Murepavadin (POL7080) represents the first member of a novel class of outer membrane protein-targeting antibiotics. It specifically interacts with LptD and inhibits lipopolysaccharide (LPS) transport.
KEYWORDS: murepavadin, pharmacodynamics, pharmacokinetics
ABSTRACT
Murepavadin (POL7080) represents the first member of a novel class of outer membrane protein-targeting antibiotics. It specifically interacts with LptD and inhibits lipopolysaccharide (LPS) transport. Murepavadin is being developed for the treatment of serious infections by Pseudomonas aeruginosa. We determined the plasma protein binding and the pharmacokinetics of murepavadin in plasma and epithelial lining fluid (ELF; pulmonary) in infected animals, and we determined the exposure-response relationship. Treatment of CD-1 neutropenic mice was started 2 h after infection using murepavadin at different dosing frequencies for 24 h, and the number of CFU per lung was determined. The sigmoid maximum-effect model was used to fit the dose-response, and the pharmacodynamic index (PDI) response was used to determine the PDI values, resulting in a static effect and 1-log kill reduction. Using R2 as an indicator of the best fit, the area under the concentration-time curve for the unbound fraction of the drug (fAUC)/MIC ratio correlated best with efficacy. The mean AUC required to provide a static effect was 36.83 mg h/liter (fAUC = 8.25 mg h/liter), and that to provide a 1-log reduction was 44.0 mg h/liter (fAUC = 9.86 mg h/liter). The mean static fAUC/MIC was determined to be 27.78, and that for a 1-log reduction was 39.85. These data may serve to determine doses in humans that are likely to be efficacious.
INTRODUCTION
Hospital-acquired bacterial pneumonia (HABP) and ventilator-associated bacterial pneumonia (VABP) are some of the most common nosocomial infections and leading causes of death despite improvements in both prevention and supportive care (1, 2). Moreover, when the infection is due to Pseudomonas aeruginosa, it is often associated with significant morbidity and increased costs and mortality (3). P. aeruginosa is one of the most common causes of VABP, with an occurrence of approximately 25% (4, 5). When an inappropriate initial antibiotic therapy (IIAT) is prescribed, mortality increases, and this risk is further enhanced due to the presence of multidrug-resistant (MDR) pathogens. Therefore, an increase in the frequency of MDR pathogens results in increased mortality in these patients (6–9). Progressively more isolates of P. aeruginosa have become resistant to many antibiotics, and the incidence of MDR P. aeruginosa infections is very concerning in the intensive care unit (ICU) (10). It is estimated that about 30% of the P. aeruginosa strains isolated from patients with HABP and VABP are MDR (4). Thus, additional options are needed for the treatment of HABP/VABP due to P. aeruginosa (11).
Murepavadin is the first outer membrane protein-targeting antibiotic (OMPTA) with a novel mode of action being developed (12, 13). The molecule targets the lipopolysaccharide transport protein D (LptD), an outer membrane protein involved in lipopolysaccharide biogenesis in Gram-negative bacteria (14, 15). The compound demonstrates selective and potent bactericidal antimicrobial activity against P. aeruginosa in vitro. Moreover, when evaluated against over 1,200 P. aeruginosa isolates from the United States, Europe, and China, or more recently against a panel of 785 extensive drug-resistant (XDR) isolates, the MIC90 was 0.12 to 0.25 mg/liter (16, 17).
An important issue that needs attention is the site of infection, in particular, the lung. Several compounds developed recently have failed in patients with ventilator-associated pneumonia (18, 19). It is not clear what the exact reasons for failure of treatment were, but relatively low concentrations at the site of infection are thought to be one of the possibilities. It is therefore critical to determine the effect of murepavadin on P. aeruginosa in a pulmonary model of infection, including the concentration-effect relationships in epithelial lining fluid (ELF). Given this, the goal of our experiments was to characterize the in vivo pharmacokinetic/pharmacodynamic (PK/PD) properties of murepavadin in plasma and in ELF and the magnitude of the PK/PD index required for efficacy against P. aeruginosa in the murine lung infection model. Since it is unclear whether inflammation in the lung affects the PK/PD properties in ELF, all PK analyses were performed in infected animals. The murine thigh model was also employed to confirm the pharmacodynamic driver in dose-fractionation studies.
RESULTS
In vitro susceptibility studies.
The MICs to murepavadin of the strains ranged from 0.125 to 1.0 mg/liter (Table 1). The majority (11/15) of the isolates tested were MDR/XDR to known antipseudomonal antibiotics, all of which were nonsusceptible to carbapenems, and 2 which were only susceptible to polymyxins, according to EUCAST breakpoints (20).
TABLE 1.
MIC characterization of the P. aeruginosa isolates used in the experiments
| Isolate IDa | MIC (mg/liter) forb
: |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Murepavadin | CST | ATM | CAZ | FEP | IPM | MEM | DOR | TZP | CIP | LVX | GEN | TOB | AKN | |
| ATCC 27853 | 0.125 | 1 | 4 | 2 | 2 | 2 | 0.5 | 0.25 | 2 | 0.5 | 0.5 | 1 | 0.5 | 2 |
| ATCC BAA 2113 | 0.125 | 2 | 16 | 4 | 4 | 0.5 | 0.5 | 0.25 | 16 | 0.12 | 0.5 | 1 | 0.5 | 2 |
| NCTC 13437 | 0.25 | 1 | >16 | >32 | >16 | >8 | 32 | >4 | 32 | >4 | >4 | >8 | >8 | 32 |
| 5 | 1 | 2 | >16 | >32 | 16 | >8 | 8 | >4 | >64 | 2 | 4 | 8 | 2 | 32 |
| 9 | 0.25 | 1 | 4 | 1 | 4 | 0.5 | 0.25 | 0.25 | 4 | 0.25 | 0.5 | 2 | 0.5 | 4 |
| 6 | 0.5 | 2 | 8 | 8 | 4 | 8 | 8 | 4 | 32 | 0.12 | 0.5 | 2 | 0.5 | 4 |
| 11 | 0.5 | 2 | >16 | >32 | 16 | >8 | 16 | >4 | >64 | >4 | >4 | >8 | >8 | 16 |
| 12 | 0.25 | 1 | 16 | 16 | 8 | 8 | 4 | 4 | 64 | >4 | >4 | >8 | >8 | 4 |
| 15 | 0.25 | 1 | >16 | 32 | 8 | 8 | 4 | 4 | >64 | >4 | >4 | >8 | >8 | 4 |
| 16 | 0.25 | 1 | 16 | 4 | 4 | 8 | 8 | >4 | 32 | >4 | >4 | ≤0.5 | ≤0.12 | 0.5 |
| 18 | 0.25 | 1 | >16 | 32 | 16 | 8 | 4 | 4 | >64 | >4 | >4 | 2 | 0.5 | 4 |
| 19 | 0.25 | 1 | >16 | 32 | 16 | 8 | 4 | 4 | 64 | >4 | >4 | 2 | 0.5 | 4 |
| 21 | 0.25 | 1 | 8 | >32 | 16 | 8 | 4 | 2 | 32 | >4 | >4 | >8 | >8 | >32 |
| 22 | 0.25 | 1 | 8 | 32 | 16 | 4 | 4 | 4 | 16 | >4 | >4 | >8 | >8 | >32 |
| X11045 | 0.25 | 2 | 2 | 1 | 2 | 2 | 1 | 0.25 | 2 | 0.25 | 0.5 | 4 | 1 | 16 |
ID, identifier.
CST, colistin; ATM, aztreonam; CAZ, ceftazidime; FEP, cefepime; IPM, imipenem; MEM, meropenem; DOR, doripenem; TZP, piperacillin-tazobactam; CIP, ciprofloxacin; LVX, levofloxacin; GEN, gentamicin; TOB, tobramycin; AKN, amikacin.
Plasma protein binding.
Plasma protein binding (PPB) was determined in two external labs, giving similar results. Over the entire concentration range tested and in all plasma samples from the different species, no concentration dependence of PPB was observed. The %fu (percent fraction unbound; mean ± standard deviation [SD]) for mice was determined to be 22.6% ± 5.95% based on 82 observations, and that from humans was 22.4% ± 1.96% based on 84 observations. For all the species tested, the mean %fu was determined to be 22.4% ± 6.4% based on 256 observations. Thus, for the calculation of exposures to unbound compound (maximum concentration in serum of the free fraction of drug [fCmax] and area under the concentration-time curve for the unbound fraction of the drug [fAUC]), a single factor of 22.4% was used across all tested species over the concentration range of 0.1 to 20.0 mg/liter.
Pharmacokinetics.
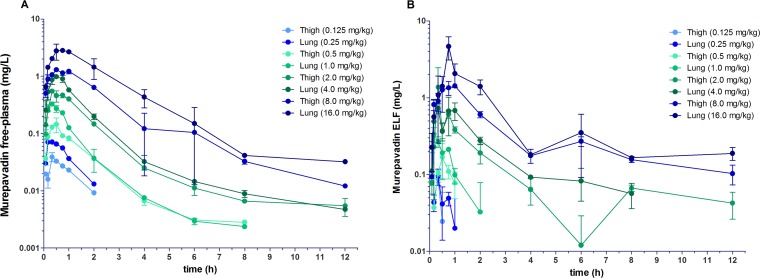
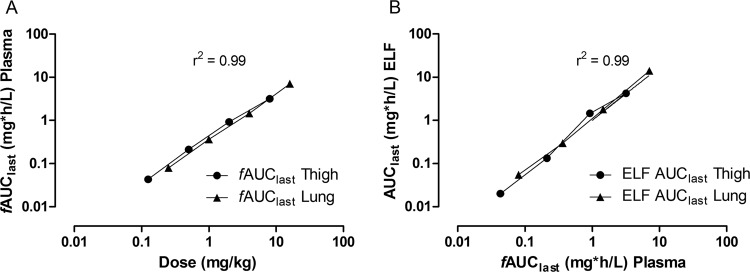
The time-concentration data of murepavadin in mice after subcutaneous doses are shown in Fig. 1. The peak plasma levels ranged from 0.175 to 12.6 mg/liter, and the AUC for the period from t = 0 to the last quantifiable concentration level (AUC0–last) values ranged from 0.186 to 30.0 mg h/liter (Table 2). At low-dose ranges (0.125 to 0.25 mg/kg of body weight), plasma levels were no longer quantifiable after 2 h, whereas at the medium ranges (0.5 to 1 mg/kg), the last measurable plasma level was at 8 h postdose. Time to Cmax (Tmax) parameters were consistently observed in the 20- to 45-min postadministration period, with Tmax rising with increasing dose. The AUCs were determined for each dose level and plotted as log fAUC versus log dose in a dose-proportionality plot for both the lung and thigh models (Fig. 2a). The relationship was linear, and there were no significant differences in pharmacokinetics between thigh and lung models (F = 0.500781, P = 0.5182 for differences in slope and F = 3.56582, P = 0.1176 for differences in intercept when analyzed using linear regression). Over the dose range, the relationship was linear (fAUClast R2 = 0.99, fCmax R2 = 0.99); thus, exposure levels may be calculated using dose levels multiplied by the respective slope factors. The overall relationship between dose and fAUC could therefore be described by the slope factor fAUC = dose (mg/kg) × 0.4078 ± 0.0087 (h mg kg/liter mg). The fCmax can be described by fCmax = dose (mg/kg) × 0.1789 ± 0.0081 (mg kg/liter mg).
FIG 1.
Single dose time-concentration profiles of eight 2-fold increasing doses of murepavadin by s.c. injection. Four doses were applied in the thigh infection model (0.125, 0.5, 2, and 8 mg/kg) and four in the lung infection model (0.25, 1, 4, and 16 mg/kg). Shown are the free murepavadin time-concentration profile in plasma (A) and the ELF time-concentration profile (B). Each symbol represents the mean ± SD of the results from two mice.
TABLE 2.
Pharmacokinetic parameter estimates for murepavadin in plasma of neutropenic mice following s.c. administration
| Pharmacokinetic parametera | Value for model at a dose (mg/kg) of: |
|||||||
|---|---|---|---|---|---|---|---|---|
| 0.125 (thigh) | 0.25 (lung) | 0.5 (thigh) | 1 (lung) | 2 (thigh) | 4 (lung) | 8 (thigh) | 16 (lung) | |
| Tmax (h) | 0.3 | 0.3 | 0.5 | 0.3 | 0.3 | 0.5 | 0.5 | 0.8 |
| Cmax (ng/ml) | 175 | 319 | 650 | 1,470 | 2,460 | 4,400 | 5,820 | 12,600 |
| Tlast (h) | 2 | 2 | 8 | 8 | 12 | 12 | 12 | 12 |
| Clast (ng/ml) | 41.2 | 58.8 | 12.6 | 10.6 | 24.5 | 21.2 | 54.2 | 144 |
| AUClast (h ng/ml) | 186 | 349 | 922 | 1,570 | 3,860 | 6,100 | 13,400 | 30,000 |
| AUCINF (h ng/ml) | 234 | 403 | 946 | 1,580 | 3,920 | 6,210 | 13,500 | 30,400 |
| CL/F (ml/h/kg) | 533 | 620 | 529 | 632 | 511 | 644 | 592 | 527 |
| Vz/F (ml/kg) | 622 | 566 | 1,000 | 947 | 1,160 | 3,520 | 1,400 | 1,230 |
| t1/2 (h) | 0.8 | 0.6 | 1.3 | 1 | 1.6 | 3.8 | 1.6 | 1.6 |
| R2 | 0.997 | 0.992 | 0.918 | 0.919 | 0.86 | 0.986 | 0.954 | 0.916 |
Tlast, time of last quantifiable concentration; Clast, last quantifiable concentration; AUCINF, AUC to infinity; CL/F, apparent total clearance of drug after oral administration; Vz/F, apparent volume of distribution during terminal phase after oral administration; t1/2, half-life.
FIG 2.
Dose-proportionality plot of murepavadin (A) and relationship of free AUC (plasma) to the ELF AUC of murepavadin (B). Eight different doses of murepavadin were administered to mice by s.c. injection, with four doses in the thigh infection model and four doses in the lung infection model. Each symbol represents the exposure per dose. The linear regression line is indicated, and R2 represents the coefficient of determination. Analysis was conducted using GraphPad Prism version 5.03 for Windows (GraphPad Software, La Jolla, CA, USA).
The profile for ELF concentrations followed that of plasma, with a slight time delay, in that concentrations were initially lower but then increased relative to those in plasma over time. The peak levels ranged from 0.088 to 4.67 mg/liter, and the AUC0–last values ranged from 0.020 to 14.0 mg h/liter (Table 3). The penetration of murepavadin in ELF was estimated by comparing the AUCs and fAUC in plasma to those in ELF. The ELF AUCs for each dose level were plotted as log AUC versus log fAUC (plasma) for both the lung and thigh models (Fig. 2b). The relationship was linear, and there were no significant differences in pharmacokinetics between the thigh and lung models (F = 0.212514, P = 0.6688 for differences in slope and F = 0.0841804, P = 0.7834 for differences in intercept). The ELF/plasma AUC ratio indicates that murepavadin penetrates ELF well (Table 3). Over the dose range, the relationship was linear (AUClast R2 =0.99, Cmax R2 = 0.99); thus, exposure levels may be calculated using dose levels multiplied by the respective slope factors. The overall relationship between fAUC (plasma) and ELF AUC could therefore be described by the slope factor AUC = fAUC (plasma) × 1.240 ± 0.045 (h mg kg/liter mg). The Cmax can be described by Cmax = fCmax (plasma) × 0.929 ± 0.109 (mg/liter). The concentration of drug in the ELF was thus approximately equal to the free fraction in plasma.
TABLE 3.
Penetration of murepavadin in ELF
| Dose (mg/kg) | Values by sample type |
|||||
|---|---|---|---|---|---|---|
| ELF |
Plasma |
ELF/fplasmaa |
||||
| Cmax (mg/liter) | AUC (h mg/liter) | fCmax (mg/liter) | fAUC (h mg/liter) | Cmax (%) | AUC (%) | |
| 0.125 | 0.088 | 0.02 | 0.0392 | 0.042 | 224.5 | 48 |
| 0.25 | 0.095 | 0.055 | 0.0715 | 0.078 | 132.9 | 70.4 |
| 0.5 | 0.169 | 0.133 | 0.146 | 0.207 | 115.8 | 64.4 |
| 1 | 0.336 | 0.296 | 0.329 | 0.352 | 102.1 | 84.2 |
| 2 | 1.37 | 1.455 | 0.551 | 0.865 | 248.6 | 168.3 |
| 4 | 0.726 | 1.795 | 0.986 | 1.366 | 73.6 | 131.4 |
| 8 | 1.43 | 4.239 | 1.304 | 3.002 | 109.7 | 141.2 |
| 16 | 4.76 | 14.001 | 2.82 | 6.72 | 168.7 | 208.3 |
Values for ELF/fplasma (ratio of ELF exposure to free plasma exposure expressed as percent) Cmax (%) and AUC (%) were geometric mean, 136.3 and 102.2; mean, 147.0 and 114.5; and SD, 31.4 and 56.7, respectively.
Determination of the pharmacodynamic index linked to efficacy.
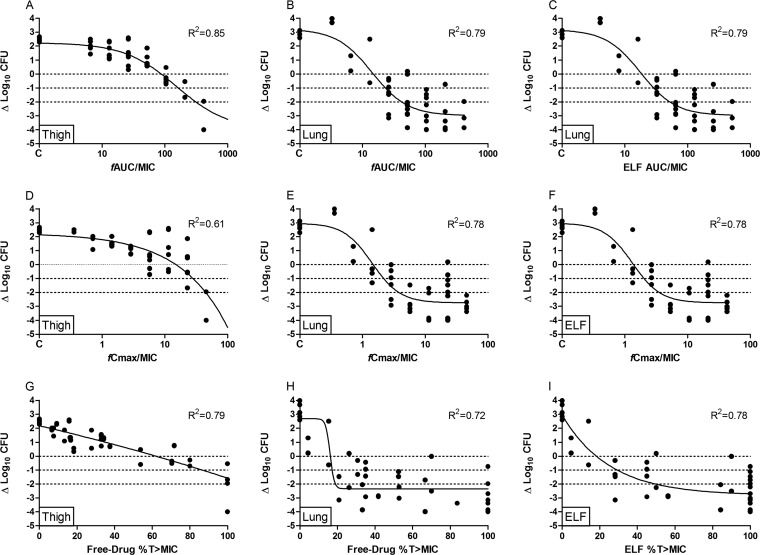
Dose-fractionation studies were performed in both the thigh and lung models with two strains, P. aeruginosa ATCC 27853 and the multidrug-resistant (XDR) clinical isolate 18. The exposure-response curves of ATCC 27853 in the thigh and lung models are shown in Fig. 3. The dose frequency did not have a significant effect on overall efficacy. Since the compound shows linear pharmacokinetics for the total drug, this indicates that AUC is the dominant pharmacokinetic parameter driving the pharmacodynamic effect. This is further substantiated by the R2 values of the model fits that were the highest for fAUC/MIC (R2 = 0.85 and 0.79 for thigh and lung models, respectively).
FIG 3.
Relationships of murepavadin 24-h AUC/MIC (A to C), Cmax/MIC (D to F), and the percentage of time above the MIC (%T>MIC) (G to I) for P. aeruginosa ATCC 27853 in the neutropenic thigh and lung infection models with change in CFU per thigh or lung from start of treatment and after 24 h of therapy. Each symbol represents a therapy response in one mouse thigh or lung. The line is the best-fit line based on the sigmoid Emax model.
Determination of the magnitude of the PD target.
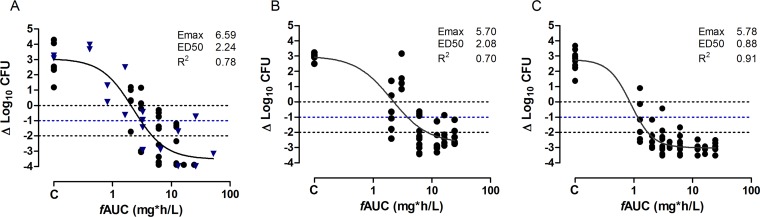
Since AUC appears to be the main driver of effect, experiments were carried out with 15 P. aeruginosa strains with a wide range of MIC values (0.125 mg/liter to 1 mg/liter) all equal to or above the MIC90 (Table 1) to estimate the fAUC to result in a static, 1-log drop, and 2-log drop effects (Table 4) in the lung model. The dose frequencies chosen were every 24 h (q24h), every 12 h (q12h), and every 6 h (q6h). For all isolates, a static effect was achieved; for 14 of 15 isolates, a 1-log drop was observed, and for 8 of 15 isolates, a 2-log drop was observed at the doses tested. The exposure-response relationships of 15 strains were well described by the sigmoid maximum effect (Emax) model, and a static and 1-log reduction effects were calculated. An example of responses is shown in Fig. 4. For 7/15 of these strains, a 2-log reduction was not observed, but this could be due to organism-specific characteristics, as it does not seem to be related to the MIC. Strain 5 only reached a static effect, and this may be due to the high MIC toward this isolate (1 mg/liter); also, since murepavadin, like many peptide drugs, is less tolerated by mice (which is attributed to a nonimmunogenic histamine release from mast cells), the maximum dose was given and did not allow higher drug doses to be investigated (13, 21). In addition, for this isolate, the homogenates of the lung were streaked on nonselective agar to determine the CFU load; the MICs of the subsequent colonies were tested, and no increase in MIC was observed, suggesting that resistance to murepavadin was not the reason for treatment failure. The mean plasma fAUC required to provide a static effect was 8.25 mg h/liter, and that to provide a 1-log reduction was 9.86 mg h/liter. If normalized to the MIC, the mean static fAUC/MIC was determined to be 27.8, and that for a 1-log reduction was 39.9 (Table 2). For the ELF, the values were determined to be 34.5 and 49.4 for the static and 1-log reduction effects, respectively.
TABLE 4.
In vivo efficacies of murepavadin against P. aeruginosa isolates in the neutropenic lung infection models
| P. aeruginosa isolate or data measure | MIC (mg/liter) | Bacterial burden at start of therapy (log10 CFU/lung) | Bacterial burden with 24 h growth in untreated control animals (log10 CFU/lung) | No. of CFU/lung for maximum kill at 24 h from 0 h (Δlog10 CFU/lung) | Values resulting in static effect |
Values resulting in 1-log reductiona
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| fAUC | fAUC/MIC | ELF AUC/MIC | fAUC | fAUC/MIC | ELF AUC/MIC | |||||
| ATCC 27853 | 0.125 | 5.62 | 2.94 | −3.98 | 2.03 | 16.25 | 20.15 | 3.09 | 24.74 | 30.67 |
| ATCC BAA2113 | 0.125 | 5.48 | 2.3 | −2.03 | 0.76 | 6.10 | 7.57 | 4.68 | 37.43 | 46.41 |
| NCTC 13437 | 0.25 | 5.25 | 2.97 | −2.98 | 2.24 | 8.94 | 11.09 | 3.63 | 14.54 | 18.03 |
| 5 | 1 | 6.39 | 3.42 | −0.25 | 25.72 | 25.72 | 31.89 | NR | NR | NR |
| 9 | 0.25 | 5.72 | 3.65 | −1.75 | 7.77 | 31.07 | 38.52 | 12.89 | 51.56 | 63.93 |
| 6 | 0.5 | 4.58 | 3.42 | −2.93 | 2.51 | 5.03 | 6.23 | 5.01 | 10.03 | 12.43 |
| 11 | 0.5 | 4.48 | 1.64 | −2.78 | 3.63 | 7.27 | 9.01 | 7.77 | 15.53 | 19.26 |
| 12 | 0.25 | 4.56 | 3.21 | −1.97 | 9.33 | 37.33 | 46.29 | 14.80 | 59.20 | 73.40 |
| 15 | 0.25 | 6.33 | 2.05 | −1.98 | 6.31 | 25.26 | 31.32 | 6.61 | 26.45 | 32.80 |
| 16 | 0.25 | 4.89 | 2.42 | −3.25 | 0.84 | 3.34 | 4.15 | 1.15 | 4.60 | 5.71 |
| 18 | 0.25 | 6.06 | 2.81 | −3.05 | 6.17 | 24.67 | 30.59 | 11.23 | 44.90 | 55.68 |
| 19 | 0.25 | 5.87 | 3.15 | −1.76 | 21.89 | 87.55 | 108.57 | 23.30 | 93.18 | 115.55 |
| 21 | 0.25 | 4.99 | 2.94 | −2.44 | 6.92 | 27.69 | 34.33 | 7.08 | 28.31 | 35.10 |
| 22 | 0.25 | 4.93 | 3.45 | −1.15 | 13.50 | 53.99 | 66.95 | 20.91 | 83.65 | 103.73 |
| X11045 | 0.25 | 4.99 | 1.32 | −1.95 | 14.13 | 56.54 | 70.10 | 15.95 | 63.81 | 79.13 |
| Geometric mean | 5.31 | 2.68 | −2.00 | 5.20 | 18.98 | 23.53 | 7.52 | 30.06 | 37.28 | |
| Mean | 5.34 | 2.78 | −2.28 | 8.25 | 27.78 | 34.45 | 9.86 | 39.85 | 49.42 | |
| Median | 5.25 | 2.94 | −2.03 | 6.31 | 25.26 | 31.32 | 7.42 | 32.87 | 40.76 | |
| SD | 0.64 | 0.69 | 0.92 | 7.58 | 23.46 | 29.08 | 6.84 | 27.44 | 34.02 | |
NR, not reported.
FIG 4.
In vivo dose response of murepavadin against three P. aeruginosa isolates in the neutropenic murine lung infection model. (A) P. aeruginosa ATCC 27853. (B) NCTC 13437. (C) Isolate 16. Each symbol represents a therapy response in one mouse lung. The x axis is the murepavadin exposure expressed as AUC of the unbound fraction of the drug from 0 to 24 h (fAUC0–24 h). The y axis is the change in log10 of bacterial burden from the start of treatment. (A) The results from the study conducted at Radboud University are indicated with a blue triangle, and those from the study conducted at Evotec (UK) Ltd. are indicated with a black dot. The ED50 represents the AUC associated with 50% of the maximal effect (Emax). The line is the best fit based on the sigmoid Emax model.
DISCUSSION
In this study, we found that the pharmacodynamic index of murepavadin that best correlated to efficacy was the fAUC/MIC ratio. Estimates of the free drug exposures for stasis and a 1-log reduction were 27.8 and 39.9, respectively. As expected, and concurrent with the exposure-response relationship of most antibiotics, with the notable exception of beta-lactams, the efficacy of murepavadin showed the strongest dependence on fAUC/MIC. In addition, the fAUC/MIC response relationship was consistent in dose-fractionation studies in both the thigh and lung models, further supporting the importance of this relationship. The dependence on AUC has also been observed for other peptide antibiotics with overall positive charge, such as colistin (22). In the dose-fractionation studies, however, lower doses and fAUC/MIC exposures were required to achieve antibacterial activity in the lung similar to that in the thigh. This may be a result of the high penetration of murepavadin into ELF compared to other tissues (data not shown). This is very much in contrast to colistin, where in the lung infection model, colistin was substantially less effective in killing the bacteria (22).
The ultracentrifugation method was chosen, for it is well suited for the determination of plasma protein binding of drugs that exhibit extensive nonspecific binding to laboratory equipment (22, 23). Overall, systemic exposure to murepavadin was considered generally comparable between thigh- and lung-infected mice. Murepavadin was rapidly distributed, and concentrations peaked in the period of 20 to 45 min postdose, and then concentrations declined in a generally biexponential manner. Murepavadin showed linear pharmacokinetics in plasma for total drug and was dose proportional, which is similar to observations in humans (24). There was no significant difference in PK between the thigh and lung models; thus, we assume that inflammation in the lung due to infection does not play a significant role in the exposure of murepavadin in the ELF. Murepavadin had good penetration into ELF, with a mean penetration (AUC) ELF/plasma ratio of 24.4% for total drug and 108.9% for free drug. However, there was a tendency toward increased ELF exposure at the higher doses. The ELF penetration was approximately equivalent to the plasma free drug concentration, and as such, the pharmacodynamics of the ELF can be estimated from the free plasma drug concentrations.
In conclusion, this study provides information on the pharmacokinetic and pharmacodynamic relationship of murepavadin in neutropenic mouse models. The main PK/PD index that correlated with an effect was the fAUC/MIC ratio, with a mean fAUC/MIC ratio of 39.9 required to achieve a 1-log reduction in the lung infection model against a panel of difficult-to-treat organisms. Importantly, the efficacy of murepavadin was not affected by resistance mechanisms, including carbapenem resistance. Many of the organisms tested were XDR and by definition have limited treatment options. Murepavadin displays both potent in vitro and in vivo activities against P. aeruginosa strains, including extensively drug-resistant pathogens; as such, murepavadin has the potential to address an unmet medical need. The data provided here may serve to determine doses in humans that are likely to be effective.
MATERIALS AND METHODS
Organisms, media, and antibiotics.
Fifteen P. aeruginosa isolates were used for these studies (Table 1). The strains were chosen to span the MIC range of murepavadin but also to include a majority of carbapenem-nonsusceptible and XDR isolates. They were grown, subcultured, and quantified using Mueller-Hinton broth (MHB) and agar (Difco Laboratories, Detroit, MI) or Pseudomonas selective agar (Oxoid, Basingstoke, United Kingdom). Murepavadin was supplied by Polyphor, Ltd., Allschwil, Switzerland.
In vitro susceptibility testing.
The MICs for murepavadin and reference antibiotics for the various isolates were determined using Clinical and Laboratory Standards Institute (CLSI) microdilution methods (25, 26). All MIC assays for murepavadin were performed 5 times, and the median MIC from replicate assays is reported and was utilized in the PK/PD analysis.
Murine thigh and lung infection models.
The experiments were carried out in three separate facilities in the Central Animal Facility (Centraal Dierenlab) at Radboud University Nijmegen Medical Center, in Evotec (UK) Ltd., and in Statens Serum Institut (SSI). Pharmacodynamic studies were performed at three separate facilities following the same protocol for the neutropenic lung infection model, except for the number of animals used per dosing regimen (Radboud University, Evotec [UK] Ltd., and SSI used 2, 6, and 6 animals/dosing regimen, respectively). Three isolates (ATCC 27853, 12, and 22) were investigated at both Radboud University Nijmegen Medical Center and Evotec Ltd., and the data were combined for analysis. The animal studies were conducted in accordance with the recommendations of the 2010/63/EU 22 September 2010 directive (27). Outbred CD-1 mice (±8 weeks) weighing 20 to 25 g at the day of infection were used for all studies (Charles River, United Kingdom or Germany). Mice were rendered neutropenic by immunosuppression with cyclophosphamide at 200 mg/kg at 4 days before infection and 100 mg/kg at 1 day before infection by intraperitoneal injection. The inoculum with fresh broth was diluted to an optimal concentration with phosphate-buffered saline (PBS). On the day of infection, isolates grown in fresh broth were diluted with PBS to a final inoculum of approximately 107 CFU/ml. On day 0, each mouse was inoculated with 50 μl of approximately 106 to 107 CFU per thigh by intramuscular injection. For lung infection, the animals were instilled intranasally. Murepavadin powder was dissolved in saline for injection for a stock solution of 4 mg/ml. Murepavadin therapy was initiated 2 h (t = 0 h) after the infection and continued for 24 h (t = 24 h). At 0 h, a group of mice were humanely euthanized to determine the initial bacterial burden just before treatment. All other animals were terminated at 24 h unless the welfare of the animals necessitated earlier termination, in accordance with animal welfare regulations. After 24 h of treatment, the animals were euthanized, and the thighs or lungs were aseptically removed, homogenized, and plated for determination of the number of CFU. The lower limit of quantification was 1.7 log CFU. No-treatment (vehicle) and 0-h controls were included in all experiments.
For pharmacokinetic experiments, animals were separated into two groups, with four dose levels studied in a thigh model and four dose levels studied in a lung model. Murepavadin was administered subcutaneously (s.c.) in mice at doses of 0.125, 0.25, 0.5, 1, 2, 4, 8, and 16 mg/kg. A single dose of murepavadin was administered through 0.1-ml s.c. injection 2 h after infection. Blood was collected in 1-ml K3EDTA tubes through orbital sinus bleeding under isoflurane sedation; subsequently, the mice were killed through cervical dislocation. Time points for sampling were before t = 0 (predose) and 0.083, 0.167, 0.333, 0.5, 0.75, 1, 2, 4, 6, 8, and 12 h after administration. Blood samples were centrifuged immediately in a precooled centrifuge, and plasma samples were stored at −80°C until murepavadin plasma concentrations were determined. One sample was taken per mouse, and every time point was sampled in duplicate (2 mice).
Bronchoalveolar lavage (BAL) was performed immediately after mice were euthanized humanely, subsequent to previous blood collection. The concentrations in ELF were determined by taking BAL fluid samples and a concomitant plasma sample, as previously described (28). In short, after being sacrificed under isoflurane anesthesia followed by cervical dislocation, mice were secured on a plastic platform, and the trachea was exposed by a 1-cm incision on the ventral neck skin for insertion of the cannula. Lungs were instilled 4 times with 0.5 ml of sterile saline, and the fluid was aspirated immediately. The aspirates recovered were pooled, directly placed on ice, and subsequently stored at −80°C.
Protein binding and concentration determinations in plasma and ELF.
The binding of murepavadin to plasma proteins of different species was tested concurrently in two independent contract laboratories using ultracentrifugation (Microconstants, Inc., San Diego, CA, and Alliance Pharma, Malvern, PA). The two facilities followed the same methods, and the data were evaluated separately. Plasma protein binding experiments with mice, rats, monkeys, and humans were tested at each test site over an extended concentration range from 0.1 to 20.0 mg/liter (0.1, 0.2, 0.4, 1.0, 4.0, 10.0, 15.0, and 20.0 mg/liter). A stock solution of murepavadin was prepared at 1 mg/ml using dimethyl sulfoxide (DMSO)-water (9:1 [vol/vol]), and a positive control (warfarin) was prepared at 5 mM in DMSO. Working solutions of test compound were prepared by diluting stock solution (1 mg/ml) using DMSO-water (9:1 [vol/vol]). The working solution of positive control was prepared by diluting the stock solution to 1 mM using methanol-water (1:1 [vol/vol]). Spiked plasma samples were prepared by adding 36 μl of working solution to 3.6 ml of blank plasma. Half a milliliter of spiked plasma samples was transferred to ultracentrifuge tubes in triplicate, followed by incubation at 37°C with 5% carbon dioxide for 45 min. After 45 min of incubation, the triplicates of test compound and positive control were transferred into the ultracentrifuge and spun at 120,000 rpm for 3 h at 37°C. After centrifugation, 80 μl of centrifuged plasma was transferred from the ultracentrifuge tube to low-protein-binding library tubes containing 20 μl of plasma. Meanwhile, recovery samples were made via a mixture of 20 μl of spiked samples incubated at 37°C with 80 μl of plasma water, or 20 μl of spiked samples incubated at 0°C (on ice) with 80 μl of plasma water extracted from mouse and human blank plasma, respectively. A 200-μl aliquot of stop solution containing internal standard (0.2 μg/ml internal standard in acetonitrile-DMSO [3:1 {vol/vol}]) was added to each tube. The low-protein-binding library tubes were vortexed for 3 min at 1,700 rpm and then centrifuged at 3,500 rpm for 15 min at 4°C. A 100-μl aliquot of supernatant from each tube was transferred to a clean low-protein-binding 96-well plate and further mixed with 100 μl of Milli-Q (ultrapure) water. The samples were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The plasma samples were extracted by protein precipitation in 0.1% formic acid in acetonitrile-DMSO (75/25 [vol/vol]). After 5 min of centrifugation at 3,500 rpm, 10 μl supernatant was injected on a Waters ultrahigh-performance liquid chromatography (UPLC) system at 65°C equipped with a Waters Acquity BEH C18 (2.1 by 50 mm, 1.7-μm particle size) column, running at a flow rate of 500 μl/min. An AB Sciex API 5000 mass spectrometer with a TurboIonSpray source (positive mode) and the Analyst (version 1.6.2) software was used to monitor mass transitions (±0.2 for each mass) by selected-reaction monitoring (m/z 518.9 → m/z 115.1 [M+3H]3+). The method was validated in the range of 10 to 2,000 ng/ml, with a lower limit of quantification (LLOQ) of 10 ng/ml using a 0.1-ml volume. The assay accuracy and precision values for murepavadin were 3.3 to 9.5%, and urine concentrations of murepavadin were determined by LC-MS/MS. Murepavadin was extracted from plasma into 2% formic acid in DMSO-acetonitrile (25:75 [vol/vol]) by protein precipitation. The supernatant was diluted 1:1 with 0.1% formic acid, injected onto an allure biphenyl column, and detected by MS/MS with positive electrospray ionization. The method was validated in the range of 10 to 2,000 ng/ml, with a lower limit of quantification of 10 ng/ml using a 0.1-ml volume. The assay accuracy and precision values for murepavadin were 3.3 to 9.5%.
The calculation of antibiotic concentrations in ELF was performed as follows: concentrations of murepavadin in ELF were determined by using the ratio of the urea concentration in BAL fluid compared to that in plasma, as measured with a modified enzymatic assay (QuantiChrom urea assay kit [DIUR-500]; BioAssay Systems). As urea concentrations are normally the same in plasma and ELF because of rapid diffusion and an equilibrium across the capillary-alveolar membrane, the apparent ELF volume was estimated by using urea as an endogenous marker of ELF dilution and was calculated as follows: drug concentration in ELF = drug concentration in BAL fluid × urea plasma/urea BAL fluid (reference standards ranged from 0.5 to 125 ng/ml and were linear [R2 = 0.989] over the concentration range). The lower limit of detection was 0.25 ng/ml. In ELF, protein binding of antibiotics is expected to be negligible; thus, drug concentration in ELF was assumed to equal the free (unbound) concentration (29, 30).
Pharmacokinetic evaluation.
PK parameters were estimated using Phoenix/WinNonlin pharmacokinetic software version 6.4 (Certara USA, Princeton, NJ) using a noncompartmental approach consistent with the s.c. route of administration of murepavadin. Plasma concentration values reported as not quantifiable were excluded from the evaluation. The area under the murepavadin concentration versus time curve was calculated using the linear up/log down interpolation method for the period from t = 0 to the last quantifiable concentration level (AUC0–last). Evaluation of the terminal elimination phase was not practical, as terminal phase concentration data were either not available (low doses) or sparse.
Pharmacodynamic studies in mice.
Dose-fractionation studies were undertaken for 2 P. aeruginosa strains (clinical isolate 18 and ATCC 27853). A dose range of 0.25 to 32 mg/kg was used 1, 2, 4, or 8 times in 24 h. Dose-response experiments were performed for an additional 13 P. aeruginosa strains (5, 6, 9, 11, 12, 15, 16, 19, 21, 22, X11045, ATCC BAA 2113, and NCTC 13437). For dose-response models, murepavadin was administered as single (q24h), b.i.d. (twice a day [q12h]), or q.i.d. (4 times a day [q6h]) doses for 24 h in 2-fold increasing dosages (range, 0.5 to 30 mg/kg). In two experiments, 32 mg/kg was given, and in one experiment, 40 mg/kg was given. Effect relationships were determined for both total drug and free drug concentrations. Protein binding and PK parameters used to study the relationship between pharmacokinetic and pharmacodynamic indices were obtained from the PK study and the protein binding studies. The sigmoid Emax model with variable slope was used fit to the dose and PK/PD index (PDI) responses to determine the PDI values of murepavadin resulting in a static, 1-log drop, and 2-log drop effects using GraphPad Prism version 5.03 (GraphPad, Inc., San Diego, CA). When the data were combined from separate studies, all data were aggregated and included in a single model. Models were fitted without constraint, and the fits were judged by R2 values and visual inspection in most instances. In some cases, if no clear fit was obtained, the lowest effect was used as a constraint or the log10 CFU at t = 0. Since data are expressed as Δlog10 CFU, the actual value used was the lower limit of detection minus the log10 CFU at the start of treatment (t = 0).
ACKNOWLEDGMENTS
J.W.M. has received research funding from Adenium, Astra-Zeneca, Basilea, Cubist, Polyphor, Roche, Eumedica, Basilea, VenatoRx, AiCuris, Gilead, and Wockhardt. M.J.M. declares no conflicts of interest. P.W. and J.T. are employees of Evotec (UK) Ltd. J.H. is an employee of Statens Serum Institut. F.B., A.W., D.O., and G.E.D. are employees of Polyphor Ltd.
This study was funded by Polyphor Ltd.
REFERENCES
- 1.Klevens RM, Edwards JR, Richards CL Jr, Horan TC, Gaynes RP, Pollock DA, Cardo DM. 2007. Estimating health care-associated infections and deaths in U.S. hospitals, 2002. Public Health Rep 122:160–166. doi: 10.1177/003335490712200205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tumbarello M, De Pascale G, Trecarichi EM, Spanu T, Antonicelli F, Maviglia R, Pennisi MA, Bello G, Antonelli M. 2013. Clinical outcomes of Pseudomonas aeruginosa pneumonia in intensive care unit patients. Intensive Care Med 39:682–692. doi: 10.1007/s00134-013-2828-9. [DOI] [PubMed] [Google Scholar]
- 3.Micek ST, Wunderink RG, Kollef MH, Chen C, Rello J, Chastre J, Antonelli M, Welte T, Clair B, Ostermann H, Calbo E, Torres A, Menichetti F, Schramm GE, Menon V. 2015. An international multicenter retrospective study of Pseudomonas aeruginosa nosocomial pneumonia: impact of multidrug resistance. Crit Care 19:219. doi: 10.1186/s13054-015-0926-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones RN. 2010. Microbial etiologies of hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin Infect Dis 51:S81–S87. doi: 10.1086/653053. [DOI] [PubMed] [Google Scholar]
- 5.Kollef MH, Chastre J, Fagon JY, Francois B, Niederman MS, Rello J, Torres A, Vincent JL, Wunderink RG, Go KW, Rehm C. 2014. Global prospective epidemiologic and surveillance study of ventilator-associated pneumonia due to Pseudomonas aeruginosa. Crit Care Med 42:2178–2187. [DOI] [PubMed] [Google Scholar]
- 6.Ramírez-Estrada S, Borgatta B, Rello J. 2016. Pseudomonas aeruginosa ventilator-associated pneumonia management. Infect Drug Resist 20:7–18. doi: 10.2147/IDR.S50669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 8.American Thoracic Society, Infectious Diseases Society of America. 2005. Guidelines for the management of adults with hospital-acquired, ventilator‐associated, and healthcare‐associated pneumonia. Am J Respir Crit Care Med 171:388–416. [DOI] [PubMed] [Google Scholar]
- 9.Spellberg B, Talbot G. 2010. Recommended design features of future clinical trials of antibacterial agents for hospital-acquired bacterial pneumonia and ventilator-associated bacterial pneumonia. Clin Infect Dis 51:S150–S170. doi: 10.1086/653065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin-Loeches I, Torres A, Rinaudo M, Terraneo S, de Rosa F, Ramirez P, Diaz E, Fernandez-Barat L, Li Bassi GL, Ferrer M. 2015. Resistance patterns and outcomes in intensive care unit (ICU)-acquired pneumonia. Validation of European Centre for Disease Prevention and Control (ECDC) and the Centers for Disease Control and Prevention (CDC) classification of multidrug resistant organisms. J Infect 70:213–222. doi: 10.1016/j.jinf.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 11.Bassetti M, Welte T, Wunderink RG. 2016. Treatment of Gram-negative pneumonia in the critical care setting: is the beta-lactam antibiotic backbone broken beyond repair? Crit Care 20:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luther A, Moehle K, Chevalier E, Dale G, Obrecht D. 2017. Protein epitope mimetic macrocycles as biopharmaceuticals. Curr Opin Chem Biol 38:45–51. doi: 10.1016/j.cbpa.2017.02.004. [DOI] [PubMed] [Google Scholar]
- 13.Martin-Loeches I, Dale GE, Torres A. 2018. Murepavadin: a new antibiotic class in the pipeline. Expert Rev Anti Infect Ther 16:259–268. doi: 10.1080/14787210.2018.1441024. [DOI] [PubMed] [Google Scholar]
- 14.Srinivas N, Jetter P, Ueberbacher BJ, Werneburg M, Zerbe K, Steinmann J, Van der Meijden B, Bernardini F, Lederer A, Dias RLA, Misson PE, Henze H, Zumbrunn J, Gombert FO, Obrecht D, Hunziker P, Schauer S, Ziegler U, Kach A, Eberl L, Riedel K, DeMarco SJ, Robinson JA. 2010. Peptidomimetic antibiotics target outer-membrane biogenesis in Pseudomonas aeruginosa. Science 327:1010–1013. doi: 10.1126/science.1182749. [DOI] [PubMed] [Google Scholar]
- 15.Andolina G, Bencze L-C, Zerbe K, Müller M, Steinmann J, Kocherla H, Monda M, Sobek J, Moehle K, Malojčić G, Wollscheid B, Robinson JA. 2018. A peptidomimetic antibiotic interacts with the periplasmic domain of LptD from Pseudomonas aeruginosa. ACS Chem Biol 13:666–675. doi: 10.1021/acschembio.7b00822. [DOI] [PubMed] [Google Scholar]
- 16.Sader HS, Dale GE, Rhomberg PR, Flamm RK. 2018. Antimicrobial activity of murepavadin tested against clinical isolates of Pseudomonas aeruginosa collected in the United States. Europe, and China Antimicrob Agents Chemother 62:e00311-18. doi: 10.1128/AAC.00311-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sader HS, Flamm RK, Dale GE, Rhomberg PR, Castanheira M. 2018. Murepavadin activity tested against contemporary (2016–17) clinical isolates of XDR Pseudomonas aeruginosa. J Antimicrob Chemother 73:2400–2404. doi: 10.1093/jac/dky227. [DOI] [PubMed] [Google Scholar]
- 18.Awad SS, Rodriguez AH, Chuang Y-C, Marjanek Z, Pareigis AJ, Reis G, Scheeren TW, Sanchez AS, Zhou X, Saulay M, Engelhardt M. 2014. A phase 3 randomized double-blind comparison of ceftobiprole medocaril versus ceftazidime plus linezolid for the treatment of hospital-acquired pneumonia. Clin Infect Dis 59:51–61. doi: 10.1093/cid/ciu219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freire AT, Melnyk V, Kim MJ, Datsenko O, Dzyublik O, Glumcher F, Chuang YC, Maroko RT, Dukart G, Cooper CA, Korth-Bradley JM, Dartois N, Gandjini H, 311 Study Group. 2010. Comparison of tigecycline with imipenem/cilastatin for the treatment of hospital-acquired pneumonia. Diagn Microbiol Infect Dis 68:140–151. doi: 10.1016/j.diagmicrobio.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 20.European Committee on Antimicrobial Susceptibility Testing. 2017. Breakpoint tables for interpretation of MICs and zone diameters. Version 7.1. European Committee on Antimicrobial Susceptibility Testing, Växjö, Sweden: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_7.1_Breakpoint_Tables.pdf. [Google Scholar]
- 21.Murakami T, Suzuki K, Niyonsaba F, Tada H, Reich J, Tamura H, Nagaoka I. 2018. MrgX2‑mediated internalization of LL‑37 and degranulation of human LAD2 mast cells. Mol Med Rep 18:4951–4959. doi: 10.3892/mmr.2018.9532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheah SE, Wang J, Nguyen VT, Turnidge JD, Li J, Nation RL. 2015. New pharmacokinetic/pharmacodynamic studies of systemically administered colistin against Pseudomonas aeruginosa and Acinetobacter baumannii in mouse thigh and lung infection models: smaller response in lung infection. J Antimicrob Chemother 70:3291–3297. doi: 10.1093/jac/dkv267. [DOI] [PubMed] [Google Scholar]
- 23.Legg B, Rowland M. 1987. Cyclosporin: measurement of fraction unbound in plasma. J Pharm Pharmacol 39:599–603. [DOI] [PubMed] [Google Scholar]
- 24.Wach A, Dembowsky K, Dale GE. 2018. Pharmacokinetics and safety of intravenous murepavadin infusion in healthy adult subjects administered single and multiple ascending doses. Antimicrob Agents Chemother 62:e02355-17. doi: 10.1128/AAC.02355-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clinical and Laboratory Standards Institute. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 8th ed CLSI document M07-A10. Clinical and Laboratory Standards Institute (CLSI), Wayne, PA. [Google Scholar]
- 26.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing; 24th informational supplement. CLSI document M100-S25. Clinical and Laboratory Standards Institute (CLSI), Wayne, PA. [Google Scholar]
- 27.European Union. 2010. Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes. European Union, Brussels, Belgium: https://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=OJ:L:2010:276:0033:0079:en:PDF. [Google Scholar]
- 28.Seyedmousavi S, Brüggemann RJM, Melchers WJG, Verweij PE, Mouton JW. 2014. Intrapulmonary penetration of posaconazole at the infection site in an immunosuppressed murine model of invasive pulmonary aspergillosis receiving oral prophylactic regimens. Antimicrob Agents Chemother 58:2964–2967. doi: 10.1128/AAC.00053-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodvold KA, Yoo L, George JM. 2011. Penetration of anti-infective agents into pulmonary epithelial lining fluid: focus on antifungal, antitubercular and miscellaneous anti-infective agents. Clin Pharmacokinet 50:689–704. doi: 10.2165/11592900-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 30.Kiem S, Schentag JJ. 2008. Interpretation of antibiotic concentration ratios measured in epithelial lining fluid. Antimicrob Agents Chemother 52:24–36. doi: 10.1128/AAC.00133-06. [DOI] [PMC free article] [PubMed] [Google Scholar]