with significantly decreased (Z-score < −2, i.e. the codon’s log2(FD) value is more than two standard deviations lower than the mean; 651 individual codons) or increased (Z-score > 2; 856 individual codons) FD in both biological replicates (see Supplementary Dataset S3). For these codons we extracted the sequence starting from the A-site (+1 position), P-site (0 position), and E-site (−1 position), as well as for the positions of the amino acids in the polypeptide tunnel (from −2 to −5). To compute amino acid overrepresentation scores we used pLogo39 that outputs log10-odds of over- and underrepresentation in relation to the background (see Supplementary Methods section in Supplementary information). The horizontal red bar in Fig. 5b–e represents the statistical significance threshold of the multiple test adjusted p-value (0.05).
Figure 5.
Amino acid- and position-specific redistribution of ribosomal density upon depletion of eEF3 or eIF5A. (a) Distribution of genomic sites (codons) according to the relative fold difference (FD) in the ribosome density between eEF3-deficient and eEF3-proficient cells. Positive FD values (right side of the distribution) indicate the sites where depletion of the factor leads to an increase in ribosome density, and negative FD values correspond to a relative decrease in ribosome density upon depletion of the factor. pLogo39 was used to calculate overrepresentation of specific amino acids at positions relative to the P-site codon using codons with Z-score <-2 (eEF3: panel b, 651 codon positions; eIF5A32: panel d, 510 codon positions) and >2 (eEF3: panel c, 856 codon positions; eIF5A32: panel e, 998 codon positions) that are common for both biological replicates. Horizontal red lines on the pLogos represent significance threshold (the log10-odds 3.45) corresponding to a Bonferroni corrected p-value of 0.05.
We detected no dramatic overrepresentation of specific amino acids in the E-site, indicating that the identity of the E-site tRNA does not influence the redistribution of ribosomal densities upon eEF3 depletion (Fig. 5a,c; Supplementary Table S2). This suggests that eEF3’s function in translation elongation is not E-site codon specific.
In the A-site, the strongest effect is overrepresentation of histidine-encoding codons amongst those with increased overage upon eEF3 depletion (Fig. 5c). Histidine is overrepresented 5.4 times (log10-odds of 19) in comparison to the background, followed by phenylalanine (2.6 times; log10-odds 9) and asparagine (2.5 times; log10-odds 6.6) in eEF3-deficient conditions. In the P-site, eEF3 deficiency leads to a moderate increase in ribosome density for hydrophobic amino acids, with the strongest signal being methionine (5 times; log10-odds 25) and valine (2.1 times; log10-odds 13).
The strongest amino acid-specific effect that we observe over all is a relative underrepresentation of proline codons in the P-site under eEF3-deficient conditions (Fig. 5b). That is, ribosomal density corresponding to P-site prolines is relatively higher in eEF3-proficient conditions. As this is a non-trivial result, we performed several additional analyses all of which support the P-site-specific proline effect (Supplementary Fig. S7). To test reproducibility, we performed separate analyses of the two replicates (Supplementary Fig. S7a and b). To ensure that the effect is not caused by mis-assignment of ribosomal P-site positions by Rp-Bp26, we inspected the metagene plots (Supplementary Fig. S5) to select high-quality read lengths. First, we used the reads that have good periodicity in both eEF3-proficient and eEF3-deficient conditions (Supplementary Fig. S7c: 33 nt-long reads, replicate 2). Since in the case of replicate 1 we could not choose good periodicity reads of the same length for both eEF3-proficient and eEF3-deficient conditions, we used, similarly to Fig. 5, ribosomal positions that are common for both biological replicates (Supplementary Fig. S7d; replicate 1: 28 nt reads in the case of eEF3-proficient conditions and 33 nt reads in the case of eEF3-deficient conditions, replicate 2: 33 nt reads for both conditions). Finally, to deconvolute the codon-specific effects caused by eEF3 depletion from changes in gene expression driven by methionine, we omitted differentially expressed genes, that is, those affected on the transcriptional level by methionine addition (Supplementary Fig. S7e).
To put the P-site proline effect into perspective, we re-analyzed the Ribo-Seq dataset of Schuller and colleagues, who profiled a yeast strain depleted in the translation factor eIF5A32. Polyproline motifs are well-known to cause ribosomal pausing40, and eIF5A promotes translation of these stalling-inducing sequences33,41. Upon eIF5A depletion, ribosome pause sites (high FD; Z-score > 2) are enriched with proline and aspartic acid (Fig. 5e; Supplementary Table S3). While proline codons are overrepresented in all of the three ribosomal sites, the E-site effect is the most dramatic (7.9 times; log10-odds 274). The next-strongest effect upon eIF5A depletion is overrepresentation of proline in the P-site (4 times; log10-odds 91). The strength of this effect is very similar to the proline overrepresentation in this position that we observe in eEF3-proficient conditions in relation to eEF3-depeleted (5.5 times; log10-odds of 93.5).
As in the case of eEF3, we detect a clear signal of relative underrepresentation of specific amino acids in the eIF5A-deficient strain, although the nature of amino acids is different. Positively charged amino acids lysine and arginine – well-known ribosomal stallers42–44 – are overrepresented 3.3–5.9 times in A- and P-sites (Fig. 5d, Supplementary Fig. 3; log10-odds of 13–39).
Discussion
In this study, we have applied ribosome profiling to an eEF3-depleted S. cerevisiae strain under conditions of balanced growth. Surprisingly, the depletion of eEF3 leads to decreased occupancy of elongating ribosomes on P-site proline residues (Fig. 5b). The opposite effect, i.e. increased occupancy of elongating ribosomes on P-site proline residues is brought about by depletion of another E-site binder – translation factor eIF5A which promotes translation of polyprolines32 (Fig. 5e). Therefore, we hypothesise that lowering eEF3 concentration increases the E-site availability for eIF5A binding, increasing the efficiency of transpeptidation in the case of prolines. An alternative explanation is that the general reduction in the efficiency of translation elongation as observed in eEF3-deficient cells may simply mask the pauses that normally occur at proline codons, thus leading to an observed relative decrease in the ribosome density of P-site proline residues.
While eEF3 deficiency specifically decreases occupancy of elongating ribosomes on prolines, our re-analysis of the ribosome profiling dataset of the eIF5A-deficient strain32 has identified specifically decreased occupancy on lysine and arginine (Fig. 5d). It is instructive to contrast this result with that of Pelechano and Alepuz who performed 5Pseq, a genome-wide method of analysing translation by sequencing 5′-phosphorylated mRNA degradation intermediates45. This work used an eIF5A temperature-sensitive mutant33 rather than the degrone fusion used by Schuller and colleagues32. While Pelechano and Alepuz also detected lysine- and arginine-specific effects of eIF5A depletion, the effect is the opposite, i.e. lysine and arginine are overrepresented amongst the codons with increased ribosome occupancy in the eIF5A-depleted strain.
Since we detect no pile up of the ribosome protected fragments at the stop codon – a characteristic sign of defective ribosome recycling36 – we conclude that ribosome recycling is not the primary function of eEF3. We also fail to detect any ribosomal stalling signature specific to the nature of E-site codon. From this we conclude that the identity of the E-site codon (and, hence the identity of deacylated E-site tRNA) does not seem to have a significant role in eEF3′s function as a general elongation factor.
Methods
Growth media, strains and genetic procedures
Yeast media were prepared as described46 with the difference that the composition of drop-out mix was as per Johansson47. Difco Yeast Nitrogen Base w/o Amino Acids was purchased from Becton Dickinson (291940), amino acids and other supplements from Sigma-Aldrich. YEPD medium supplemented with 200 µg/ml GeneticinTM (Gibco 11811-023) was used to select for transformants containing the kanMX6 marker48.
To construct a strain in which the expression of eEF3 (YEF3) is under control of the methionine repressible MET25 promoter (PMET25), we first transformed a diploid strain, formed between BY4728 and BY474249, with a kanMX6-PMET25 DNA fragment with appropriate homologies. The kanMX6-PMET25 DNA fragment was generated in three steps. The kanMX6 marker was amplified from pFA6a-kanMX648 using the primers V3 (5′-TATCCGGCCCCACCCATGCATAACCCTAAATTATTAGATCCGGATCCCCGGGTTAATTAA-3′), which introduces 40 bp homology to sequences ≈300 bp upstream of the YEF3 ORF, and V4 (5′-GAATTCGAGCTCGTTTAAAC-3′). The PMET25 fragment was amplified from genomic DNA from the BY4742 strain49 using primers V5 (5′-GTTTAAACGAGCTCGAATTCGGATGCAAGGGTTCGAATC-3′), which introduces 20 bp homolgy to 3′ end of kanMX6 fragment, and V6 (5′-GTTCTTCTAGAACCTTAATGGATTGCTGGGAATCAGACATTGTATGGATGGGGGTAATAGA-3′), which introduces 40 bp homology to the 5′ end of YEF3 open reading frame. The kanMX6 and PMET25 DNA fragments were then fused by overlap extension PCR50, generating the kanMX6-PMET25 DNA fragment. After transformation, purifications by single cell streaks and PCR confirmation, a heterozygous PYEF3::kanMX6-PMET25/YEF3 strain was allowed to sporulate and the VKY8 (MATα ura3Δ0 his3 leu2Δ0 PYEF3::kanMX6-PMET25) and VKY9 (MATa ura3Δ0 his3 leu2Δ0) strains were obtained from a tetrad on SC-met-cys medium. The PYEF3::kanMX6-PMET25 allele in VKY8 was confirmed by PCR, using primers that annealed outside of sequences in the transformed DNA fragment, and subsequent DNA sequencing of the PCR product.
Growth assays
The PMET25-YEF3 (VKY8) strain was grown overnight in SC-met-cys medium at 30 °C, diluted to an optical density at 600 nm (OD600) of 0.05 in SC-met-cys medium supplemented with different L-methionine concentrations. Cultures were grown at 30 °C in a shaking water bath (New Brunswick™ Innova® 3100) at 195 rpm. After 8 hours, the cultures were diluted to OD600≈0.05 in the same medium and grown overnight. In the morning, the cultures were re-diluted to OD600≈0.05 and growth was monitored through hourly OD600 measurements. Growth rates (µ2) were calculated as slopes of linear regression lines through log2-transformed OD600 data points.
The effect of the methionine concentration on the growth of wild-type (VKY9) and PMET25-YEF3 (VKY8) cells on solid medium was determined from cultures grown overnight in liquid SC-met-cys medium. Cells were harvested, washed, serially diluted and spotted51 onto plates containing different methionine concentrations.
Polysome profile analysis
The PMET25-YEF3 (VKY8) strain was grown overnight in SC-met-cys medium at 30 °C and diluted to an OD600 of 0.05 in SC-met-cys and SC-cys (0.5 mM Met) medium. After 8 h of growth at 30 °C, the cultures were re-diluted in 150 ml of the same medium thereby ensuring that the OD600 of the cultures was below 0.6 the next morning (15–17 h). Growth was then monitored until the OD600 reached 0.8–1. A 50 ml aliquot of each culture was transferred into a pre-warmed flask and treated with 100 µg/ml cycloheximide, CHX, (C7698, Sigma-Aldrich) for 10 min under continuous shaking. Cells from both untreated and CHX treated cultures were pelleted by centrifugation at 3,000 × g for 5 minutes at room temperature, placed on ice, washed with 5 ml of ice cold Breaking buffer (20 mM Tris-HCl pH 7.4, 10 mM MgCl2, 100 mM KCl) with or without 100 µg/ml CHX, and pelleted again at 4 °C. The pellet was resuspended in 250 µl of the respective Breaking buffer containing 1 mM DTT and 1x EDTA-free protease inhibitor cocktail and transferred to a 2 ml FastPrep-24 compatible microcentrifuge tube. Cells were lysed using 0.25 g of glass beads (0.5 mm diameter) and the FastPrep-24 for two 20 sec cycles at a speed setting of 4 m/sec with 1 min on ice between the steps. Lysates were cleared by centrifugation at max speed for 15 min at 4 °C and 5 A260 units loaded on precooled 7–45% linear sucrose gradients in SW41 tubes (made in Breaking buffer +/− CHX supplemented with 1 mM DTT and using a Biocomp Gradient Master instrument). Following centrifugation at 35,000 rpm for 3 hours at 4 °C, the gradients were analyzed by measuring continuous absorbance at 260 nm using a Piston Gradient Fractionator (Biocomp Instruments).
Preparation of NGS libraries (Ribo-Seq and RNA-Seq) and data analysis
The PMET25-YEF3 (VKY8) strain was grown as described for the polysome profile analyses with the difference that the final culture volume was 750 ml and the cells harvested at OD600≈0.6 without prior CHX-treatment. Cells were harvested by rapid vacuum filtration onto a 0.45 µm nitrocellulose membrane, scraped off using a spatula, and frozen in liquid nitrogen. Cells were lysed by cryogenic milling in the presence of CHX-containing lysis buffer, and RNA-Seq and Ribo-Seq libraries were prepared from the cell extracts. RNA-Seq libraries were prepared using ScripSeq Complete Gold Yeast Kit (Epicentre). Ribo-Seq libraries were prepared essentially as per Ingolia and colleagues18 with modifications on the rRNA removal and sample purification procedures. Detailed protocols can be found in our GitHub repository https://github.com/GCA-VH-lab/Lab_protocols.
Multiplexed Ribo-Seq and RNA-Seq libraries were sequenced for 51 cycles (single read) on an Illumina HiSeq 2500 platform. Quality of Illumina reads was controlled using FastQC52, and low quality reads (Phred score below 20) were discarded. The adaptor sequence (5′-CTGTAGGCACCATCAAT-3′) was removed using Cutadapt53. After removing reads mapping to non-coding RNA, reads were mapped to S. cerevisiae reference genome R64-1-1.85 using HISAT254. In the case of Ribo-Seq out of 132.5–143.5 million unprocessed reads, 46.7–68.9 million remained after removal of non-coding RNA and reads mapped multiple times. RNA-Seq reads were processed similarly, omitting the Cutadapt step; out of 78.5–87.3 million unprocessed reads, 51.2–59.0 million remained after removing non-coding RNA and reads mapped multiple times. Ribo-Seq data analysis was performed using custom software written in Python 3, available in GitHub at https://github.com/GCA-VH-lab/RiboSeqPy. For more detail see Supplementary information.
Supplementary information
Acknowledgements
We are grateful to Akira Kaji for sharing anti-eEF3 antibodies and Nicholas Ingolia for sharing protocols for Ribo-Seq library preparation and help with data analysis. This work was supported by the funds from European Regional Development Fund through the Centre of Excellence for Molecular Cell Technology (V.H.); the Molecular Infection Medicine Sweden (MIMS) (V.H.); Swedish Research council (grant 2017-03783 to VH, 2015-04746 to GCA); Ragnar Söderberg foundation (V.H.); Magnus Bergvalls Foundation (2017-02098 to M.J.); Åke Wibergs Foundation (M14-0207 to M.J.); and Kempestiftelsernas grants (JCK-1627 to GCA and SMK-1349 to V.H.).
Author Contributions
V.H. conceived the study, coordinated the study, and drafted the manuscript together with V.K., T.M., G.C.A. and M.J. V.K., M.J. and V.H. designed experiments and analyzed the data. T.M. and G.C.A. analysed the ribosome profiling data. All authors have read and approved the manuscript as submitted.
Data Availability
Ribo-Seq and RNA-Seq sequencing data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-6938.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Villu Kasari and Tõnu Margus contributed equally.
Contributor Information
Gemma C. Atkinson, Email: gemma.atkinson@umu.se
Marcus J. O. Johansson, Email: marcus.johansson@umu.se
Vasili Hauryliuk, Email: vasili.hauryliuk@umu.se.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-019-39403-y.
References
- 1.Atkinson GC. The evolutionary and functional diversity of classical and lesser-known cytoplasmic and organellar translational GTPases across the tree of life. BMC Genomics. 2015;16:78. doi: 10.1186/s12864-015-1289-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hopfner KP. Invited review: Architectures and mechanisms of ATP binding cassette proteins. Biopolymers. 2016;105:492–504. doi: 10.1002/bip.22843. [DOI] [PubMed] [Google Scholar]
- 3.Murina V, Kasari M, Reith M, Hauryliuk V, Atkinson GC. ABCF ATPases involved in protein synthesis, ribosome assembly and antibiotic resistance: structural and functional diversification across the tree of life. bioRxiv. 2017 doi: 10.1101/220046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ross-Smith N, Tan P, Belfield G, Tuite MF. Translational elongation factor 3 (EF-3): a study of its structural and functional divergence in fungi. Biochem Soc Trans. 1995;23:132S. doi: 10.1042/bst023132s. [DOI] [PubMed] [Google Scholar]
- 5.Mateyak MK, et al. Demonstration of translation elongation factor 3 activity from a non-fungal species, Phytophthora infestans. PLoS One. 2018;13:e0190524. doi: 10.1371/journal.pone.0190524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandbaken MG, Lupisella JA, DiDomenico B, Chakraburtty K. Protein synthesis in yeast. Structural and functional analysis of the gene encoding elongation factor 3. J Biol Chem. 1990;265:15838–15844. [PubMed] [Google Scholar]
- 7.Hutchison JS, Feinberg B, Rothwell TC, Moldave K. Monoclonal antibody specific for yeast elongation factor 3. Biochemistry. 1984;23:3055–3063. doi: 10.1021/bi00308a032. [DOI] [PubMed] [Google Scholar]
- 8.Uritani M, Miyazaki M. Role of yeast peptide elongation factor 3 (EF-3) at the AA-tRNA binding step. J Biochem. 1988;104:118–126. doi: 10.1093/oxfordjournals.jbchem.a122405. [DOI] [PubMed] [Google Scholar]
- 9.Kamath A, Chakraburtty K. Role of yeast elongation factor 3 in the elongation cycle. J Biol Chem. 1989;264:15423–15428. [PubMed] [Google Scholar]
- 10.Anand M, Balar B, Ulloque R, Gross SR, Kinzy TG. Domain and nucleotide dependence of the interaction between Saccharomyces cerevisiae translation elongation factors 3 and 1A. J Biol Chem. 2006;281:32318–32326. doi: 10.1074/jbc.M601899200. [DOI] [PubMed] [Google Scholar]
- 11.Anand M, Chakraburtty K, Marton MJ, Hinnebusch AG, Kinzy TG. Functional interactions between yeast translation eukaryotic elongation factor (eEF) 1A and eEF3. J Biol Chem. 2003;278:6985–6991. doi: 10.1074/jbc.M209224200. [DOI] [PubMed] [Google Scholar]
- 12.Kurata S, et al. Ribosome recycling step in yeast cytoplasmic protein synthesis is catalyzed by eEF3 and ATP. Proc Natl Acad Sci USA. 2010;107:10854–10859. doi: 10.1073/pnas.1006247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ling C, Ermolenko DN. Structural insights into ribosome translocation. Wiley Interdiscip Rev RNA. 2016;7:620–636. doi: 10.1002/wrna.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hirokawa G, Demeshkina N, Iwakura N, Kaji H, Kaji A. The ribosome-recycling step: consensus or controversy? Trends Biochem Sci. 2006;31:143–149. doi: 10.1016/j.tibs.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 15.Kurata S, et al. Possible steps of complete disassembly of post-termination complex by yeast eEF3 deduced from inhibition by translocation inhibitors. Nucleic Acids Res. 2013;41:264–276. doi: 10.1093/nar/gks958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen CB, et al. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature. 2006;443:663–668. doi: 10.1038/nature05126. [DOI] [PubMed] [Google Scholar]
- 17.Triana-Alonso FJ, Chakraburtty K, Nierhaus KH. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J Biol Chem. 1995;270:20473–20478. doi: 10.1074/jbc.270.35.20473. [DOI] [PubMed] [Google Scholar]
- 18.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brar GA, Weissman JS. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol. 2015;16:651–664. doi: 10.1038/nrm4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreev DE, et al. Insights into the mechanisms of eukaryotic translation gained with ribosome profiling. Nucleic Acids Res. 2017;45:513–526. doi: 10.1093/nar/gkw1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kotter P, Weigand JE, Meyer B, Entian KD, Suess B. A fast and efficient translational control system for conditional expression of yeast genes. Nucleic Acids Res. 2009;37:e120. doi: 10.1093/nar/gkp578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sangsoda S, Cherest H, Surdin-Kerjan Y. The expression of the MET25 gene of Saccharomyces cerevisiae is regulated transcriptionally. Mol Gen Genet. 1985;200:407–414. doi: 10.1007/BF00425724. [DOI] [PubMed] [Google Scholar]
- 24.Hussmann JA, Patchett S, Johnson A, Sawyer S, Press WH. Understanding Biases in Ribosome Profiling Experiments Reveals Signatures of Translation Dynamics in Yeast. PLoS Genet. 2015;11:e1005732. doi: 10.1371/journal.pgen.1005732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerashchenko, M. V. & Gladyshev, V. N. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res42 (2014). [DOI] [PMC free article] [PubMed]
- 26.Malone B, et al. Bayesian prediction of RNA translation from ribosome profiling. Nucleic Acids Res. 2017;45:2960–2972. doi: 10.1093/nar/gkw1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- 28.Fares MA, Keane OM, Toft C, Carretero-Paulet L, Jones GW. The roles of whole-genome and small-scale duplications in the functional specialization of Saccharomyces cerevisiae genes. PLoS Genet. 2013;9:e1003176. doi: 10.1371/journal.pgen.1003176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Samra N, Atir-Lande A, Pnueli L, Arava Y. The elongation factor eEF3 (Yef3) interacts with mRNA in a translation independent manner. BMC Mol Biol. 2015;16:17. doi: 10.1186/s12867-015-0045-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balakrishnan R, et al. YeastMine–an integrated data warehouse for Saccharomyces cerevisiae data as a multipurpose tool-kit. Database (Oxford) 2012;2012:bar062. doi: 10.1093/database/bar062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bush KT, Goldberg AL, Nigam SK. Proteasome inhibition leads to a heat-shock response, induction of endoplasmic reticulum chaperones, and thermotolerance. J Biol Chem. 1997;272:9086–9092. doi: 10.1074/jbc.272.14.9086. [DOI] [PubMed] [Google Scholar]
- 32.Schuller AP, Wu CC, Dever TE, Buskirk AR, Green R. eIF5A Functions Globally in Translation Elongation and Termination. Mol Cell. 2017;66:194–205 e195. doi: 10.1016/j.molcel.2017.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pelechano V, Alepuz P. eIF5A facilitates translation termination globally and promotes the elongation of many non polyproline-specific tripeptide sequences. Nucleic Acids Res. 2017;45:7326–7338. doi: 10.1093/nar/gkx479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pelechano V, Wei W, Steinmetz LM. Widespread Co-translational RNA Decay Reveals Ribosome Dynamics. Cell. 2015;161:1400–1412. doi: 10.1016/j.cell.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu W, Sweet TJ, Chamnongpol S, Baker KE, Coller J. Co-translational mRNA decay in Saccharomyces cerevisiae. Nature. 2009;461:225–229. doi: 10.1038/nature08265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young DJ, Guydosh NR, Zhang F, Hinnebusch AG, Green R. Rli1/ABCE1 Recycles Terminating Ribosomes and Controls Translation Reinitiation in 3′UTRs In Vivo. Cell. 2015;162:872–884. doi: 10.1016/j.cell.2015.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marks J, et al. Context-specific inhibition of translation by ribosomal antibiotics targeting the peptidyl transferase center. Proc Natl Acad Sci USA. 2016;113:12150–12155. doi: 10.1073/pnas.1613055113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kannan K, et al. The general mode of translation inhibition by macrolide antibiotics. Proc Natl Acad Sci USA. 2014;111:15958–15963. doi: 10.1073/pnas.1417334111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Shea JP, et al. pLogo: a probabilistic approach to visualizing sequence motifs. Nat Methods. 2013;10:1211–1212. doi: 10.1038/nmeth.2646. [DOI] [PubMed] [Google Scholar]
- 40.Artieri CG, Fraser HB. Accounting for biases in riboprofiling data indicates a major role for proline in stalling translation. Genome Res. 2014;24:2011–2021. doi: 10.1101/gr.175893.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gutierrez E, et al. eIF5A promotes translation of polyproline motifs. Mol Cell. 2013;51:35–45. doi: 10.1016/j.molcel.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lu J, Deutsch C. Electrostatics in the ribosomal tunnel modulate chain elongation rates. J Mol Biol. 2008;384:73–86. doi: 10.1016/j.jmb.2008.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sabi R, Tuller T. Computational analysis of nascent peptides that induce ribosome stalling and their proteomic distribution in Saccharomyces cerevisiae. RNA. 2017;23:983–994. doi: 10.1261/rna.059188.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sabi R, Tuller T. A comparative genomics study on the effect of individual amino acids on ribosome stalling. BMC Genomics. 2015;16(Suppl 10):S5. doi: 10.1186/1471-2164-16-S10-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pelechano V, Wei W, Steinmetz LM. Genome-wide quantification of 5′-phosphorylated mRNA degradation intermediates for analysis of ribosome dynamics. Nat Protoc. 2016;11:359–376. doi: 10.1038/nprot.2016.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amberg, D. C., Burke, D. & Strathern, J. N. Methods in yeast genetics: a cold spring harbor laboratory course manual. (CSHL Press, 2005).
- 47.Johansson MJ. Determining if an mRNA is a Substrate of Nonsense-Mediated mRNA Decay in Saccharomyces cerevisiae. Methods Mol Biol. 2017;1507:169–177. doi: 10.1007/978-1-4939-6518-2_13. [DOI] [PubMed] [Google Scholar]
- 48.Longtine MS, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 49.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 50.Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He F, Amrani N, Johansson MJ, Jacobson A. Chapter 6. Qualitative and quantitative assessment of the activity of the yeast nonsense-mediated mRNA decay pathway. Methods Enzymol. 2008;449:127–147. doi: 10.1016/S0076-6879(08)02406-3. [DOI] [PubMed] [Google Scholar]
- 52.Andrews, S. FastQC: a quality control tool for high throughput sequence data, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (2010).
- 53.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 54.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Ribo-Seq and RNA-Seq sequencing data have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-6938.