Abstract
Nosocomial infections with Pseudomonas aeruginosa (PA) are difficult to treat due to the low outer membrane permeability of the bacterium and the development of resistance. In the present study, the anti-microbial peptide (AMP) mutant chensinin-1 (MC1) was revealed to exhibit anti-bacterial activity against a multidrug-resistant PA (MRPA) strain in vitro, and the minimum inhibitory concentration was 25 µM, which was 4-fold higher than that of the native strain. MC1 was able to disrupt the integrity of the cytoplasmic membrane in the native PA strain and MRPA and had a similar membrane depolarization ability in these strains, but the outer membrane permeability of MRPA cells was lower than that of native PA cells, as determined by a 1-N-phenylnaphthylamine assay. In addition, the abundance of the gene Psl encoding for biofilm-associated polysaccharides was detected using Congo red, and a high concentration of MC1 inhibited the formation of MRPA biofilms. Furthermore, the expression levels of biofilm-associated genes affected by the AMP, MC1, were quantified by polymerase chain reaction analysis. The results indicated that MC1 induced biofilm inhibition by downregulating the relative expression of specific biofilm polysaccharide-associated genes, including pelA, algD and pslA. The present results indicated that the AMP MC1 may be an effective antibiotic against MRPA strains.
Keywords: mutant chensinin-1 peptide, multiple drug resistance, P. aeruginosa, biofilm, permeability, polymerase chain reaction
Introduction
The rise of multidrug-resistant bacteria poses a severe threat to human health and is a major cause of death in the clinical setting (1,2). Among such bacteria, Pseudomonas aeruginosa (PA) is an opportunistic pathogen and a leading cause of nosocomial infections (3). Numerous infections caused by PA occur in immunocompromised patients and this species is the major cause of ventilator-associated pneumonia (4). Pneumonia caused by PA frequently leads to acute lung injury and secondary sepsis, resulting in high infection-associated mortality. Antibiotics are effective for the majority of the infections caused by PA; however, PA has a natural resistance and the capacity for exposure-induced resistance against antibiotics with various mechanisms of action, leading to an increasing rate of resistance (5). Therefore, developing antibiotics with novel mechanisms of action has attracted the interest of researchers.
Anti-microbial peptides (AMPs), which are encoded by specific genes, are a class of low-molecular-weight polypeptides with potent anti-microbial activity against a broad spectrum of microorganisms (6). They are a fundamental component of innate immunity and are nontoxic to mammalian cells. AMPs initially bind to the negatively charged bacterial cell membrane by electrostatic interactions and then insert into the hydrophobic core and perturb its structure to increase the bacterial membrane permeability, which induces the leakage of cytoplasmic components and ultimately the death of the microorganism (7). Due to the unique bactericidal mechanism of AMPs, it is not easy for bacteria to develop resistance through evolution, and the entire evolutionary process is slow; therefore, AMPs are among the most promising candidates for novel antibiotics for the treatment of multidrug-resistant bacteria (8).
In a previous study, the AMP chensinin-1 was isolated from the skin secretions of the Chinese brown frog, Rana chensinensis, comprising 18 amino acid residues in the sequence SAVGRHGRRFGLRKHRKH (9,10). Chensinin-1 exhibited a moderate anti-microbial activity against gram-positive bacteria, but no activity against gram-negative bacteria, which may be due to its low hydrophobicity, amphipathicity and random coil conformation in the membrane environment. Chensinin-1 is able to form aggregates when it attaches to the outer cell membrane, as indicated by the quenching of the fluorescence intensity of rhodamine-labeled chensinin-1 in the presence of lipopolysaccharides, which serve as the major component of the outer membrane of gram-negative bacteria. Previous research has indicated that in AMP sequences, bulky and hydrophobic Trp residues are not present in a large proportion, but that such residues may facilitate the anchoring and insertion of the peptide into the bilayer surface of the cell membrane (11,12). Therefore, to improve the broad-spectrum anti-microbial activity of chensinin-1, a novel mutant analog of chensinin-1, MC1, was designed by replacing three Gly residues with Trp residues. MC1 is an 18-amino acid peptide with the sequence SAVWRHWRRFWLRKHRKH (13). MC1 exhibits potent anti-microbial activity against selected gram-positive bacteria and gram-negative bacteria, including PA cells. Mechanistically, the action of the AMP MC1 is initiated through electrostatic interactions, causing the adsorption of AMPs onto the surface of the negatively charged cell membrane. The majority of AMPs then perform membrane permeabilization by inserting into the hydrophobic core of the outer membrane and disrupting the bacterial membrane, leading to cell death. Of note, MC1 has no hemolytic activity and is therefore suitable as a novel antibiotic. However, it remains elusive whether MC1 possesses bactericidal activity against multidrug-resistant pathogens encountered in the clinic. In the present study, the anti-bacterial activity of MC1 against multi-drug resistant PA (MRPA), which was isolated from a clinical setting, was investigated in vitro. In particular, the ability of PA to grow in a biofilm may enhance its aversion of host defenses and resistance to chemotherapy, and therefore, the ability of MC1 to inhibit biofilm formation was also examined in the present study. For this, the effect of MC1 on the relative expression of specific biofilm-associated genes in multi-drug resistant PA was investigated.
Materials and methods
Peptide synthesis
The AMP MC1 was synthesized by KareBay Biochem Inc. (Ningbo, China) using a standard Fmoc solid-phase peptide synthesis protocol. The peptides were purified to near homogeneity (95%) by reverse-phase high-performance liquid chromatography using a Vydac 218TP1022 C-18 column (2.2 × 25 cm; Separations Group, Hesperis, CA, USA) with a mobile phase of acetonitrile/water/trifluoroacetic acid. The relative mass of the peptide was determined using matrix-assisted laser desorption ionization-time of flight mass spectrometry (Shimadzu, Kyoto, Japan).
Bacterial strains
PA was acquired from the China General Microbiological Culture Collection Centre (Beijing, China). The MRPA strain was obtained from the Department of Central Laboratory of Hunan Cancer Hospital (Changsha, China) (14) and exhibited multidrug resistance to amikacin, cefepime, aztreonam, ciprofloxacin and piperacillin. Antibiotic susceptibility testing of the strains had been performed by the Department of Central Laboratory of Hunan Cancer Hospital (15), the precautions taken with regard to biosafety when handling the pathogens were described previously (16).
Anti-microbial assay
The minimum inhibitory concentration (MIC) of the MC1 peptide for the multidrug-resistant strain MRPA and the susceptible strain PA was determined using the two-fold dilution method (17). The peptide was two-fold serially diluted to achieve concentrations between 1.56 and 200 µM. Subsequently, 50 µl of the peptide solution mixed with 50 µl of a log-phase bacterial inoculum [2×105 colony-forming units (CFU)/ml] in PBS was added to the wells of a 96-well microtiter plate. The cultures were incubated for 24 h at 37°C in air. The absorbance at 600 nm for each sample was recorded using a microtiter plate reader. The MIC was defined as the lowest peptide concentration that inhibited 95% of bacterial growth.
Bactericidal kinetics assay
The bactericidal kinetics of the peptide against PA and MRPA were assessed by generating time-kill curves according to a previously described method (9). Log-phase bacterial cultures were incubated with the peptide at its MIC at 37°C for 0–180 min. After being washed twice with sterile Meuller-Hinton broth (MHB; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) and centrifuged at 4°C and 1,064 × g for 10 min, the surviving bacteria were diluted 102- or 105-fold and then spread on agar plates (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China). Bacterial colonies were counted after the plates were incubated for 24 h at 37°C. Polymyxin B (PMB), the positive control drug, was tested under the same conditions. Bactericidal kinetics were determined by plotting the number of surviving bacteria against the time.
Post-antibiotic effect (PAE)
PA and MRPA bacteria were grown until they reached the log phase and then diluted to 2×106 CFU/ml. The bacteria were incubated with MC1 at its MIC. After being washed twice with sterile MHB, the cultures were centrifuged at 4,000 × g for 10 min at 4°C, and the pellets were re-suspended and incubated with shaking at 37°C for 8 h. The cell numbers were determined using the spiral-plating method (18). PMB-treated bacteria were included as the positive control. The time required for the colonies to reach 1.0 log10 CFU was recorded. The post-antibiotic effect (PAE) was defined as the time difference between an experimental culture and the control culture to achieve an increase of 1.0 log10 CFU/ml.
Membrane depolarization assay
Bacterial membrane depolarization was measured using the previously reported method (19). Mid-log phase bacterial cells were centrifuged, washed twice with 5 mM HEPES containing 20 mM glucose and 100 mM KCl and then re-suspended in HEPES buffer at a final concentration of 2×106 CFU/ml. After EDTA was added at a final concentration of 0.5 mM, the bacterial suspensions were incubated with 3,3′-dipropylthiadicarbocyanine iodide (DiSC3-5; 4 µM) to allow for the uptake of the DiSC3-5 probe in a 96-well microtiter plate. Once DiSC3-5 was taken up by the bacteria, MC1 was added to the bacterial samples at a final concentration of 1-, 2- or 4-fold of its MIC, and the change in fluorescence intensity was recorded.
Bacterial outer membrane permeability
The outer membrane permeability was analyzed using the 1-N-phenylnaphthylamine (NPN) dye (20). The bacterial cells were grown to mid-log phase, harvested by centrifugation and then washed and re-suspended in buffer (5 mM HEPES, 1 mM NaN3) at a density of 2×106 CFU/ml. NPN was added to 500 µl of the diluted bacterial cells at a final concentration of 10 µM, and the peptide was then added at increasing concentrations. After 1 h, the basal fluorescence intensity was recorded with an excitation wavelength of 350 nm and an emission maximum of 420 nm using a microplate reader. Gentamicin with MC1 (the two were incubated at the following concentrations: 1.56, 3.13, 6.25, 12.5, 25 and 50µM) served as positive controls.
Biofilm susceptibility assay
Biofilm formation was detected using a previously published method (17). The bacterial cells were grown to mid-log phase and diluted to 2×105 CFU/ml. The peptide was two-fold diluted from 50 to 1.56 µM and added to 50 µl of the bacterial suspension in a 96-well microtiter plate. The untreated control groups were setup at the same time. The planktonic cells were removed after incubation at 37°C for 24 h, and the biofilms in the wells were washed two times with PBS. The adherent bacteria were fixed with methanol at room temperature for 15 min, and each well was stained with 0.1% (w/v) crystal violet (CV) dye at room temperature for 5 min and washed with water. Subsequently, 200 µl of 95% ethanol was added to each CV-stained well. The absorbance of the biofilm biomass was measured at 600 nm. PMB was used as a positive control. The percentage of inhibition for each sample concentration was calculated according to the following equation: Inhibition (%)=1-(Absorbancesample/Absorbancecontrol)×100% (21,22).
Activity against 1-day-old biofilms was determined according to a previously described method (10). The bacterial cells were grown to mid-log phase and diluted to 2×105 CFU/ml. Equal volumes of water and bacteria (50 µl) were added to a 96-well microtiter plate and then incubated at 37°C for 24 h. The biofilms were washed with PBS and incubated with different concentrations of peptide for 24 h. Subsequently, the adherent bacteria were fixed with methanol at room temperature for 15 min and each well was stained with 0.1% (w/v) CV dye at room temperature for 5 min and washed with water prior to addition of 200 µl of 95% ethanol to each CV-stained well. After agitation for 30 min, the absorbance at 600 nm was measured with a microtiter plate reader. The minimum biofilm reduction concentration was defined as the minimum concentration of the peptide required to reduce the biofilm by >50%. PMB was used as a positive control.
Polysaccharide Psl assay
Psl is a crucial adhesive scaffolding component of the biofilm matrix, promoting cell-cell interactions and surface attachment (23,24), which can be examined by using the liquid Congo red (CR) method according to a published protocol (5). The mid-log phase bacterial suspension [optical density at 600 nm (OD600), ~1.0] was inoculated with the MC1 peptide at a final concentration of 1-, 4-, 8- or 16-fold of its MIC in unsalted Luria-Bertani medium (Beijing Solarbio Science & Technology Co., Ltd., Beijing, China) containing 40 µg/ml CR. Subsequently, the sample was incubated with agitation overnight at 37°C. To determine the OD600, 1 ml of the culture liquid was removed. The remaining bacterial cells were centrifuged for 10 min at 4°C and 4,000 × g, and the supernatant was measured at 490 nm to determine the binding ability of bacterial cells to unbound CR, a dye that detects neutral polysaccharides or polysaccharides (25).
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Mid-log phase bacterial cells were divided into two groups: One group was treated with AMPs at a final concentration equal to the MIC for 24 h and the other group was incubated without the peptides. One milliliter of sample was removed and centrifuged at 13,000 × g for 1 min at 4°C. The supernatant was removed, 10 mg/ml lysozyme (Takara Biotechnology Co., Ltd., Dalian, China) was added to the sample and the sample was incubated at 4°C for 10 min. Subsequently, 1 ml TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) was added, and the samples were vortexed for 20 sec and incubated at 4°C for 5 min. The supernatant was transferred into a fresh 1.5 ml Eppendorf tube, chloroform was added, and the tube was vortexed for 1 min. The sample was incubated for 15 min to develop a milky appearance and centrifuged at 4°C. Subsequently, an equal volume of isopropanol was added to the supernatant, and the sample was incubated for 4 min and centrifuged at 12,000 × g for 15 min at 4°C. Anhydrous ethanol was added for precipitation, the supernatant was removed by centrifugation, and 50 µl DEPC-treated water was added. The RNA bands were separated by agarose gel electrophoresis.
The extracted RNA was reverse-transcribed into complementary DNA and qPCR was performed using a Super ScriptÔ III One-Step RT-PCR System with Platinum™ Taq DNA Polymerase (cat. no. 12574026; Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol and an ABI Prism 7000 sequence detection system (Thermo Fisher Scientific, Inc.). The primers were designed and synthesized by Takara Biotechnology Co., Ltd. The following primers were used: PlsA (gene ID, 879717) forward, 5′-AAACGCTACGGCTACAACAACC-3′ and reverse, 5′-TATTCGCTGACCGCCTCCT-3′; PelA (gene ID, 878833) forward, 5′-ACGCCCTTCGCCTATCTGT-3′ and reverse, 5′-GAGGTCCATTACCTGGCTGTTC-3′; alginate (Alg)D (gene ID, 879004) forward, 5′-CTCATCACCAGCCACGACA-3′ and reverse, 5′-AGCACCAGCACATCGGAAC-3′; and GAPDH forward, 5′-ACCACAGTCCATGCCATCAC-3′ and reverse, 5′-TCCACCACCCTGTTGCTGTA-3′. GAPDH expression was used as an internal control. The reaction conditions were set as follows: 94°C for 30 sec, followed by 40 cycles at 94°C for 5 sec, 55°C for 15 sec and 72°C for 10 sec. After normalization to the internal control GAPDH, fold changes were calculated using the comparative cycle threshold method (26).
Statistical analysis
All experiments were repeated three times, independently. Values are expressed as the mean ± standard deviation. Differences between groups were assessed using one-way analysis of variance followed by the Student-Newman-Keuls post-hoc test in SAS 9.2 software (SAS Institute Inc., Shanghai, China). P<0.05 was considered to indicate a statistically significant difference.
Results
Anti-bacterial activity of MC1 against multidrug-resistant bacteria
The AMP MC1 exhibited anti-bacterial activity against the tested multidrug-resistant bacteria. In detail, MC1 exhibited a marked anti-bacterial activity against PA, with an MIC of 6.25 µM, while an MIC of 25 µM was obtained for the MRPA strain, which was significantly higher than that for the susceptible PA strain.
Bactericidal kinetics and PAE of MC1
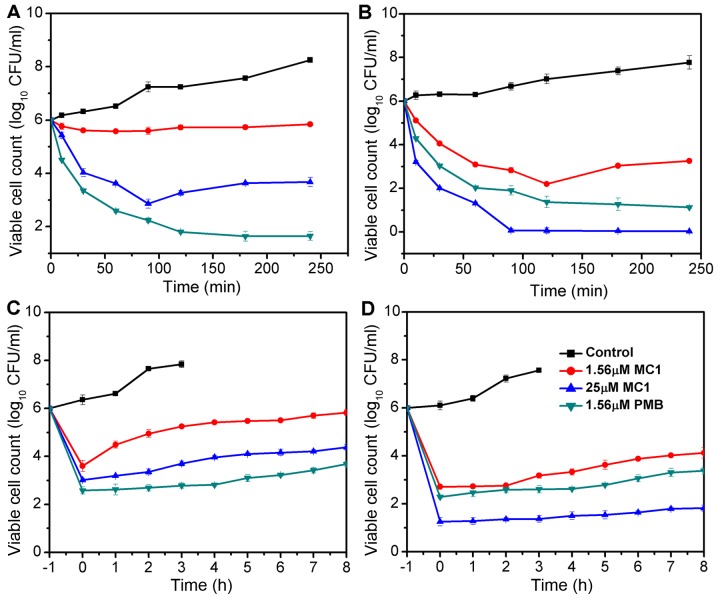
The bactericidal kinetics curves revealed that at its MIC, MC1 killed the MRPA cells in a time-dependent manner; the growth of MRPA cells was completely inhibited at 90 min when the concentration of MC1 was 25 µM and a reduction of >4 log10 CFU/ml was observed. MC1 had no effect on the growth of MRPA cells at a concentration of 1.56 µM (Fig. 1A), at which the growth of PA cells was inhibited, but the exponentially growing PA cells were completely eliminated after incubation with 25 µM MC1 for 90 min (Fig. 1B). PMB served as the positive control and exhibited a similar anti-bacterial activity against PA and MRPA. The PAE is defined as persistent suppression of bacterial growth after a brief exposure (1–2 h) of bacteria to an antibiotic (27). No PAE was observed when MRPA cells were treated with 1.56 µM MC1, but when the concentration was increased to 25 µM, an effect was detected at 3 h (Fig. 1C). A PAE for PA was observed at peptide concentrations of 1.56 and 25 µM at 4–6 h (Fig. 1D).
Figure 1.
Antibacterial activity of MC1. Killing kinetics of agents against (A) MRPA and (B) PA. Post-antibiotic effect of agents on (C) MRPA and (D) PA. Each data-point represents an average of six independent experiments. PA, Pseudomonas aeruginosa; MRPA, multidrug-resistant PA; CFU, colony-forming units; PMB, polymyxin B; MC1, mutant chensinin-1 peptide.
MC1 affects the inner membrane permeability of PA cells
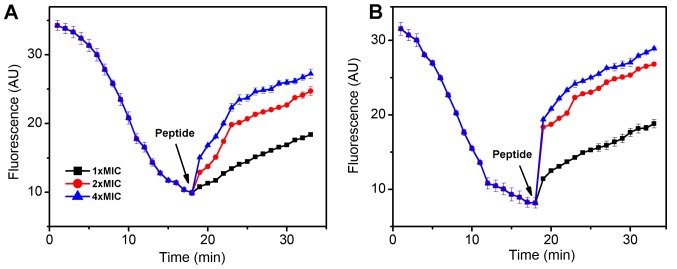
The fluorescent probe DiSC3-5 was used to determine the effect of the AMP MC1 on the cytoplasmic membrane depolarization of PA and MRPA. As presented in Fig. 2, when the dye was added to the bacterial cells, its fluorescence decreased rapidly as the dye self-quenched in the membranes. At 18 min, the fluorescence reached a steady state. The peptide was then added and the fluorescence increased rapidly. As depolarization was completed, the maximum fluorescence intensity was 27 absorption units (AU) for MRPA and 29 AU for PA. The slope change of the susceptible PA strain was clearly steeper than that of the MRPA strain, indicating that the capacity of MC1 to change the inner membrane permeability of the MRPA was relatively low. The slope changes were positively associated with the concentration of the peptide.
Figure 2.
Cytoplasmic membrane depolarization of (A) multidrug-resistant PA and (B) PA by mutant chensinin-1 peptide. The fluorescence of the membrane potential-sensitive dye diSC3-5 was detected. PA, Pseudomonas aeruginosa; MIC, minimum inhibitory concentration.
Outer membrane permeability is affected by MC1
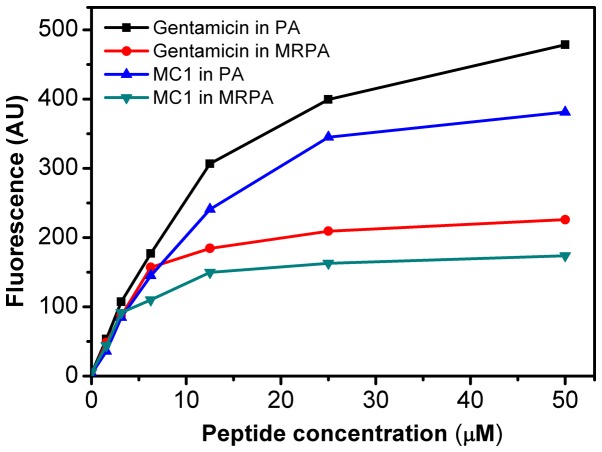
As presented in Fig. 3, the AMP MC1 dose-dependently permeabilized the outer membrane of MRPA, reflected by the change in the fluorescence intensity of the NPN dye. Initially, the fluorescence intensity increased gradually. At the peptide concentration of 25 µM, a fluorescence intensity of 165 AU was reached and the increase stagnated at concentrations beyond this. However, at the same concentration, the ability of the peptide to permeabilize the outer membrane of the susceptible PA strain was relatively higher, as the maximum fluorescence intensity reached 350 AU, suggesting that the ability of the AMP MC1 to permeabilize the outer membrane of PA is greater than that to permeabilize the outer membrane of the drug-resistant strain. However, the permeability of the sensitive and drug-resistant strains also depended on the concentration of the peptide. The permeability of MC1 in the drug-resistant strain was lower than that in the sensitive strain. Furthermore, the ability of the AMP MC1 to permeabilize the outer membrane of the bacteria was inferior to that of the positive control gentamicin.
Figure 3.
MC1-induced 1-N-phenylnaphthylamine uptake in MRPA and PA cells. The fluorescence intensity of the dye that entered the hydrophobic interior of the outer membrane was quantified. PA, Pseudomonas aeruginosa; MRPA, multidrug-resistant PA; MC1, mutant chensinin-1 peptide.
Anti-biofilm activity of the AMP MC1
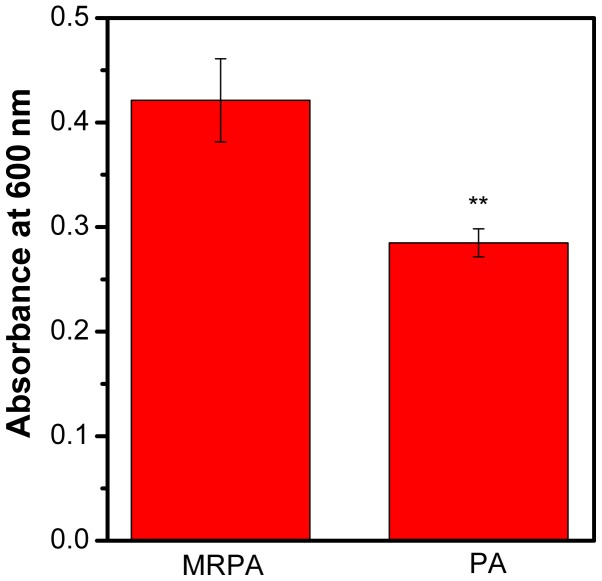
The biomass of PA and MRPA was quantified by CV. As presented in Fig. 4, compared to the sensitive strain, the multidrug-resistant strain had a greater biofilm biomass after 24 h of incubation. It may be concluded that the ability of MRPA to form a biofilm is stronger than that of the susceptible strain under the same culture conditions.
Figure 4.
Biofilm formation of MRPA and PA cells. The biofilm biomass was stained with crystal violet dye and quantified by measuring the absorbance at 600 nm without mutant chensinin-1 peptide. **P<0.01 vs. MRPA. PA, Pseudomonas aeruginosa; MRPA, multidrug-resistant PA.
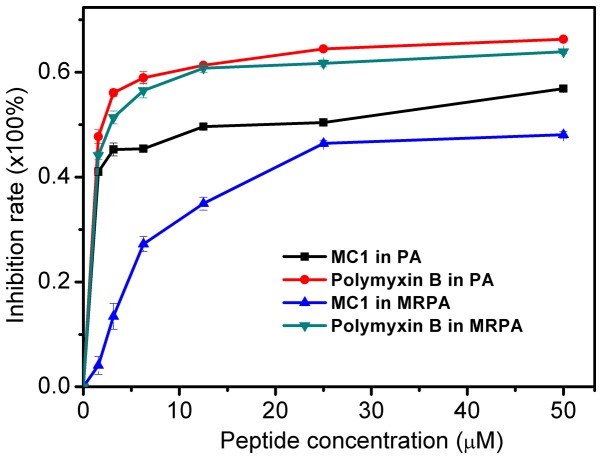
The inhibitory effect of the AMP on the MRPA biofilm was determined by measuring the OD600. As presented in Fig. 5, the inhibition of biofilm formation by the peptide occurred in a dose-dependent manner. For MRPA, with MC1 at a concentration of 1.56 µM, the inhibition rate was only ~4%. However, for the susceptible strain, the inhibition rate was ~41%. For MC1 at a concentration of 25 µM, the inhibition rate of MRPA was ~46%, which was similar to that observed for biofilms of the sensitive strain with 1.56 µM peptide. The results suggested that MC1 inhibits biofilm formation, and as the concentration of the AMP increases, the inhibition rate of the biofilm also increases, but the inhibition was less distinct for MRPA than for PA. Furthermore, the positive control PMB more effectively inhibited the biofilm formation of PA and MRPA than MC1, and its effects on these two strains were similar.
Figure 5.
Inhibitory effect of MC1 on bacterial biofilm formation. Values are expressed the mean ± standard deviation (n=3). PA, Pseudomonas aeruginosa; MRPA, multidrug-resistant PA; MC1, mutant chensinin-1 peptide.
Effect of MC1 on the biofilm polysaccharide Psl
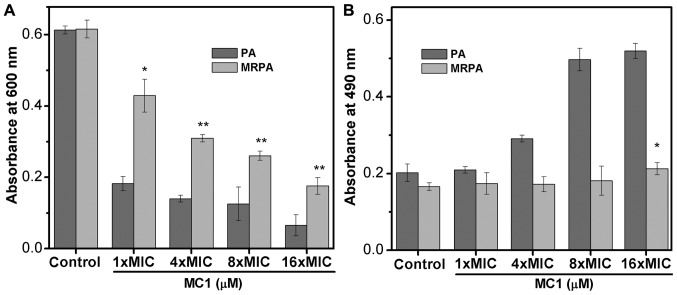
Previous studies have indicated that at least three types of polysaccharide, namely Psl, Pel and AlgD, are produced by PA to constitute the biofilm (28,29). The biofilms of the sensitive strain and the multidrug-resistant strain mainly contain two polysaccharides, Psl and Pel, which are important factors in preventing antibiotics from entering drug-resistant cells. Therefore, the effect of AMPs on the synthesis of the biofilm polysaccharide Psl reflects the effect of AMPs on biofilms (Fig. 6A and B). In the present study, the OD600 represented suspended cells and the OD490 indicated that the dye did not bind to the bacteria. When Psl is overproduced, the binding of Psl to the dye CR increases the OD600 value but decreases the absorbance at 490 nm. As the number of bacteria decreases, the synthesis of Psl is also inhibited, which decreases the binding capacity of the dye, and the OD490 increases. The experimental results indicate that the OD600 values of the multidrug-resistant strain were larger than those of the sensitive strain, indicating that the multidrug-resistant strain exhibited less cell death compared with the PA strain. With the addition of the AMP MC1, the OD600 value decreased and the OD490 value increased, suggesting that MC1 inhibited Psl synthesis. This inhibition was dependent on the peptide concentration. In addition, at a peptide concentration of 1X MIC (6.25 µM for PA and 25 µM for MRPA strain), the OD490 value of the multidrug-resistant bacteria was less than that of the sensitive bacteria, indicating that the multidrug-resistant strain produced more biofilm, which suggested that MRPA had a relatively greater capacity to synthesize Psl.
Figure 6.
Polysaccharide biosynthesis of PA biofilm inhibited by MC1. (A) OD600 of the cultures and (B) OD490 of the supernatant were measured to determine the binding of Congo red to bacterial cells. Values are expressed the mean ± standard deviation (n=3). *P<0.05; **P<0.01 vs. control MRPA cells. PA, Pseudomonas aeruginosa; MRPA, multidrug-resistant PA; OD600, optical density at 600 nm; MIC, minimum inhibitory concentration; MC1, mutant chensinin-1 peptide.
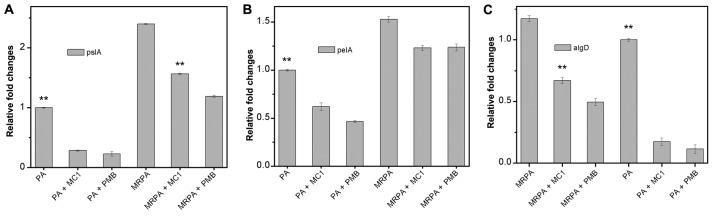
Gene expression of the biofilm components PslA, PelA and AlgD is affected by MC1
To examine the relative expression of polysaccharide-associated genes in the multidrug-resistant strain during biofilm formation, RT-qPCR was used to determine the effect of MC1 on the relative expression of the genes PelA, PslA and AlgD, which encode for biofilm components. PMB was used as a positive control drug. As presented in Fig. 7, the level of transcription of the PslA and PelA genes in the MRPA cells was almost 2-fold higher than that observed in the PA cells. In the absence of MC1 and PMB, the three genes were stably expressed, and the gene expression levels of MRPA were higher than those in the sensitive strain. When MC1 was added, all three genes were inhibited, and the relative expression of the genes was downregulated, particularly that of PslA.
Figure 7.
Effect of MC1 on the expression of biofilm polysaccharide-associated genes determined by quantitative polymerase chain reaction. (A) pslA, (B) pelA and (C) algD. Fold changes of genes were normalized to the housekeeping gene GAPDH and the results are expressed as the average of three independent experiments. **P<0.01 vs. MRPA cells. PA, Pseudomonas aeruginosa; MRPA, multidrug-resistant PA; PMB, polymyxin B; MC1, mutant chensinin-1 peptide.
Discussion
P. aeruginosa is resistant to most antibiotics due to its low outer membrane permeability (30). The present study focused on the anti-microbial activity of the AMP MC1 against MRPA and its effect on biofilm formation. The membrane permeability of pathogenic microorganisms affects whether AMP molecules enter pathogenic microorganisms and kill them. Therefore, the effect of the MC1 peptide on the permeability of the internal and external membranes of a multidrug-resistant strain and a sensitive strain was tested. Depolarization of the bacterial plasma membrane provides a direct assessment of effects on membrane permeability (31). A depolarization experiment demonstrated that the degree of depolarization of the multidrug-resistant strain and the sensitive strain by MC1 were similar at 30 min, but the slope of the depolarization trend was clearly different as the fluorescence intensity was increased from 9.9 AU to 15.06 for MRPA, while the fluorescence intensity was sharply increased from 8.15 AU to 18.35 AU in 1 min for PA in the presence of 4 × MIC at the interval between 17 and 18 min. However, as the peptide concentration increased, there was no significant increase in depolarization for PA and MRPA. An outer membrane penetration test indicated that the permeability of the AMP MC1 in the sensitive strain was significantly higher than that observed in the multidrug-resistant strain, which was also dependent on the peptide concentration. However, with increasing peptide concentrations, the permeability of MRPA exhibited relatively lesser increases and approached a maximum, which may be due to the low permeability of the outer membrane.
The mechanism of drug resistance in MRPA is associated with its biofilm formation ability. By quantifying the biofilm formed by the bacteria within 24 h, it was determined that the multidrug-resistant strain MRPA was able to produce more biofilm under normal conditions. By comparing the inhibitory effect of the AMP MC1 on the biofilm formation of the sensitive strain PA and the multidrug-resistant strain MRPA, it was revealed that MC1 inhibited the biofilm formation of the sensitive strain PA at a low concentration, while the inhibition rate in the MRPA groups was relatively low, which indicated that MRPA produced more biofilm biomass and was resistant to the inhibitory effects on biofilm formation. In the biomass inhibition experiment using biofilms produced by the sensitive strain PA and the multidrug-resistant strain MRPA over 24 h, MC1 had a comparatively greater effect on the decomposition of the biofilm from the sensitive strain. In addition, as MRPA produced more biofilm than the sensitive strain, the effect on biofilm decomposition was low at the same peptide concentration.
With respect to biofilm components, the effect of the AMP MC1 on the amount of the biofilm polysaccharides, Psl and Pel, and the expression of the genes PslA, PelA and AlgD, which encode for biofilm components, was assessed. It was revealed that, compared to the susceptible strain PA, the multidrug-resistant strain MRPA contained a larger amount of PslA, PelA and AlgD, and the polysaccharide Psl in each strain was reduced with the addition of AMP. The RT-qPCR results suggested that the AMP MC1 inhibited the relative expression of polysaccharide-associated genes. In general, the multidrug-resistant strain MRPA and the susceptible strain PA produce biofilm; however, the biofilm formation by MRPA is faster, and the structure is denser, so the biofilms are more difficult to decompose (32,33). P. aeruginosa produces at least three polysaccharides (alginate, Pel, and Psl) to stabilize the biofilm structure (28,29). The data in the current study also demonstrated that MC1 significantly inhibited Pel and Psl synthesis in MRPA cells as the transcription levels of the algD and PslA genes were significantly downregulated following the treatment of MRPA 0108 cells with MC1. Therefore, a peptide-driven downregulation of polysaccharide biosynthesis may occur as MC1 inhibited the expression of the algD and PslA genes to decrease the structural stability of biofilms and interfere with the formation of MRPA-containing biofilms.
For the development of novel antibiotics against multi-drug-resistant pathogens, the bacterial cell membrane may serve as the major target, as the evolution of membrane composition changes may be a slow process. Therefore, membrane-targeted AMPs are expected to be effective compared with traditional antibiotics with a single target. MC1 exhibited a similar ability to permeate the cell membrane of the susceptible and the MRPA strain, suggesting that it has the potential to be developed as an anti-microbial agent with membrane-perforating activity against MRPA.
In summary, the present study demonstrates that the AMP MC1 decreases the drug resistance of MRPA by reducing the outer membrane permeability and the production of biofilm biomass. The AMP MC1 inhibited biofilm formation and exhibited an anti-biofilm effect. In addition, MC1 potently inhibited the expression of biofilm-associated genes. These results indicated that MC1 may serve as an effective antibiotic against multidrug-resistant bacterial strains.
Acknowledgements
Not applicable.
Funding
The present study was supported by Hunan Cancer Hospital (grant no. 303050).
Availability of data and materials
Data are available from the corresponding authors on reasonable request.
Authors' contributions
ZY and JL conceived the study, designed the experiments, analysed the data and wrote the manuscript. ZY, YK, ZL and TL performed the experiments.
Ethical approval and informed consent
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Lyczak JB, Cannon CL, Pier GB. Establishment of Pseudomonas aeruginosa infection: Lessons from a versatile opportunist. Microbes Infect. 2000;2:1051–1060. doi: 10.1016/S1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 2.Rowe SM, Miller S, Sorscher EJ. Cystic Fibrosis. N Engl J Med. 2005;352:1992–2001. doi: 10.1056/NEJMra043184. [DOI] [PubMed] [Google Scholar]
- 3.Nicas TI, Hancock RE. Pseudomonas aeruginosa outer membrane permeability: Isolation of a porin protein F-deficient mutant. J Bacteriol. 1983;153:281–285. doi: 10.1128/jb.153.1.281-285.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breidenstein EB, de la Fuente-Núñez C, Hancock RE. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011;19:419–426. doi: 10.1016/j.tim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 5.Pasupuleti M, Schmidtchen A, Malmsten M. Antimicrobial peptides: Key component of the innate immune system. Crit Rev Biotechnol. 2012;32:143–171. doi: 10.3109/07388551.2011.594423. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Xiang Q, Zhang Q, Huang Y, Su Z. Overview on the recent study of antimicrobial peptides: Origins, functions, relative mechanisms and application. Peptides. 2012;37:207–215. doi: 10.1016/j.peptides.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- 8.Shang D, Sun Y, Wang C, Wei S, Ma L, Sun L. Membrane interaction and antibacterial properties of chensinin-1, an antimicrobial peptide with atypical structural features from the skin of Rana chensinensis. Appl Microbiol Biotechnol. 2012;96:1551–1560. doi: 10.1007/s00253-012-4148-3. [DOI] [PubMed] [Google Scholar]
- 9.Shang D, Yu F, Li J, Zheng J, Li Y. Molecular cloning of cDNAs encoding antimicrobial peptide precursors from the skin of the Chinese brown frog, Rana chensinensis. Zoolog Sci. 2009;26:220–226. doi: 10.2108/zsj.26.220. [DOI] [PubMed] [Google Scholar]
- 10.Chan DI, Prenner EJ, Vogel HJ. Tryptophan- and arginine-rich antimicrobial peptides: Structures and mechanisms of action. Biochim Biophys Acta. 2006;1758:1184–1202. doi: 10.1016/j.bbamem.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 11.Yau WM, Wimley WC, Gawrisch K, White SH. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 12.Dong W, Mao X, Guan Y, Kang Y, Shang D. Antimicrobial and anti-inflammatory activities of three chensinin-1 peptides containing mutation of glycine and histidine residues. Sci Rep. 2017;7:40228. doi: 10.1038/srep40228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pal T, Abraham B, Sonnevend A, Jumaa P, Conlon JM. Brevinin-1BYa: A naturally occurring peptide from frog skin with broad-spectrum antibacterial and antifungal properties. Int J Antimicrob Agents. 2006;27:525–529. doi: 10.1016/j.ijantimicag.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 14.Saiman L, Mehar F, Niu WW, Neu HC, Shaw KJ, Miller G, Prince A. Antibiotic susceptibility of multiply resistant Pseudomonas aeruginosa isolated from patients with cystic fibrosis, including candidates for transplantation. Clin Infect Dis. 1996;23:532–537. doi: 10.1093/clinids/23.3.532. [DOI] [PubMed] [Google Scholar]
- 15.Reller LB, Schoenknecht FD, Kenny MA, Sherris JC. Antibiotic susceptibility testing of Pseudomonas aeruginosa: Selection of a control strain and criteria for magnesium and calcium content in media. J Infect Dis. 1974;130:454–463. doi: 10.1093/infdis/130.5.454. [DOI] [PubMed] [Google Scholar]
- 16.Working party on antibiotic sensitivity testing of the britishsociety for antimicrobial chemotherapy. report of the working party on antibiotic sensitivity testing of the British Society for Antimicrobial Chemotherapy. A guide to sensitivity testing. J Antimicrob Chemother. 1991;27:41–43. [PubMed] [Google Scholar]
- 17.Sun Y, Dong W, Sun L, Ma L, Shang D. Insights into the membrane interaction mechanism and antibacterial properties of chensinin-1b. Biomaterials. 2015;37:299–311. doi: 10.1016/j.biomaterials.2014.10.041. [DOI] [PubMed] [Google Scholar]
- 18.Gilchrist JE, Campbell JE, Donnelly CB, Peeler JT, Delaney JM. Spiral plate method for bacterial determination. Appl Microbiol. 1973;25:244–252. doi: 10.1128/am.25.2.244-252.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saravanan R, Mohanram H, Joshi M, Domadia PN, Torres J, Ruedl C, Bhattacharjya S. Structure, activity and interactions of the cysteine deleted analog of tachyplesin-1 with lipopolysaccharide micelle: Mechanistic insights into outer-membrane permeabilization and endotoxin neutralization. Biochim Biophys Acta. 2012;1818:1613–1624. doi: 10.1016/j.bbamem.2012.03.015. [DOI] [PubMed] [Google Scholar]
- 20.Beckloff N, Laube D, Castro T, Furgang D, Park S, Perlin D, Clements D, Tang H, Scott RW, Tew GN, Diamond G. Activity of an antimicrobial peptide mimetic against planktonic and biofilm cultures of oral pathogens. Antimicrob Agents Chemother. 2007;51:4125–4132. doi: 10.1128/AAC.00208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shang D, Zhang Q, Dong W, Liang H, Bi X. The effects of LPS on the activity of Trp-containing antimicrobial peptides against Gram-negative bacteria and endotoxin neutralization. Acta Biomater. 2016;33:153–165. doi: 10.1016/j.actbio.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 22.Ma L, Jackson KD, Landry RM, Parsek MR, Wozniak DJ. Analysis of Pseudomonas aeruginosa conditional psl variants reveals roles for the psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J Bacteriol. 2006;188:8213–8221. doi: 10.1128/JB.01202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol. 2004;186:4457–4465. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsukawa M, Greenberg EP. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186:4449–4456. doi: 10.1128/JB.186.14.4449-4456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei GX, Campaqna AN, Bobek LA. Effect of MUC7 peptides on the growth of bacteria and on Streptococcus mutans biofilm. J Antimicrob Chemother. 2006;57:1100–1009. doi: 10.1093/jac/dkl120. [DOI] [PubMed] [Google Scholar]
- 26.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma KK, Sangraulah H, Mediratta PK. Some new concepts in antibacterial drug therapy. Indian J Pharm. 2002;34:390–396. [Google Scholar]
- 28.Ryder C, Byrd M, Wozniak DJ. Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol. 2007;10:644–648. doi: 10.1016/j.mib.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghafoor A, Hay ID, Rehm BH. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl Environ Microbiol. 2011;77:5238–5246. doi: 10.1128/AEM.00637-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie J, Gou Y, Zhao Q, Wang K, Yang X, Yan J, Zhang W, Zhang B, Ma C, Wang R. Antimicrobial activities and membrane-active mechanism of CPF-C1 against multidrug-resistant bacteria, a novel antimicrobial peptide derived from skin secretions of the tetraploid frog Xenopus clivii. J Pept Sci. 2014;20:876–884. doi: 10.1002/psc.2679. [DOI] [PubMed] [Google Scholar]
- 31.Torrent M, Navarro S, Moussaoui M, Nogués MV, Boix E. Eosinophil cationic protein high-affinity binding to bacteria wall lipopolysaccharides and peptidoglycans. Biochemistry. 2008;47:3544–3555. doi: 10.1021/bi702065b. [DOI] [PubMed] [Google Scholar]
- 32.Li XZ, Zhang L, Poole AK. Interplay between the MexA-MexB-OprM multidrug efflux system and the outer membrane barrier in the multiple antibiotic resistance of Pseudomonas aeruginosa. J Antimicrob Chemother. 2000;45:433–436. doi: 10.1093/jac/45.4.433. [DOI] [PubMed] [Google Scholar]
- 33.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. Int J Antimicrob Agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available from the corresponding authors on reasonable request.