Abstract
MicroRNAs (miRs), a class of small non-coding RNAs, have been demonstrated to be involved in the development and progression of human malignancies, including cutaneous squamous cell carcinoma (CSCC). miR-186 serves a suppressive role in certain common types of human cancer; however, its exact function in CSCC has not been reported previously. In the present study, the expression of miR-186 was significantly increased in CSCC tissues compared with adjacent non-tumour tissues. Overexpression of miR-186 significantly promoted CSCC cell proliferation while inhibiting cell apoptosis. Reticulophagy regulator 1 (RETREG1), a gene that is significantly downregulated in CSCC tissues and cell lines, was identified as a novel target of miR-186. In addition, the expression of RETREG1 was inversely correlated with miR-186 expression in CSCC tissues. Furthermore, the expression of RETREG1 was negatively regulated by miR-186 in CSCC cells, and restoration of RETREG1 attenuated the effects of miR-186 on CSCC cells. Taken together, the results of the current study suggest that miR-186 serves an oncogenic role in CSCC and may be used as a potential therapeutic target for the treatment of this disease.
Keywords: cutaneous squamous cell carcinoma, microRNA, reticulophagy regulator 1, oncogene
Introduction
The occurrence of cutaneous squamous cell carcinoma (CSCC), one of the most common non-melanoma skin cancers, is increasing mainly due to the damage of the ozonosphere and the lack of risk awareness (1,2). Certain factors, including infection, chemical substances, inheritance and immune responses, have been implicated in the development and progression of CSCC (3). High rates of recurrence and metastasis contribute to the poor prognosis of patients with CSCC (4). The one-year survival rate of patients with CSCC with recurrence and metastasis is approximately 50% (4). Therefore, it is necessary to investigate the molecular mechanisms underlying the growth and metastasis of CSCC to support the development of novel therapeutic targets for this disease.
MicroRNAs (miRs), a class of small non-coding RNAs 22–25 nucleotides in length, can regulate gene expression at the post-transcriptional level by directly binding to the 3′UTR of their target mRNAs (5,6). It has been well established that numerous miRs participate in the regulation of various biological processes, including cell survival, proliferation, differentiation, motility and tumorigenesis (5–7). Furthermore, certain miRs, including miR-31, miR-142-5p and miR-365, have been reported to serve oncogenic or tumour suppressive roles in CSCC (8–11). For instance, miR-125b inhibits the expression of MMP13 and suppresses CSCC cell proliferation, migration and invasion (11).
miR-186 is an important member of cancer-associated miRs and has been reported to generally serve a tumour suppressive role in various types of human cancer, including chronic myeloid leukaemia (12), colorectal cancer (13), gastrointestinal stromal tumour (14), lung cancer (15), liver cancer (16), cervical cancer (17), and gastric cancer (18). miR-186 inhibits the proliferation, metastasis and epithelial-to-mesenchymal transition of colorectal cancer cells by targeting ZEB1 (13). However, the exact function of miR-186 in CSCC has not been previously studied.
The current study aimed to determine the expression and regulatory role of miR-186 in CSCC.
Materials and methods
Clinical tissue samples
The current study was approved by the Ethics Committee of the First Affiliated Hospital of Hunan College of Traditional Chinese Medicine (Zhuzhou, China). A total of 32 CSCC tissues and matched adjacent non-tumour tissues were collected from patients with primary CSCC (18 males and 14 females; age range, 35–68 years; mean age, 53.6 years) at First Affiliated Hospital of Hunan College of Traditional Chinese Medicine between March 2015 and May 2017, and informed consent was obtained from all patients. Exclusion criteria included radiotherapy or chemotherapy before surgical resection. After surgical resection, the tissues were immediately immersed in liquid nitrogen and stored until further use.
Cell lines
The human normal skin cell line HaCaT (cat. no. CL0114) and the CSCC cell lines A431 and SCL-1 were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China). Cells were cultured in Dulbecco's modified Eagle's Medium (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% foetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and maintained in a humidified incubator containing 5% CO2 and 95% O2 at 37°C.
Cell transfection
The miR-186 mimic, miR-186 inhibitor and negative control (NC) were all purchased from Shanghai GenePharma Co., Ltd. (Shanghai, China). miRs were produced from GMR-miR microRNA single-stranded mimics (cat. no. B01001), which are proprietary. pcDNA3.1 vector and pcDNA3.1-RETREG1 expression plasmid were purchased from Hunan Yearth Biotechnology Co., Ltd. (Changsha, China). For the in vitro functional study of miR-186 in CSCC, SCL-1 and A431 cells were transfected with 100 nM NC, 100 nM miR-186 mimics or 100 nM miR-186 inhibitor, or co-transfected with 100 nM miR-186 mimics and 1 µg pcDNA3.1 vector (the miR-186 + blank group) or pcDNA3.1-RETREG1 expression plasmid (the miR-186 + RETREG1 group) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. Following 48 h of transfection, the subsequent experiments were conducted.
RNA extraction and reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues or cell lines using the TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The RNA was subsequently reverse transcribed into cDNA using the PrimeScript RT reagent kit with gDNA Eraser (Takara Bio, Inc., Otsu, Japan), according to the manufacturer's protocol. The qPCR was performed using the SYBR Green Realtime PCR Master Mix (Toyobo Life Science, Osaka, Japan), according to the manufacturer's protocol. The thermocycling conditions were as follows: Initial denaturation at 95°C for 5 min; 35 cycles of 95°C for 15 sec and 60°C for 30 sec. U6 was used as the internal control of miR-186 and GAPDH was used as the internal control of reticulophagy regulator 1 (RETREG1). The primers for RETREG1 (cat. no. HQP013485), GAPDH (cat. no. HQP006940), miR-186 (cat. no. HmiRQP0248) and U6 (cat. no. HmiRQP9001) were all purchased from Guangzhou Fulengen (GeneCopoeia, Inc., Rockville, MD, USA). The relative expression levels were calculated using the 2−ΔΔCt method (19).
Cell proliferation assay
Cell proliferation was studied using Cell Counting kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto, Japan). At 48 h after transfection, the cells were seeded into 96-well plates (3×103 cells/well) and cultured for 0, 24, 48 and 72 h. The optical density value at 450 nm was measured using a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Cell apoptosis analysis
Cell apoptosis was analyzed by using an Annexin V-fluorescein isothiocyanate (FITC) Apoptosis kit (BD Biosciences, San Diego, CA, USA) in accordance with the manufacturer's protocol. Transfected SCL-1 and A431 cells were seeded in 24-well plates (1×105 cells/well) and cultured in a humidified incubator containing 5% CO2 and 95% O2 at 37°C for 24 h. Subsequently, the cells were re-suspended in 500 µl binding buffer containing 1% FITC-labelled Annexin-V and propidium iodide (which is in the Annexin V-FITC Apoptosis kit). After incubation in the dark for 30 min, the apoptosis levels were evaluated using BD Accuri CFlow software 1.0 on a BD C6 FACSVerse™ flow cytometer (both BD Biosciences).
Western blots
Total protein was extracted using RIPA lysis buffer (Beyotime Institute of Biotechnology, Beijing, China). The protein concentration was measured using a bicinchoninic acid protein assay kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Equal amounts of protein (50 µg/lane) were isolated using 10% SDS-PAGE and transferred onto polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). The membranes were blocked with 5% skimmed milk at 4°C overnight and incubated with rabbit anti-human RETREG1 antibodies (cat. no. ab151755) and GAPDH antibodies (cat. no. ab9485; both 1:500; Abcam, Cambridge, MA, USA) at room temperature for 3 h. Subsequently, the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit secondary antibodies (1:5,000; cat. no. ab6721; Abcam) at room temperature for 40 min. The protein bands were detected using an Enhanced Chemiluminescence Western Blotting kit (Pierce; Thermo Fisher Scientific, Inc.) and quantified using Image Lab analysis software version 3.1 (Bio-Rad Laboratories, Inc.).
Bioinformatics analysis and luciferase reporter gene assay
TargetScan software 7.2 (www.targetscan.org/) was used to predict the potential target genes of miR-186. The wild-type (WT) and mutant (MT) sequences containing the putative binding sites of miR-186 in the 3′-UTR of the RETREG1 mRNA transcript were cloned into separate pMIR-REPORT luciferase reporter plasmids (Promega Corporation, Madison, WI, USA). Cells (5×104 cells per well) in 24-well plates were transfected with 10 nM miR-186 mimics (or miR-NC) and 500 ng WT RETREG1 (or MT RETREG1) plasmid using Lipofectamine 2000. After 48 h of transfection, the relative luciferase activity was detected using a Dual-Luciferase® Reporter Assay System (Promega Corporation). The activity of firefly luciferase was normalized to the activity of Renilla luciferase.
Statistical analysis
Data are presented as the mean ± standard deviation. SPSS 20.0 software (IBM Corp., Armonk, NY, USA) was used for the statistical analyses. Comparisons were performed with one-way analysis of variance followed with Tukey's post hoc test or Student's t test. A Pearson correlation test was used to determine the association between miR-186 and RETREG1 expression in CSCC tissues. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-186 is upregulated in CSCC
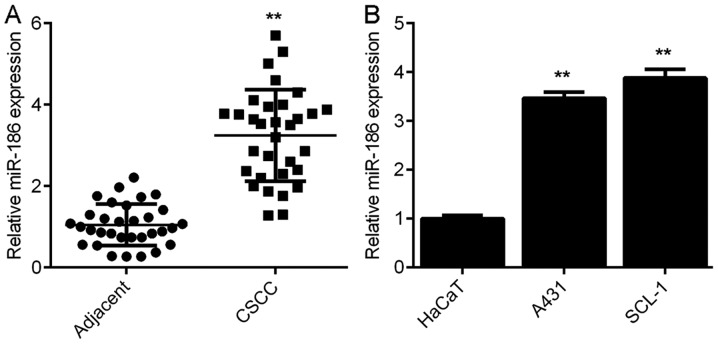
To investigate the role of miR-186 in CSCC, the expression levels of miR-186 were determined in 32 CSCC tissues and their matched adjacent non-tumour tissues. miR-186 was significantly upregulated in the CSCC tissues compared with the matched adjacent non-tumour tissues (Fig. 1A). Using the mean expression level of miR-186 as a cut-off value (3.26), the authors divided the patients into high and low miR-186 groups. The expression of miR-186 in tumour tissues exhibited no difference between the male and female patients with CSCC (P=0.735; Table I). To further confirm these results, miR-186 expression was determined in the normal human skin cell line HaCaT and the CSCC cell lines A431 and SCL-1. miR-186 expression levels increased in the A431 and SCL-1 cell lines compared with the HaCaT cells (Fig. 1B). The above results indicated that miR-186 was upregulated in CSCC.
Figure 1.
miR-186 is upregulated in CSCC. (A) RT-qPCR was used to determine the expression of miR-186 in CSCC tissues and matched adjacent non-tumour tissues. **P<0.01 vs. Adjacent. (B) RT-qPCR was used to determine the expression of miR-186 in CSCC cell lines and the human normal skin cell line HaCaT. **P<0.01 vs. HaCaT. CSCC, cutaneous squamous cell carcinoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; miR, micro RNA.
Table I.
Association between miR-186 expression and sex of patients with cutaneous squamous cell carcinoma.
| Sex | Low miR-186 (n=15) | High miR-186 (n=17) | P-value |
|---|---|---|---|
| Male | 9 | 9 | 0.735 |
| Female | 6 | 8 |
miR, micro RNA.
miR-186 affects CSCC cell proliferation and apoptosis
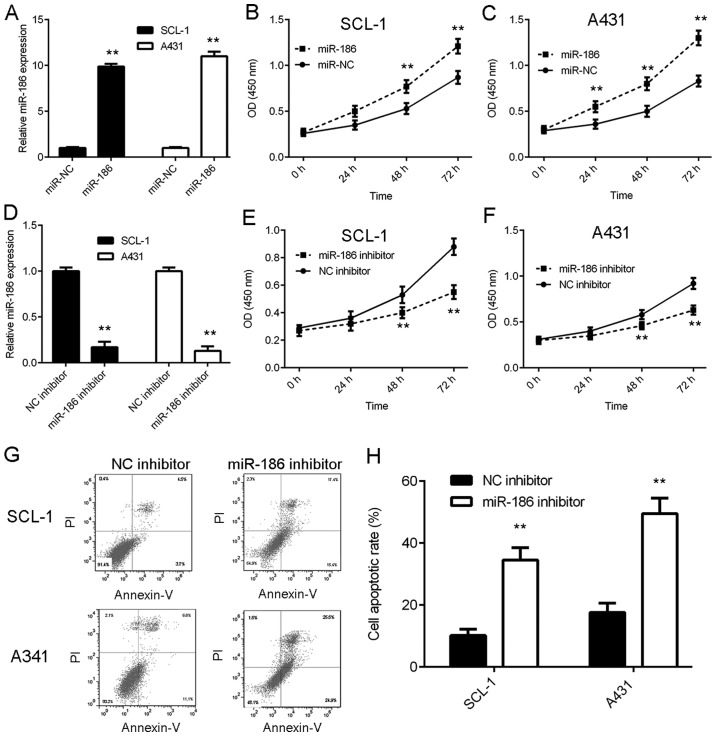
The function of miR-186 in CSCC was investigated. SCL-1 and A431 cells were transfected with miR-186 mimics and qPCR data indicated that the miR-186 levels were significantly increased in the miR-186 group compared with the miR-NC group in both cell lines (Fig. 2A). A CCK-8 assay was subsequently used to study the effects of miR-186 upregulation on CSCC cell proliferation. The results suggested that overexpression of miR-186 caused a significant increase in the proliferation of SCL-1 and A431 cells compared with the miR-NC group (Fig. 2B and C). Flow cytometry was subsequently conducted to study cell apoptosis and it was not significantly affected by miR-186 overexpression (data not shown).
Figure 2.
miR-186 affects CSCC cell proliferation and apoptosis. SCL-1 and A431 cells were transfected with miR-186 mimics or miR-NC. Following transfection, (A) RT-qPCR was used to examine the expression of miR-186, and CCK-8 assay was used to examine cell proliferation in (B) SCL-1 and (C) A431 cell lines. **P<0.01 vs. miR-NC. Subsequently, SCL-1 and A431 cells were transfected with NC inhibitor or miR-186 inhibitor. Following transfection, (D) RT-qPCR was used to examine the expression of miR-186, and CCK-8 assay was used to examine cell proliferation in (E) SCL-1 and (F) A431 cell lines. (G) Flow cytometry was used to study cell apoptosis. (H) Quantitative analysis of cell apoptotic rate. **P<0.01 vs. NC inhibitor. Nc, negative control; miR, micro RNA; CCK-8, cell counting kit-8; CSCC, cutaneous squamous cell carcinoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; PI, propidium iodide.
SCL-1 and A431 cells were transfected with miR-186 inhibitor to knockdown miR-186 expression. qPCR data indicated that the miR-186 levels were significantly reduced in the miR-186 inhibitor group compared with the NC inhibitor group (Fig. 2D). Data from the CCK-8 assay indicated that inhibition of miR-186 expression caused a significant reduction in CSCC cell proliferation (Fig. 2E and F). Furthermore, apoptosis of CSCC cells markedly increased following miR-186 inhibition (Fig. 2G and H). Therefore, the results of the current study indicated that miR-186 overexpression promoted CSCC cell proliferation, and knockdown of miR-186 induced CSCC cell apoptosis.
RETREG1 is a target gene of miR-186
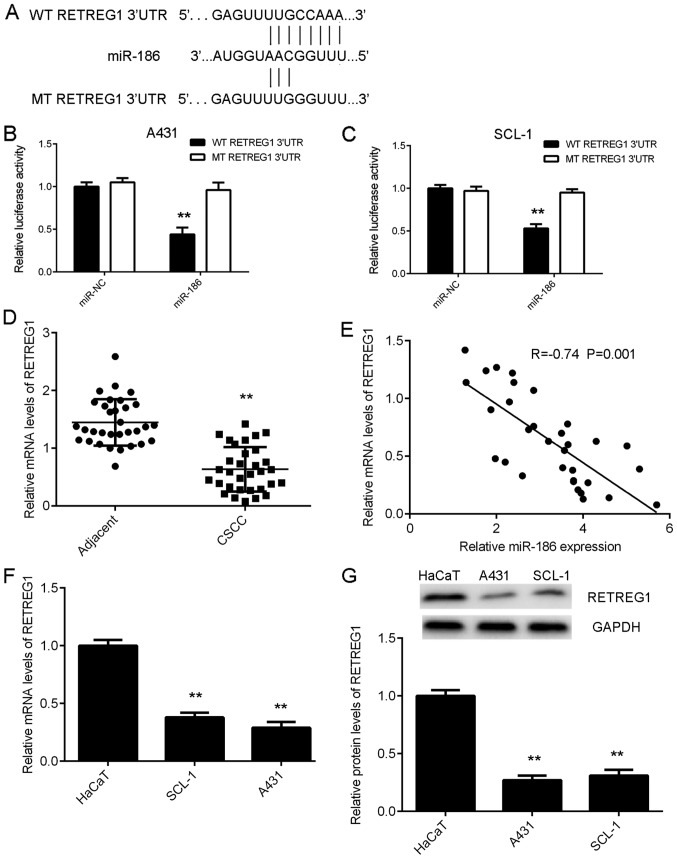
TargetScan software 7.2 was used to predict the potential target genes of miR-186, and the data indicated that there were predicted binding sites for miR-186 in the 3′UTR of the RETREG1 mRNA (Fig. 3A). A dual-luciferase assay was performed to verify whether RETREG1 was a target gene of miR-186. The results indicated that overexpression of miR-186 significantly downregulated the luciferase activity of the WT 3′-UTR of RETREG1; however, the luciferase activity of the MT 3′-UTR of RETREG1 was not affected in CSCC cells (Fig. 3B and C). Therefore, the results indicate that RETREG1 is a direct target gene of miR-186.
Figure 3.
RETREG1 is a target gene of miR-186. (A) WT and MT sequences of the RETREG1 3′ UTR. A dual-luciferase assay was performed to verify whether RETREG1 was a target gene of miR-186 in (B) A431 and (C) SCL-1 cells. **P<0.01 vs. miR-NC. (D) RT-qPCR was used to determine the mRNA levels of RETREG1 in CSCC tissues and matched adjacent non-tumour tissues. **P<0.01 vs. Adjacent. (E) RETREG1 expression levels were inversely correlated with the expression levels of miR-186 in CSCC tissues. (F) RT-qPCR and (G) western blotting were used to examine the mRNA and protein levels of RETREG1 in CSCC cell lines and the human normal skin cell line HaCaT. **P<0.01 vs. HaCaT. WT, wild type; MT, mutant; UTR, untranslated region; CSCC, cutaneous squamous cell carcinoma; miR, micro RNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; RETREG1, reticulophagy regulator 1.
Expression level of RETREG1 is reduced in CSCC tissues and inversely correlated to the miR-186 expression
RT-qPCR data indicated that RETREG1 expression was significantly lower in the CSCC tissues compared with the matched adjacent non-tumour tissues (Fig. 3D). Using the mean expression value of RETREG1 as a cut-off value (0.64), the authors divided the patients into high and low RETREG1 groups. The expression of RETREG1 in the tumour tissues exhibited no difference between male and female patients with CSCC (P=0.473; Table II). Furthermore, the RETREG1 expression levels were inversely correlated with the expression levels of miR-186 in CSCC tissues (Fig. 3E). Taken together, these data suggest that the reduced expression of RETREG1 in CSCC tissues may be due to the increased expression of miR-186. Furthermore, the mRNA and protein levels of RETREG1 were lower in CSCC cells compared with HaCaT cells (Fig. 3F and G).
Table II.
Association between RETREG1 expression and sex of patients with cutaneous squamous cell carcinoma.
| Sex | Low RETREG1 (n=19) | High RETREG1 (n=13) | P-value |
|---|---|---|---|
| Male | 12 | 6 | 0.473 |
| Female | 7 | 7 |
RETREG1, reticulophagy regulator 1.
miR-186 negatively regulates the expression of RETREG1 in CSCC cells
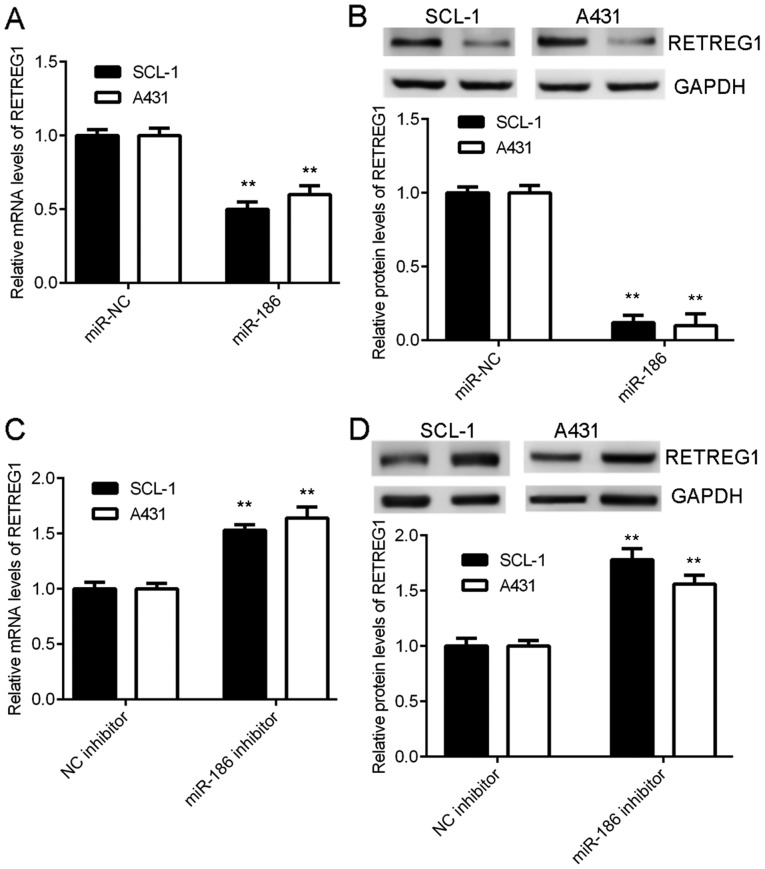
The effects of miR-186 on the expression levels of RETREG1 in CSCC cells were subsequently studied. Data indicated that overexpression of miR-186 significantly reduced the mRNA and protein expression of RETREG1 in CSCC cells (Fig. 4A and B). By contrast, knockdown of miR-186 significantly increased the expression of RETREG1 in CSCC cells (Fig. 4C and D). The above results indicated that RETREG1 expression was negatively regulated by miR-186 in CSCC.
Figure 4.
miR-186 negatively regulates the expression of RETREG1 in CSCC cells. SCL-1 and A431 cells were transfected with miR-186 mimics or miR-NC. Following transfection, (A) RT-qPCR and (B) western blotting were used to determine the mRNA and protein levels of RETREG1. **P<0.01 vs. miR-NC. Subsequently, SCL-1 and A431 cells were transfected with NC inhibitor or miR-186 inhibitor. Following transfection, (C) RT-qPCR and (D) western blotting were used to determine the mRNA and protein levels of RETREG1. **P<0.01 vs. NC inhibitor. NC, negative control; miR, micro RNA; RETREG1, reticulophagy regulator 1; CSCC, cutaneous squamous cell carcinoma; RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
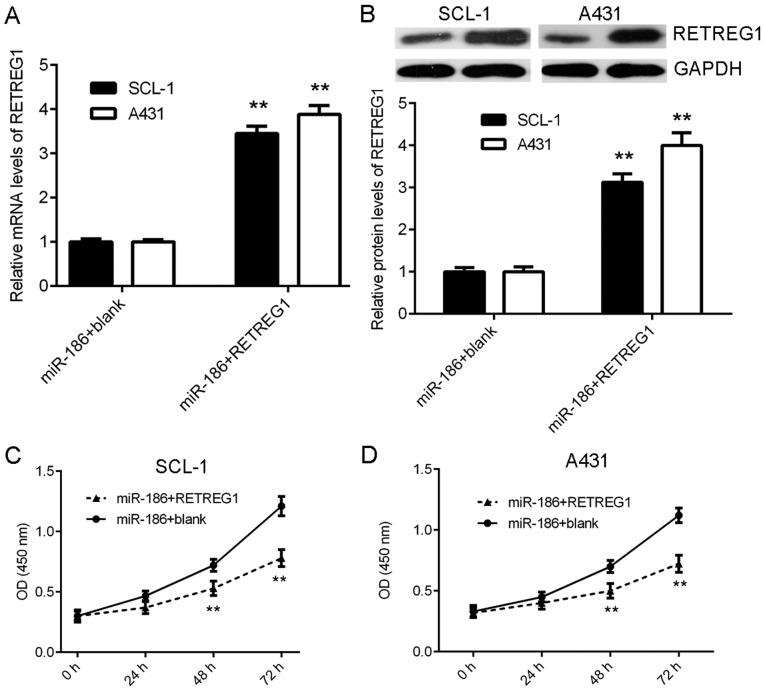
Overexpression of RETREG1 attenuates the effects of miR-186 on CSCC cell proliferation
Finally, the current study examined whether RETREG1 served a role in the miR-186-mediated proliferation of CSCC cells. The miR-186-overexpressing CSCC cells were transfected with a RETREG1 expression plasmid to restore its expression. Following transfection, the mRNA and protein levels of RETREG1 were significantly increased in the miR-186 + RETREG1 group compared with the miR-186 + blank group (Fig. 5A and B). Further investigation indicated that the proliferation of CSCC cells was significantly reduced in the miR-186 + RETREG1 group compared with the miR-186 + blank group (Fig. 5C and D). These results suggest that RETREG1 overexpression attenuated the effects of miR-186 on CSCC cell proliferation.
Figure 5.
Overexpression of RETREG1 attenuates the promoting effects of miR-186 on CSCC cell proliferation. The miR-186-overexpressing CSCC cells were transfected with a RETREG1 expression plasmid or a control vector. Following transfection, (A) reverse transcription-quantitative polymerase chain reaction and (B) western blotting were used to determine the mRNA and protein levels of RETREG1. Subsequently, cell counting kit-8 assays were used to study cell proliferation in (C) SCL-1 and (D) A431 cells. **P<0.01 vs. miR-186 + blank. miR, micro RNA; RETREG1, reticulophagy regulator 1; CSCC, cutaneous squamous cell carcinoma.
Discussion
miR-186 serves a suppressive role in certain common types of human cancer; however, its exact function in CSCC has not been previously reported. In the current study, the expression of miR-186 was significantly increased in CSCC tissues compared with adjacent non-tumour tissues. Overexpression of miR-186 significantly promoted CSCC cell proliferation and inhibited cell apoptosis. RETREG1, which is significantly downregulated in CSCC tissues and cell lines, was identified as a novel target gene of miR-186. In addition, the expression of RETREG1 was inversely correlated with the expression level of miR-186 in CSCC tissues. Furthermore, the expression of RETREG1 was negatively regulated by miR-186 in CSCC cells, and restoration of RETREG1 attenuated the effects of miR-186 on CSCC cells.
In recent decades, a large number of miRs, including, miR-125b (11), miR-365 (20), miR-205 (21), miR-199a-5p (22), and miR-203 (23) have been identified as key regulators of the development and progression of CSCC and numerous other types of human cancer. For instance, miR-199a-5p induces CSCC cell invasion by suppressing E-cadherin expression (22). Among the cancer-associated miRs, miR-186 has been reported to generally function as a tumour suppressor (24,25). For instance, Cai et al (24) demonstrated that miR-186 served as a tumour suppressor in oral squamous cell carcinoma by negatively regulating the expression of tyrosine-protein phosphatase non-receptor type 11. Ruan et al (25) revealed that miR-186 suppressed lung cancer progression by targeting SIRT6. However, whether miR-186 serves a role in CSCC remains unclear. The current study indicated that the expression of miR-186 was significantly increased in CSCC tissues compared to adjacent non-tumour tissues. However, the expression of miR-186 in tumour tissues exhibited no difference between male and female patients. The present study further revealed that overexpression of miR-186 significantly promoted CSCC cell proliferation, and knockdown of miR-186 significantly inhibited CSCC cell proliferation and induced cell apoptosis. These results suggest that miR-186 serves an oncogenic role in CSCC.
RETREG1, also known as FAM134B, is a reticulophagy receptor that regulates the turnover of the endoplasmic reticulum (ER) by selective phagocytosis (26). Inhibition of RETREG1 contributes to impaired proteostasis in the ER due to the accumulation of misfolded or aggregated proteins, leading to compromised neuronal survival and progressive neuronal degenerative diseases (26). In previous studies, RETREG1 was downregulated in colorectal cancer, and its low expression was associated with younger age, larger tumour size, advanced cancer stages, higher recurrence rate, lymphovascular invasion and poor prognosis of patients (27,28). Furthermore, overexpression of RETREG1 inhibited the proliferation of colorectal cancer cells in vitro, and tumour growth in vivo (29). However, the function of RETREG1, and the regulatory mechanism of RETREG1 expression in CSCC remains unclear. The current study demonstrated that RETREG1 was a target gene of miR-186 in CSCC cells and the expression of RETREG1 was reduced in CSCC tissues and inversely correlated with miR-186 expression. These results suggest that the reduced expression of RETREG1 in CSCC may be due to the upregulation of miR-186. Similar observations were also reported in colorectal cancer. Islam et al (30) demonstrated that miR-186 overexpression significantly decreased the expression of RETREG1 in colorectal cancer cells, and the expression levels of RETREG1 and miR-186 were inversely correlated in colorectal cancer tissues and cells. The current study expanded the understanding of the association between miR-186 and RETREG1 in human cancer. In addition, the expression of miR-186 in tumour tissues exhibited no difference between male and female patients with CSCC. Furthermore, overexpression of RETREG1 attenuated the effects of miR-186 on CSCC cell proliferation, suggesting that RETREG1 may act as a downstream signalling molecule of miR-186 in CSCC cells.
In conclusion, the present study demonstrated that miR-186 may promote CSCC cell proliferation and inhibit cell apoptosis at least partly through direct targeting of RETREG1, suggesting that targeting the miR-186/RETREG1 axis may be used as a potential strategy for the treatment of CSCC. Future studies should further determine the function of the miR-186/RETREG1 axis in CSCC growth and metastasis in vivo.
Acknowledgements
Not applicable.
Funding
Not applicable.
Availability of data and materials
All data generated or analysed during this study are included in this published article.
Authors' contributions
SH designed the study. XH collected clinical tissues and wrote the manuscript. YL, YT, TW, LL and CC performed experiments. PA conducted statistical analysis.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of First Affiliated Hospital of Hunan College of Traditional Chinese Medicine. Written informed consent to participate in the current study were obtained from all patients.
Patient consent for publication
Written informed consent to publish the current study were obtained from all patients.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Gordon R. Skin cancer: An overview of epidemiology and risk factors. Semin Oncol Nurs. 2013;29:160–169. doi: 10.1016/j.soncn.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 2.Linares MA, Zakaria A, Nizran P. Skin cancer. Prim Care. 2015;42:645–659. doi: 10.1016/j.pop.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Prieto-Granada C, Rodriguez-Waitkus P. Cutaneous squamous cell carcinoma and related entities: Epidemiology, clinical and histological features, and basic science overview. Curr Probl Cancer. 2015;39:206–215. doi: 10.1016/j.currproblcancer.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Burton KA, Ashack KA, Khachemoune A. Cutaneous squamous cell carcinoma: A review of high-risk and metastatic disease. Am J Clin Dermatol. 2016;17:491–508. doi: 10.1007/s40257-016-0207-3. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 7.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bai X, Zhou Y, Chen P, Yang M, Xu J. MicroRNA-142-5p induces cancer stem cell-like properties of cutaneous squamous cell carcinoma via inhibiting PTEN. J Cell Biochem. 2018;119:2179–2188. doi: 10.1002/jcb.26379. [DOI] [PubMed] [Google Scholar]
- 9.Lin N, Zhou Y, Lian X, Tu Y. MicroRNA-31 functions as an oncogenic microRNA in cutaneous squamous cell carcinoma cells by targeting RhoTBT1. Oncol Lett. 2017;13:1078–1082. doi: 10.3892/ol.2017.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou M, Zhou L, Zheng L, Guo L, Wang Y, Liu H, Ou C, Ding Z. miR-365 promotes cutaneous squamous cell carcinoma (CSCC) through targeting nuclear factor I/B (NFIB) PLoS One. 2014;9:e100620. doi: 10.1371/journal.pone.0100620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu N, Zhang L, Meisgen F, Harada M, Heilborn J, Homey B, Grandér D, Ståhle M, Sonkoly E, Pivarcsi A. MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J Biol Chem. 2012;287:29899–29908. doi: 10.1074/jbc.M112.391243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin J, Ma JC, Yang J, Yin JY, Chen XX, Guo H, Wen XM, Zhang TJ, Qian W, Qian J, Deng ZQ. Arresting of miR-186 and releasing of H19 by DDX43 facilitate tumorigenesis and CML progression. Oncogene. 2018;37:2432–2443. doi: 10.1038/s41388-018-0146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, Xia L, Zhou Z, Zuo Z, Xu C, Song H, Cai J. MiR-186-5p upregulation inhibits proliferation, metastasis and epithelial-to-mesenchymal transition of colorectal cancer cell by targeting ZEB1. Arch Biochem Biophys. 2018;640:53–60. doi: 10.1016/j.abb.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Niinuma T, Kai M, Kitajima H, Yamamoto E, Harada T, Maruyama R, Nobuoka T, Nishida T, Kanda T, Hasegawa T, et al. Downregulation of miR-186 is associated with metastatic recurrence of gastrointestinal stromal tumors. Oncol Lett. 2017;14:5703–5710. doi: 10.3892/ol.2017.6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang T, Wang G, Yang L, Peng B, Wen Y, Ding G, Wang Z. MiR-186 inhibits proliferation, migration, and invasion of non-small cell lung cancer cells by downregulating Yin Yang 1. Cancer Biomark. 2017;21:221–228. doi: 10.3233/CBM-170670. [DOI] [PubMed] [Google Scholar]
- 16.Gou Y, Zhai F, Zhang L, Cui L. RUNX3 regulates hepatocellular carcinoma cell metastasis via targeting miR-186/E-cadherin/EMT pathway. Oncotarget. 2017;8:61475–61486. doi: 10.18632/oncotarget.18424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang JJ, Wang DD, Du CX, Wang Y. Long noncoding RNA ANRIL promotes cervical cancer development by acting as a sponge of miR-186. Oncol Res. 2018;26:345–352. doi: 10.3727/096504017X14953948675449. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Liu L, Wang Y, Bai R, Yang K, Tian Z. MiR-186 inhibited aerobic glycolysis in gastric cancer via HIF-1α regulation. Oncogenesis. 2016;5:e224. doi: 10.1038/oncsis.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Zhou L, Gao R, Wang Y, Zhou M, Ding Z. Loss of BAX by miR-365 promotes cutaneous squamous cell carcinoma progression by suppressing apoptosis. Int J Mol Sci. 2017;18:E1157. doi: 10.3390/ijms18061157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stojadinovic O, Ramirez H, Pastar I, Gordon KA, Stone R, Choudhary S, Badiavas E, Nouri K, Tomic-Canic M. MiR-21 and miR-205 are induced in invasive cutaneous squamous cell carcinomas. Arch Dermatol Res. 2017;309:133–139. doi: 10.1007/s00403-016-1705-0. [DOI] [PubMed] [Google Scholar]
- 22.Wang S, Cao KE, He Q, Yin Z, Zhou J. miR-199a-5p induces cell invasion by suppressing E-cadherin expression in cutaneous squamous cell carcinoma. Oncol Lett. 2016;12:97–101. doi: 10.3892/ol.2016.4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Canueto J, Cardenoso-Alvarez E, Garcia-Hernandez JL, Galindo-Villardón P, Vicente-Galindo P, Vicente-Villardón JL, Alonso-López D, De Las Rivas J, Valero J, Moyano-Sanz E, et al. MicroRNA (miR)-203 and miR-205 expression patterns identify subgroups of prognosis in cutaneous squamous cell carcinoma. Br J Dermatol. 2017;177:168–178. doi: 10.1111/bjd.15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai Z, Hao XY, Liu FX. MicroRNA-186 serves as a tumor suppressor in oral squamous cell carcinoma by negatively regulating the protein tyrosine phosphatase SHP2 expression. Arch Oral Biol. 2018;89:20–25. doi: 10.1016/j.archoralbio.2018.01.016. [DOI] [PubMed] [Google Scholar]
- 25.Ruan L, Chen J, Ruan L, Yang T, Wang P. MicroRNA-186 suppresses lung cancer progression by targeting SIRT6. Cancer Biomark. 2018;21:415–423. doi: 10.3233/CBM-170650. [DOI] [PubMed] [Google Scholar]
- 26.Islam F, Gopalan V, Lam AK. RETREG1 (FAM134B): A new player in human diseases: 15 years after the discovery in cancer. J Cell Physiol. 2018;233:4479–4489. doi: 10.1002/jcp.26384. [DOI] [PubMed] [Google Scholar]
- 27.Kasem K, Gopalan V, Salajegheh A, Lu CT, Smith RA, Lam AK. The roles of JK-1 (FAM134B) expressions in colorectal cancer. Exp Cell Res. 2014;326:166–173. doi: 10.1016/j.yexcr.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 28.Islam F, Gopalan V, Pillai S, Lu CT, Kasem K, Lam AK. Promoter hypermethylation inactivate tumor suppressor FAM134B and is associated with poor prognosis in colorectal cancer. Genes Chromosomes Cancer. 2018;57:240–251. doi: 10.1002/gcc.22525. [DOI] [PubMed] [Google Scholar]
- 29.Islam F, Gopalan V, Wahab R, Smith RA, Qiao B, Lam AK. Stage dependent expression and tumor suppressive function of FAM134B (JK1) in colon cancer. Mol Carcinog. 2017;56:238–249. doi: 10.1002/mc.22488. [DOI] [PubMed] [Google Scholar]
- 30.Islam F, Gopalan V, Vider J, Wahab R, Ebrahimi F, Lu CT, Kasem K, Lam AKY. MicroRNA-186-5p overexpression modulates colon cancer growth by repressing the expression of the FAM134B tumour inhibitor. Exp Cell Res. 2017;357:260–270. doi: 10.1016/j.yexcr.2017.05.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.