Human parvovirus B19 (B19V), a member of the genus Erythroparvovirus of the family Parvoviridae, is a small nonenveloped virus that has a single-stranded DNA (ssDNA) genome of 5.6 kb with two inverted terminal repeats (ITRs). B19V infection often results in severe hematological disorders and fetal death in humans.
KEYWORDS: in vitro nicking assay, parvovirus B19, antivirals
ABSTRACT
Human parvovirus B19 (B19V), a member of the genus Erythroparvovirus of the family Parvoviridae, is a small nonenveloped virus that has a single-stranded DNA (ssDNA) genome of 5.6 kb with two inverted terminal repeats (ITRs). B19V infection often results in severe hematological disorders and fetal death in humans. B19V replication follows a model of rolling hairpin-dependent DNA replication, in which the large nonstructural protein NS1 introduces a site-specific single-strand nick in the viral DNA replication origins, which locate at the ITRs. NS1 executes endonuclease activity through the N-terminal origin-binding domain. Nicking of the viral replication origin is a pivotal step in rolling hairpin-dependent viral DNA replication. Here, we developed a fluorophore-based in vitro nicking assay of the replication origin using the origin-binding domain of NS1 and compared it with the radioactive in vitro nicking assay. We used both assays to screen a set of small-molecule compounds (n = 96) that have potential antinuclease activity. We found that the fluorophore-based in vitro nicking assay demonstrates sensitivity and specificity values as high as those of the radioactive assay. Among the 96 compounds, we identified 8 which have an inhibition of >80% at 10 µM in both the fluorophore-based and radioactive in vitro nicking assays. We further tested 3 compounds that have a flavonoid-like structure and an in vitro 50% inhibitory concentration that fell in the range of 1 to 3 µM. Importantly, they also exhibited inhibition of B19V DNA replication in UT7/Epo-S1 cells and ex vivo-expanded human erythroid progenitor cells.
INTRODUCTION
Human parvovirus B19 (B19V) was identified in 1975 when Cossart and colleagues screened a panel of human serum samples for hepatitis B virus (HBV) (1). B19V is a small, nonenveloped single-stranded DNA (ssDNA) virus belonging to the genus Erythroparvovirus within the family Parvoviridae (2). B19V exhibits a remarkable tropism for human erythroid progenitor cells (EPCs) in the bone marrow and fetal liver (3–7). B19V most commonly causes fifth disease or slapped cheek syndrome in children (8, 9); however, B19V infection can cause a series of severe hematological disorders (10). B19V infection of the fetus can cause severe fetal anemia, resulting in nonimmune hydrops fetalis and fetal death (11–14). In certain circumstances, B19V infection often results in bone marrow failure, most notably, transient aplastic crisis in patients with increased red blood cell turnover (e.g., sickle cell disease patients) and pure red cell aplasia in immunodeficient and immunocompromised patients (e.g., HIV/AIDS patients and organ transplant recipients) (11, 15). The clinical manifestations of B19V infection, as seen in hydrops fetalis, transient aplastic crisis, and pure red cell aplasia, are due to direct cytotoxicity resulting from virus infection, which results in the death of the EPCs in which B19V replicates (5, 16–22).
B19V has a linear ssDNA genome of about 5.6 kb, which has identical inverted terminal repeats (ITRs) of 383 nucleotides at both ends. The double-stranded replicative-form (RF) DNA of the B19V genome contains a P6 promoter at the left hand (10). The left side of the RF genome encodes a large nonstructural protein (NS1) and a small nonstructural 7.5-kDa protein, whereas the right side of the genome encodes two capsid proteins (VP1 and VP2), along with a small nonstructural 11-kDa protein, encoded using a different open reading frame (23). B19V NS1, 671 amino acids (aa) long, has a molecular weight of approximate 78 kDa (Fig. 1A) (24, 25). NS1 predominantly localizes in the nucleus of infected cells, as it contains nuclear localization signals at amino acid residues 177 to 180 (KKPR) and 316 to 321 (KKCGKK) (25, 26). The N terminus (aa 1 to 176) of NS1 contains a DNA replication origin-binding domain (OBD) that also exhibits endonuclease activity (27, 28), the central region contains ATPase and nucleoside triphosphate binding motifs (20), and the C terminus contains transactivation domains (20, 29). NS1 is essential for the replication of viral DNA through its endonuclease and helicase activities (30). NS1 also binds the P6 promoter of the viral RF genome to regulate viral gene expression (31). In addition, NS1 has been reported to transactivate several other host genes (29, 32, 33).
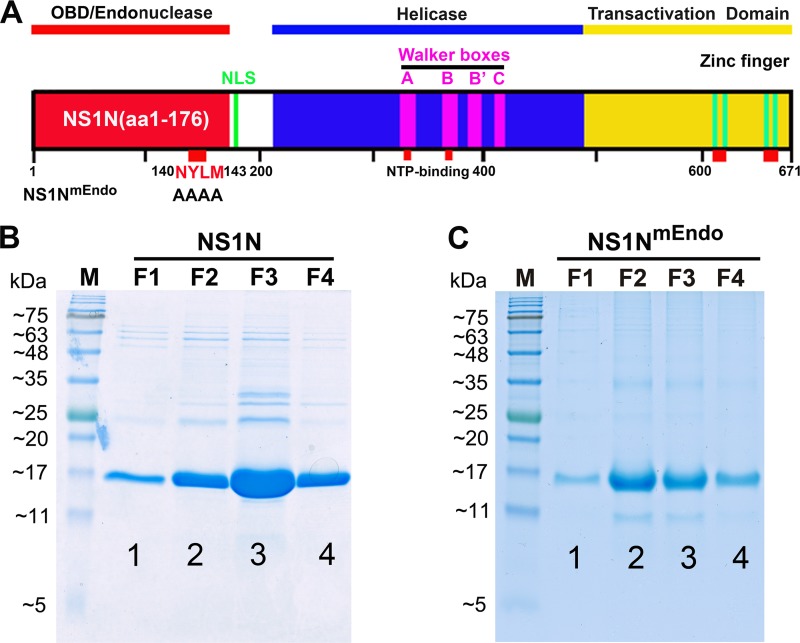
FIG 1.
Functional domains of B19V NS1 and purification of the NS1N and NS1NmEndo proteins. (A) Schematic diagram of the B19V NS1 protein. B19V NS1 is depicted with the Ori-binding (OBD/Endonuclease), helicase, and transactivation domains. NS1N has NS1 (aa 1 to 176), and NS1NmEndo has alanine substitutions in the endonuclease motif (aa 140 to 143) (20). The Walker boxes, nucleoside triphosphate (NTP)-binding sites, and zinc finger motifs are indicated. The C-terminal region (shown in yellow) contains a transactivation domain 2 (TAD2; 523SSFFNLITP531) (20). NLS, nuclear localization signal. (B and C) Purification of NS1N and NS1NmEndo proteins. One liter of IPTG-induced bacteria was collected, and the bacteria were sonicated and lysed. The cleared lysate was mixed with ∼1 ml of NTA beads (Qiagen) and loaded onto a column. The beads were then washed with wash buffer, followed by elution buffer. About 1 ml of each fraction was collected, and 20 µl was loaded for SDS-15% PAGE. The gels were stained with Coomassie blue. Dialyzed fraction F2 or F3 was used in the in vitro nicking assay. Lanes M, molecular weight markers.
B19V is an autonomous parvovirus, replicating itself in the host cells without a helper virus (10), as are the majority of the members in the Parvoviridae family, except for adeno-associated viruses (AAVs) (34). In contrast, AAVs, whose genome also contains their unique ITRs of 144 nucleotides, requires coinfection with a helper virus, such as adenovirus, herpesvirus, or human bocavirus, for replication (35, 36). B19V replication arrests the cell cycle at late S phase, hijacks cellular DNA replication factors present in the S phase, and replicates its genome following a rolling hairpin model of DNA replication (37–39). In principle, the B19V ssDNA genome uses the 3′-end hairpin as a self-primer (3′OH) to extend viral ssDNA into the double-stranded DNA (dsDNA) genome by cellular replication proteins (40), a step called first-strand DNA synthesis (39). The extended 3′ end is presumably ligated to the 5′ end of the genome to form a partial circular DNA genome. NS1, which has site-specific endonuclease activity and DNA helicase activity, nicks ssDNA at the terminal resolution site (trs) between the 5′-end hairpins and the newly synthesized viral DNA to form a novel 3′ primer that initiates hairpin transfer, followed by strand displacement (39). The elongated viral genomes (both RF and double-RF intermediates) are also resolved by NS1 nicking to release ssDNA, which is finally packaged into the capsid.
The minimal replication origin (Ori) of B19V DNA has been identified to be 67 nucleotides (nt), which includes a STAT5-binding element (STAT5BE), a trs or nicking site, two copies of the NS1-binding element (NS1BE), and the putative cellular factor-binding site (CFBE) (Fig. 2A) (25, 27, 41, 42). Specific cleavage of the ssDNA Ori has been demonstrated in an in vitro nicking assay using the purified B19V NS1 N terminus (aa 1 to 176) and a 5′-end 32P-labeled oligonucleotide that contain the trs (28). In this study, we further demonstrate the specificity of the radioactive nicking assay and developed a nonradioactive nicking assay using an oligonucleotide of 20 nucleotides labeled with 6-carboxyfluorescein (FAM) and the quencher Iowa Black FQ (IBFQ) at the 5′ and 3′ ends, respectively. The fluorophore-based nicking assay recapitulates the same sensitivity and specificity found in the radioactive assay. We further validated the assay using a small library of 96 compounds, where 8 compounds inhibited NS1 nicking activity by over 80%. Of these 8 compounds, 3 compounds also inhibited viral DNA replication in B19V RF DNA-transfected UT7/Epo-S1 cells and B19V-infected CD36+ EPCs.
FIG 2.
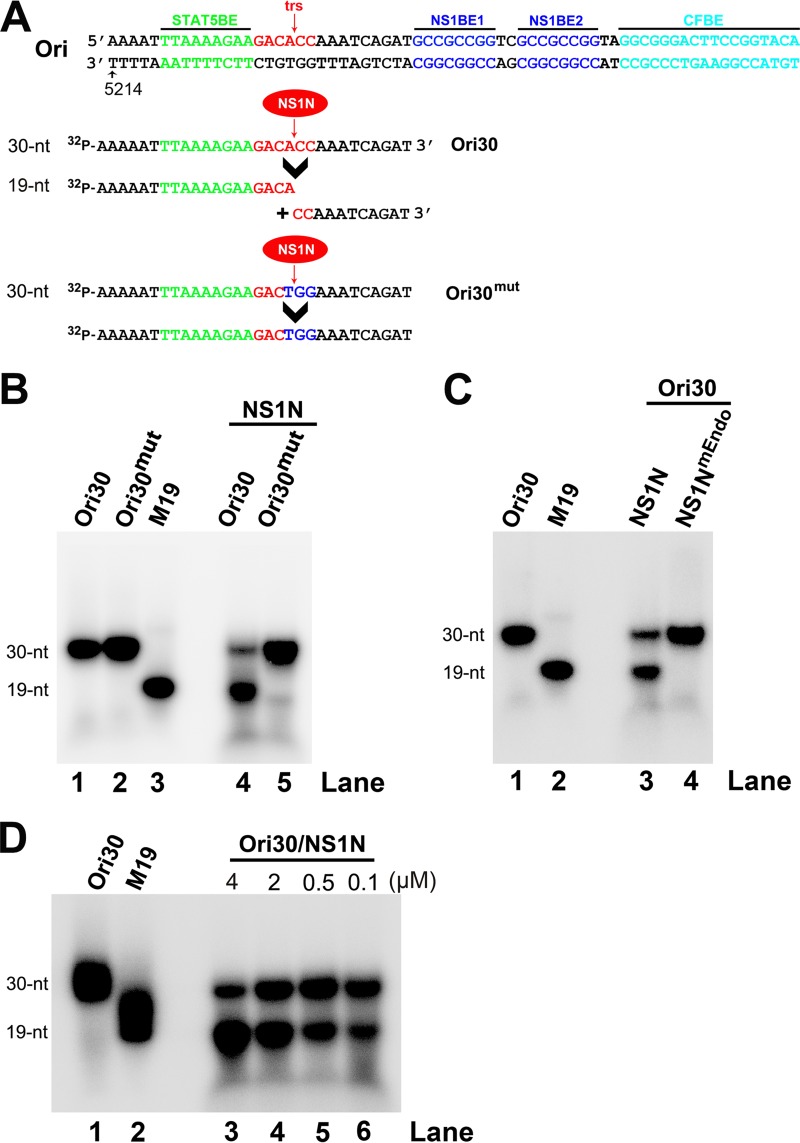
Purified B19V NS1N specifically cleaves the B19V ssDNA Ori at the terminal resolution site. (A) Sequences of Ori30, Ori30mut, and a size marker of 19 nt (M19). The core sequence of Ori is shown with STAT5BE, terminal resolution site (trs), and NS1-binding elements (NS1BEs). The Ori30 probe and the Ori30mut probe, which has three mutations (in blue) at the trs site, are shown. The nicked DNA fragments from Ori30 are shown under the arrowhead. (B, C) Radioactive in vitro nicking assay. The reaction mixtures for in vitro nicking were analyzed on a denaturing polyacrylamide gel, together with labeled probes alone and a labeled oligonucleotide of 19 nt as a size marker for the cleaved band. (B) NS1N cleaved Ori30 but not Ori30mut. 32P-labeled Ori30 or Ori30mut was incubated with NS1N in the nicking buffer. (C) NS1N, but not NS1NmEndo, cleaved Ori30. 32P-labeled Ori30 was incubated with NS1N or NS1NmEndo in the nicking buffer. (D) Determination of a minimal concentration of NS1N used in the nicking reactions. 32P-labeled Ori30 oligonucleotide (2 nM) was incubated with NS1N at a final concentration of 0.1, 0.5, 2, or 4 µM in the nicking buffer.
RESULTS
Purified NS1N, but not the endonuclease motif mutant, cleaves B19V Ori ssDNA and does not cleave Orimut.
B19V NS1 has endonuclease activity at N terminus aa 1 to 176 (NS1N; Fig. 1A). NS1N has been proved to bind the minimal replication origin (Ori) and executes its cleavage in vitro (27, 28). We first tested the specific Ori cleavage of NS1N. To this end, we expressed and purified both wild-type NS1N and the endonuclease motif mutant (NS1NmEndo), which has alanine substitutions in the endonuclease motif (residues 140 to 143) (20) (Fig. 1A). Purified NS1N and NS1NmEndo, which contain a 6× histidine tag at the C terminus, showed a purity of >90% (Fig. 1B and C). We used a short version of the B19V ssDNA from the Ori (Ori30), which is 30 nt, as a template for cleavage, because the NS1-binding elements (NS1BEs) are not required for nicking of the ssDNA Ori under the conditions of the in vitro nicking assay, which contains high concentrations of NS1N (28). Ori30 and the Ori30 mutant (Ori30mut) (Fig. 2A) were 5′ end labeled with 32P and then used at a final concentration of 2 nM in the nicking buffer (see Materials and Methods) with 2 μM the purified NS1N. After incubation for 16 to 18 h at 37°C, the reaction mixtures were analyzed in a denaturing urea polyacrylamide gel. We found that NS1N cleaved Ori30 at the trs and produced a cleaved DNA band at 19 nt (Fig. 2B, lane 4), while NS1N did not cleave the trs mutant, Ori30mut (Fig. 2B, lane 5). Moreover, we asked whether the NS1N nick is endonuclease motif specific. We used purified NS1NmEndo in the nicking assay. The result showed that while wild-type NS1N was capable of cleaving Ori30 at trs and released a band of 19 nt, the NS1NmEndo did not (Fig. 2C, lanes 3 and 4). We next determined the minimal concentration of NS1N required for the nicking of the Ori. We found that there was a dose-dependent increase in the level of the nicked band (19 nt) (Fig. 2D). At 2 μM NS1N, greater than 70% of the probe was cleaved. We chose to use NS1N at 2 μM in subsequent experiments.
Taken together, these results confirm that NS1N specifically and effectively cleaves the Ori (at 2 nM) at trs at a concentration as low as 2 μM and that the endonuclease motif is important for the nicking activity.
Small-molecule compounds inhibit cleavage of the ssDNA Ori by NS1N.
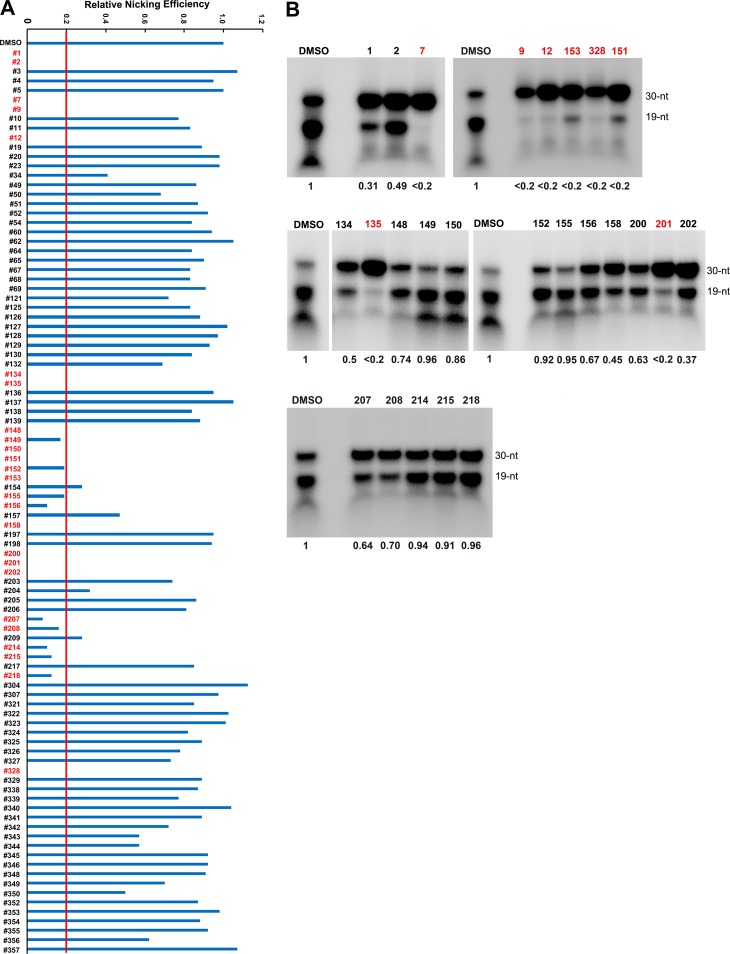
To identify small-molecule compounds that can inhibit the NS1N-mediated Ori cleavage, we used 96 compounds (see Table S1 in the supplemental material), which we have used previously to screen inhibitors for the RNase H nuclease activity of the human hepatitis B virus (HBV) polymerase (43–47), to test them for inhibitory effects on the NS1N-mediated Ori cleavage using a 32P-labeled Ori30 template. We first tested them at 100 µM and found that 25 compounds inhibited the nicking of NS1N by greater than 80%, as shown in Fig. 3A. Next, we tested the 25 positive compounds at 10 µM. Eight compounds (compounds 7, 9, 12, 135, 151, 153, 201, and 328) appeared to nearly abolish cleavage of the Ori by NS1N, a >80% inhibition compared with that for the dimethyl sulfoxide (DMSO) control (Fig. 3B).
FIG 3.
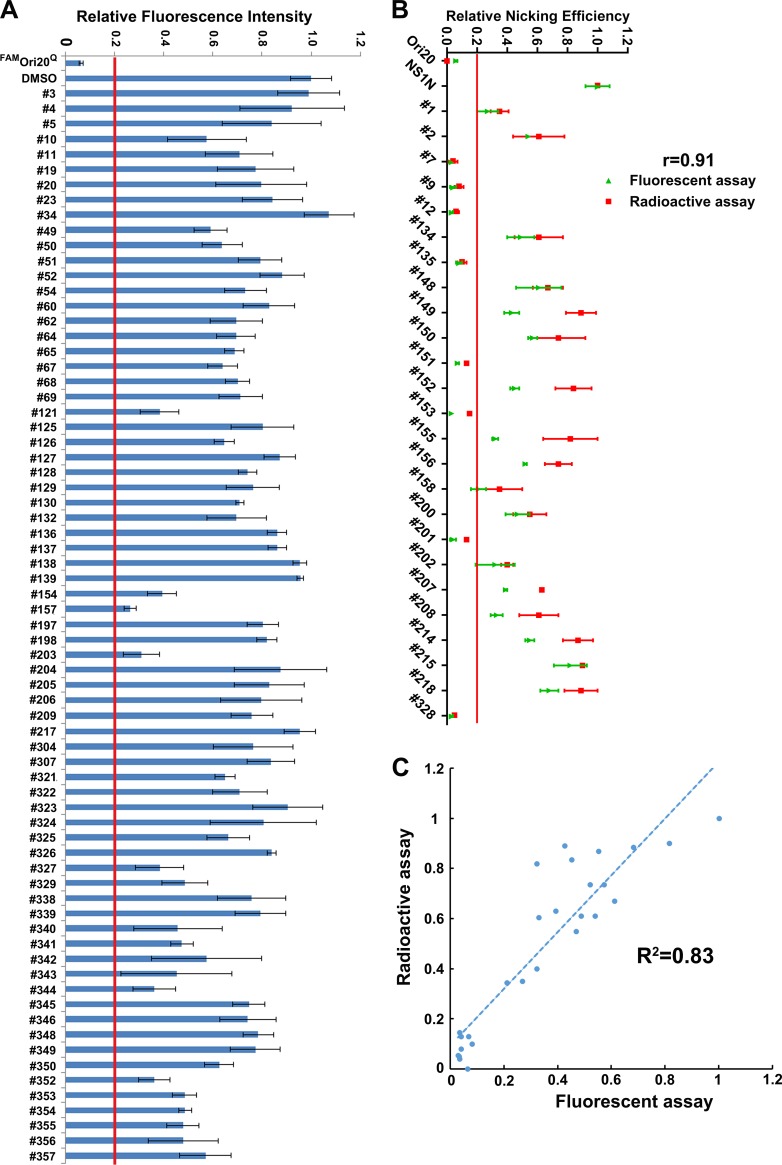
Compound screening for inhibition of cleavage of the 32P-labeled B19V Ori30 oligonucleotide. A total of 96 small-molecule compounds at 100 µM (A) or the chosen 25 compounds at 10 µM (B) were incubated with 32P-labeled Ori30 and NS1N in the nicking buffer. The reaction mixtures were analyzed on a denaturing polyacrylamide gel. The relative nicking efficiencies are shown beneath the gels. The ratio of the signals at 19 nt versus those at 19 nt plus 30 nt in the DMSO vehicle control group is set equal to 1. (A) The nicking efficiency for compounds that inhibited all nicking activity is shown as a blank, as no obvious nicked bands were detected at 19 nt. (B) Autoradiography gel images for the 25 compounds assessed at 10 µM are shown.
The fluorophore FAM-based nicking assay reproduces the sensitivity and specificity of the radioactive nicking assay.
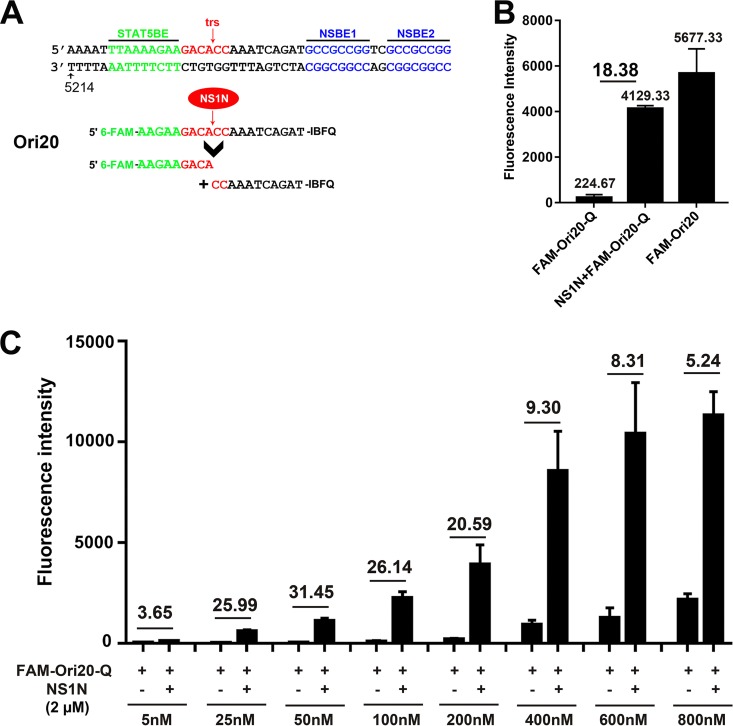
We next developed a method that could easily screen a large number of compounds. We synthesized one fluorescent Ori oligonucleotide, FAMOri20Q, a 20-nt ssDNA oligonucleotide in which the 5′ end is labeled with FAM and the 3′ end is labeled with a quencher, IBFQ (Fig. 4A). The quencher dye quenches the fluorescence emitted by the fluorophore via Förster resonance energy transfer (FRET) when the two dyes are in close proximity, and the intact FAMOri20Q did not show significant fluorescence. When NS1N was added, cleavage occurred at the trs, which resulted in an increase in fluorescence to a level of 72% of the fluorescence from the same oligomer but without a quencher present (FAMOri20), and the fluorescence intensity difference with and without NS1N reached 18.38-fold (Fig. 4B). We further tested the optimal concentration of the oligonucleotide. We found that at 25 to 200 nM FAMOri20Q, the assay displayed a >20-fold difference in the fluorescence intensity with and without NS1N (Fig. 4C). Therefore, we chose FAMOri20Q at 200 nM for use in subsequent experiments, based on the high fold change (20-fold) and the maximum fluorescence intensity (reading). We noticed that 2 µM NS1N was enough to saturate the nicking of the Ori at concentrations ranging from 25 to 800 nM, as increased fluorescence intensities were detected (Fig. 4C).
FIG 4.
Establishment of a 6-carboxyfluorescein (FAM)-based in vitro nicking assay. (A) Diagram of the FAM-labeled oligonucleotides. The sequences of Ori20 are shown with FAM and the Iowa Black FQ quencher (Q) at the 5′ and 3′ ends, respectively. After incubation with NS1N, Ori20 is cleaved into two shorter oligonucleotides, and then a FAM-linked short oligonucleotide of 9 nt is released for fluorescence detection. (B) FAMOri20Q-based nicking assay. FAMOri20Q (200 nM) was incubated with 2 µM NS1N protein in the nicking buffer. The fluorescence intensity of each sample was detected on a microplate reader. FAMOri20Q without NS1N and FAMOri20 without a quencher were used as controls. (C) Optimization of the probe concentration. Various concentrations of the FAMOri20Q probe were used in the nicking assay. Fluorescence intensity was determined with or without NS1N, as indicated. The fold changes in fluorescence intensity in the presence of NS1N from the fluorescence intensity with no NS1N are shown.
We next used the FAMOri20Q probe to repeat screening of the 71 compounds (at 10 µM) which did not show >80% inhibition at 100 µM by the radioactive assay (Fig. 3A). We set up a cutoff value of 80% inhibition, as in the radioactive assay, compared with that achieved with the DMSO control. We found none of the 71 compounds showed an inhibition of >80% at 10 µM (Fig. 5A). We next carefully tested the 25 compounds which showed an inhibition of >80% at 100 µM in the radioactive assay (Fig. 3B) at 10 µM with the FAM-based nicking assay using the FAMOri20Q probe. We found that 8 compounds (compounds 7, 9, 12, 135, 151, 153, 201, and 328) were positive (>80% inhibition) for the inhibition of NS1N nicking activity (Fig. 5B). The results confirmed that the fluorescent assay detected all 8 positive compounds from the radioactive assay.
FIG 5.
Screening of compounds for inhibition of cleavage of the B19V Ori by NS1N using FAM-labeled B19V Ori20. (A) FAM-based in vitro nicking assay. Each of the 71 compounds was incubated with NS1N at 10 µM in the nicking buffer at 37°C for 16 to 18 h. The fluorescence intensity of each sample was detected on a microplate reader. Ori20 without NS1N was set up as the background, and Ori20 with NS1N and DMSO were set up as positive controls. We set up inhibition of 80% of the relative fluorescence intensity as a cutoff value for the inhibition of NS1N nicking. (B) Comparison of the 25 chosen compounds between the radioactive and fluorescent nicking assays. At a final concentration of 10 µM, 8 compounds showed inhibition of >80% in the NS1N nicking of the 32P-labeled Ori30 (in red), and 8 compounds were positive for the NS1N nicking of the FAM-labeled Ori20 (in green). The correlation coefficient (r) between the radioactive and fluorescent nicking assays was calculated using SPSS software. (C) Scatter plot and trend line of the radioactive nicking assay with the fluorescent nicking assay. The scatter plot, trend line, and R2 value for the two assays were calculated using SPSS software.
We established the correlation coefficient value (r) between the two assays, which was 0.91 (Fig. 5B), indicating that the results of the two methods matched well. We also generated a scatter plot and trend line for the two assays (Fig. 5C). The coefficient R2 was 0.83, representing smaller differences between the two assays.
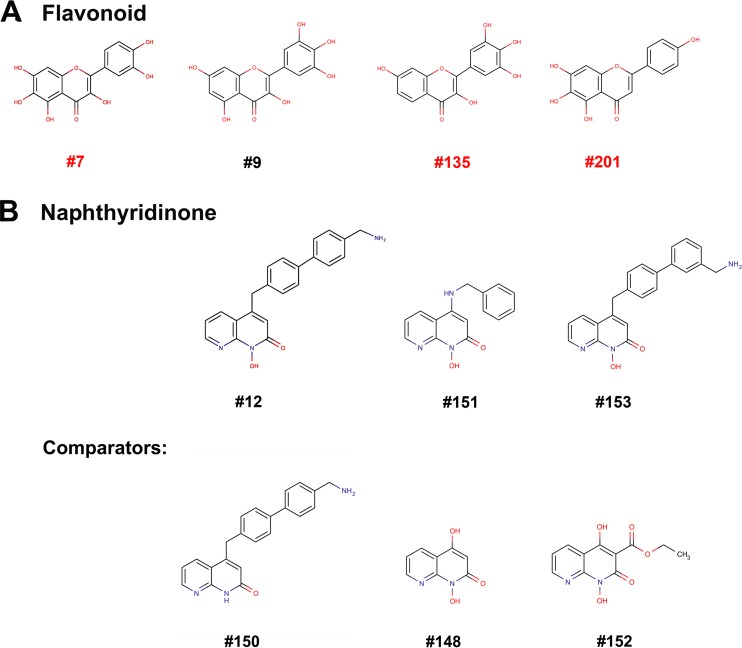
Cheminformatics analysis showed that compounds 7, 9, 135, 201, and 328 have flavonoid structures, while compounds 12, 151, and 153 possess naphthyridinone structures (Fig. 6). We chose flavonoid compounds 7, 135, and 201 for further study in cells, as described below.
FIG 6.
Chemical structures of the eight screened compounds. (A) Flavonoids. (B) Naphthyridinones.
Flavonoid compounds inhibit B19V DNA replication in UT7/Epo-S1 cells.
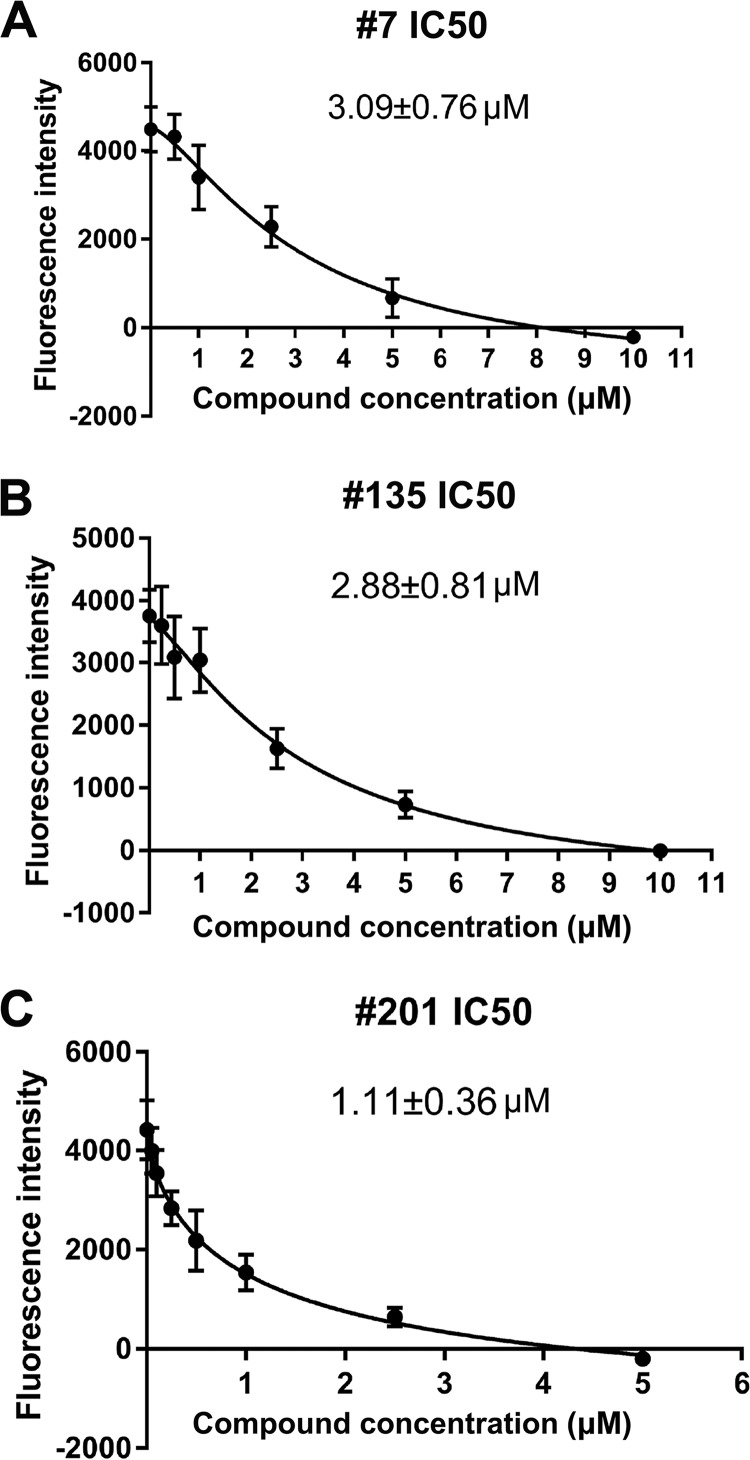
We first determined the 50% inhibitory concentration (IC50; the concentration for half-maximal inhibition of in vitro nicking) of the three flavonoids compounds, compounds 7, 135, and 201, which were 3.1 ± 0.8 µM, 2.9 ± 0.8 µM, and 1.1 ± 0.4 µM, respectively (Fig. 7). We next asked whether they inhibited B19V DNA replication in B19V-permissive cells. For a direct result of inhibition of viral DNA replication, we tested them in UT7/Epo-S1 cells electroporated with RF DNA of the B19V genome, the B19V infectious M20 DNA (48). B19V infects UT7/Epo-S1 cells poorly (41, 49), and electroporation using Nucleofector (Lonza) directly delivers the viral double-stranded RF DNA into the nucleus, where the RF DNA is nicked by NS1 to initiate viral DNA replication (39). After electroporation of the cells with linearized M20 DNA, a compound was added at various concentrations. At 2 days postelectroporation under hypoxic conditions, cells were collected for flow cytometry using an anti-B19V capsid antibody to select capsid-expressing cells.
FIG 7.
Determination of IC50s of compounds 7, 135, and 201. In the FAM-based nicking assay, various concentrations of the compound were added to the nicking assay mixture with FAMOri20Q and NS1N protein. The fluorescence intensity of each reaction was detected. FAMOri20Q without NS1N was set up as the background. The fluorescence intensity obtained at each concentration was compared with that obtained with DMSO. The IC50 of each inhibitor was calculated using GraphPad Prism software.
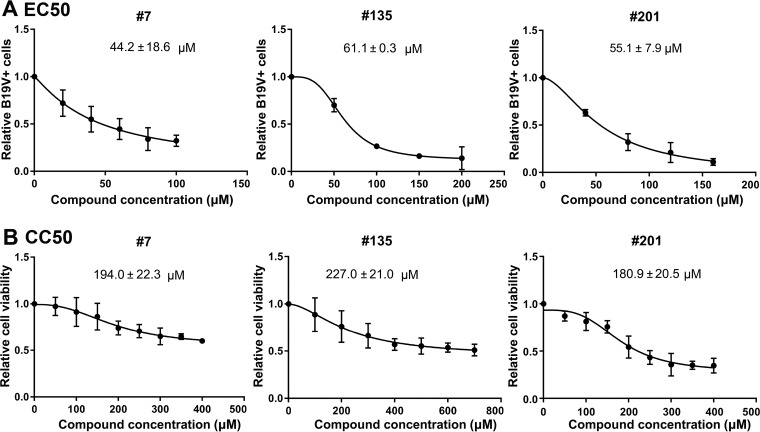
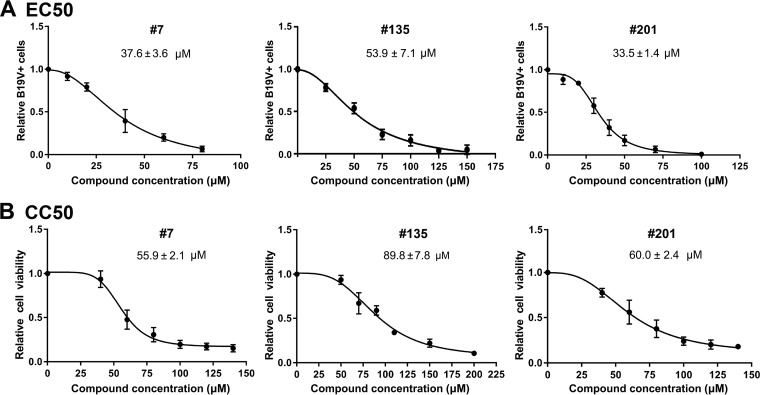
The half-maximal (50%) effective concentration (EC50) for B19V replication inhibition was calculated by comparison with the results for the vehicle control (DMSO). Compounds 7, 135, and 201 had EC50 values of 44.2 ± 18.6 µM, 61.1 ± 0.3 µM, and 55.1 ± 7.9 µM, respectively (Fig. 8A). The drug concentrations that affected the viability of 50% of cells in culture (the 50% cytotoxic concentrations [CC50]) for compounds 7, 135, and 201, as determined in UT7/Epo-S1 cells, were 194.0 ± 22.3 µM, 227.0 ± 21.0 µM, and 180.9 ± 20.5 µM, respectively (Fig. 8B). Therefore, the selective indexes (SI; which is equal to CC50/EC50) for compounds 7, 135, and 201 were 4.4, 3.7, and 3.3, respectively.
FIG 8.
Determination of EC50 and CC50 of compounds 7, 135, and 201. (A) EC50 determination. UT7/Epo-S1 cells were transfected with B19V duplex genome M20. Each inhibitor was added at different concentrations, as indicated. After 2 days, the cells were collected for flow cytometry to detect the capsid-expressing cells. DMSO was used as a vehicle control. The final EC50 was calculated with GraphPad Prism software. (B) CC50 determination. The compounds were added at various concentrations, as shown, to UT7/Epo-S1 cells in 96-well plates. After 2 days, the percentage of viable cells were determined using a CytoTox-Glo cytotoxicity assay kit (Promega), and the CC50 was calculated using GraphPad Prism software.
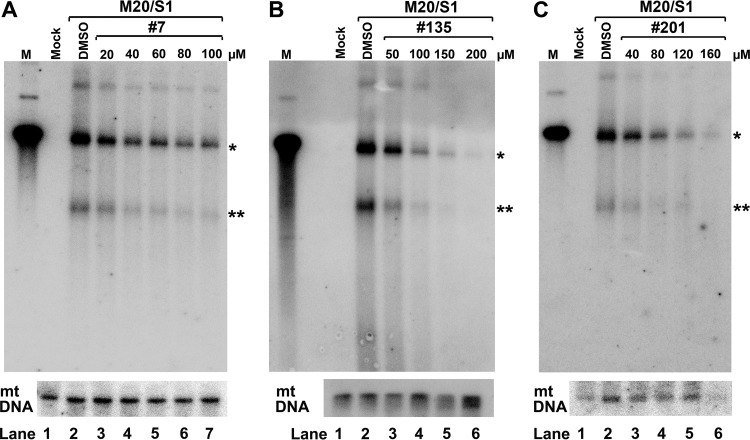
We further analyzed viral DNA replication in UT7/Epo-S1 cells using Southern blotting. The results showed that at concentrations close to their EC50s, compounds 7, 135, and 201 inhibited at least half of the level of the RF DNA (Fig. 9). These results suggest that the inhibition of B19V replication is likely due to the inhibition of viral DNA replication.
FIG 9.
Compounds 7, 135, and 201 inhibit B19V DNA replication in UT7/Epo-S1 cells. UT7/Epo-S1 cells were electroporated with B19V duplex genome M20, followed by addition of compound 7 (A), compound 135 (B), or compound 201 (C) at various concentrations, as shown. At 2 days posttransfection, Hirt DNA samples prepared from treated cells were digested with DpnI and subjected to Southern blotting with the M20 probe. DMSO was used as a vehicle control. UT7/Epo-S1 cells without transfection were set up as mock-transfected controls. pM20 digested with SalI was loaded as a size marker (lanes M). Mitochondrial (mt) DNA was detected as a loading control using a specific mitochondrial DNA probe (91). *, monomer replicative-form DNA; **, single-stranded DNA.
Flavonoid compounds inhibit B19V infection in CD36+ EPCs.
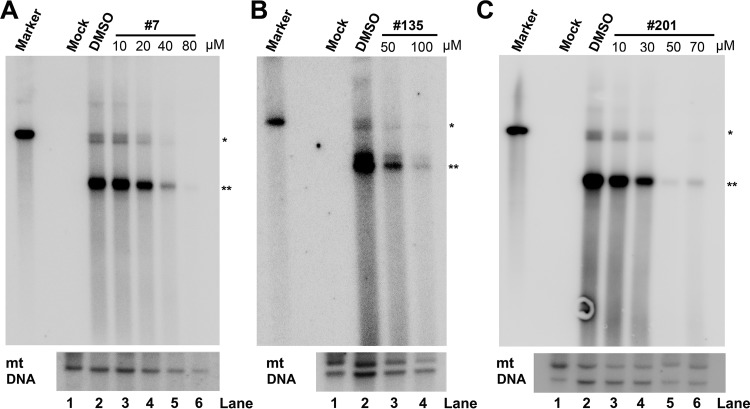
To mimic the B19V infection in human bone marrow, we used ex vivo-expanded primary CD36+ EPCs, which were cultured under hypoxic conditions and which were differentiated from CD34+ hematopoietic stem cells isolated from human bone marrow. We infected the CD36+ EPCs with B19V-infected plasma, and at the same time, the cells were treated with various concentrations of the compounds. At 48 h postinfection, the cells were collected for determining B19V infection by flow cytometry using an anticapsid antibody. We found that for compounds 7, 135, and 201, the EC50s were 37.6 ± 3.6, 53.9 ± 7.1, and 33.5 ± 1.4 µM, respectively (Fig. 10A); the CC50s were 55.9 ± 2.1, 89.8 ± 7.8, and 60.0 ± 2.4 µM, respectively (Fig. 10B). Therefore, the SI for compounds 7, 135, and 201 were 1.5, 1.7, and 1.8, respectively. For compound 7 at 40 µM, there were >95% living cells, whereas B19V infection was inhibited at an efficiency of >60%. For compound 135, at 50 µM there were >95% living cells, whereas B19V infection was inhibited at an efficiency of 50%. For compound 201, at 40 µM, there were >80% living cells, whereas B19V infection was inhibited at an efficiency of 70% (Fig. 10). We next analyzed viral DNA replication in drug-treated infected cells at 48 h postinfection using Southern blotting. It was obvious that compounds 7, 135, and 201 inhibited viral DNA replication by over 10-fold at concentrations of 40, 50, and 50 µM, respectively (Fig. 11).
FIG 10.
EC50 and CC50 determination of compound 7, compound 135, and compound 201. (A) EC50 determination. CD36+ EPCs were infected with B19V. Each compound was added at different concentrations, as shown. At 2 days postinfection, the cells were collected for flow cytometry to detect the capsid-expressing cells. DMSO was used as a vehicle control. The final EC50 was calculated with GraphPad Prism software. (B) CC50 determination. EPCs in 96-well plates were treated with each compound at various concentrations, as shown. At 2 days postinfection, the percentages of viable cells were determined, and the CC50 was calculated using GraphPad Prism software.
FIG 11.
Compounds 7, 135, and 201 inhibit B19V replication in CD36+ EPCs. CD36+ EPCs were infected with B19V, followed by addition of each compound, compound 7 (A), compound 135 (B), and compound 201 (C), at various concentrations, as shown. At 2 days postinfection, Hirt DNA samples were prepared from infected cells and were subjected to Southern blotting using the M20 probe. DMSO was used as a vehicle control. CD36+ EPCs without infection were set up as mock-infected cells (Mock). pM20 digested with SalI was loaded as a size marker (Marker). Mitochondrial (mt) DNA was detected as a loading control using a specific mt DNA probe (91). *, monomer replicative-form DNA; **, ssDNA, single-stranded DNA.
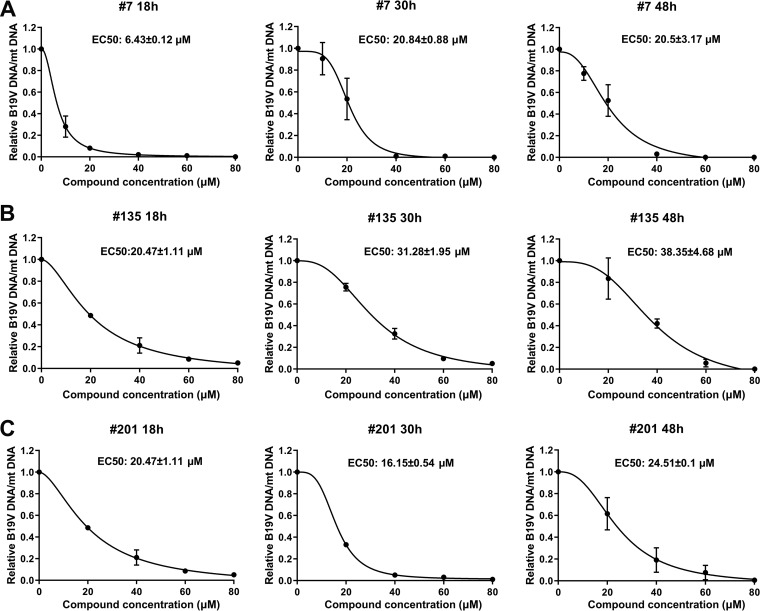
Furthermore, using quantitative PCR (qPCR), we were able to determine the inhibition of viral DNA replication by the compounds at an early time (18 h) postinfection compared to that at middle (30 h) and late (48 h) times during infection. At 18 h, 30 h, and 48 h postinfection, cells were collected for extraction of total DNA, which was subjected to qPCR for determination of the number of copies of the viral genome relative to that of mitochondrial (mt) DNA, which was used to calculate the EC50. Compound 7 had EC50 values of 6.43 ± 0.12 µM, 20.84 ± 0.88 µM, and 20.5 ± 3.17 µM at 18 h, 30 h, and 48 h postinfection, respectively (Fig. 12A); compound 135 had EC50 values of 20.47 ± 1.11 µM, 31.28 ± 1.95 µM, and 38.35 ± 4.68 µM at 18 h, 30 h, and 48 h postinfection, respectively (Fig. 12B); and compound 201 had EC50 values of 20.47 ± 1.11 µM, 16.15 ± 0.54 µM, and 24.51 ± 0.1 µM at 18 h, 30 h, and 48 h postinfection, respectively (Fig. 12C). These results suggest that all the three compounds showed a level of inhibition of B19V DNA replication during early infection similar to that during the middle and late time points of infection. Of note, compound 7 had a relatively effective EC50 of 6.4 µM at 18 h postinfection. Therefore, the selective indexes for compounds 7, 135, and 201, as determined by quantification of viral DNA at 48 h postinfection, were 2.7, 2.4, and 2.5, respectively.
FIG 12.
Compounds 7, 135, and 201 inhibit B19V DNA replication in CD36+ EPCs at 18 h, 30 h, and 48 h postinfection. CD36+ EPCs were infected with B19V, followed by addition of compound 7 (A), compound 135 (B), and compound 201 (C) at various concentrations, as shown. At 18 h, 30 h, and 48 h postinfection, total DNA was extracted from each sample and subjected to multiplex qPCR for detection of viral DNA and cellular (mt) DNA. DMSO was used as a vehicle control. The ratios of B19V DNA/mt DNA were calculated and are shown in relation to the ratio for the vehicle control (which is set equal to 1). EC50 was calculated using GraphPad Prism software.
Collectively, these results demonstrate that compounds 7, 135, and 201 inhibited B19V infection and DNA replication in CD36+ EPCs at a concentration that does not significantly kill cells.
DISCUSSION
In B19V infection-caused transient aplastic crisis, pure red cell aplasia, chronic anemia, and hydrops fetalis, the clinical manifestations are due to the direct cytotoxicity of the virus infection (11, 15, 21), a direct outcome of the cell cycle arrest and cell death of human erythroid progenitor cells (EPCs) that host B19V replication (5, 16, 18, 19). To date, a vaccine against B19V infection and specific treatments for B19V infection-caused transient aplastic crisis, chronic anemia, pure red cell aplasia, hydrops fetalis, and congenital anemia in infants are not available. Although intravenous immunoglobulin (IVIG) can be administered to patients with B19V-associated chronic anemia and pure red cell aplasia (50–52), the symptoms often recur when IVIG treatment is interrupted (53–56). The repeated applications of IVIG and the maintenance therapy that are required to fully eliminate B19V-associated pure red cell aplasia are cost prohibitive (51, 57–63). As all these complications of hematological disorders caused by B19V infection are the direct outcome of B19V infection of EPCs, diminishing B19V replication in EPCs is expected to be an effective approach for treating B19V infection-induced hematological disorders. Cidofovir, an acyclic nucleoside phosphonate broadly active against dsDNA viruses, has shown some promise in having an antiviral effect on B19V infection (64, 65). Small-molecule compounds that can specifically inhibit B19V replication in ex vivo-expanded CD36+ EPCs will be ideal candidates for testing as drugs for the treatment of hematological disorders caused by B19V infection.
In the current study, we identified 8 compounds that exhibit inhibition of NS1 nicking of the B19V Ori from the screening of 96 small-molecule compounds, which have been used to screen inhibitors of the RNase H nuclease activity of the HBV polymerase (43–47), using an in vitro nicking assay. We chose three compounds, compounds 7, 135, and 201, that share a similar flavonoid chemical structure for detailed study. All three flavonoid compounds inhibited B19V DNA replication in UT7/Epo-S1 cells and exhibited a selective index of 3 to 4. However, the compounds showed higher cytotoxicity in primary CD36+ EPCs with a therapeutic index of >1.5, although they inhibited B19V infection at concentrations that did not show significant cytotoxicity. These data indicate that the 8 screening hits identified here are candidates for future medicinal chemistry campaigns to improve efficacy and reduce cytotoxicity.
Flavonoids are a class of ubiquitous secondary plant metabolites. They have been reported to have functions against a wide range of biological activities, including antimicrobe, antioxidant, anticancer, anti-inflammatory, and eukaryotic enzyme inhibition properties (66, 67). As antivirals, flavonoids have been shown to execute antiviral activities against many human viruses, including DNA viruses, RNA viruses, and retroviruses, and these have been observed in cell culture and animals (68). Mechanistically, flavonoids function in various pathways of the virus life cycle, including virus entry (69), RNA transcription of the viral genome (70, 71), and viral RNA translation (72, 73). Flavonoids can inhibit viral protease activity (74, 75), viral helicase activity (76, 77), and viral (HIV) reverse transcriptase (78, 79). They have also been shown to suppress cellular pathways that are essential for virus replication (80). Our studies expand the range of antiviral mechanisms possessed by the flavonoids to include the endonuclease activity of viral proteins (68). The flavonoids inhibited B19V infection, determined by assessment of viral capsid expression, and B19V DNA replication in UT7/Epo-S1 cells transfected using nucleofection (nucleofected) with M20 B19V infectious DNA (Fig. 8 and 9), as well as B19V DNA replication during early infection of CD36+ EPCs (Fig. 12), suggesting that the inhibition likely takes place in steps of viral DNA replication. As the nicking of the Ori by NS1 is an essential step for B19V DNA replication (81), we believe that flavonoids specifically target the nicking step. Another important class of small molecules that inhibits NS1N nicking are naphthyridinones. Compounds 12 and 153 have been shown to inhibit the RNase H activity of the HBV polymerase (45, 82). Naphthyridinones have also been shown to inhibit the nuclease activity of HIV reverse transcriptase (83). As a next step, we will test compounds 12, 151, and 153 for their activity against B19V infection in cells.
Identification of multiple hits in two chemotypes provides clues regarding the structure-activity relationships of these compounds. All of the flavonoid and naphthyridinone hits share a hydrophilic two-ring heteroatom aromatic structure with a single appendage containing one or more benzyl moieties extending from similar spots on the bi-ring structure. Converting the benzylic extensions seen in naphthyridinones 151 and 153 to a hydroxyl in compounds 152 and 148 (Fig. 6) ablated the activity against NS1N (Fig. 3 and 5). Similarly, ablating the hydroxyl on the nitrogen heteroatom in compound 12 to create compound 150 (Fig. 6) also ablated the activity, possibly by removing the potential for chelation of the Mg2+ ions in the NS1N active site, which is known to be important, as the naphthyridinones bind in this way to the HIV RNase H (84). These similarities will be valuable during future medicinal chemistry optimization of both the flavonoid and the naphthyridinone scaffolds.
An important contribution of this study is the establishment of a fluorescent in vitro nicking assay. We have identified the 67-nt Ori that supports B19V DNA replication in B19V-permisive UT7/Epo-S1 cells (41) and characterized the NS1BEs of the Ori in vitro (27). Based on this information, an in vitro nicking assay using a 32P-labeled DNA probe, which spans the trs in the Ori, and purified B19V NS1N was established (28). This radioactive nicking assay is not appropriate for high-throughput screening of small-molecule compounds that inhibit the B19V NS1N-driven nicking of the Ori. NS1N efficiently cleaves the FAMOri20Q probe and releases a significant fluorescent signal. Compared with the fluorescence intensity of the non-NS1N control, NS1N increases the fluorescence intensity by >18-fold, achieving a signal ∼72% of the signal from a nonquenched probe, FAMOri20. Notably, the FAM-based nicking assay possesses the same sensitivity and specificity for the detection of inhibition of NS1N cleavage of the Ori as the radioactive nicking assay. The two assays exhibited a good correlation and minimal difference in detection of the NS1N-effected nicking of the B19V Ori. We expect that this FAM-based fluorescent nicking assay could be utilized in high-throughput screening of large libraries of small-molecule compounds for anti-B19V drug candidates.
MATERIALS AND METHODS
Compounds.
The compounds used in this study were as follows: 1 to 5, 7, 9 to 12, 19, 20, 23, 34 49 to 52, 54, 60, 62, 64, 65, 67 to 69, 121, 125 to 130, 132, 134 to 139, 148 to 158, 197, 198, 200 to 209, 214, 215, 217, 218, 304, 307, 321 to 329, 338 to 346, 348 to 350, and 352 to 357. Their structures are shown in Table S1 in the supplemental material. They were acquired commercially or synthesized (as described in the methods in the supplemental material) (85–87). The synthesized compounds were purified to ≥95% purity, as analyzed by high-performance liquid chromatography. All compounds were dissolved in DMSO (Sigma, St. Louis, MO) and stored at −80°C.
Ethics statement.
Primary human CD34+ hematopoietic stem cells (HSCs) were isolated from the bone marrow of a healthy human donor. We purchased them from AllCells LLC (Alameda, CA), and B19V-containng plasma samples were obtained from ViraCor Eurofins Laboratories (Lee’s Summit, MO). Both the cell and virus samples were deidentified, and therefore institutional review board (IRB) reviews were waived.
Cell line and primary cells.
(i) Primary human CD36+ EPCs. We expanded CD34+ hematopoietic stem cells ex vivo under normoxic conditions (21% O2, 5% CO2) at 37°C until day 4 (the day of procurement was designated day 0) (49, 88). On day 4, the expanded cells were frozen in liquid nitrogen and are referred to as “day 4 cells.” For each experiment, day 4 cells were thawed and cultured for 2 to 3 days in Wong expansion medium (88) under normoxic conditions. On day 6 or 7, the cells were cultured under hypoxic conditions (1% O2 and 5% CO2) for 2 days; these cells expressed CD36+ (a marker for erythroid cells), and so we named them CD36+ erythroid progenitor cells (CD36+ EPCs).
(ii) UT7/Epo-S1 cells. UT7/Epo-S1 cells, which are cells of a human megakaryoblastoid cell line obtained from Kevin Brown with permission from Kazuo Sugamura (89), were cultured under normoxic conditions in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum and erythropoietin (2 U/ml; Amgen, Thousand Oaks, CA).
Virus and infection. Plasma sample 404, which contained B19V at 1 × 1012 viral genomic copies (vgc) per ml, as quantified by qPCR, was provided by ViraCor Eurofins Laboratories (Lee’s Summit, MO). CD36+ EPCs were infected with B19V at a multiplicity of infection (MOI) of 1,000 vgc per cell. After 2 days of incubation under hypoxic conditions, the infected cells were collected for flow cytometry and extraction of low-molecular-weight (Hirt) DNA (41). In addition, at 18 h, 30 h, and 48 h postinfection, total DNA was extracted from the cells for qPCR.
Electroporation.
Two million UT7/Epo-S1 cells were electroporated with 3 µg of SalI-linearized B19V infectious clone pM20 (48) in solution V using the Amaxa Nucleofector technology (Lonza, Basel, Switzerland), as described previously (29). After electroporation, the cells were cultured under hypoxic conditions (1% O2 and 5% CO2) for 2 days.
Southern blot analysis.
Hirt DNA was extracted from either B19V-infected CD36+ EPCs or B19V duplex genome (M20)-transfected UT7/Epo-S1 cells and was analyzed by Southern blotting, as described previously (41). M20 excised from SalI-digested pM20 was used as a probe. The blots were reprobed for mitochondrial (mt) DNA using a specific probe (90).
Protein expression and purification.
NS1 N-terminal aa 1 to 176 (NS1N)- and NS1N endonuclease motif mutant (NS1NmEndo)-coding sequences were ligated into the vector pET-30a (GE Healthcare Life Sciences) through NdeI and XhoI sites. Both plasmids were transformed into Escherichia coli BL21(DE3)pLyss competent cells. Protein expression was induced by addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to the bacterial culture at 1 mM. Protein was purified with Ni-nitrilotriacetic acid (NTA) affinity agarose (Qiagen) as described previously (42).
In vitro DNA nicking assays.
(i) Radioactive nicking assay. The oligonucleotide was 5′ end labeled with [γ-32P]ATP (PerkinElmer, Inc.) using T4 polynucleotide kinase (NEB). The labeled oligonucleotide was purified using MicroSpin G-50 columns (GE Healthcare). 32P-labeled oligonucleotide and NS1N at final concentrations of 2 nM and 2 µM, respectively, were added to the nicking buffer (50 mM HEPES-NaOH, pH 7.0, 150 mM NaCl, 10 mM CoCl2) in a total volume of 20 μl. The reaction mixture was incubated at 37°C for 16 to 18 h and was quenched by the addition of 20 μl of quenching solution (80% formamide, 50 mM EDTA, 1 mg/ml xylene cyanol FF dye, 1 mg/ml bromophenol blue dye), before electrophoresis on denaturing polyacrylamide (20% acrylamide-bisacrylamide [19:1 ratio], 4 M urea, 89 mM Tris base, 89 mM boric acid, 2 mM EDTA) gels. The gels were autoradiographed without drying against a phosphor screen at 4°C for 12 to 18 h. Images were obtained by scanning the phosphor screen on a GE Typhoon FLA 9000 scanner (GE Healthcare) and processed with ImageQuant TL (GE Healthcare) software for quantification.
(ii) Fluorophore-based nicking assay. Fluorescein (FAM)-labeled oligonucleotides were synthesized with 6-carboxyfluorescein (FAM) at the 5′ end and Iowa Black FQ quencher at the 3′ end at Integrated DNA Technologies, Inc. (Coralville, IA). The labeled oligonucleotide was diluted to 100 µM as a stock solution. The in vitro nicking reaction mixture was set up at 60 µl and was composed of labeled oligonucleotide at 200 nM and purified NS1N at 2 µM in the nicking buffer. After incubation for 16 to 18 h, the samples were transferred to a black 96-well plate (catalog number 3991; Corning). The fluorescence intensity of each sample was detected with excitation at 492 nm and emission at 518 nm on a microplate reader (Synergy H1 BioTek).
IC50, EC50, and CC50 determinations.
(i) IC50. In the fluorophore-based nicking assay, various concentrations of each inhibitor were used in the in vitro nicking reaction with the Ori20 oligonucleotide. The fluorescence intensity of reactions with FAMOri20Q but without NS1N was set up as the background. The fluorescence readings of the reactions with each concentration of the compound were compared with those for the DMSO (1%) control. The half-maximal inhibitory concentration (IC50) of each compound was calculated using GraphPad Prism software.
(ii) EC50 based on viral capsid expression. The half-maximal effective concentration (EC50) is the concentration at which half of virus replication is inhibited in cells. For UT7/Epo-S1 cells, the cells were cultured under hypoxic conditions for 2 days, and then the cells were transfected with B19V duplex genome M20. Transfected cells were diluted with medium to 0.4 million cells/ml, and 1 ml was transferred to 12-well plates, followed by addition of a compound at various concentrations. After 2 days under hypoxic conditions, the cells were collected for flow cytometry to detect the capsid-expressing cells. For CD36+ EPCs, compounds were added to B19V-infected CD36+ EPCs at various concentrations. After 2 days, the infected cells were collected for flow cytometry to detect the capsid-positive cells. The final EC50 was calculated using GraphPad Prism software. DMSO (0.1%) was used as a vehicle control.
(iii) EC50 determined based on levels of viral DNA. Day 6 CD36+ EPCs were cultured under hypoxic condition for 2 days, and then the cells were infected with B19V for 1 h and washed three times with the culture medium. The infected cells were diluted with the medium to 0.4 million cells/ml, and 0.1 ml of cells was transferred to the wells of a 96-well plate, followed by addition of the compounds at various concentrations. At 18 h, 30 h, and 48 h postinfection, infected cells were collected for extraction of total DNA using a DNeasy blood and tissue kit (Qiagen) following the manufacturer’s instructions. A multiplex qPCR assay using a set of probe and primers targeting the B19V VP1 unique region-coding sequence and a set of probe and primers targeting cellular mitochondrial DNA was used to quantify the viral DNA and cellular DNA, respectively. The final EC50 was calculated using GraphPad Prism software as the ratio of B19V DNA/mt DNA for treated cells relative to that for the DMSO (0.1%)-treated control group.
(iv) Drug concentration that affects the viability of 50% of cells in culture (CC50). UT7/Epo-S1 cells or day 6 CD36+ EPCs were cultured under hypoxic condition for 2 days, and then the cells were diluted with medium to 0.4 million cells/ml and 0.1 ml was transferred to 96-well plates. The compounds were added at different concentrations. After 2 days, cell viability was determined using a CytoTox-Glo cytotoxicity assay kit (Promega) by following the manufacturer’s instructions. We used GraphPad Prism software to calculate the CC50.
Flow cytometry.
To detect B19V capsid-expressing cells, infected or transfected cells were collected, fixed, and permeabilized as described previously (29). A mouse anti-B19V capsid monoclonal antibody (catalog no. MAB8292; clone 521-5D; MilliporeSigma) was used to detect capsid-expressing cells on a 3-laser flow cytometer (LSR II; BD Biosciences, San Jose, CA). All flow data were analyzed using FACSDIVA software (BD Biosciences).
Quantification of viral and mt DNA using multiplex qPCR.
The primers and FAM-labeled probe used for quantification of B19V DNA were VP1u forward primer (5′-CCT GGG CAA GTT AGC GTA C-3′; nt 2975 to 2993), VP1u reverse primer (5′-ATG AAT CCT TGC AGC ACT GTC A-3′; nt 3082-3061), and the VP1u probe (5′–6-FAM-CCG GTA CTA/ZEN/ACT ATG TTG GGC CTG GCA A-3IABkFQ–3′; nt 3000 to 3027). The primers and 6-carboxy-4',5'-dichloro-2',7'-dimethoxyfluorescein (JOE)-labeled probe used for quantification of mt DNA were the mt forward primer (5′-TCA AAC TCA AAC TAC GCC CTG-3′; nt 3673 to 3693), the mt reverse primer (5′-GTT GTG ATA AGG GTG GAG AGG-3′; nt 3809 to 3789), and the mt probe (5′–6-JOEN/TGC GAG CAG/ZEN/TAG CCC AAA CAA TCT-3IABkFQ–3′; nt 3704 to 3727). Multiple PCR was performed as described previously (41, 49) on a 7500 Fast real-time PCR system (Applied Biosystems). For the nucleotide sequences of B19V and mitochondrial DNA, refer to GenBank accession numbers AY386330.1 and GU170821.1, respectively.
Statistical analyses.
GraphPad Prism (version 8) software was used to calculate the IC50, EC50, and CC50 using the inhibitor-versus-response variable slope (four parameters).
The correlation (r value) between data obtained by two methods (the two assays) was analyzed by use of the Pearson correlation and SPSS software (IBM Corp, Armonk, NY). When comparing two methods through SPSS analysis, the coefficient of correlation was a P value of <0.01, which was considered significant. The two methods were considered to be highly correlated as long as r was ≥0.8. A scatter plot, the trend line, and R2 were calculated at the same time.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the J. Qiu and J. E. Tavis labs for technical support and valuable discussions. We acknowledge the Flow Cytometry Core Laboratory, University of Kansas Medical Center, for help in flow cytometry analysis. We also thank Sarah McNitt and Max Bourdillon for the synthesis of compounds.
This study was supported by PHS grant R01 AI070723 (to J.Q.), R01 AI104494 (to J.E.T.), and R21 AI124672 (to J.E.T. and M.J.M.) from the National Institute of Allergy and Infectious Diseases.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AAC.01879-18.
REFERENCES
- 1.Cossart YE, Field AM, Cant B, Widdows D. 1975. Parvovirus-like particles in human sera. Lancet i:72–73. [DOI] [PubMed] [Google Scholar]
- 2.Cotmore SF, Agbandje-McKenna M, Chiorini JA, Mukha DV, Pintel DJ, Qiu J, Söderlund-Venermo M, Tattersall P, Tijssen P, Gatherer D, Davison AJ. 2014. The family Parvoviridae. Arch Virol 159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ozawa K, Kurtzman G, Young N. 1986. Replication of the B19 parvovirus in human bone marrow cell cultures. Science 233:883–886. doi: 10.1126/science.3738514. [DOI] [PubMed] [Google Scholar]
- 4.Srivastava A, Lu L. 1988. Replication of B19 parvovirus in highly enriched hematopoietic progenitor cells from normal human bone marrow. J Virol 62:3059–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yaegashi N, Niinuma T, Chisaka H, Uehara S, Moffatt S, Tada K, Iwabuchi M, Matsunaga Y, Nakayama M, Yutani C, Osamura Y, Hirayama E, Okamura K, Sugamura K, Yajima A. 1999. Parvovirus B19 infection induces apoptosis of erythroid cells in vitro and in vivo. J Infect 39:68–76. doi: 10.1016/S0163-4453(99)90105-6. [DOI] [PubMed] [Google Scholar]
- 6.Morey AL, Fleming KA. 1992. Immunophenotyping of fetal haemopoietic cells permissive for human parvovirus B19 replication in vitro. Br J Haematol 82:302–309. doi: 10.1111/j.1365-2141.1992.tb06422.x. [DOI] [PubMed] [Google Scholar]
- 7.Anderson MJ, Khousam MN, Maxwell DJ, Gould SJ, Happerfield LC, Smith WJ. 1988. Human parvovirus B19 and hydrops fetalis. Lancet i:535. [DOI] [PubMed] [Google Scholar]
- 8.Nunoue T, Okochi K, Mortimer PP, Cohen BJ. 1985. Human parvovirus (B19) and erythema infectiosum. J Pediatr 107:38–40. doi: 10.1016/S0022-3476(85)80610-7. [DOI] [PubMed] [Google Scholar]
- 9.Chorba T, Coccia P, Holman RC, Tattersall P, Anderson LJ, Sudman J, Young NS, Kurczynski E, Saarinen UM, Moir R. 1986. The role of parvovirus B19 in aplastic crisis and erythema infectiosum (fifth disease). J Infect Dis 154:383–393. doi: 10.1093/infdis/154.3.383. [DOI] [PubMed] [Google Scholar]
- 10.Qiu J, Söderlund-Venermo M, Young NS. 2017. Human parvoviruses. Clin Microbiol Rev 30:43–113. doi: 10.1128/CMR.00040-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young NS, Brown KE. 2004. Parvovirus B19. N Engl J Med 350:586–597. doi: 10.1056/NEJMra030840. [DOI] [PubMed] [Google Scholar]
- 12.de Jong EP, de Haan TR, Kroes AC, Beersma MF, Oepkes D, Walther FJ. 2006. Parvovirus B19 infection in pregnancy. J Clin Virol 36:1–7. doi: 10.1016/j.jcv.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 13.Woolf AD, Campion GV, Chishick A, Wise S, Cohen BJ, Klouda PT, Caul O, Dieppe PA. 1989. Clinical manifestations of human parvovirus B19 in adults. Arch Intern Med 149:1153–1156. doi: 10.1001/archinte.1989.00390050111022. [DOI] [PubMed] [Google Scholar]
- 14.Al-Khan A, Caligiuri A, Apuzzio J. 2003. Parvovirus B-19 infection during pregnancy. Infect Dis Obstet Gynecol 11:175–179. doi: 10.1080/10647440300025518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chisaka H, Morita E, Yaegashi N, Sugamura K. 2003. Parvovirus B19 and the pathogenesis of anaemia. Rev Med Virol 13:347–359. doi: 10.1002/rmv.395. [DOI] [PubMed] [Google Scholar]
- 16.Sol N, Le JJ, Vassias I, Freyssinier JM, Thomas A, Prigent AF, Rudkin BB, Fichelson S, Morinet F. 1999. Possible interactions between the NS-1 protein and tumor necrosis factor alpha pathways in erythroid cell apoptosis induced by human parvovirus B19. J Virol 73:8762–8770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morita E, Sugamura K. 2002. Human parvovirus B19-induced cell cycle arrest and apoptosis. Springer Semin Immunopathol 24:187–199. doi: 10.1007/s00281-002-0099-6. [DOI] [PubMed] [Google Scholar]
- 18.Chen AY, Zhang EY, Guan W, Cheng F, Kleiboeker S, Yankee TM, Qiu J. 2010. The small 11kDa non-structural protein of human parvovirus B19 plays a key role in inducing apoptosis during B19 virus infection of primary erythroid progenitor cells. Blood 115:1070–1080. doi: 10.1182/blood-2009-04-215756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moffatt S, Yaegashi N, Tada K, Tanaka N, Sugamura K. 1998. Human parvovirus B19 nonstructural (NS1) protein induces apoptosis in erythroid lineage cells. J Virol 72:3018–3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lou S, Luo Y, Cheng F, Huang Q, Shen W, Kleiboeker S, Tisdale JF, Liu Z, Qiu J. 2012. Human parvovirus B19 DNA replication induces a DNA damage response that is dispensable for cell cycle arrest at G2/M phase. J Virol 86:10748–10758. doi: 10.1128/JVI.01007-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown KE, Young N. 1997. Human parvovirus B19: pathogenesis of disease, p 105–119. In Anderson LJ, Young N (ed), Human parvovirus B19, vol 20 Karger, Basel, Switzerland. [Google Scholar]
- 22.Chen AY, Qiu J. 2010. Parvovirus infection-induced cell death and cell cycle arrest. Future Virol 5:731–741. doi: 10.2217/fvl.10.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Qiu J, Cheng F, Chu Y, Yoto Y, O'Sullivan MG, Brown KE, Pintel DJ. 2004. Comparison of the transcription profile of simian parvovirus with that of the human erythrovirus B19 reveals a number of unique features. J Virol 78:12929–12939. doi: 10.1128/JVI.78.23.12929-12939.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cotmore SF, McKie VC, Anderson LJ, Astell CR, Tattersall P. 1986. Identification of the major structural and nonstructural proteins encoded by human parvovirus B19 and mapping of their genes by procaryotic expression of isolated genomic fragments. J Virol 60:548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ganaie SS, Qiu J. 2018. Recent advances in replication and infection of human parvovirus B19. Front Cell Infect Microbiol 8:166. doi: 10.3389/fcimb.2018.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wan Z, Zhi N, Wong S, Keyvanfar K, Liu D, Raghavachari N, Munson PJ, Su S, Malide D, Kajigaya S, Young NS. 2010. Human parvovirus B19 causes cell cycle arrest of human erythroid progenitors via deregulation of the E2F family of transcription factors. J Clin Invest 120:3530–3544. doi: 10.1172/JCI41805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tewary SK, Zhao H, Deng X, Qiu J, Tang L. 2014. The human parvovirus B19 non-structural protein 1 N-terminal domain specifically binds to the origin of replication in the viral DNA. Virology 449:297–303. doi: 10.1016/j.virol.2013.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez JL, Romero Z, Quinones A, Torgeson KR, Horton NC. 2016. DNA binding and cleavage by the human parvovirus B19 NS1 nuclease domain. Biochemistry 55:6577–6593. doi: 10.1021/acs.biochem.6b00534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu P, Zhou Z, Xiong M, Zou W, Deng X, Ganaie SS, Kleiboeker S, Peng J, Liu K, Wang S, Ye SQ, Qiu J. 2017. Parvovirus B19 NS1 protein induces cell cycle arrest at G2-phase by activating the ATR-CDC25C-CDK1 pathway. PLoS Pathog 13:e1006266. doi: 10.1371/journal.ppat.1006266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhi N, Mills IP, Lu J, Wong S, Filippone C, Brown KE. 2006. Molecular and functional analyses of a human parvovirus B19 infectious clone demonstrates essential roles for NS1, VP1, and the 11-kilodalton protein in virus replication and infectivity. J Virol 80:5941–5950. doi: 10.1128/JVI.02430-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gareus R, Gigler A, Hemauer A, Leruez-Ville M, Morinet F, Wolf H, Modrow S. 1998. Characterization of cis-acting and NS1 protein-responsive elements in the p6 promoter of parvovirus B19. J Virol 72:609–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moffatt S, Tanaka N, Tada K, Nose M, Nakamura M, Muraoka O, Hirano T, Sugamura K. 1996. A cytotoxic nonstructural protein, NS1, of human parvovirus B19 induces activation of interleukin-6 gene expression. J Virol 70:8485–8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakashima A, Morita E, Saito S, Sugamura K. 2004. Human parvovirus B19 nonstructural protein transactivates the p21/WAF1 through Sp1. Virology 329:493–504. doi: 10.1016/j.virol.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Cotmore SF, Tattersall P. 2014. Parvoviruses: small does not mean simple. Annu Rev Virol 1:517–537. doi: 10.1146/annurev-virology-031413-085444. [DOI] [PubMed] [Google Scholar]
- 35.Weitzman MD, Linden RM. 2011. Adeno-associated virus biology. Methods Mol Biol 807:1–23. doi: 10.1007/978-1-61779-370-7_1. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Deng X, Zou W, Engelhardt JF, Yan Z, Qiu J. 2017. Human bocavirus 1 is a novel helper for adeno-associated virus replication. J Virol 91:e00710-17. doi: 10.1128/JVI.00710-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo Y, Lou S, Deng X, Liu Z, Li Y, Kleiboeker S, Qiu J. 2011. Parvovirus B19 infection of human primary erythroid progenitor cells triggers ATR-Chk1 signaling, which promotes B19 virus replication. J Virol 85:8046–8055. doi: 10.1128/JVI.00831-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo Y, Kleiboeker S, Deng X, Qiu J. 2013. Human parvovirus B19 infection causes cell cycle arrest of human erythroid progenitors at late S phase that favors viral DNA replication. J Virol 87:12766–12775. doi: 10.1128/JVI.02333-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Luo Y, Qiu J. 2015. Human parvovirus B19: a mechanistic overview of infection and DNA replication. Future Virol 10:155–167. doi: 10.2217/fvl.14.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zou W, Wang Z, Xiong M, Chen AY, Xu P, Ganaie SS, Badawi Y, Kleiboeker S, Nishimune H, Ye SQ, Qiu J. 2018. Human parvovirus B19 utilizes cellular DNA replication machinery for viral DNA replication. J Virol 92:e01881-17. doi: 10.1128/JVI.01881-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guan W, Wong S, Zhi N, Qiu J. 2009. The genome of human parvovirus B19 virus can replicate in non-permissive cells with the help of adenovirus genes and produces infectious virus. J Virol 83:9541–9553. doi: 10.1128/JVI.00702-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ganaie SS, Zou W, Xu P, Deng X, Kleiboeker S, Qiu J. 2017. Phosphorylated STAT5 directly facilitates parvovirus B19 DNA replication in human erythroid progenitors through interaction with the MCM complex. PLoS Pathog 13:e1006370. doi: 10.1371/journal.ppat.1006370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cai CW, Lomonosova E, Moran EA, Cheng X, Patel KB, Bailly F, Cotelle P, Meyers MJ, Tavis JE. 2014. Hepatitis B virus replication is blocked by a 2-hydroxyisoquinoline-1,3(2H,4H)-dione (HID) inhibitor of the viral ribonuclease H activity. Antiviral Res 108:48–55. doi: 10.1016/j.antiviral.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tavis JE, Cheng X, Hu Y, Totten M, Cao F, Michailidis E, Aurora R, Meyers MJ, Jacobsen EJ, Parniak MA, Sarafianos SG. 2013. The hepatitis B virus ribonuclease H is sensitive to inhibitors of the human immunodeficiency virus ribonuclease H and integrase enzymes. PLoS Pathog 9:e1003125. doi: 10.1371/journal.ppat.1003125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tavis JE, Lomonosova E. 2015. The hepatitis B virus ribonuclease H as a drug target. Antiviral Res 118:132–138. doi: 10.1016/j.antiviral.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lomonosova E, Daw J, Garimallaprabhakaran AK, Agyemang NB, Ashani Y, Murelli RP, Tavis JE. 2017. Efficacy and cytotoxicity in cell culture of novel alpha-hydroxytropolone inhibitors of hepatitis B virus ribonuclease H. Antiviral Res 144:164–172. doi: 10.1016/j.antiviral.2017.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edwards TC, Lomonosova E, Patel JA, Li Q, Villa JA, Gupta AK, Morrison LA, Bailly F, Cotelle P, Giannakopoulou E, Zoidis G, Tavis JE. 2017. Inhibition of hepatitis B virus replication by N-hydroxyisoquinolinediones and related polyoxygenated heterocycles. Antiviral Res 143:205–217. doi: 10.1016/j.antiviral.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhi N, Zadori Z, Brown KE, Tijssen P. 2004. Construction and sequencing of an infectious clone of the human parvovirus B19. Virology 318:142–152. doi: 10.1016/j.virol.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 49.Chen AY, Kleiboeker S, Qiu J. 2011. Productive parvovirus B19 infection of primary human erythroid progenitor cells at hypoxia is regulated by STAT5A and MEK signaling but not HIF alpha. PLoS Pathog 7:e1002088. doi: 10.1371/journal.ppat.1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kurtzman G, Frickhofen N, Kimball J, Jenkins DW, Nienhuis AW, Young NS. 1989. Pure red-cell aplasia of 10 years' duration due to persistent parvovirus B19 infection and its cure with immunoglobulin therapy. N Engl J Med 321:519–523. doi: 10.1056/NEJM198908243210807. [DOI] [PubMed] [Google Scholar]
- 51.Mouthon L, Guillevin L, Tellier Z. 2005. Intravenous immunoglobulins in autoimmune- or parvovirus B19-mediated pure red-cell aplasia. Autoimmun Rev 4:264–269. doi: 10.1016/j.autrev.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 52.Young NS. 1996. Parvovirus infection and its treatment. Clin Exp Immunol 104:26–30. [PubMed] [Google Scholar]
- 53.Hayakawa F, Imada K, Towatari M, Saito H. 2002. Life-threatening human parvovirus B19 infection transmitted by intravenous immune globulin. Br J Haematol 118:1187–1189. doi: 10.1046/j.1365-2141.2002.03741.x. [DOI] [PubMed] [Google Scholar]
- 54.Egbuna O, Zand MS, Arbini A, Menegus M, Taylor J. 2006. A cluster of parvovirus B19 infections in renal transplant recipients: a prospective case series and review of the literature. Am J Transplant 6:225–231. doi: 10.1111/j.1600-6143.2005.01139.x. [DOI] [PubMed] [Google Scholar]
- 55.Renoult E, Bachelet C, Krier-Coudert M-J, Diarrassouba A, André J-L, Kessler M. 2006. Recurrent anemia in kidney transplant recipients with parvovirus B19 infection. Transplant Proc 38:2321–2323. doi: 10.1016/j.transproceed.2006.06.116. [DOI] [PubMed] [Google Scholar]
- 56.Ogawa E, Otaguro S, Murata M, Kainuma M, Sawayama Y, Furusyo N, Hayashi J, Furusyo N, Hayashi J. 2008. Intravenous immunoglobulin therapy for severe arthritis associated with human parvovirus B19 infection. J Infect Chemother 14:377–382. doi: 10.1007/s10156-008-0636-X. [DOI] [PubMed] [Google Scholar]
- 57.Shen Q, Xu H, Cao Q, Zhou LJ, Xu J, Fang XY, Ge J. 2011. Long-term remission of recurrent severe anemia as a result of parvovirus B19 infection in a pediatric renal transplant recipient. Pediatr Transplant 15:E76–E79. doi: 10.1111/j.1399-3046.2010.01291.x. [DOI] [PubMed] [Google Scholar]
- 58.Mrzljak A, Kardum-Skelin I, Cvrlje VC, Kanizaj TF, Sustercic D, Gustin D, Kocman B. 2010. Parvovirus B 19 (PVB19) induced pure red cell aplasia (PRCA) in immunocompromised patient after liver transplantation. Coll Antropol 34:271–274. [PubMed] [Google Scholar]
- 59.Moudgil A, Shidban H, Nast CC, Bagga A, Aswad S, Graham SL, Mendez R, Jordan SC. 1997. Parvovirus B19 infection-related complications in renal transplant recipients: treatment with intravenous immunoglobulin. Transplantation 64:1847–1850. doi: 10.1097/00007890-199712270-00037. [DOI] [PubMed] [Google Scholar]
- 60.Koduri PR, Kumapley R, Khokha ND, Patel AR. 1997. Red cell aplasia caused by parvovirus B19 in AIDS: use of i.v. immunoglobulin. Ann Hematol 75:67–68. doi: 10.1007/s002770050315. [DOI] [PubMed] [Google Scholar]
- 61.Ramratnam B, Gollerkeri A, Schiffman FJ, Rintels P, Flanigan TP. 1995. Management of persistent B19 parvovirus infection in AIDS. Br J Haematol 91:90–92. doi: 10.1111/j.1365-2141.1995.tb05250.x. [DOI] [PubMed] [Google Scholar]
- 62.Koduri PR, Kumapley R, Valladares J, Teter C. 1999. Chronic pure red cell aplasia caused by parvovirus B19 in AIDS: use of intravenous immunoglobulin—a report of eight patients. Am J Hematol 61:16–20. doi:. [DOI] [PubMed] [Google Scholar]
- 63.Hung CC, Lee KL, Chen MY. 2001. Increase in B19 viral load prior to relapse of anaemia in an AIDS patient with persistent B19 infection. J Infect 43:150–152. doi: 10.1053/jinf.2001.0882. [DOI] [PubMed] [Google Scholar]
- 64.Bonvicini F, Bua G, Manaresi E, Gallinella G. 2015. Antiviral effect of cidofovir on parvovirus B19 replication. Antiviral Res 113:11–18. doi: 10.1016/j.antiviral.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 65.Bonvicini F, Bua G, Manaresi E, Gallinella G. 2016. Enhanced inhibition of parvovirus B19 replication by cidofovir in extendedly exposed erythroid progenitor cells. Virus Res 220:47–51. doi: 10.1016/j.virusres.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 66.Cushnie TP, Lamb AJ. 2005. Antimicrobial activity of flavonoids. Int J Antimicrob Agents 26:343–356. doi: 10.1016/j.ijantimicag.2005.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mansuri ML, Parihar P, Solanki I, Parihar MS. 2014. Flavonoids in modulation of cell survival signalling pathways. Genes Nutr 9:400. doi: 10.1007/s12263-014-0400-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zakaryan H, Arabyan E, Oo A, Zandi K. 2017. Flavonoids: promising natural compounds against viral infections. Arch Virol 162:2539–2551. doi: 10.1007/s00705-017-3417-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hung PY, Ho BC, Lee SY, Chang SY, Kao CL, Lee SS, Lee CN. 2015. Houttuynia cordata targets the beginning stage of herpes simplex virus infection. PLoS One 10:e0115475. doi: 10.1371/journal.pone.0115475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Argenta DF, Silva IT, Bassani VL, Koester LS, Teixeira HF, Simoes CM. 2015. Antiherpes evaluation of soybean isoflavonoids. Arch Virol 160:2335–2342. doi: 10.1007/s00705-015-2514-z. [DOI] [PubMed] [Google Scholar]
- 71.Qian K, Gao AJ, Zhu MY, Shao HX, Jin WJ, Ye JQ, Qin AJ. 2014. Genistein inhibits the replication of avian leucosis virus subgroup J in DF-1 cells. Virus Res 192:114–120. doi: 10.1016/j.virusres.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 72.Ganesan S, Faris AN, Comstock AT, Wang Q, Nanua S, Hershenson MB, Sajjan US. 2012. Quercetin inhibits rhinovirus replication in vitro and in vivo. Antiviral Res 94:258–271. doi: 10.1016/j.antiviral.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Min N, Leong PT, Lee RCH, Khuan JSE, Chu JJH. 2018. A flavonoid compound library screen revealed potent antiviral activity of plant-derived flavonoids on human enterovirus A71 replication. Antiviral Res 150:60–68. doi: 10.1016/j.antiviral.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 74.Lin YJ, Chang YC, Hsiao NW, Hsieh JL, Wang CY, Kung SH, Tsai FJ, Lan YC, Lin CW. 2012. Fisetin and rutin as 3C protease inhibitors of enterovirus A71. J Virol Methods 182:93–98. doi: 10.1016/j.jviromet.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 75.Yang L, Lin J, Zhou B, Liu Y, Zhu B. 2017. Activity of compounds from Taxillus sutchuenensis as inhibitors of HCV NS3 serine protease. Nat Prod Res 31:487–491. doi: 10.1080/14786419.2016.1190719. [DOI] [PubMed] [Google Scholar]
- 76.Bachmetov L, Gal-Tanamy M, Shapira A, Vorobeychik M, Giterman-Galam T, Sathiyamoorthy P, Golan-Goldhirsh A, Benhar I, Tur-Kaspa R, Zemel R. 2012. Suppression of hepatitis C virus by the flavonoid quercetin is mediated by inhibition of NS3 protease activity. J Viral Hepat 19:e81–e88. doi: 10.1111/j.1365-2893.2011.01507.x. [DOI] [PubMed] [Google Scholar]
- 77.Yu MS, Lee J, Lee JM, Kim Y, Chin YW, Jee JG, Keum YS, Jeong YJ. 2012. Identification of myricetin and scutellarein as novel chemical inhibitors of the SARS coronavirus helicase, nsP13. Bioorg Med Chem Lett 22:4049–4054. doi: 10.1016/j.bmcl.2012.04.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Behbahani M, Sayedipour S, Pourazar A, Shanehsazzadeh M. 2014. In vitro anti-HIV-1 activities of kaempferol and kaempferol-7-O-glucoside isolated from Securigera securidaca. Res Pharm Sci 9:463–469. [PMC free article] [PubMed] [Google Scholar]
- 79.Nakane H, Ono K. 1989. Differential inhibition of HIV-reverse transcriptase and various DNA and RNA polymerases by some catechin derivatives. Nucleic Acids Symp Ser 1989:115–116. [PubMed] [Google Scholar]
- 80.Wang C, Wang P, Chen X, Wang W, Jin Y. 2015. Saururus chinensis (Lour.) Baill blocks enterovirus 71 infection by hijacking MEK1-ERK signaling pathway. Antiviral Res 119:47–56. doi: 10.1016/j.antiviral.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 81.Cotmore SF, Tattersall P. 2005. A rolling-hairpin strategy: basic mechanisms of DNA replication in the parvoviruses, p 171–181. In Kerr J, Cotmore SF, Bloom ME, Linden RM, Parrish CR(ed), Parvoviruses. Hodder Arnold, London, United Kingdom. [Google Scholar]
- 82.Lu G, Villa JA, Donlin MJ, Edwards TC, Cheng X, Heier RF, Meyers MJ, Tavis JE. 2016. Hepatitis B virus genetic diversity has minimal impact on sensitivity of the viral ribonuclease H to inhibitors. Antiviral Res 135:24–30. doi: 10.1016/j.antiviral.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meck C, D'Erasmo MP, Hirsch DR, Murelli RP. 2014. The biology and synthesis of alpha-hydroxytropolones. Medchemcomm 5:842–852. doi: 10.1039/C4MD00055B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Su HP, Yan Y, Prasad GS, Smith RF, Daniels CL, Abeywickrema PD, Reid JC, Loughran HM, Kornienko M, Sharma S, Grobler JA, Xu B, Sardana V, Allison TJ, Williams PD, Darke PL, Hazuda DJ, Munshi S. 2010. Structural basis for the inhibition of RNase H activity of HIV-1 reverse transcriptase by RNase H active site-directed inhibitors. J Virol 84:7625–7633. doi: 10.1128/JVI.00353-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Summa V, Petrocchi A, Bonelli F, Crescenzi B, Donghi M, Ferrara M, Fiore F, Gardelli C, Gonzalez PO, Hazuda DJ, Jones P, Kinzel O, Laufer R, Monteagudo E, Muraglia E, Nizi E, Orvieto F, Pace P, Pescatore G, Scarpelli R, Stillmock K, Witmer MV, Rowley M. 2008. Discovery of raltegravir, a potent, selective orally bioavailable HIV-integrase inhibitor for the treatment of HIV-AIDS infection. J Med Chem 51:5843–5855. doi: 10.1021/jm800245z. [DOI] [PubMed] [Google Scholar]
- 86.Masaoka T, Zhao H, Hirsch DR, D’Erasmo MP, Meck C, Varnado B, Gupta A, Meyers MJ, Baines J, Beutler JA, Murelli RP, Tang L, Le Grice SFJ. 2016. Characterization of the C-terminal nuclease domain of herpes simplex virus pUL15 as a target of nucleotidyltransferase inhibitors. Biochemistry 55:809–819. doi: 10.1021/acs.biochem.5b01254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Williams PD, Venkatraman S, Langford HM, Kim B, Booth TM, Grobler JA, Staas D, Ruzek RD, Embrey MW, Wiscount CM, Lyle TA. July 2007. 1-Hydroxy naphthyridine compounds as anti-HIV agents. US patent PCT/US2007/016052.
- 88.Wong S, Zhi N, Filippone C, Keyvanfar K, Kajigaya S, Brown KE, Young NS. 2008. Ex vivo-generated CD36+ erythroid progenitors are highly permissive to human parvovirus B19 replication. J Virol 82:2470–2476. doi: 10.1128/JVI.02247-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morita E, Nakashima A, Asao H, Sato H, Sugamura K. 2003. Human parvovirus B19 nonstructural protein (NS1) induces cell cycle arrest at G(1) phase. J Virol 77:2915–2921. doi: 10.1128/JVI.77.5.2915-2921.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sowd GA, Li NY, Fanning E. 2013. ATM and ATR activities maintain replication fork integrity during SV40 chromatin replication. PLoS Pathog 9:e1003283. doi: 10.1371/journal.ppat.1003283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Deng X, Yan Z, Cheng F, Engelhardt JF, Qiu J. 2016. Replication of an autonomous human parvovirus in non-dividing human airway epithelium is facilitated through the DNA damage and repair pathways. PLoS Pathog 12:e1005399. doi: 10.1371/journal.ppat.1005399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.