The potential spread of antimalarial drug resistance to Africa, in particular for artemisinins and key partner drugs, is a major concern. We surveyed Plasmodium falciparum genetic markers associated with drug sensitivity on 3 occasions at ∼6-month intervals in 2016 and 2017 at 10 sites representing a range of epidemiological settings in Uganda.
KEYWORDS: Uganda, antimalarial drug sensitivity, molecular markers
ABSTRACT
The potential spread of antimalarial drug resistance to Africa, in particular for artemisinins and key partner drugs, is a major concern. We surveyed Plasmodium falciparum genetic markers associated with drug sensitivity on 3 occasions at ∼6-month intervals in 2016 and 2017 at 10 sites representing a range of epidemiological settings in Uganda. For putative drug transporters, we found continued evolution toward wild-type sequences associated with increased sensitivity to chloroquine. For pfcrt K76T, by 2017 the prevalence of the wild type was >60% at all sites and >90% at 6 sites. For the pfmdr1 N86Y and D1246Y alleles, wild type prevalence ranged from 80 to 100%. We found low prevalence of K13 propeller domain mutations, which are associated with artemisinin resistance in Asia, but one mutation previously identified in northern Uganda, 675V, was seen in 2.0% of samples, including 5.5% of those from the 3 northernmost sites. Amplification of the pfmdr1 and plasmepsin2 genes, associated elsewhere with decreased sensitivity to lumefantrine and piperaquine, respectively, was seen in <1% of samples. For the antifolate targets pfdhfr and pfdhps, 5 mutations previously associated with resistance were very common, and the pfdhfr 164L and pfdhps 581G mutations associated with higher-level resistance were seen at multiple sites, although prevalence did not clearly increase over time. Overall, changes were consistent with the selective pressure of the national treatment regimen, artemether-lumefantrine, with increased sensitivity to chloroquine, and with poor efficacy of antifolates. Strong evidence for resistance to artemisinins was not seen. Continued surveillance of markers that predict antimalarial drug sensitivity is warranted.
INTRODUCTION
Antimalarial drug resistance is a major concern. In Africa, resistance to chloroquine and antifolates has been widespread for many years (1). In Uganda, the standard therapy for uncomplicated malaria changed from chloroquine to chloroquine plus sulfadoxine-pyrimethamine (SP) in 2000 and then to the artemisinin-based combination therapy (ACT) artemether-lumefantrine in 2004, although implementation of the new regimen was slow (2). Many countries in Africa utilize the ACT artesunate-amodiaquine as first-line therapy. Dihydroartemisinin-piperaquine is an alternative ACT for uncomplicated malaria in some countries and is under evaluation for chemoprevention (3). Amodiaquine plus SP is used for seasonal malaria chemoprevention in parts of west and central Africa (4). SP is the standard for intermittent preventive therapy during pregnancy (IPTp) (5). Resistance to each component of these regimens has been detected (6) and the spread of resistance to Africa, in particular that to artemisinins and key partner drugs, may have devastating consequences.
Mechanisms of altered sensitivity to a number of antimalarial drugs are quite well understood. Mutations in genes encoding two putative drug transporters, Plasmodium falciparum multidrug resistance protein-1 (pfmdr1) and P. falciparum chloroquine resistance transporter (pfcrt), impact sensitivities to a number of drugs (1). Three mutations that have been common in Uganda, pfcrt 76T, pfmdr1 86Y, and pfmdr1 1246Y (7), are selected by therapy with artesunate-amodiaquine (8, 9) and associated with decreased sensitivity to aminoquinolines (10). Wild-type sequences at these same alleles are selected by prior therapy with artemether-lumefantrine (11–13), associated with decreased lumefantrine sensitivity (10), and, in a pooled analysis, predicted recrudescence after treatment with artemether-lumefantrine (14).
P. falciparum resistance to artemisinin derivatives, defined as delayed parasite clearance either clinically (15) or in the in vitro ring stage survival assay (16), is now widespread in southeast Asia (17, 18). This phenotype is associated with a number of different mutations in the propeller domain of the kelch13 (K13) protein (19). To date, 20 K13 propeller domain mutations in Asia have been associated with delayed clearance (20). K13 mutations have also been seen in P. falciparum isolates from Africa; most of these differ from the mutations associated with delayed clearance, and, based on clinical, parasitological, and molecular data, artemisinin resistance has not clearly been identified in Africa (21–25). Resistance to ACT partner drugs is also of great concern. In southeast Asia resistance to mefloquine, mediated by increased pfmdr1 copy number (26), and to piperaquine, mediated by increased plasmepsin2 copy number (27, 28), has been noted. Furthermore, combined resistance to artemisinins and piperaquine that has led to very high rates of treatment failure for dihydroartemisinin-piperaquine in Cambodia is a concern (29, 30).
P. falciparum resistance to SP is mediated by mutations in the target dihydrofolate reductase (pfdhfr) and dihydropteroate synthase (pfdhps) genes (31). The combination of three mutations in pfdhfr (51I, 59R, and 108N) and two in pfdhps (437G and 540E) leads to an intermediate level of SP resistance (32). This genotype is common in Uganda and other parts of Africa, although the pfdhps 540E mutation is absent in much of west and central Africa (33). Addition of either pfdhfr 164L or pfdhps 581G leads to higher-level SP resistance (31). These additional mutations have been uncommon in Africa, but some reports have noted moderate prevalence of the pfdhfr 164L mutation in parasites from southwestern Uganda (34–36) and of the pfdhps 581G mutation in parasites from Uganda and Tanzania (35–37).
Changes in treatment practices appear to have impacted antimalarial drug sensitivity in Africa. In the clearest demonstration of this phenomenon, nonuse of chloroquine in Malawi in the 1990s was followed by reversion of parasites to the wild-type pfcrt K76 genotype, with subsequent demonstration of improved chloroquine sensitivity in vitro (38) and in vivo (39). In Uganda, with increased use of artemether-lumefantrine and decreased use of chloroquine to treat malaria, parasite genotypes have been changing, with increased prevalence of pfcrt and pfmdr1 wild-type alleles (7, 35). Recently, high prevalence of wild-type pfcrt K76 parasites and remarkable improvement in in vitro sensitivity to chloroquine was observed in Tororo in eastern Uganda (10). Changing parasite sensitivities appear to have clinical consequences, with the efficacy of artesunate-amodiaquine being better than that of artemether-lumefantrine in recent trials, a change from the results of trials conducted about a decade ago (21, 40). For antifolates, resistance-associated mutations are widespread, but SP remains the standard of care for IPTp, and there is concern for selection of more highly resistant parasites. With this background, careful surveillance for established markers of antimalarial drug resistance is a high priority, and so we surveyed the prevalence of key polymorphisms over time at 10 sites across Uganda.
RESULTS
Collection of samples.
We set out to collect blood samples from 50 children presenting with malaria at each of 10 sites (Fig. 1) on 3 occasions during 2016 and 2017. A total of 1,459 samples were collected, as follows: 499 from April to June 2016 (collection 1), 491 from November 2016 to January 2017 (collection 2), and 469 from May to June 2017 (collection 3; Table S1); for Kabale, the third collection continued to October 2017 due to the low number of available samples. Fewer than 50 samples per collection were available from Kabale and Tororo during some collection periods due to low incidence of malaria (Table S1). Due to anticipated low prevalence of polymorphisms, sequencing of K13 and assays for amplification of pfmdr1 and plasmepsin2 were only performed on samples from 2017; due to slow collection, these assays were not performed on samples from Kabale. Reported results are for samples that yielded data for at least one polymorphism.
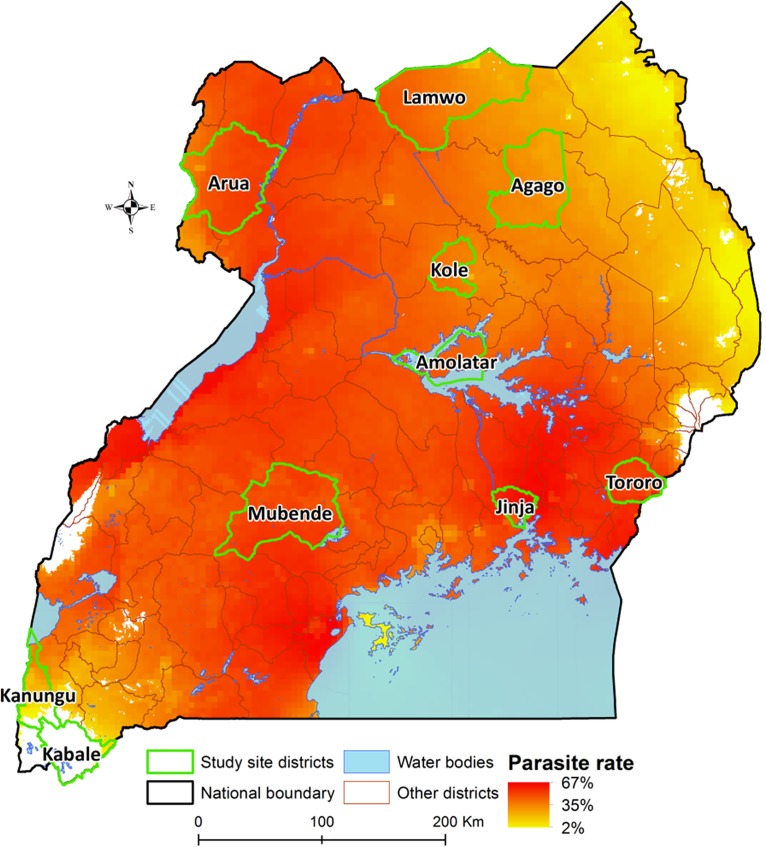
FIG 1.
Surveillance sites in Uganda. The map shows study sites and estimated parasite prevalence for children 2 to 10 years of age. Estimates are based on community surveys between 1985 and 2010 under the Malaria Atlas Project. White locations have indeterminate prevalence based on insufficient data; these are primarily high-elevation areas with known low malaria transmission intensity.
Prevalence of drug resistance-mediating polymorphisms in putative transporters.
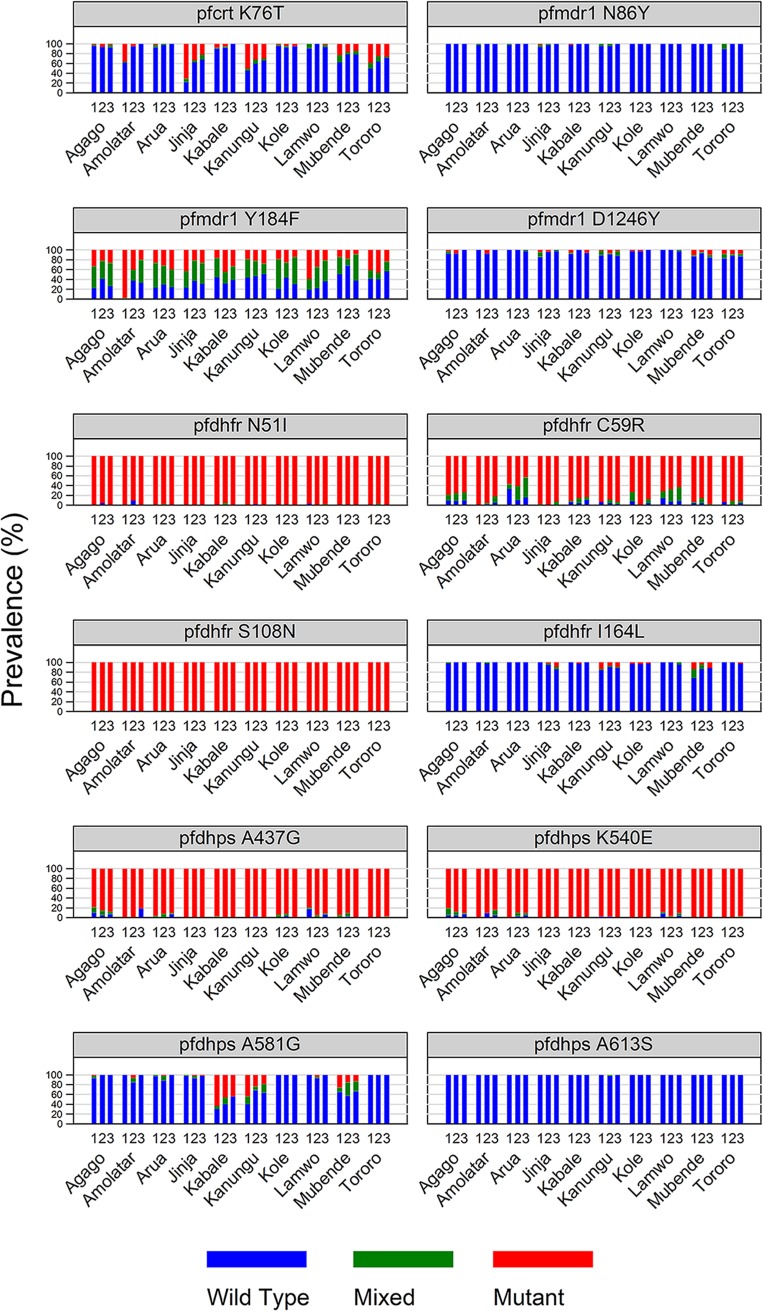
The prevalence of key pfcrt and pfmdr1 polymorphisms varied across the country, but the temporal trends were similar (Fig. 2). For the pfcrt K76T polymorphism, for which the mutant is associated with resistance to chloroquine, wild-type prevalence was higher than that reported in earlier surveys performed at 3 of the sites (7, 35), and this prevalence increased over time, such that, in 2017, >60% of samples were wild type at all sites and >90% were wild type at 6 of the sites. For pfmdr1 N86Y and pfmdr1 D1246Y, 80 to 100% of parasites were wild type at all study sites. As seen previously, the pfmdr1 Y184F allele was polymorphic, but the prevalence of wild-type alleles did not change notably over time.
FIG 2.
Prevalence of wild-type, mixed, and mutant alleles at the named sites over the indicated survey periods. The numbers above the site names represent survey periods (1, April 2016 to June 2016; 2, November 2016 to January 2017; 3, May 2017 to June 2017).
Prevalence of K13 polymorphisms.
Of the 412 samples with successful sequencing of the K13 propeller domain, 25 carried nonsynonymous polymorphisms, with 7 different mutations observed (Tables S2 and S3). Two single-nucleotide polymorphisms (SNPs) had a prevalence of >1%, A675V (2.0%) and C469Y (1.4%); both alleles were seen primarily at sites in northern Uganda (Table 1). The 675V mutation was seen in 7/128 (5.5%) samples from the 3 northernmost sites. The most common mutation previously reported in Africa, 578S, was seen in 3 (0.7%) samples.
TABLE 1.
Prevalence of K13 and copy number polymorphisms at the different sites in 2017
| Site | Na | K13 candidate artemisinin resistance markers |
Increased copy number |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
PfK13 A675V |
PfK13 C469Y |
|||||||||
| Wild type (%) | Mixed (%) | Mutant (%) | Wild type (%) | Mixed (%) | Mutant (%) | N | pfmdr1 (%) | plasmepsin2 (%) | ||
| Agago | 42 | 40 (95.2) | 1 (2.4) | 1 (2.4) | 40 (95.2) | 1 (2.4) | 1 (2.4) | 30 | 2 (6.7) | 1 (3.3) |
| Amolatar | 48 | 48 (100) | 0 | 0 | 48 (100) | 0 | 0 | 41 | 0 | 0 |
| Arua | 43 | 42 (97.7) | 0 | 1 (2.3) | 43 (100) | 0 | 0 | 37 | 0 | 0 |
| Jinja | 48 | 48 (100) | 0 | 0 | 48 (100) | 0 | 0 | 37 | 0 | 1 (2.7) |
| Kanungu | 48 | 48 (100) | 0 | 0 | 48 (100) | 0 | 0 | 45 | 0 | 0 |
| Kole | 47 | 46 (97.9) | 0 | 1 (2.1) | 46 (97.9) | 0 | 1 (2.1) | 31 | 0 | 0 |
| Lamwo | 43 | 39 (90.7) | 0 | 4 (9.3) | 41 (95.4) | 1 (2.3) | 1 (2.3) | 35 | 0 | 0 |
| Mubende | 45 | 45 (100) | 0 | 0 | 44 (97.8) | 1 (2.2) | 0 | 39 | 0 | 0 |
| Tororo | 48 | 48 (100) | 0 | 0 | 48 (100) | 0 | 0 | 40 | 0 | 0 |
N, number of samples successfully evaluated.
Prevalence of parasites with increased pfmdr1 and plasmepsin2 gene copy numbers.
Of 335 samples successfully assessed for copy number variation, only 2 (0.6%), both from Agago, had an increased pfmdr1 copy number and 2 (0.6%; one each from Agago and Jinja) had increased plasmepsin2 copy numbers (Table 1). Thus, consistent with prior reports (10), increased pfmdr1 and plasmepsin2 copy numbers were uncommon in Ugandan isolates.
Prevalence of drug resistance-mediating polymorphisms in folate pathway enzymes.
As seen previously (7, 35), the prevalences of 5 mutations in pfdhfr (51I, 59R, and 108N) and pfdhps (437G and 540E) were high across Uganda (Fig. 2). Additional mutations associated with higher-level antifolate resistance (pfdhfr 164L and pfdhps 581G) were seen, with the prevalence of mixed or mutant pfdhps 581G at ∼25 to 60% at sites in central and southwestern Uganda. However, the prevalences of pfdhfr 164L and pfdhps 581G mutant parasites did not increase over time at most sites.
DISCUSSION
In Uganda, treatment of malaria primarily with artemether-lumefantrine for the last decade has been associated with marked changes in P. falciparum genetic markers associated with drug sensitivity. To gain insight into recent trends across the country, we performed surveillance for relevant markers on 3 occasions in 2016 and 2017 at 10 sites representing a range of epidemiological settings. We found continued evolution toward wild-type transporter sequences, low prevalence of K13 mutations or amplified pfmdr1 or plasmepsin2, and high prevalence of antifolate mutations. These results suggest increasing sensitivity of P. falciparum to chloroquine, a lack of resistance to artemisinins or major ACT partner drugs, and continued poor antimalarial efficacy of SP. Thus, in Uganda, P. falciparum appears to remain sensitive to the ACTs available to treat malaria, but the utility of antifolates to prevent malaria is in question.
The current evolution of transporter gene sequences in Uganda is not surprising. In Malawi, withdrawal of chloroquine in the 1990s was followed by increased prevalence of parasites with the wild-type pfcrt K76 allele, accompanied by improved chloroquine sensitivity (38, 39). Similar changes have been documented in other African countries, including Kenya (41) and Tanzania (42). In Uganda, reversion to wild-type pfcrt K76 and pfmdr1 N86 and D1246 alleles was initially slow following chloroquine withdrawal, perhaps due to continued usage of chloroquine in the community and reasonably strong fitness of chloroquine-resistant parasites. Prior analyses showed a <10% prevalence of parasites with wild-type pfcrt K76 until 2012 in Tororo (7), but a steady increase in prevalence of the wild type in Tororo and two other sites since that time (35). Reversion to wild-type pfmdr1 N86 and D1246 alleles was also seen, with changes more rapid than those for pfcrt K76T (7, 35). Our new results show continued selection of parasites that have wild-type sequences at key transporter alleles. Consistent with this finding, parasites collected in Tororo demonstrated increasing chloroquine ex vivo sensitivity over time (10). Remarkably, recent results suggest that chloroquine may soon again be a highly effective antimalarial in Uganda, although widespread use would likely reselect for resistant parasites.
Resistance to artemisinins, manifested as delayed parasite clearance after therapy or in vitro, is associated with polymorphisms in the K13 gene. A total of 20 different K13 propeller domain mutations have been associated with delayed clearance in southeast Asia (20), with resistance documented across the Greater Mekong Subregion (17, 18). In Africa, delayed clearance after therapy with ACTs (43) or when measured in vitro (22) appears to be very uncommon. Multiple K13 mutations have been seen at low prevalence in African parasites, but many, including the most common polymorphism reported in Africa, A578S, have not been associated with delayed clearance (20). One mutation that has been associated with delayed clearance in southeast Asia, 675V, was seen in 2.0% of our study samples. This mutation was also noted in one sample collected in Rwanda in 2015 (44) and in one sample that showed delayed clearance in vitro in a recent study from northern Uganda (45). The clinical significance of this finding is uncertain, but in our study the polymorphism was geographically clustered in northern Uganda.
Amplification of pfmdr1 has been associated with decreased sensitivity of P. falciparum to mefloquine (26) and lumefantrine (46) and amplification of plasmepsin2 with decreased sensitivity to piperaquine (27, 28). Amplification of pfmdr1 (47, 48) and plasmepsin2 (10) has been uncommon in previous studies from Uganda, as also seen in our new results. These results are reassuring, as they suggest continued strong efficacy of important ACT partner drugs, consistent with excellent efficacy for leading ACTs in recent trials (21, 49).
SP was abandoned as a treatment for malaria due to widespread resistance in P. falciparum, mediated by well-characterized mutations in the pfdhfr and pfdhps genes (31), however, SP remains the standard of care for IPTp in areas of Africa where malaria is endemic (5). SP is also increasingly used for seasonal malaria chemoprophylaxis, whereby treatment courses of SP plus amodiaquine are provided monthly during the rainy season in parts of west and central Africa (4). We found that all 5 pfdhfr and pfdhps mutations commonly associated with SP resistance in Africa remain widespread in Uganda. In addition, the pfdhfr 164L and pfdhps 581G mutations, which predict higher-level resistance, were seen in samples from many sites. These results suggest that, in Uganda, the antimalarial efficacy of SP for IPTp or other indications is poor, consistent with results of recent clinical studies (5, 36, 50). Furthermore, while SP plus amodiaquine appears to be efficacious for malaria chemoprevention in areas where the pfdhps 540E mutation is absent (4), this regimen is unlikely to be effective in Uganda. Other regimens for chemoprevention, in particular the ACT dihydroartemisinin-piperaquine, are under study for intermittent preventive therapy (IPT) in pregnancy (50, 51) and in children (3, 52). Our data suggest continued good antimalarial activity of dihydroartemisinin-piperaquine in Uganda, although loss of activity of this regimen in Cambodia (29, 30) is concerning.
Our study had important limitations. First, we studied convenience samples collected across Uganda; we cannot be sure that our results are representative of all parasites from the study areas. Second, we assessed only polymorphisms already associated with resistance to antimalarials. Additional genetic changes in P. falciparum likely impact sensitivity to various antimalarial agents. Although it is difficult to identify new resistance mediators in highly diverse clinical isolates, broader deep sequencing approaches should shed light on additional polymorphisms contributing to drug sensitivity. Third, for some uncommon markers we only evaluated the most recent available samples, so were unable to characterize temporal trends.
In summary, in studies from a range of sites in Uganda, we identified consistent changes in P. falciparum genetic markers associated with drug sensitivity over time. Importantly, markers associated with resistance to artemisinins or key ACT partner drugs were not seen. Markers indicating resistance to antifolates had continued high prevalence. These findings suggest that continued use of leading ACTs to treat malaria in Uganda is warranted, but that continued surveillance for markers associated elsewhere with ACT resistance is a high priority.
MATERIALS AND METHODS
We selected 10 sites to represent different regions of Uganda with varied malaria transmission intensity and epidemiology (Fig. 1). At each site, as part of routine care, health care personnel evaluated children 6 months to 10 years of age with clinical syndromes suggestive of malaria using either Giemsa-stained blood smears or histidine-rich protein 2 (HRP2)-based rapid diagnostic tests, following national guidelines and depending on local availability of these tests. Consecutive children diagnosed with malaria and their parents or guardians were approached for enrollment, and, if consent was obtained, blood was collected as 4 blood spots dried on Whatman 3MM filter paper. Filter paper samples were stored in zipper storage bags with desiccant at room temperature and transported to our laboratory in Kampala for evaluation. This study was approved by the Makerere University Research and Ethics Committee, the Uganda National Council of Science and Technology, and by the University of California, San Francisco Committee on Human Research.
Genomic DNA was extracted from blood spots using Chelex 100, as previously described (53). Pfmdr1, pfcrt, pfdhfr, and pfdhps polymorphisms of interest were characterized by PCR and ligase detection reaction-fluorescent microsphere assays, as previously described (54), with minor modification to incorporate nested PCR (48). Copy number variations for pfmdr1 and plasmepsin2 were assessed by quantitative PCR (qPCR), and the K13 propeller domain was amplified and sequenced, all as previously described (10).
Data availability.
Nucleotide sequences are available in the GenBank database under the accession numbers MH788997 to MH789408.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by funding from the National Institutes of Health (grants AI089674 and AI075045).
We thank the study participants, their parents and guardians, and the clinical staffs of the health care facilities for their cooperation.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01818-18.
REFERENCES
- 1.Rosenthal PJ. 2013. The interplay between drug resistance and fitness in malaria parasites. Mol Microbiol 89:1025–1038. doi: 10.1111/mmi.12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yeka A, Gasasira A, Mpimbaza A, Achan J, Nankabirwa J, Nsobya S, Staedke SG, Donnelly MJ, Wabwire-Mangen F, Talisuna A, Dorsey G, Kamya MR, Rosenthal PJ. 2012. Malaria in Uganda: challenges to control on the long road to elimination: I. Epidemiology and current control efforts. Acta Trop 121:184–195. doi: 10.1016/j.actatropica.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nankabirwa JI, Wandera B, Amuge P, Kiwanuka N, Dorsey G, Rosenthal PJ, Brooker SJ, Staedke SG, Kamya MR. 2014. Impact of intermittent preventive treatment with dihydroartemisinin-piperaquine on malaria in Ugandan schoolchildren: a randomized, placebo-controlled trial. Clin Infect Dis 58:1404–1412. doi: 10.1093/cid/ciu150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cissé B, Ba EH, Sokhna C, NDiaye JL, Gomis JF, Dial Y, Pitt C, NDiaye M, Cairns M, Faye E, NDiaye M, Lo A, Tine R, Faye S, Faye B, Sy O, Konate L, Kouevijdin E, Flach C, Faye O, Trape J-F, Sutherland C, Fall FB, Thior PM, Faye OK, Greenwood B, Gaye O, Milligan P. 2016. Effectiveness of seasonal malaria chemoprevention in children under ten years of age in Senegal: a stepped-wedge cluster-randomised trial. PLoS Med 13:e1002175. doi: 10.1371/journal.pmed.1002175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desai M, Hill J, Fernandes S, Walker P, Pell C, Gutman J, Kayentao K, Gonzalez R, Webster J, Greenwood B, Cot M, ter Kuile FO. 2018. Prevention of malaria in pregnancy. Lancet Infect Dis 18:e119. doi: 10.1016/S1473-3099(18)30064-1. [DOI] [PubMed] [Google Scholar]
- 6.Blasco B, Leroy D, Fidock DA. 2017. Antimalarial drug resistance: linking Plasmodium falciparum parasite biology to the clinic. Nat Med 23:917. doi: 10.1038/nm.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mbogo GW, Nankoberanyi S, Tukwasibwe S, Baliraine FN, Nsobya SL, Conrad MD, Arinaitwe E, Kamya M, Tappero J, Staedke SG, Dorsey G, Greenhouse B, Rosenthal PJ. 2014. Temporal changes in prevalence of molecular markers mediating antimalarial drug resistance in a high malaria transmission setting in Uganda. Am J Trop Med Hyg:54–56. doi: 10.4269/ajtmh.13-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nsobya SL, Dokomajilar C, Joloba M, Dorsey G, Rosenthal PJ. 2007. Resistance-mediating Plasmodium falciparum pfcrt and pfmdr1 alleles after treatment with artesunate-amodiaquine in Uganda. Antimicrob Agents Chemother 51:3023–3025. doi: 10.1128/AAC.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humphreys G, Merinopoulos I, Ahmed J, Whitty C, Mutabingwa T, Sutherland C, Hallett R. 2007. Amodiaquine and artemether-lumefantrine select distinct alleles of the Plasmodium falciparum mdr1 gene in Tanzanian children treated for uncomplicated malaria. Antimicrob Agents Chemother 51:991–997. doi: 10.1128/AAC.00875-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rasmussen SA, Ceja FG, Conrad MD, Tumwebaze PK, Byaruhanga O, Katairo T, Nsobya SL, Rosenthal PJ, Cooper RA. 2017. Changing antimalarial drug sensitivities in Uganda. Antimicrob Agents Chemother 61:e01516-17. doi: 10.1128/AAC.01516-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sisowath C, Strömberg J, Mårtensson A, Msellem M, Obondo C, Björkman A, Gil JP. 2005. In vivo selection of Plasmodium falciparum pfmdr1 86N coding alleles by artemether-lumefantrine (Coartem). J Infect Dis 191:1014–1017. doi: 10.1086/427997. [DOI] [PubMed] [Google Scholar]
- 12.Zongo I, Dorsey G, Rouamba N, Tinto H, Dokomajilar C, Guiguemde RT, Rosenthal PJ, Ouedraogo JB. 2007. Artemether-lumefantrine versus amodiaquine plus sulfadoxine-pyrimethamine for uncomplicated falciparum malaria in Burkina Faso: a randomised non-inferiority trial. Lancet 369:491–498. doi: 10.1016/S0140-6736(07)60236-0. [DOI] [PubMed] [Google Scholar]
- 13.Baliraine FN, Rosenthal PJ. 2011. Prolonged selection of pfmdr1 polymorphisms after treatment of falciparum malaria with artemether-lumefantrine in Uganda. J Infect Dis 204:1120–1124. doi: 10.1093/infdis/jir486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Venkatesan M, Gadalla NB, Stepniewska K, Dahal P, Nsanzabana C, Moriera C, Price RN, Mårtensson A, Rosenthal PJ, Dorsey G, Sutherland CJ, Guérin P, Davis TME, Ménard D, Adam I, Ademowo G, Arze C, Baliraine FN, Berens-Riha N, Björkman A, Borrmann S, Checchi F, Desai M, Dhorda M, Djimdé AA, El-Sayed BB, Eshetu T, Eyase F, Falade C, Faucher JF, Fröberg G, Grivoyannis A, Hamour S, Houzé S, Johnson J, Kamugisha E, Kariuki S, Kiechel JR, Kironde F, Kofoed PE, LeBras J, Malmberg M, Mwai L, Ngasala B, Nosten F, Nsobya SL, Nzila A, Oguike M, Otienoburu SD, Ogutu B, Ouédraogo JB, Piola P, Rombo L, Schramm B, Somé AF, Thwing J, Ursing J, Wong RPM, Zeynudin A, Zongo I, Plowe CV, Sibley CH. 2014. Polymorphisms in Plasmodium falciparum chloroquine resistance transporter and multidrug resistance 1 genes: parasite risk factors that affect treatment outcomes for P. falciparum malaria after artemether-lumefantrine and artesunate-amodiaquine. Am. J Trop Med Hyg 91:833–843. doi: 10.4269/ajtmh.14-0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NPJ, Lindegardh N, Socheat D, White NJ. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, Lim P, Mao S, Sopha C, Sam B, Anderson JM, Duong S, Chuor CM, Taylor WRJ, Suon S, Mercereau-Puijalon O, Fairhurst RM, Menard D. 2013. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis 13:1043–1049. doi: 10.1016/S1473-3099(13)70252-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han KT, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz AB, Ghose A, Hossain MA, Samad R, Rahman MR, Hasan MM, Islam A, Miotto O, Amato R, MacInnis B, Stalker J, Kwiatkowski DP, Bozdech Z, Jeeyapant A, Cheah PY, Sakulthaew T, Chalk J, Intharabut B, Silamut K, Lee SJ, Vihokhern B, Kunasol C, Imwong M, Tarning J, Taylor WJ, Yeung S, Woodrow CJ, Flegg JA, Das D, Smith J, Venkatesan M, Plowe CV, Stepniewska K, Guerin JP, Dondorp AM, Day NP, White NJ. 2014. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371:411–423. doi: 10.1056/NEJMoa1314981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imwong M, Suwannasin K, Kunasol C, Sutawong K, Mayxay M, Rekol H, Smithuis FM, Hlaing TM, Tun KM, van der Pluijm RW, Tripura R, Miotto O, Menard D, Dhorda M, Day NPJ, White NJ, Dondorp AM. 2017. The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong subregion: a molecular epidemiology observational study. Lancet Infect Dis 17:491–497. doi: 10.1016/S1473-3099(17)30048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois A-C, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Ménard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Bras JL, Berry A, Barale CJ, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Ménard M. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.WWARN K13 Genotype-Phenotype Study Group. Association of mutations in the Plasmodium falciparum Kelch13 gene (Pf3D7_1343700) with parasite clearance rates after artemisinin-based treatments—a WWARN individual patient data meta-analysis. BMC Infect Dis, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeka A, Kigozi R, Conrad MD, Lugemwa M, Okui P, Katureebe C, Belay K, Kapella BK, Chang MA, Kamya MR, Staedke SG, Dorsey G, Rosenthal PJ. 2016. Artesunate/amodiaquine versus artemether/lumefantrine for the treatment of uncomplicated malaria in Uganda: a randomized trial. J Infect Dis 213:1134–1142. doi: 10.1093/infdis/jiv551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cooper RA, Conrad MD, Watson QD, Huezo SJ, Ninsiima H, Tumwebaze P, Nsobya SL, Rosenthal PJ. 2015. Lack of artemisinin resistance in Plasmodium falciparum in Uganda based on parasitological and molecular assays. Antimicrob Agents Chemother 59:5061–5064. doi: 10.1128/AAC.00921-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conrad MD, Bigira V, Kapisi J, Muhindo M, Kamya MR, Havlir DV, Dorsey G, Rosenthal PJ. 2014. Polymorphisms in K13 and falcipain-2 associated with artemisinin resistance are not prevalent in Plasmodium falciparum isolated from Ugandan children. PLoS One 9:e105690. doi: 10.1371/journal.pone.0105690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor SM, Parobek CM, Deconti DK, Kayentao K, Coulibaly SO, Greenwood BM, Tagbor H, Williams J, Bojang K, Njie F, Desai M, Kariuki S, Gutman J, Mathanga DP, Mårtensson A, Ngasala B, Conrad MD, Rosenthal PJ, Tshefu AK, Moormann AM, Vulule JM, Doumbo OK, Ter Kuile FO, Meshnick SR, Bailey JA, Juliano JJ. 2015. Absence of putative artemisinin resistance mutations among Plasmodium falciparum in Sub-Saharan Africa: a molecular epidemiologic study. J Infect Dis 211:680–688. doi: 10.1093/infdis/jiu467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Talundzic E, Ndiaye YD, Deme AB, Olsen C, Patel DS, Biliya S, Daniels R, Vannberg FO, Volkman SK, Udhayakumar VJA, Ndiaye D. 2017. The molecular epidemiology of Plasmodium falciparum kelch13 mutations in Senegal using targeted amplicon deep sequencing. Antimicrob Agents Chemother 61:e02116-16. doi: 10.1128/AAC.02116-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price RN, Uhlemann A-C, Brockman A, McGready R, Ashley E, Phaipun L, Patel R, Laing K, Looareesuwan S, White NJ, Nosten F, Krishna S. 2004. Mefloquine resistance in Plasmodium falciparum and increased Pfmdr1 gene copy number. Lancet 364:438–447. doi: 10.1016/S0140-6736(04)16767-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Amato R, Lim P, Miotto O, Amaratunga C, Dek D, Pearson RD, Almagro-Garcia J, Neal AT, Sreng S, Suon S, Drury E, Jyothi D, Stalker J, Kwiatkowski DP, Fairhurst RM. 2017. Genetic markers associated with dihydroartemisinin–piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype–phenotype association study. Lancet Infect Dis 17:164–173. doi: 10.1016/S1473-3099(16)30409-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Witkowski B, Duru V, Khim N, Ross LS, Saintpierre B, Beghain J, Chy S, Kim S, Ke S, Kloeung N, Eam R, Khean C, Ken M, Loch K, Bouillon A, Domergue A, Ma L, Bouchier C, Leang R, Huy R, Nuel G, Barale CJ, Legrand E, Ringwald P, Fidock DA, Mercereau-Puijalon O, Ariey F, Ménard D. 2017. A surrogate marker of piperaquine-resistant Plasmodium falciparum malaria: a phenotype–genotype association study. Lancet Infect Dis 17:174–183. doi: 10.1016/S1473-3099(16)30415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spring MD, Lin JT, Manning JE, Vanachayangkul P, Somethy S, Bun R, Se Y, Chann S, Ittiverakul M, Sia-ngam P, Kuntawunginn W, Arsanok M, Buathong N, Chaorattanakawee S, Gosi S, Ta-aksorn W, Chanarat N, Sundrakes S, Kong N, Heng TK, Nou S, Teja-isavadharm P, Pichyangkul S, Phann ST, Balasubramanian S, Juliano SJ, Meshnick SR, Chour CM, Prom S, Lanteri CA, Lon C, Saunders DL. 2015. Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: an observational cohort study. Lancet Infect Dis 15:683–691. doi: 10.1016/S1473-3099(15)70049-6. [DOI] [PubMed] [Google Scholar]
- 30.Amaratunga C, Lim P, Suon S, Sreng S, Mao S, Sopha C, Sam B, Dek D, Try V, Amato R, Blessborn D, Song L, Tullo GS, Fay MP, Anderson JM, Tarning J, Fairhurst RM. 2016. Dihydroartemisinin–piperaquine resistance in Plasmodium falciparum malaria in Cambodia: a multisite prospective cohort study. Lancet Infect Dis 16:357–365. doi: 10.1016/S1473-3099(15)00487-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gregson A, Plowe CV. 2005. Mechanisms of resistance of malaria parasites to antifolates. Pharmacol Rev 57:117–145. doi: 10.1124/pr.57.1.4. [DOI] [PubMed] [Google Scholar]
- 32.Naidoo I, Roper C. 2013. Mapping ‘partially resistant’, ‘fully resistant’, and ‘super resistant’ malaria. Trends Parasitol 29:505–515. doi: 10.1016/j.pt.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Pearce RJ, Pota H, Evehe M-SB, Bâ E-H, Mombo-Ngoma G, Malisa AL, Ord R, Inojosa W, Matondo A, Diallo DAJ, Mbacham W, Van den Broek I, Swarthout TD, Getachew A, Dejene S, Grobusch MP, Njie F, Dunyo S, Kweku M, Owusu-Agyei S, Chandramohan D, Bonnet M, Guthmann JP, Clarke S, Barnes KI, Streat E, Katokele ST, Uusiku P, Agboghoroma CO, Elegba OY, Cissé B, A-Elbasit IE, Giha HA, Kachur SP, Lynch C, Rwakimari JB, Chanda P, Hawela M, Sharp B, Naidoo I, Roper C. 2009. Multiple origins and regional dispersal of resistant dhps in African Plasmodium falciparum malaria. PLoS Med 6:e1000055. doi: 10.1371/journal.pmed.1000055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lynch C, Pearce R, Pota H, Cox J, Abeku TA, Rwakimari J, Naidoo I, Tibenderana J, Roper C. 2008. Emergence of a dhfr mutation conferring high-level drug resistance in Plasmodium falciparum populations from southwest Uganda. J Infect Dis 197:1598–1604. doi: 10.1086/587845. [DOI] [PubMed] [Google Scholar]
- 35.Tumwebaze P, Tukwasibwe S, Taylor A, Conrad M, Ruhamyankaka E, Asua V, Walakira A, Nankabirwa J, Yeka A, Staedke SG, Greenhouse B, Nsobya SL, Kamya MR, Dorsey G, Rosenthal PJ. 2016. Changing antimalarial drug resistance patterns identified by surveillance at three sites in Uganda. J Infect Dis 215:631–635. doi: 10.1093/infdis/jiw614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Braun V, Rempis E, Schnack A, Decker S, Rubaihayo J, Tumwesigye NM, Theuring S, Harms G, Busingye P, Mockenhaupt FPJM. 2015. Lack of effect of intermittent preventive treatment for malaria in pregnancy and intense drug resistance in western Uganda. Malar J 14:372. doi: 10.1186/s12936-015-0909-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gesase S, Gosling RD, Hashim R, Ord R, Naidoo I, Madebe R, Mosha JF, Joho A, Mandia V, Mrema H, Mapunda E, Savael Z, Lemnge M, Mosha FW, Greenwood B, Roper C, Chandramohan D. 2009. High resistance of Plasmodium falciparum to sulphadoxine/pyrimethamine in northern Tanzania and the emergence of dhps resistance mutation at Codon 581. PLoS One 4:e4569. doi: 10.1371/journal.pone.0004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kublin JG, Cortese JF, Njunju EM, Mukadam RAG, Wirima JJ, Kazembe PN, Djimdé AA, Kouriba B, Taylor TE, Plowe CV. 2003. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect DIS 187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- 39.Laufer MK, Hesing PC, Eddington ND, Masonga R, Dzinjalamala FK, Takala SL, Taylor TE, Plowe CV. 2006. Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med 355:1959–1966. doi: 10.1056/NEJMoa062032. [DOI] [PubMed] [Google Scholar]
- 40.Bukirwa H, Yeka A, Kamya MR, Talisuna A, Banek K, Bakyaita N, Rwakimari JB, Rosenthal PJ, Wabwire-Mangen F, Dorsey G, Staedke SG. 2006. Artemisinin combination therapies for treatment of uncomplicated malaria in Uganda. PLoS Clin Trial 1:e7. doi: 10.1371/journal.pctr.0010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eyase FL, Akala HM, Ingasia L, Cheruiyot A, Omondi A, Okudo C, Juma D, Yeda R, Andagalu B, Wanja E, Kamau E, Schnabel D, Bulimo W, Waters NC, Walsh DS, Johnson JD. 2013. The role of pfmdr1 and pfcrt in changing chloroquine, amodiaquine, mefloquine and lumefantrine susceptibility in western-Kenya P. falciparum samples during 2008–2011. PLoS One 8:e64299. doi: 10.1371/journal.pone.0064299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Malmberg M, Ngasala B, Ferreira PE, Larsson E, Jovel I, Hjalmarsson A, Petzold M, Premji Z, Gil JP, Björkman A, Martensson A. 2013. Temporal trends of molecular markers associated with artemether-lumefantrine tolerance/resistance in Bagamoyo district, Tanzania. Malar J 12:103. doi: 10.1186/1475-2875-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.WWARN Artemisinin Based Combination Therapy (ACT) Africa Baseline Study Group, Dahal P, d’Alessandro U, Dorsey G, Guerin PJ, Nsanzabana C, Price RN, Sibley CH, Stepniewska K, Talisuna AO. 2015. Clinical determinants of early parasitological response to ACTs in African patients with uncomplicated falciparum malaria: a literature review and meta-analysis of individual patient data. BMC Med 13:212. doi: 10.1186/s12916-015-0445-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tacoli C, Gai PP, Bayingana C, Sifft K, Geus D, Ndoli J, Sendegeya A, Gahutu JB, Mockenhaupt FP. 2016. Artemisinin resistance-associated K13 polymorphisms of Plasmodium falciparum in southern Rwanda, 2010–2015. Am J Trop Med Hyg 95:1090–1093. doi: 10.4269/ajtmh.16-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ikeda M, Kaneko M, Tachibana S-I, Balikagala B, Sakurai-Yatsushiro M, Yatsushiro S, Takahashi N, Yamauchi M, Sekihara M, Hashimoto M, Katuro OT, Olia A, Obwoya PS, Auma MA, Anywar DA, Odongo-Aginya EI, Okello-Onen J, Hirai M, Ohashi J, Palacpac NMQ, Kataoka M, Tsuboi T, Kimura E, Horii T, Mita T. 2018. Artemisinin-resistant Plasmodium falciparum with high survival rates, Uganda, 2014–2016. Emerg Infect Dis 24:718. doi: 10.3201/eid2404.170141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Price RN, Uhlemann A-C, van Vugt M, Brockman A, Hutagalung R, Nair S, Nash D, Singhasivanon P, Anderson TJ, Krishna S, White NJ, Nosten F. 2006. Molecular and pharmacological determinants of the therapeutic response to artemether-lumefantrine in multidrug-resistant Plasmodium falciparum malaria. Clin Infect Dis 42:1570–1577. doi: 10.1086/503423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dokomajilar C, Nsobya SL, Greenhouse B, Rosenthal PJ, Dorsey G. 2006. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob Agents Chemother 50:1893–1895. doi: 10.1128/AAC.50.5.1893-1895.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conrad MD, LeClair N, Arinaitwe E, Wanzira H, Kakuru A, Bigira V, Muhindo M, Kamya MR, Tappero JW, Greenhouse B, Dorsey G, Rosenthal PJ. 2014. Comparative impacts over 5 years of artemisinin-based combination therapies on Plasmodium falciparum polymorphisms that modulate drug sensitivity in Ugandan children. J Infect Dis 210:344–353. doi: 10.1093/infdis/jiu141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sagara I, Beavogui AH, Zongo I, Soulama I, Borghini-Fuhrer I, Fofana B, Traore A, Diallo N, Diakite H, Togo AH, Koumare S, Keita M, Camara D, Somé AF, Coulibaly AS, Traore OB, Dama S, Goita S, Djimde M, Bamadio A, Dara N, Maiga H, Sidibe B, Dao F, Coulibaly M, Alhousseini ML, Niangaly H, Sangare B, Diarra M, Coumare S, Kabore MJT, Ouattara SM, Barry A, Kargougou D, Diarra A, Henry N, Soré H, Bougouma EC, Thera I, Compaore YD, Sutherland CJ, Sylla MM, Nikiema F, Diallo MS, Dicko A, Picot S, Borrmann S, Duparc S, Miller RM, Doumbo OK, Shin J, Gil PJ, Björkman A, Ouedraogo JB, Sirima SB, Djimde AA. 2018. Pyronaridine-artesunate or dihydroartemisinin-piperaquine versus current first-line therapies for repeated treatment of uncomplicated malaria: a randomised, multicentre, open-label, longitudinal, controlled, phase 3b/4 trial. Lancet 391:1378–1390. doi: 10.1016/S0140-6736(18)30291-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Desai M, Gutman J, L’lanziva A, Otieno K, Juma E, Kariuki S, Ouma P, Were V, Laserson K, Katana A, Williamson J, ter Kuile FO. 2015. Intermittent screening and treatment or intermittent preventive treatment with dihydroartemisinin–piperaquine versus intermittent preventive treatment with sulfadoxine–pyrimethamine for the control of malaria during pregnancy in western Kenya: an open-label, three-group, randomised controlled superiority trial. Lancet 386:2507–2519. doi: 10.1016/S0140-6736(15)00310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kakuru A, Jagannathan P, Muhindo MK, Natureeba P, Awori P, Nakalembe M, Opira B, Olwoch P, Ategeka J, Nayebare P, Clark TD, Feeney ME, Charlebois ED, Rizzuto G, Muehlenbachs A, Havlir DV, Kamya MR, Dorsey G. 2016. Dihydroartemisinin-piperaquine for the prevention of malaria in pregnancy. N Engl J Med 374:928–939. doi: 10.1056/NEJMoa1509150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bigira V, Kapisi J, Clark TD, Kinara S, Mwangwa F, Muhindo MK, Osterbauer B, Aweeka FT, Huang L, Achan J, Havlir DV, Rosenthal PJ, Kamya MR, Dorsey G. 2014. Protective efficacy and safety of three antimalarial regimens for the prevention of malaria in young Ugandan children: a randomized controlled trial. PLoS Med 11:e1001689. doi: 10.1371/journal.pmed.1001689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Asua V, Tukwasibwe S, Conrad M, Walakira A, Nankabirwa JI, Mugenyi L, Kamya MR, Nsobya SL, Rosenthal PJ. 2017. Plasmodium species infecting children presenting with malaria in Uganda. Am J Trop Med Hyg 97:753–757. doi: 10.4269/ajtmh.17-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.LeClair NP, Conrad MD, Baliraine FN, Nsanzabana C, Nsobya SL, Rosenthal PJ. 2013. Optimization of a ligase detection reaction-fluorescent microsphere assay for characterization of resistance-mediating polymorphisms in African samples of Plasmodium falciparum. J Clin Microbiol 51:2564–2570. doi: 10.1128/JCM.00904-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Nucleotide sequences are available in the GenBank database under the accession numbers MH788997 to MH789408.