Abstract
Mitochondria have originated in eukaryotic cells by endosymbiosis of a specialized prokaryote approximately 2 billion years ago. They are essential for normal cell function by providing energy through their role in oxidizing carbon substrates. Glutathione (GSH) is a major thiol-disulfide redox buffer of the cell including the mitochondrial matrix and intermembrane space. We have generated cardiomyocyte-specific Grx1-roGFP2 GSH redox potential (EGSH) biosensor mice in the past, in which the sensor is targeted to the mitochondrial matrix. Using this mouse model a distinct EGSH of the mitochondrial matrix (−278.9 ± 0.4 mV) in isolated cardiomyocytes is observed. When analyzing the EGSH in isolated mitochondria from the transgenic hearts, however, the EGSH in the mitochondrial matrix is significantly oxidized (−247.7 ± 8.7 mV). This is prevented by adding N-Ethylmaleimide during the mitochondria isolation procedure, which precludes disulfide bond formation. A similar reducing effect is observed by isolating mitochondria in hypoxic (0.1–3% O2) conditions that mimics mitochondrial pO2 levels in cellulo. The reduced EGSH is accompanied by lower ROS production, reduced complex III activity but increased ATP levels produced at baseline and after stimulation with succinate/ADP. Altogether, we demonstrate that oxygenation is an essential factor that needs to be considered when analyzing mitochondrial function ex vivo.
Keywords: Glutathione redox potential, Hypoxia, Mitochondrial matrix, Grx1-roGFP
Abbreviations: EGSH, glutathione redox potential; GSH, glutathione; mtOxD, mitochondrial oxidation difference; ROS, reactive oxygen species
Highlights
-
•
We identified that mitochondria isolated in room air at 20.9% O2 exhibit a strong oxidation of the EGSH in the matrix.
-
•
Isolation of mitochondria in hypoxic conditions mimicking their in cellulo conditions prevents oxidation of the EGSH.
-
•
Normoxic and hypoxic isolated mitochondria differ in ROS production, complex III activity and ATP levels.
-
•
Oxygenation needs to be considered when analyzing mitochondrial function ex vivo.
1. Introduction
Mitochondria have originated in eukaryotic cells by endosymbiosis of a specialized prokaryote approximately 2 billion years ago [1]. They are essential for normal cell function, particularly by providing energy through their role in oxidative phosphorylation. Mitochondrial ATP synthesis is driven by a proton gradient across the inner mitochondrial membrane, which is generated through electron transport reactions that are linked to the oxidation of carbon substrates. However, the energy production in form of ATP is also accompanied by the production of reactive oxygen species (ROS) [2]. While ROS production plays an important role in cell signaling, excessive ROS levels can result in oxidative damage [3]. Glutathione (GSH) is considered to be one major thiol-disulfide redox buffer of the cell including the mitochondrial intermembrane space and matrix [4]. Mitochondria are devoid of GSH synthesis. Instead, GSH is mainly produced in the cytosol, from where it gets exchanged with the intracellular organelle compartments including the mitochondrial matrix [5]. There is increasing evidence that the cytosol and the mitochondrial matrix have separate and independent GSH redox pools, which is exemplified by distinct GSH redox potentials (EGSH) [6,7]. GSH is predominant in the reduced form, upon increased oxidative stress there is an increased oxidized GSH (GSSG) formation, which may cause glutathionylation of mitochondrial proteins [8]. The mitochondrial EGSH is routinely analyzed in isolated mitochondria by the GSH/GSSG ratio. However, the isolation method for mitochondria, i.e. by a discontinuous Percoll gradient versus differential centrifugation, affects the GSH and GSSG concentrations [9]. Moreover, respiratory substrates are important for the reduced redox environment in the mitochondria [9]. Taken together these data demonstrate that a physiological environment including a steady respiratory flux is important for the mitochondrial EGSH.
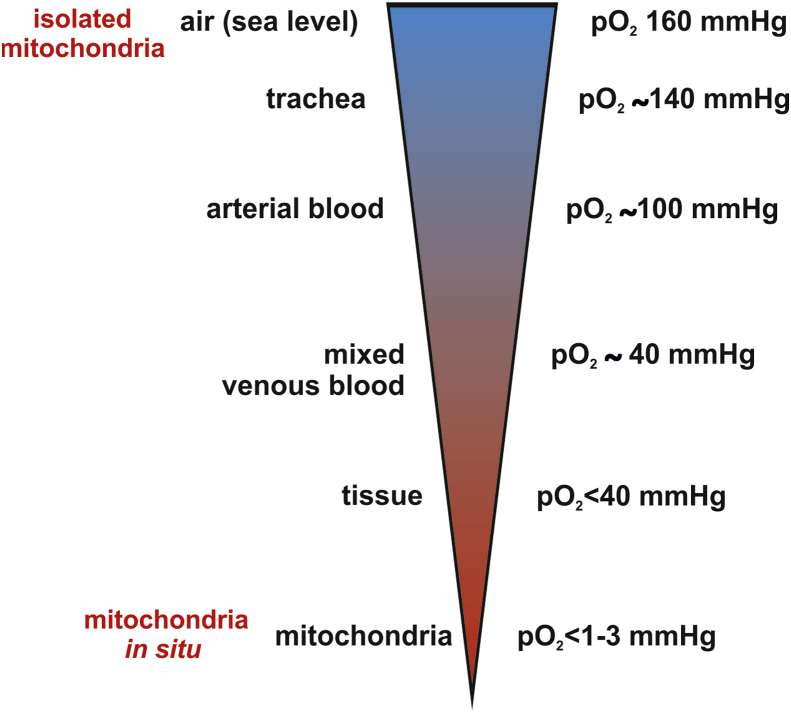
Mitochondrial bioenergetics and ROS production are highly dependent on the availability of oxygen. Atmospheric oxygen levels were probably around 0.1% at the time of mitochondria endosymbiosis [10]. The accumulation of oxygen in the atmosphere developed gradually up to the present day [11]. Mitochondria are still exposed to low oxygen-conditions within cells, in which there is a steep drop in the local pO2 around individual oxygen-consuming mitochondria [12]. Oxygen partial pressures (pO2) of <1–3 mmHg were observed at the level of mitochondria in cells exposed to normoxic (20.9% O2) culture conditions. This is most likely due to their function as oxygen sinks [13]. Isolated mitochondria are thus naturally exposed to artificially high pO2 levels, which are not found in physiological in situ conditions [14]. Therefore we examined, if the ambient oxygen concentration during the isolation process of the mitochondria influences their EGSH and mitochondrial energy production.
2. Material and methods
2.1. Grx1-roGFP2 transgenic mice
Generation and characterization of the Grx1-roGFP2 cardiomyocyte specific mice, in which the biosensor is targeted to the mitochondrial matrix, were described previously by our group [6]. Adult transgenic mice between 8 and 12 weeks of age were used for isolating hearts, cardiomyocytes or mitochondria. Wild type littermates were used for determining ATP, complex III activity and ROS levels in isolated mitochondria.
2.2. Isolation of mitochondria from tissues in normoxia and hypoxia
Organs were extracted from mice and immediately transferred to ice cold PBS at the indicated oxygen concentrations. All further steps of isolation were performed on ice. Tissue was cut into small pieces and homogenized in a potter with isolation buffer (in mM: HEPES 20, mannitol 220, sucrose 70, EDTA 1, freshly added PMSF 0.5). The homogenate was centrifuged at 800 g for 10 min. The homogenization step was repeated once. Supernatants were transferred into a fresh tube and centrifuged at 10,000 g for 30 min. The pellet was washed in 1.5 ml isolation buffer. The entire process of mitochondria isolation was done simultaneously in normoxic conditions (20.9% O2, room air) and hypoxic conditions (0.1%, 3%, 5% or 10% O2) using an in invivo 2 400 hypoxia workstation (Baker Ruskin).
2.3. EGSH measurements of isolated mitochondria
All measurements of sensor fluorescence were performed in the CLARIOstar microplate reader (BMG) with atmospheric control unit. During the measurements the atmospheric control unit ensured stable O2 concentrations between 0.1 and 20.9% O2 as indicated in the respective experiments. Excitation wavelengths were set to 488 and 405 nm with an emission wavelength of 510 nm. Baseline fluorescence ratio was recorded for all samples. For full oxidation and reduction of roGFP, 100 μM diamide and 2 mM DTT were added respectively. In some experiments 2.5 mM succinate/0.5 mM ADP were added as indicated. All measurements were carried out in sucrose buffer (in mM: EDTA 1, MOPS 10, sucrose 250).
2.4. Isolation of cardiomyocytes
Adult ventricular cardiomyocytes were isolated via the Langendorff perfusion. Mice were euthanized and the hearts were removed and quickly transferred into a chamber filled with ice-cold PBS where the aorta was tied to a 21G cannula. The heart was then perfused at 37 °C with Ca2+-free perfusion buffer (in mM: NaCl 113, KCl 4.7, KH2PO4 0.6, Na2HPO4 × 2H2O 0.6, MgSO4 × 7H2O 1.2, NaHCO3 12, KHCO3 10, HEPES 10, taurine 30, 2,3-butanedione-monoxime 10, glucose 5.5, pH 7.4) for 3 min. To digest the heart it was then perfused with 30 ml digestion buffer containing liberase DH (0.04 mg mL−1, Roche), trypsin (0.025%, Gibco) and CaCl2 12.5 μM. Afterwards, the atria were carefully excised and discarded; the digested ventricles were dissected for 30 s in 2.5 ml digestion buffer. To stop the digestion, 2.5 ml stop buffer I (perfusion buffer containing 1% BSA (Sigma) and 50 μM CaCl2) were added to the cell suspension, which was then homogenized for 3 min using a 1 ml syringe without a needle. Ten minutes after sedimentation, the cardiomyocyte pellet was transferred into stop buffer II (perfusion buffer containing 0.5% BSA and 37.5 μM CaCl2) for gradual recalcification up to 1 mM of Ca2+. The cardiomyocytes were plated onto round laminin (Sigma)-coated coverslides (24 mm, Thermo Scientific) and incubated at 37 °C and 5% CO2 until use.
2.5. EGSH measurements of isolated cardiac myocytes
Isolated cardiac myocytes plated onto laminin-coated glass coverslips were incubated for at least 45 min before imaging. Then, the coverslips were mounted in the imaging chamber and washed once with 400 μL of imaging buffer (in mmol/L: NaCl 144, KCl 5.4, MgCl2 1,CaCl2 1, HEPES 10, and pH 7.3) at room temperature. The redox measurements were performed using the inverted fluorescence microscope IX83 (Olympus) and Visiview software. The roGFP2 sensor was excited at 488 and 405 nm using a Polychrome V light source (Till Photonics). The emitted light from the sample was detected via a CCD camera (emission filter 510 ± 15 nm). Cardiomyocytes were either treated with 2 mM DTT or 100 μM diamide as soon as the 405/488 nm ratio reached a stable baseline. An exposure time of 10 ms usually led to a good signal:noise ratio, and images were acquired in GFP emission channels every 5 s.
2.6. EGSH measurements of cryosections
Redox histology on cryosections was performed as described previously by Fujikawa et al. [15].
2.7. Calculation of the mitochondrial oxidation difference (mtOxD) and EroGFP2 redox potentials
mtOxD and EGSH calculations were performed as described previously [6]. Determining the EGSH values at basal conditions requires the analyses of the fluorescence intensities at 405 nm and 488 nm after stimulation with diamide (maximum oxidation response) and DTT (maximum reduction response). Based on these values the mtOxD of the probe was calculated. The mtOxD is the ratio of the number of oxidized molecules to the total number of molecules (mtOxD roGFP2 = [roGFP2ox]/([roGFP2red + roGFP2ox])). In short the emission intensities (I) obtained from the measurements at 405 and 488 nm were used to calculate the mtOxD. The mtOxD then was applied to calculate the probe redox potential, where E`roGFP2 is −280 mV. Assuming that the probe and the glutathione redox couple are in equilibrium, the EGSH = EroGFP2. All measurements were performed with standardized microcopy settings including laser intensities and exposure times.
2.8. Analysis of superoxide anion (O2.-) levels with MitoSox
Mitochondria (25 μg) were diluted in sucrose buffer (in mM: EDTA 1, MOPS 10, sucrose 250) with or without 2.5 mM succinate/0.5 mM ADP. 5 μM of MitoSOX red mitochondrial superoxide indicator (Invitrogen, M36008) were added and O2•− levels were recorded in the CLARIOstar microplate reader (BMG) with atmospheric control unit at the indicated oxygen concentration. The MitoSOX dye was excited at 510 nm and emission filter was set to 580 nm. The measurement was allowed to run for 20 min. The average of the plateau phase of the last 10 min was considered for analysis.
2.9. Analysis of hydrogen peroxide (H2O2) levels with Amplex Red
Mitochondrial H2O2 production was measured using the H2O2-sensitive and specific fluorescent dye Amplex®UltraRed (Life technologies, Molecular Probes®). Amplex Red reacts with H2O2 to form the fluorescent product Resorufin (excitation at 535 nm, emission at 590 nm) in a 1:1 relation. The reaction is catalyzed by Horseradish Peroxidase (HRP). Assay component concentrations were 50 μM for Amplex Red, 0.5 U/ml for HRP and 100 U/ml superoxide dismutase (for conversion of O2•− to H2O2). Mitochondrial H2O2 production was measured with and without addition of 2.5 mM succinate/0.5 mM ADP. Experiments were performed in 96 well plates (black/transparent) using the CLARIOstar microplate reader (BMG) with a bottom reading setting. All experiments were conducted in sucrose buffer (in mM: EDTA 1, MOPS 10, sucrose 250) with 25 μg mitochondria per well at 37 °C.
2.10. Complex III activity assay
Cytochrome c reductase (complex III) activity was assessed by the reduction of cytochrome c after administration of reduced decylubichinone. 10 mM decylubichinone, dissolved in ethanol was reduced with 300 mM KBH4 and stabilized in 120 mM HCl. The enzyme assay was performed in assay buffer (in mM: KPi 50, n-Dodecyl β-D-maltoside 1, KCN 1, 2.5 μM rotenone and 0.1% BSA, pH 7.4) complemented with 0.1 mM of reduced decylubichinone and 15 μM cytochrome c. The reaction was started with the administration of 25 μg of isolated mitochondria and reduction of cytochrome c was followed over time at a wavelength of 550 nm.
2.11. ATP
Mitochondria (25 μg) were diluted in sucrose buffer (in mM: EDTA 0.04, MOPS 10, sucrose 250) with or without 2.5 mM succinate/0.5 mM ADP. Samples were incubated in the respective buffers for 20 min at defined oxygen concentrations as indicated. These mitochondria were then centrifuged at 10,000 g for 10 min. Supernatants were used for the ATP measurements using the Celltiter-Glo luminescent cell viability assay kit (Promega G7570/7571). The measurement of ATP in the supernatants was performed at room air condition including 20.9% O2.
2.12. Statistics
Data are shown as mean ± SEM. Statistical analyses were performed using student 2-tailed t-test. One-way ANOVA analysis (Conferroni post hoc test) was performed in cases of comparisons with more than two groups. Values of p < 0.05 were considered statistically significant.
3. Results
3.1. Isolation of mitochondria in low oxygen conditions preserves the EGSH
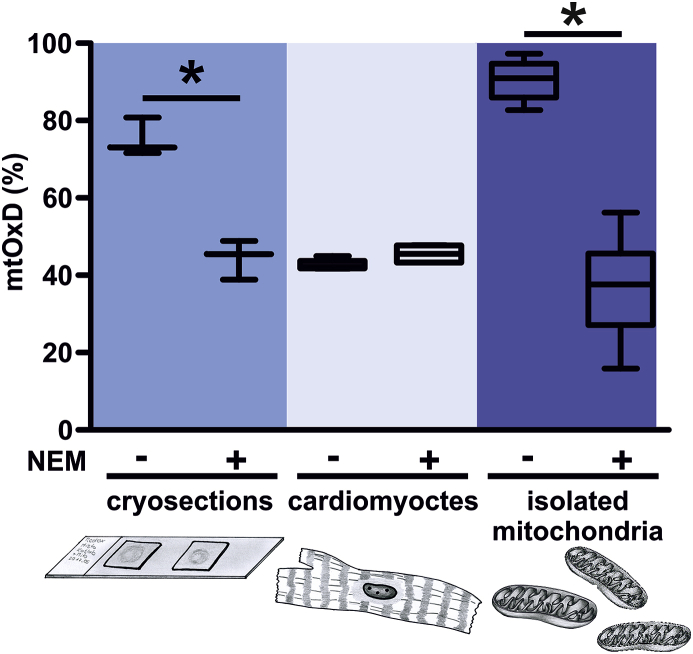
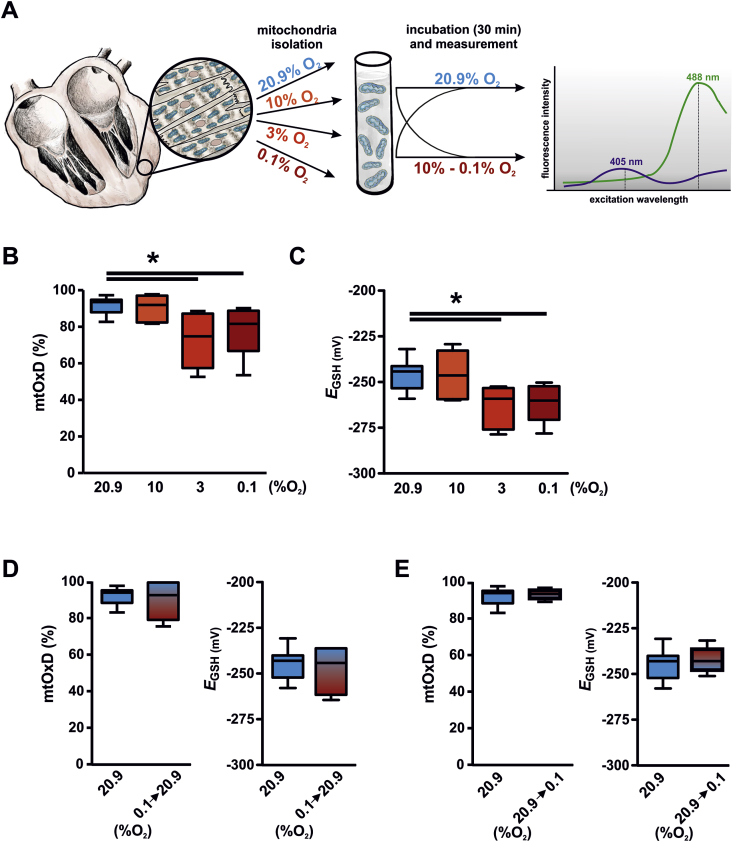
We have previously described a transgenic mouse model, which allows determining the EGSH by the use of the genetically encoded Grx1-roGFP2 biosensor [6]. The transgenic expression of the biosensor is driven by the αMHC promoter and thus results in cardiomyocyte-restricted expression. Due to the Neurospora crassa ATP9 mitochondrial targeting sequence the biosensor is located in the mitochondrial matrix. Redox biosensors including the Grx1-roGFP2 have been used in the past in cell culture but also in complex tissue preparations including tissue sections and whole organs [7,16]. Determining the EGSH at tissue level relies on procedures to prevent oxidation of the redox probes during specimen dissection and fixation [17]. We confirmed this by analyzing the mitochondrial mtOxD, defined as the ratio of oxidized roGFP2 to total roGFP2, in cryosections of mtGrx1-roGFP2 transgenic mice using a protocol described previously by Fujikawa et al. [15] (Fig. 1). Incubation of the heart sections with NEM that is protecting thiol groups prevented oxidation of the biosensor. Interestingly, incubation of isolated cardiomyocytes from Grx1-roGFP transgenic hearts with or without NEM did not alter the mtOxD indicating that the physiological intracellular milieu is sufficient for preserving the more reduced state of the EGSH of the mitochondrial matrix. Next we analyzed the mtOxD in isolated cardiomyocyte mitochondria from the Grx1-roGFP2 mice. The mtOxD of isolated mitochondria revealed a higher degree of oxidation compared to the isolated cardiomyocytes and was similar to the mtOxD of non NEM-treated cryosections (around 45%). Similar to the histology sections, the shift of the EGSH could be prevented by addition of NEM to all steps of the mitochondria isolation procedure. To analyze if mimicking the physiological pO2 of the mitochondria would prevent changes in the mtOxD and EGSH likewise, we next isolated mitochondria under defined oxygen concentrations. Preparations of the hearts as well as all steps of the mitochondria isolation, incubation and measurement were performed at oxygen concentrations ranging from 20.9% to 0.1% O2 (Fig. 2A). With decreasing oxygen concentration, i.e. 3% and 0.1% O2 the mtOxD and EGSH were significantly lower compared to 20.9% O2 (Fig. 2B and C). Next we tested, if the reduced mtOxD and EGSH in hypoxia can be reversed. Reoxygenation of mitochondria, which were isolated at 0.1% O2, for 30 min at 20.9% O2 indeed inverted the reduced mtOxD and EGSH (Fig. 2D). Incubation of mitochondria isolated in 20.9% O2 for 30 min at 0.1% O2 on the other hand could not rescue the high mtOxD of the glutathione pool indicating that the changes induced at the atmospheric oxygen levels are the result of an irreversible process (Fig. 2E).
Fig. 1.
The glutathione pool is oxidized during isolation of mitochondria. Grx1-roGFP2 cardiomyocyte specific transgenic mice were used to determine the mitochondrial Oxidation difference (mtOxD) of the glutathione pool in heart cryosection (3 mice), isolated cardiomyocytes (4 mice) or isolated cardiac mitochondria (9 mice). N-Ethylmaleimide (NEM) was either added or not during the experimental procedure as indicated. *p < 0.05.
Fig. 2.
Isolation of mitochondria in hypoxia prevents oxidation of the glutathione pool. (A) Schematic drawing of the experiments performed in B-E. (B, C) Mitochondria from Grx1-roGFP2 cardiomyocyte specific transgenic hearts were isolated, incubated and analyzed in a defined oxygen environment as indicated. 11, 6, 6, and 11 hearts were included in the analysis at 20.9, 10, 3, and 0.1% O2, respectively. Subsequently the mitochondrial Oxidation Difference (mtOxD) and the glutathione redox potential (EGSH) were determined. The defined oxygen environment was continued during the mtOxD and EGSH measurements for each sample. (D, E) Mitochondria from Grx1-roGFP2 cardiomyocyte specific transgenic hearts were isolated either at 20.9% O2 or 0.1% O2. Subsequently, the mitochondria were incubated and analyzed at the same oxygen concentration (20.9% O2, 11 hearts), reoxygenated (0.1 → 20.9% O2, 6 hearts) or exposed to hypoxia (20.9 → 0.1% O2, 4 hearts) as indicated. *p < 0.05.
3.2. Isolation of mitochondria in hypoxia affects superoxide anion and ATP production
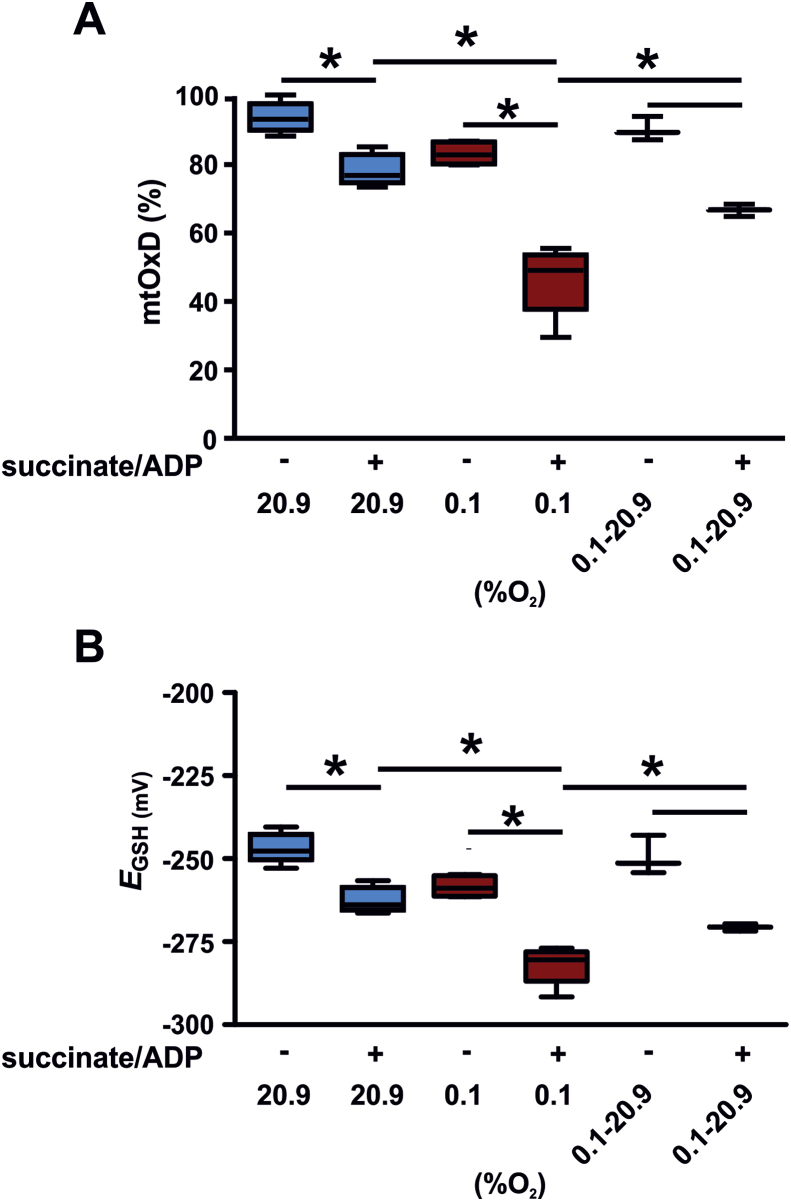
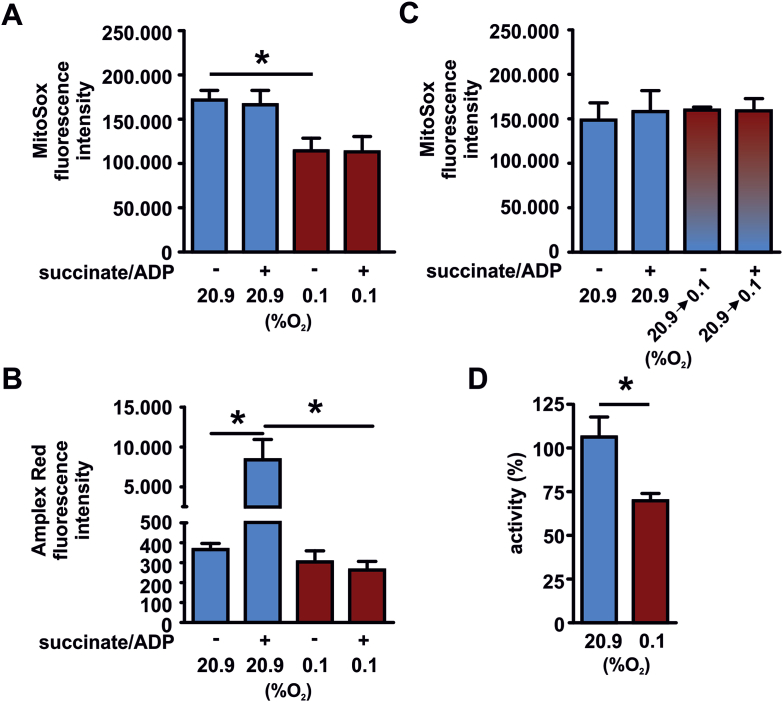
To analyze, if mitochondria either isolated in 20.9% O2 or 0.1% O2 differ in their bioenergetic profile and function, we tested the oxidation of the Grx1-roGFP2 biosensor of the mitochondria in response to succinate/ADP. The mtOxD and EGSH were significantly reduced in mitochondria isolated in 0.1% O2 confirming the earlier finding (Fig. 3A and B). Upon addition of succinate/ADP, mitochondria reduced the oxidation of the glutathione pool. This is congruent with previous reports [9]. The reduction was detectable in normoxic and in even more significant in hypoxic isolated mitochondria. To analyze how addition of succinate affects the mitochondria we next determined ROS production of mitochondria isolated in 20.9% O2 or 0.1% O2. We determined the mitochondrial O2•− levels in the mitochondrial matrix by the use of the dye MitoSOX (Fig. 4A) and H2O2 release of the mitochondria by Amplex Red (Fig. 4B). The mitochondria that were isolated in 0.1% O2 demonstrated significantly lower levels of O2•− compared to the normoxic isolated organelles. Addition of succinate did not affect the O2•− levels in the matrix upon normoxia or hypoxia. Addition of succinate, however indeed increased the release of H2O2 of the mitochondria in normoxia as detected by Amplex Red. In sharp contrast, mitochondria isolated in hypoxia lacked the succinate-induced H2O2 release. Since not only the isolation of the mitochondria but also the ROS measurements were performed in 20.9% O2 or 0.1% O2, we controlled for the effect of acute hypoxia on the O2•− readings (Fig. 4C). Exposing mitochondria isolated at 20.9% O2 during the succinate/ADP stimulation and MitoSox readings to 0.1% O2, did not affect the O2•− levels compared to mitochondria isolated at 20.9% O2 followed by the MitoSox analysis performed likewise at 20.9% O2. This excludes an impairment of O2•− production due to the hypoxic exposure during the analysis.
Fig. 3.
Addition of succinate/ADP results in a reduction of the glutathione pool in mitochondria isolated in hypoxia. Grx1-roGFP2 tg mitochondria were isolated in 20.9% O2 (4 hearts) or 0.1% O2 (4 hearts). One portion of the 0.1% O2 isolated mitochondria were incubated for 30 min at 20.9% O2 before addition of substrates (0.1%→20.9% 02). Subsequently, succinate and ADP were added for 30 min as indicated and the mitochondrial Oxidation difference (mtOxD, A) and EGSH (B) were determined. During substrate addition and mtOxD analysis the indicated oxygen concentration was continued for each sample. *p < 0.05.
Fig. 4.
Superoxide anion, hydrogen peroxide levels and complex III activity are diminished in mitochondria isolated in hypoxia. Mitochondria were isolated in 20.9% O2 or 0.1% O2. Subsequently, superoxide anion (O2•−) levels (A) and hydrogen peroxide levels (B) were analyzed at the given oxygen concentration. (C) Mitochondria were isolated in 20.9% O2. Isolated mitochondria were either analyzed with MitoSox at 20.9% O2 or at 0.1% O2 (20.9 → 0.1% 02) as indicated. (D) Mitochondria were isolated in 20.9% O2 or 0.1% O2. Subsequently, complex III activity was analyzed. For each condition 3 independent mitochondria isolations from hearts were included in the analysis.*p < 0.05.
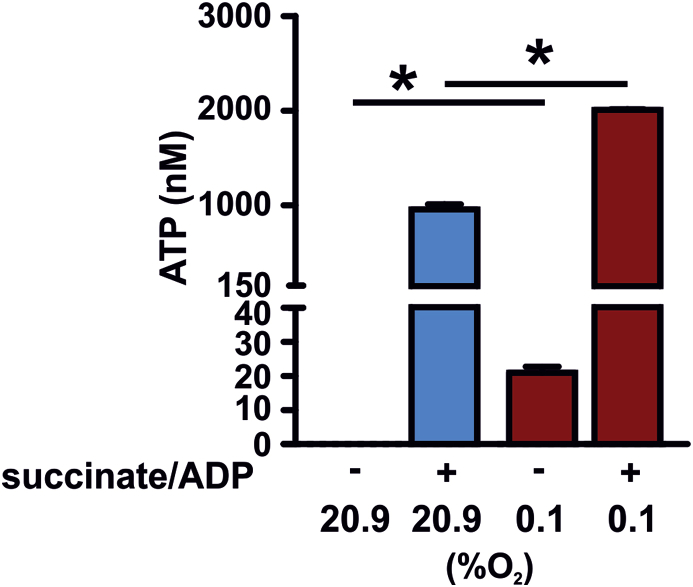
The reduced baseline release of ROS in the hypoxic isolated mitochondria was paralleled by a decreased mitochondrial complex III activity (Fig. 4D). To analyze the bioenergetics outcome we subsequently analyzed ATP levels in the supernatants of the mitochondria (Fig. 5). Even in non-energizeda mitochondria ATP levels produced by the hypoxic isolated mitochondria were significantly higher compared to the normoxic isolated mitochondria. Further ATP production was stimulated by the addition of succinate/ADP. Mitochondria isolated in 20.9% O2 and 0.1% O2 responded both with significantly increased ATP levels in response to addition of the substrate. Overall the levels produced upon stimulation however were significantly higher in the hypoxic isolated mitochondria compared to the normoxic isolated mitochondria indicating that metabolism and electron flow are altered in response to the oxygen concentration during the isolation process.
Fig. 5.
ATP production is elevated in mitochondria isolated in hypoxia. Mitochondria from in total three independent hearts per experimental condition were isolated simultaneously in 20.9% O2 or 0.1% O2. Subsequently, succinate/ADP were added for 30 min as indicated. During substrate addition the defined oxygen concentration was maintained. Subsequently, mitochondria were centrifuged, supernatants of the mitochondria were harvested and ATP levels were analyzed. *p < 0.05.
4. Discussion
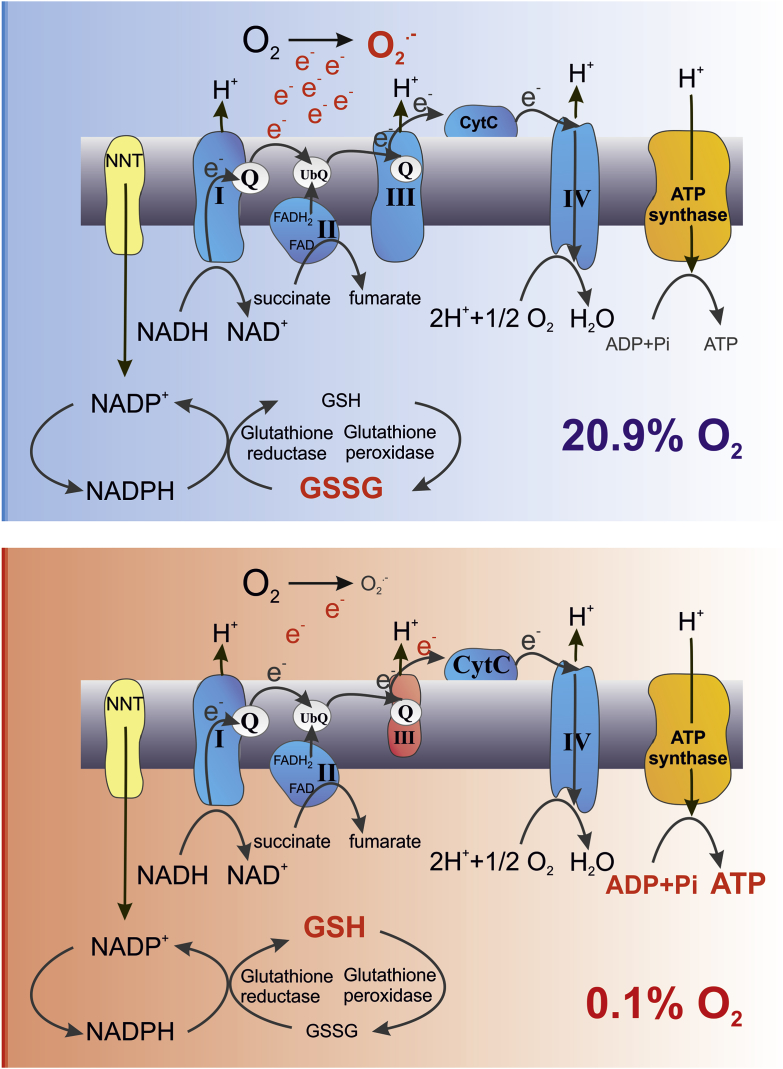
Isolated mitochondria are widely used and analyzed for studying mitochondrial biology including studies involving redox signaling pathways. Isolation of these intracellular organelles makes them accessible for example for biochemical analyses. Due to their complex morphology one major challenge during the isolation is preserving their structural integrity [18]. One main function of energized mitochondria is the production of ATP. The mitochondrial respiratory chain generates approximately 90% of the cellular ATP [19]. For balancing the oxygen consumption and production of O2•− as a side product, the mitochondria rely on the glutathione redox buffer. It is known that during sample preparation GSH can undergo oxidation [20]. There are well established protocols analyzing cultured cells and solid tissues for preventing auto-oxidation and consequently overestimation of oxidation of the glutathione pool by adding NEM during sample preparation [21]. Using NEM during isolation of mitochondria we have identified that the mitochondrial glutathione pool is prone to oxidation during the isolation process. Most interestingly adding NEM to isolated cardiomyocytes did not affect the mitochondrial EGSH indicating that the living cells per se can offer enough buffering potential to prevent oxidation. Thus, the mitochondria seem to be largely undisturbed within this intracellular physiological environment despite detaching the cardiomyocytes from their surrounding tissue. In an attempt to mimic the intracellular oxygen conditions, we were able to show that isolating mitochondria in an oxygen reduced environment prevents the oxidation of the mitochondrial glutathione pool likewise. The oxygen partial pressure shows a steep gradient from the environmental high pO2 of 160 mmHg (at sea level) to the mitochondrial pO2 of <1–3 mmHg (Fig. 6). Mimicking the physiological tissue or intracellular oxygen conditions in vitro is a major experimental challenge [22]. Despite the crucial role of O2 in cellular physiology, much of the cell culture and tissue work is still conducted under ambient air atmosphere (pO2 18.6 kPa in a humidified CO2 incubator), i.e. hyperoxic conditions compared to the in vivo situation. To mimic the in vivo conditions of the mitochondria, the isolation of the mitochondria but also the further processing and analysis of the samples need to be done in the defined oxygen environment without risking any reoxygenation. Using an experimental approach that allowed a continuous and stable oxygen environment, we demonstrate that reducing the environmental oxygen concentration prevents the oxidation of the mitochondrial glutathione pool during the isolation process (Fig. 7). This was accompanied by altered mitochondrial complex III activity and ROS production. The reduced levels of H2O2 of mitochondria in hypoxia are in line with a described linear dependence of mitochondrial H2O2 production on oxygen [23]. The isolated mitochondria likewise differed in ATP production in the sense of higher ATP levels produced in the mitochondria isolated in 0.1% O2. In this regard it is important to note that cytochrome c oxidase, which is the terminal oxidase of cellular respiration, catalyzes the transfer of electrons from ferrocytochrome c to molecular oxygen. This is the basis for generating a proton gradient, which is finally used for ATP production. The lowest oxygen concentration applied in the presented study, i.e. 0.1% O2 (equivalent to a pO2 of 0.76 mmHg or 0.1 kPa) is not limiting for cytochrome c oxidase activity considering the high affinity of the enzyme towards oxygen as reflected by KM values around 0.01 kPa measured in isolated mitochondria [24]. However, it should be noted that indeed several KM values for oxygen have been reported for the cytochrome c oxidase, which might be in part explained by the fact that for example the proton-motive force affects the oxygen affinity of the enzyme. Together our data demonstrate that despite the low ROS levels in hypoxic isolated mitochondria, more ATP was produced excluding that the electron flow in the mitochondrial respiratory chain was impaired at 0.1% O2. The reduced complex III activity together with increased ATP levels indicate that the individual complex activity is not rate limiting to the overall process but rather streamlined to match and efficiently make use of the provided oxygen by the respiratory chain. Based on these data we hypothesize that a balance is maintained between transfer of electrons to O2 to form water as opposed to premature electron transfer at complex III to form ROS. The isolation of mitochondria in 0.1% O2 thus seems to create an environment that is better suited for their function compared to the isolation in 20.9% O2. This notion is also supported by recent in vivo studies. Chronic hypoxia led to a marked improvement in survival and outcome in a genetic mouse model of Leigh syndrome, which is a mitochondrial disease [25].
Fig. 6.
pO2gradient from the ambient air to the mitochondria in situ. Oxygen partial pressures (pO2) shown are reported in the literature [14,28].
Fig. 7.
Schematic drawing of the major differences found in the mitochondria isolated in normoxia (20.9% O2) compared to hypoxia (0.1% O2).
Oxygen is a reactive chemical that oxidizes other molecules especially transition metals. As oxygen accumulated during evolution, iron became a limiting nutrient in the aerobic environment [26]. Iron is especially important for mitochondria since their function relies on iron sulfur cluster- and heme-containing proteins. Many enzymes including iron-sulfur proteins are irreversibly destroyed by oxygen. Most interestingly anaerobes have a higher content and higher variability of iron sulfur proteins than aerobes [27]. Aerobic organisms have indeed favored the use of Fe2S2 over Fe4S4, in line with the fact that superoxide anions are more damaging to Fe4S4 clusters. Therefore the preferential use of Fe2S2 clusters by mitochondrial Fe-S proteins is a logical consequence. In line it is tempting to speculate that the atmospheric oxygen concentration of 20.9% O2 is simply hyperoxic for isolated mitochondria creating a partly oxygen-toxic environment.
Taken together we found a better preserved EGSH and a more efficient electron flow in mitochondria isolated in low oxygen concentration, which might be more close to the in vivo situation. Thus, considering the oxygen concentration seems to be critical when analyzing isolated mitochondria.
Author disclosure statement
No competing financial interests exist.
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (IRTG 1816) to MN, VVB and DMK. PR is supported by the SFB1002, project A06. VVB is supported by Russian Science Foundation 17-14-01086.
References
- 1.Gray M.W. Mitochondrial evolution. Cold Spring Harb. Perspect. Biol. 2012;4:a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murphy M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kalogeris T., Bao Y., Korthuis R.J. Mitochondrial reactive oxygen species: a double edged sword in ischemia/reperfusion vs preconditioning. Redox Biol. 2014;2:702–714. doi: 10.1016/j.redox.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deponte M. The incomplete glutathione puzzle: just guessing at numbers and figures? Antioxidants Redox Signal. 2017;27:1130–1161. doi: 10.1089/ars.2017.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calabrese G., Morgan B., Riemer J. Mitochondrial glutathione: regulation and functions. Antioxidants Redox Signal. 2017;27:1162–1177. doi: 10.1089/ars.2017.7121. [DOI] [PubMed] [Google Scholar]
- 6.Swain L., Kesemeyer A., Meyer-Roxlau S., Vettel C., Zieseniss A., Guntsch A., Jatho A., Becker A., Nanadikar M.S., Morgan B., Dennerlein S., Shah A.M., El-Armouche A., Nikolaev V.O., Katschinski D.M. Redox imaging using cardiac myocyte-specific transgenic biosensor mice. Circ. Res. 2016;119:1004–1016. doi: 10.1161/CIRCRESAHA.116.309551. [DOI] [PubMed] [Google Scholar]
- 7.Swain L., Nanadikar M.S., Borowik S., Zieseniss A., Katschinski D.M. Transgenic organisms meet redox bioimaging: one step closer to physiology. Antioxidants Redox Signal. 2018;29:603–612. doi: 10.1089/ars.2017.7469. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J., Ye Z.W., Singh S., Townsend D.M., Tew K.D. An evolving understanding of the s-glutathionylation cycle in pathways of redox regulation. Free Radic. Biol. Med. 2018;120:204–216. doi: 10.1016/j.freeradbiomed.2018.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia J., Han D., Sancheti H., Yap L.P., Kaplowitz N., Cadenas E. Regulation of mitochondrial glutathione redox status and protein glutathionylation by respiratory substrates. J. Biol. Chem. 2010;285:39646–39654. doi: 10.1074/jbc.M110.164160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor C.T., McElwain J.C. Ancient atmospheres and the evolution of oxygen sensing via the hypoxia-inducible factor in metazoans. Physiology (Bethesda) 2010;25:272–279. doi: 10.1152/physiol.00029.2010. [DOI] [PubMed] [Google Scholar]
- 11.Canfield D.E., Habicht K.S., Thamdrup B. The archean sulfur cycle and the early history of atmospheric oxygen. Science. 2000;288:658–661. doi: 10.1126/science.288.5466.658. [DOI] [PubMed] [Google Scholar]
- 12.Mik E.G., Stap J., Sinaasappel M., Beek J.F., Aten J.A., van Leeuwen T.G., Ince C. Mitochondrial po2 measured by delayed fluorescence of endogenous protoporphyrin ix. Nat. Methods. 2006;3:939–945. doi: 10.1038/nmeth940. [DOI] [PubMed] [Google Scholar]
- 13.Wenger R.H. Mitochondria: oxygen sinks rather than sensors? Med. Hypotheses. 2006;66:380–383. doi: 10.1016/j.mehy.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 14.Wenger R.H., Kurtcuoglu V., Scholz C.C., Marti H.H., Hoogewijs D. Frequently asked questions in hypoxia research. Hypoxia (Auckl) 2015;3:35–43. doi: 10.2147/HP.S92198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujikawa Y., Roma L.P., Sobotta M.C., Rose A.J., Diaz M.B., Locatelli G., Breckwoldt M.O., Misgeld T., Kerschensteiner M., Herzig S., Muller-Decker K., Dick T.P. Mouse redox histology using genetically encoded probes. Sci. Signal. 2016;9:rs1. doi: 10.1126/scisignal.aad3895. [DOI] [PubMed] [Google Scholar]
- 16.Kostyuk A.I., Panova A.S., Bilan D.S., Belousov V.V. Redox biosensors in a context of multiparameter imaging. Free Radic. Biol. Med. 2018;128:23–39. doi: 10.1016/j.freeradbiomed.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Albrecht S.C., Barata A.G., Grosshans J., Teleman A.A., Dick T.P. In vivo mapping of hydrogen peroxide and oxidized glutathione reveals chemical and regional specificity of redox homeostasis. Cell Metabol. 2011;14:819–829. doi: 10.1016/j.cmet.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 18.Lehr S., Hartwig S., Kotzka J. Preparation of "functional" mitochondria: a challenging business. Methods Mol. Biol. 2015;1264:1–8. doi: 10.1007/978-1-4939-2257-4_1. [DOI] [PubMed] [Google Scholar]
- 19.Rich P. Chemiosmotic coupling: the cost of living. Nature. 2003;421:583. doi: 10.1038/421583a. [DOI] [PubMed] [Google Scholar]
- 20.Curello S., Ceconi C., Cargnoni A., Cornacchiari A., Ferrari R., Albertini A. Improved procedure for determining glutathione in plasma as an index of myocardial oxidative stress. Clin. Chem. 1987;33:1448–1449. [PubMed] [Google Scholar]
- 21.Ratnayake S., Dias I.H., Lattman E., Griffiths H.R. Stabilising cysteinyl thiol oxidation and nitrosation for proteomic analysis. J. Proteom. 2013;92:160–170. doi: 10.1016/j.jprot.2013.06.019. [DOI] [PubMed] [Google Scholar]
- 22.Keeley T.P., Mann G.E. Defining physiological normoxia for improved translation of cell physiology to animal models and humans. Physiol. Rev. 2019;99:161–234. doi: 10.1152/physrev.00041.2017. [DOI] [PubMed] [Google Scholar]
- 23.Grivennikova V.G., Kareyeva A.V., Vinogradov A.D. Oxygen-dependence of mitochondrial ros production as detected by amplex red assay. Redox Biol. 2018;17:192–199. doi: 10.1016/j.redox.2018.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gnaiger E., Lassnig B., Kuznetsov A., Rieger G., Margreiter R. Mitochondrial oxygen affinity, respiratory flux control and excess capacity of cytochrome c oxidase. J. Exp. Biol. 1998;201:1129–1139. doi: 10.1242/jeb.201.8.1129. [DOI] [PubMed] [Google Scholar]
- 25.Jain I.H., Zazzeron L., Goli R., Alexa K., Schatzman-Bone S., Dhillon H., Goldberger O., Peng J., Shalem O., Sanjana N.E., Zhang F., Goessling W., Zapol W.M., Mootha V.K. Hypoxia as a therapy for mitochondrial disease. Science. 2016;352:54–61. doi: 10.1126/science.aad9642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imlay J.A. Iron-sulphur clusters and the problem with oxygen. Mol. Microbiol. 2006;59:1073–1082. doi: 10.1111/j.1365-2958.2006.05028.x. [DOI] [PubMed] [Google Scholar]
- 27.Andreini C., Rosato A., Banci L. The relationship between environmental dioxygen and iron-sulfur proteins explored at the genome level. PLoS One. 2017;12 doi: 10.1371/journal.pone.0171279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keeley T.P., Mann G.E. Defining physiological normoxia for improved translation of cell physiology to animal models and humans. Physiol. Rev. 2019;99:161–234. doi: 10.1152/physrev.00041.2017. [DOI] [PubMed] [Google Scholar]