Abstract
Muscle-invasive and metastatic bladder cancer have an extremely poor 5-year survival rate of 5%. In comparison, all other bladder cancers (BCs) have a 5-year survival rate of 77%. This striking contrast indicates that one of the therapeutic kernels for bladder cancer is to elucidate the molecular mechanisms underlying its invasiveness and metastasis. In the current study, we demonstrated that maternally expressed gene 3 (MEG3) is significantly downregulated in human invasive bladder cancers in comparison to non-invasive bladder cancers, and that ectopic expression of MEG3 dramatically inhibits the invasiveness of human bladder cancer cells. Consistently, ectopic expression of MEG3 also attenuates metastatic ability of T24T cells, a cell line derived from T24 cells, in the lungs of nude mice. Our mechanistic studies reveal that MEG3, as a ceRNA, inhibits the invasiveness of human bladder cancer cells via negative regulation of c-Myc by competing with PHLPP2 mRNA for miR-27a. These findings not only provide a novel insight into understanding the mechanisms behind the MEG3 inhibition of bladder cancer cell invasion, but also reveal the potential for use of MEG3 as a tool for the prevention and therapy of invasive bladder cancer.
Keywords: lncRNA, miRNA, c-Myc, ceRNA, bladder cancer
Introduction
Bladder cancer (BC) is the fourth most common cancer in men in the United States and the most lethal malignancy in the urinary system.1, 2 It is estimated that, in 2017, 79,030 Americans will be diagnosed with bladder cancer and 16,870 will die of the disease.2 The occurrence of new bladder cancer cases has been rising steadily at a rate of approximately 1,000 per year. Approximately 20% to 30% of bladder cancers are diagnosed as muscle invasive and tend to be metastatic. Half of patients with muscle-invasive disease will die from metastasis within 2 years. Compared with the 77% 5-year survival rate of all stages of bladder cancer, the 5-year survival rate for metastatic bladder cancer is only 5%.1 For these reasons, to improve the clinical outcome of this disease, further understanding of the mechanisms underlying invasive bladder cancer is critical.
Long noncoding RNAs (lncRNAs) have gained widespread attention in recent years as a potential new and crucial player in biological regulation.3, 4, 5 Maternally expressed gene 3 (MEG3) is one of the lncRNAs. Loss of MEG3 expression has been found in various types of human tumors and cancer cell lines.6 Recent studies show various functions of MEG3, which include inhibition of proliferation and induction of apoptosis through MEG3’s interaction with p53 in hepatoma cells,7 suppression of migration and invasion of thyroid carcinoma by targeting Rac1,8 and promotion of osteogenic differentiation of mesenchymal stem cells by regulating BMP4.9 In addition, studies in Chinese patients suggest that the genetic variants in MEG3 contribute to the development and risk of colorectal cancer, further suggesting that MEG3 has potential as a prognostic biomarker.10, 11 In bladder cancers, downregulation of MEG3 forces increased proliferation of cancer cells,12 and it is known that low expression of serum MEG3 is associated with poor recurrence-free survival.13 However, nothing is known about the underlying mechanisms of MEG3 in bladder cancer’s invasive action.
lncRNA can function as a competing endogenous RNA (ceRNA) and indeed is a contributing factor in the onset and progression of disease.14, 15, 16 For example, MALAT1 has been reported to be a ceRNA of MCL-1 by sponging miR-363 in gallbladder cancer,17 and the NEAT1-E2F3 ceRNA pair has been demonstrated as regulating non-small-cell lung cancer’s progression by competing for miR-377.17 Our current study demonstrates that MEG3 functions as a ceRNA of PHLPP2 (PH domain and leucine-rich repeat protein phosphatase 2) mRNA in competing with miR-27a and thereby promoting PHLPP2 protein translation, which, in turn, inhibits c-Jun-mediated c-Myc mRNA transcription and bladder cancer’s invasive ability in human bladder cancer cells in vitro as well as lung metastasis in human bladder cancer cells in vivo.
Results
lncRNA MEG3 Impairs the Invasive Ability of Bladder Cancer Cells
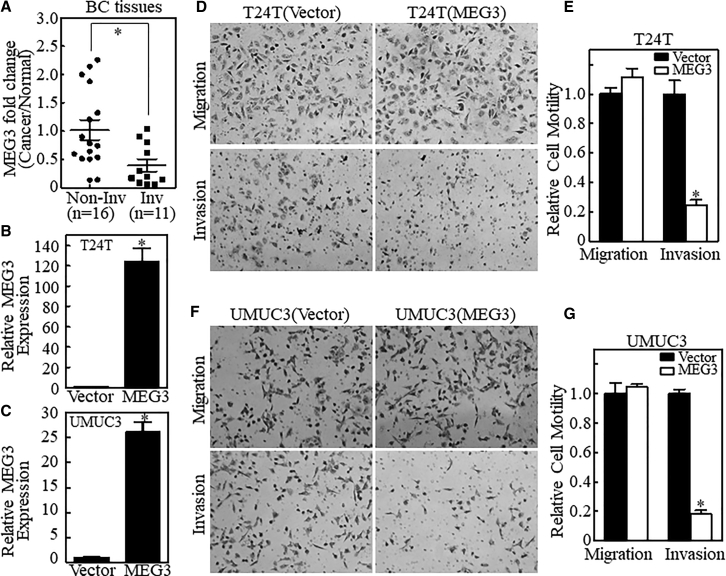
Although MEG3 has been reported to be downregulated in human bladder cancers,12 the potential association between MEG3 downregulation and bladder cancer invasion has never been explored. To investigate the function of MEG3 in bladder cancer invasion, we first observed MEG3 expression in human bladder cancer tissues. Twenty-seven pairs of bladder cancer tissue and their adjacent normal tissue, as listed in Table S1, confirmed by pathological analysis, were used to analyze MEG3 expression (Figure 1A). The pathological analysis divided the samples into two groups: non-muscle-invasive bladder cancers (Non-Inv, n = 16) and muscle-invasive bladder cancers (Inv, n = 11). As shown in Figure 1A, the MEG3 expression level was displayed as fold change (bladder cancer tissue and their adjacent normal tissue). The results showed that the expression of MEG3 in muscle-invasive bladder cancer was dramatically lower than the expression in non-muscle-invasive disease. This finding strongly indicates that MEG3 may be associated with bladder cancer invasion. To test this, we overexpressed MEG3 in two different high-grade invasive bladder cancer cell lines: T24T and UMUC3. The stable MEG3 transfectants and their vector control transfectants were established and identified, as shown in Figures 1B and 1C and Table S2. Ectopic expression of MEG3 resulted in dramatic inhibition of the invasive ability of the T24T cell line, but did not show any observable inhibition of bladder cancer cell migration (Figures 1D and 1E). Similar findings were observed in UMUC3 cells (Figures 1F and 1G). Our results strongly indicate an inhibitory effect of MEG3 on bladder cancer invasion.
Figure 1.
MEG3 Impaired the Invasive Ability of Bladder Cancer Cells
(A) lncRNA MEG3 was evaluated in human invasive bladder cancers (BCs) as compared with those in non-invasive bladder cancers. (B and C) The lncRNA MEG3 were stably transfected into T24T (B) and UMUC3 (C) cells and MEG3 expression was analyzed by qRT-PCR. GAPDH mRNA was used as the internal control. Results are presented as relative MEG3 expression. The asterisk indicates a significant change in comparison to the control (p < 0.05). Bars represent the mean ± SD from three independent experiments. The stable transfectants T24T(MEG3) (D), UMUC3(MEG3) (F), and their vector controls were used to determine their invasion ability using a BD BioCoat Matrigel Invasion Chamber. The migration ability was determined by using the empty insert membrane without the Matrigel. The corresponding quantitative analysis for T24T(MEG3) (E) and UMUC3(MEG3) (G) were presented. Results are presented as relative cell motility, and the asterisk indicates a significant change compared with the vector cells (p < 0.05). The bars indicate the mean ± SD from three independent experiments.
c-Myc Is a MEG3 Downstream Effector Responsible for Attenuation of Bladder Cancer Invasion
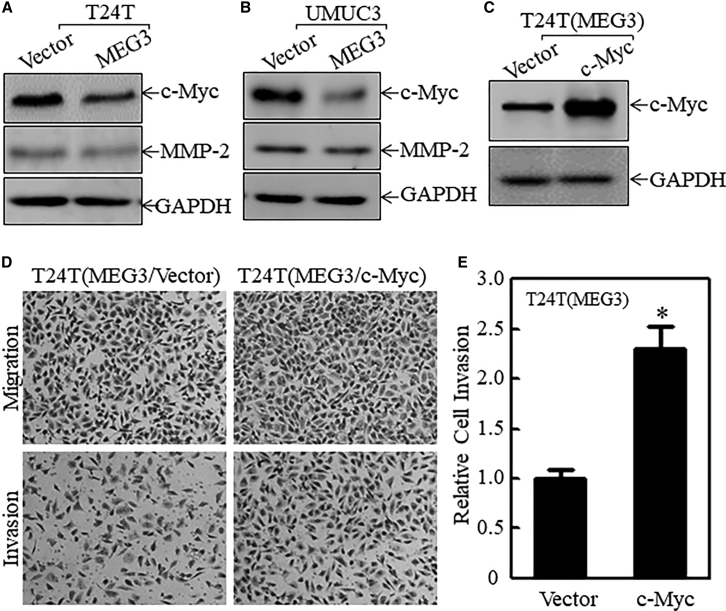
Our previous study demonstrated that MMP-2 is crucial for invasion of human bladder cancer T24T cells.18 It also has been reported that c-Myc plays a role in human bladder cancer invasion.19 To determine whether MMP-2 and c-Myc are the downstream effectors of MEG3 in its inhibition of bladder cancer invasion, the effects of MEG3 on the protein abundance of c-Myc and MMP-2 were evaluated in T24T(MEG3) versus T24T(Vector) cells and in UMUC3(MEG3) versus UMUC3(Vector) cells. As shown in Figures 2A, 2B, and S1, overexpression of MEG3 in both cell lines resulted in a reduction of c-Myc levels, but exhibited only a slight effect on MMP-2 expression. These results suggest that c-Myc is inhibited by MEG3 overexpression and may act as a MEG3 downstream effector for MEG3’s inhibition of bladder cancer invasion. To test this notion, c-Myc was introduced into T24T(MEG3) cells to see if ectopic expression of c-Myc could reverse the MEG3 inhibitory effect on bladder cancer invasion. As expected, ectopic c-Myc expression completely recovered the invasive ability of T24T(MEG3) cells (Figures 2C–2E and S2; Table S3), demonstrating that reduced c-Myc expression in T24T(MEG3) cells indeed contributes to the attenuation of the invasion of T24T cells.
Figure 2.
c-Myc Is a MEG3 Downstream Effector of Attenuation of Bladder Cancer Invasion
(A and B) As indicated, the c-Myc and MMP-2 expression levels were evaluated by western blot in the stable cell lines T24T (A) and UMUC3 (B). GAPDH was used as the internal loading control. (C) c-Myc was stably transfected into T24T(MEG3) cells, and its expression was identified by western blot. GAPDH was used as the protein loading control. (D and E) The stable transfectants, as indicated, were used to determine their invasive ability (D) and the quantitative analysis (E). The results are presented as relative cell invasion. The asterisk indicates a significant change compared with the vector transfectants (p < 0.05). The bars represent the mean ± SD of three independent experiments.
MEG3 Inhibits c-Myc Transcription by Specifically Attenuating the Activity of c-Jun Phosphorylation at Ser63/Ser73
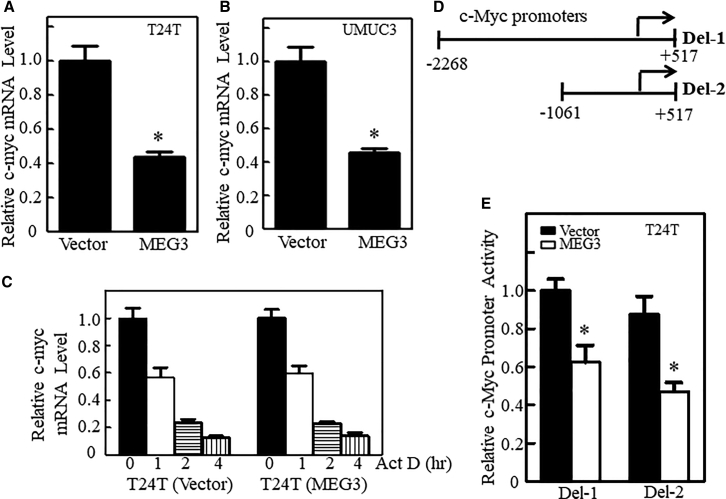
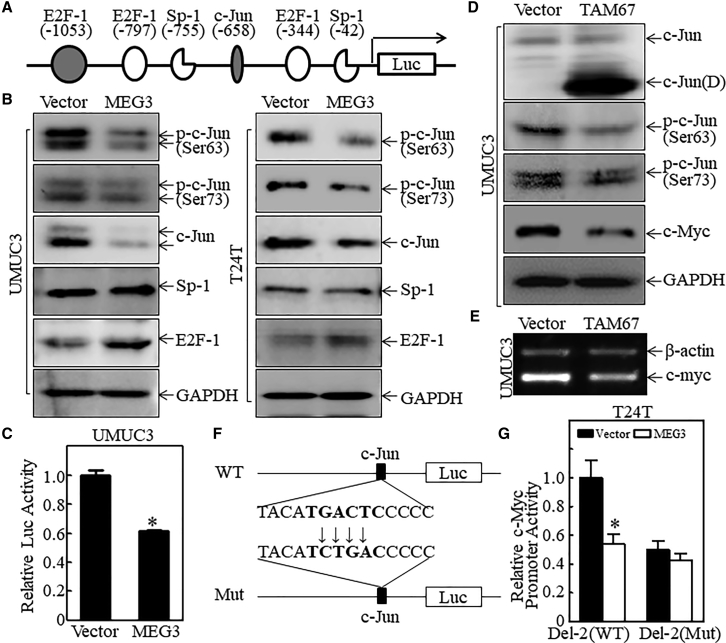
To elucidate the molecular mechanisms underlying the MEG3 regulation of c-Myc expression, we first analyzed the mRNA level of c-Myc in both T24T and UMUC3 stable transfectants expressing MEG3 in comparison to their vector control transfectants. As shown in Figures 3A and 3B, the c-Myc mRNA level was remarkably attenuated in both bladder cancer cell types with MEG3 overexpression (Table S4). To determine whether MEG3 regulates c-Myc mRNA via its RNA stability, the transcription inhibitor actinomycin D (Act D) was applied to T24T(Vector) and T24T(MEG3) cells for different time periods to observe c-Myc mRNA degradation rates. The results show that there was no observable difference in c-myc mRNA degradation between T24T(MEG3) and T24T(Vector) cells (Figure 3C), indicating that MEG3 does not affect c-Myc mRNA stability. We next examined the potential effect of MEG3 on c-Myc promoter transcription. Two different lengths of c-Myc promoter-driven luciferase reporters, Del-1(−2,268/+517 bp) and Del-2(−1,061/+517 bp), were transfected into T24T(MEG3) and T24T(Vector) cells (Figure 3D). The results show that the activities of both promoters were significantly inhibited in MEG3-overexpressing cells at a similar level (Figure 3E; Table S5), suggesting that the shared region of both c-Myc promoter reporters is regulated by the MEG3-initiated signaling axis. Thus, we performed a bioinformatics scan on the promoter region of the Del-2 prompter and several potential binding sites of transcription factors, including E2F1, c-Jun, and Sp-1, as shown in Figure 4A. To define the specific transcription factors involved in the regulation of c-Myc transcription by MEG3, we evaluated the expression of these transcription factors in T24T(MEG3) versus T24T(Vector) cells and in UMUC3(MEG3) versus UMUC3(Vector) cells. As shown in Figure 4B, among the transcription factors tested, the levels of c-Jun phosphorylation at Ser63 and Ser73 and its expression was consistently inhibited by ectopic expression of MEG3, while Sp-1 did not show an observable effect and E2F1 was increased by MEG3 overexpression. Moreover, MEG3 overexpression significantly inhibited AP-1-dependent transcriptional activity (Figure 4C). The above results suggest that c-Jun phosphorylation at Ser63/Ser73 may be downstream of MEG3 in regulating c-Myc transcription. Therefore, TAM67, a deletion mutant of c-jun that lacks amino acids 3–122 of c-jun, was transfected into UMUC3 cells to determine the potential contribution of c-Jun to c-Myc expression. As expected, ectopic expression of TAM67 successfully impaired c-Myc mRNA abundance and protein expression (Figures 4D, 4E, S3, and S4), revealing that c-Jun has an important role in the activation of MEG3 suppression of c-Myc mRNA transcription. This notion was supported by the results obtained from a point mutation of the c-Jun binding site on the c-Myc promoter-driven luciferase reporter (Del-2), as shown in Figure 4F. Ectopic expression of MEG3 dramatically inhibited the transcriptional activity of the wild-type Del-2 promoter (Del-2/WT), whereas it did not affect the transcriptional activity of the mutant promoter (Del-2/Mut) (Figure 4G). Our results demonstrate that c-Jun phosphorylation and its binding site in the c-Myc promoter region are crucial for MEG3 suppression of c-Myc transcription and protein expression.
Figure 3.
MEG3 Attenuated c-Myc Transcription by Inhibiting Its Promoter Activity
(A and B) c-Myc mRNA expression levels were analyzed by qRT-PCR in T24T(Vector) versus T24T(MEG3) cells (A) and in UMUC3(Vector) versus UMUC3(MEG3) cells (B). GAPDH mRNA was used as the internal control. Results are presented as relative c-myc mRNA levels. The asterisk indicates a significant change compared with the vector cells (p < 0.05). The bars indicate the mean ± SD of three triplicates. (C) T24T(Vector) and T24T(MEG3) were treated with actinomycin D (Act D, 15 μg/mL) for the indicated time periods, and the treated cells were extracted for total RNA. c-myc mRNA levels were determined by qRT-PCR. β-Actin mRNA was used as the internal control. (D) A diagram of two c-Myc promoter-driven luciferase reporters, Del1 and Del2. (E) c-Myc promoter-driven luciferase reporters Del1 and Del2 were used to evaluate c-Myc promoter transcription activity in the bladder cancer cells indicated. Results are presented as relative c-Myc promoter activity. The asterisk indicates a significant change compared with the vector cells (p < 0.05). The bars represent the mean ± SD of triplicates.
Figure 4.
c-Jun Was Essential for MEG3 Inhibition of c-Myc Transcription in Human Bladder Cancer Cells
(A) Diagram of the potential transcription factor binding sites in human c-Myc promoter-driven luciferase reporter. (B) The effect of MEG3 overexpression on the expression of potential transcription factors listed in (A), with GAPDH used as the internal loading control. (C) The effect of MEG3 on AP-1 activity in UMUC3 cells. The AP-1 luciferase reporter was co-transfected into UMUC3(Vector) and UMUC3(MEG3) cells with TK at a ratio of 10:1. After 24 h, the luciferase activity was determined and presented as luciferase activity relative to vector control. (D and E) Ectopic expression of TAM67 on c-Myc expression at the protein (D) and the mRNA (E) levels in UMUC3 cells. GAPDH was used as the internal loading control. (F) Diagram indicating the c-Myc promoter-driven luciferase reporter Del2 and its c-Jun binding site point mutation. (G) WT and mutant (Mut) c-Myc promoter-driven luciferase reporters were stably co-transfected with MEG3 or its scrambled vector into T24T cells, and the stable transfectants were used to evaluate the essential role of the c-Myc binding site in MEG3’s inhibition of c-Myc promoter transcription. Results are presented as relative c-Myc promoter activity. The asterisk indicates a significant inhibition compared with the vector transfectant (p < 0.05). The bars represent the mean ± SD of triplicates.
PHLPP2, but Not MAPK, Is a MEG3 Downstream Mediator for the Regulation of c-Jun Protein Phosphorylation at Ser63/Ser73
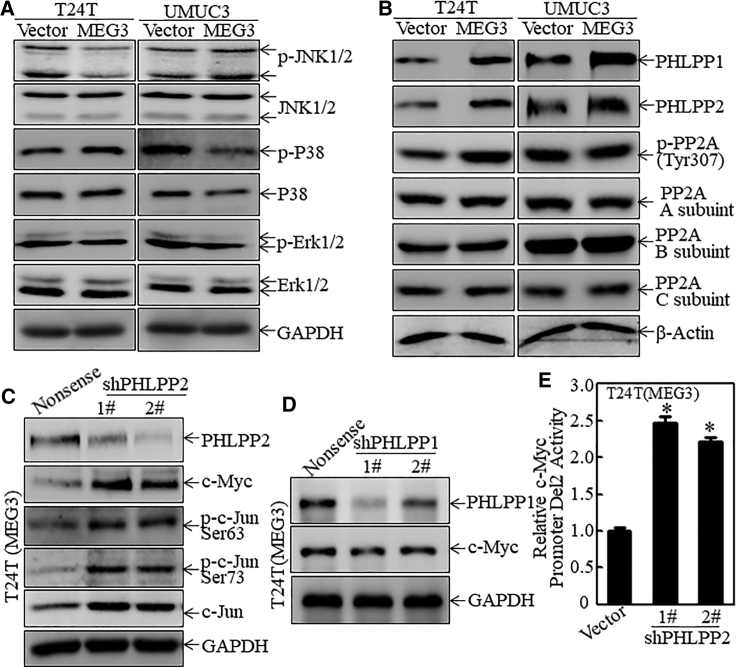
Phosphorylation of c-Jun at Ser63/Ser73 is usually regulated by mitogen-activated protein kinases (MAPKs), including JNK1/2, p38, and ERK1/2.20 To determine which MAPK is involved in MEG3-initiated c-Jun inactivation, we analyzed the levels and activation of these MAPKs in T24T(MEG3) versus T24T(Vector) cells and in UMUC3(MEG3) versus UMUC3(Vector) cells. As shown in Figure 5A, ectopic expression of MEG3 did not show a consistent effect on any of the MAPKs in both cell lines (Figures S5A and S5B), suggesting that MAPKs may not be targeted by MEG3 for inhibition of c-Jun phosphorylation at Ser63/Ser73. Our recent study demonstrated that PHLPP and PP2A can also attenuate c-Jun phosphorylation at Ser63/Ser73.21 To this end, the expressions of PHLPP1, PHLPP2, and PP2A phosphorylation were determined and compared in T24T(MEG3) versus T24T(Vector) cells and in UMUC3(MEG3) versus UMUC3(Vector) cells. As shown in Figures 5B, S5C, and S5D, PHLPP1 and PHLPP2 were consistently upregulated in both MEG3-overexpressing cell types, whereas PP2A did not show any relevant change, suggesting that upregulated PHLPP1 and PHLPP2 may participate in MEG3-induced reduction of c-Jun phosphorylation. To explore this possibility, short hairpin RNAs (shRNAs) specifically targeting human PHLPP1 and PHLPP2 were used to knock down endogenous PHLPP1 and PHLPP2 in T24T(MEG3) cells. The stable transfectants T24T(MEG3/shPHLPP1) and T24T(MEG3/shPHLPP2) and the scrambled nonsense transfectant T24T(MEG3/Nonsense) were established and identified, as shown in Figures 5C and 5D. Knockdown of PHLPP2 did elevate c-Jun phosphorylation and c-Myc expression, whereas knockdown of PHLPP1 slightly inhibited c-Myc protein abundance (Figures 5C, 5D, S5E, and S5F). These results indicate that PHLPP2 is a MEG3 downstream mediator for de-phosphorylation of c-Jun and attenuation of c-Myc transcription in MEG3-overexpresseing cells. This conclusion is also supported by the results from the c-Myc promoter-driven luciferase reporter assay, which showed a significant increase in c-Myc promoter transcription activity with the PHLPP2 knockdown transfectant (Figure 5E). These results demonstrate that PHLPP2, but not MAPK and PHLPP1, is a MEG3 downstream mediator for attenuation of c-Jun protein phosphorylation at Ser63/Ser73 and c-Myc transcription.
Figure 5.
PHLPP2 Is a MEG3 Downstream Mediator for MEG3’s Inhibition of c-Jun Activation
(A and B) Whole-cell extracts were subjected to western blot analysis to determine protein expression as indicated. GAPDH was used as the protein loading control. (C and D) shRNAs specifically targeting PHLPP1 and PHLPP2 were stably transfected into T24T(MEG3) cells to knock down PHLPP1 and PHLPP2 expression, respectively. The cell extracts from the stable transfectants were subjected to western blot analysis, as indicated. GAPDH was used as the protein loading control. (E) The c-Myc promoter-driven luciferase reporter (Del2) was stably co-transfected with PHLPP2 knockdown transfectants and the stable transfectants were used to evaluate the role of PHLPP2 on c-Myc promoter transcriptional activity. The results are presented as relative c-Myc promoter luciferase activity. The asterisk indicates a significant difference compared with the vector transfectant (p < 0.05). The bars represent the mean ± SD of triplicates.
miR-27a Plays a Crucial Role in MEG3-Mediated PHLPP2 Translational Activation
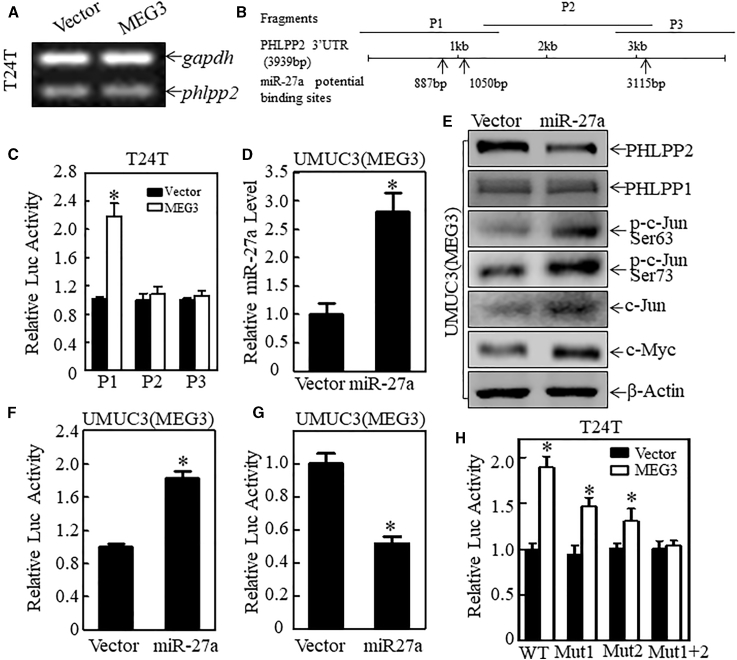
To understand how MEG3 regulates PHLPP2, we first analyzed the mRNA level of PHLPP2 in T24T stable transfectants expressing MEG3 and its vector control. As shown in Figures 6A and S6, there was no significant change in PHLPP2 mRNA between the two transfectants, suggesting that MEG3-mediated regulation of PHLPP2 expression is beyond the mRNA level. Therefore, we next examined the potential contribution of the 3′ UTR in the regulation of PHLPP2 protein expression. As shown in Figure 6B, the PHLPP2 mRNA 3′ UTR luciferase reporter was constructed in three separate parts (P1, P2, and P3). All reporters were then transfected into T24T(Vector) and T24T(MEG3) cells separately. The results in Figure 6C show that MEG3 induced a significant increase in the activity of P1, but had no effect on the activity of P2 and P3, suggesting that MEG3 regulates PHLPP2 translation through the first 1,500 bp of the 3′ UTR of PHLPP2 mRNA. Biological analysis shows that miR-27a had three potential binding sites on the PHLPP2 mRNA 3′ UTR (Figure 6B). To evaluate whether miR-27a could inhibit PHLPP2 expression and in turn increase the c-Jun phosphorylation and c-Myc expression, we overexpressed miR-27a in UMUC3(MEG3) cells. The results show that miR-27a was stably expressed in the UMUC3(MEG3) cells (Figure 6D). The whole-cell lysis was then used to analyze the expression of PHLPP2, PHLPP1, c-Jun, and c-Myc. As shown in Figures 6E and S7, overexpression of miR-27a decreased PHLPP2 protein expression, but had no effect on PHLPP1. As expected, c-Jun phosphorylation at Ser63/Ser73 and c-Myc protein was also upregulated, suggesting that miR-27a downregulates PHLPP2 expression and subsequently increases c-Jun phosphorylation, which in turn upregulates c-Myc expression. Consistently, the results from the reporter assays show that miR-27a overexpression led to an increase in c-Myc promoter-driven luciferase activity (Figure 6F) and a decrease in PHLPP2 3′ UTR luciferase activity (Figure 6G), suggesting that miR-27a downregulates PHLPP2 through the targeting of its mRNA 3′ UTR. To test whether miR-27a could bind directly to the PHLPP2 mRNA 3′ UTR, the potential miR-27a binding site point mutations were constructed based on the PHLPP2 3′ UTR luciferase reporter P1. As shown in Figures 6H and S8 and Table S6 the increase in 3′ UTR activity in the mutated reporters (Mut1 and Mut2) was significantly attenuated, compared with the WT P1 reporter, whereas the UTR activity showed no significant change when both binding sites were mutated at the same time (Mut1+2), which demonstrates that miR-27a binds directly to both sites of PHLPP2 mRNA 3′ UTR and inhibits its 3′ UTR activity, in turn inhibiting PHLPP2 protein translation. Taken together, our results demonstrate that miR-27a can bind directly to the PHLPP2 mRNA 3′ UTR and inhibit its protein translation, thus decreasing PHLPP2 phosphorylation, then upregulating expression of c-Jun and subsequently increasing c-Myc promoter transcriptional activity and then, protein expression.
Figure 6.
PHLPP2 Was Upregulated at the Translational Level through Upregulated 3′ UTR Activity
(A) RT-PCR was used to analyze the mRNA level of PHLPP2 in T24T(Vector) and T24T(MEG3) cells. GAPDH was used as the mRNA loading control. (B) Diagram of various lengths of PHLPP2 3′ UTR luciferase reporters, including P1, P2, and P3. Arrows show the potential miR-27a binding sites. (C) The PHLPP2 mRNA 3′ UTR luciferase reporters, P1, P2, and P3 were co-transfected into T24T(Vector) and T24T(MEG3) cells with TK at a ratio of 10:1. After 24 h, the luciferase activity was determined. TK was used as the internal control. The asterisk indicates a significant difference compared with the vector transfectant (p < 0.05). The bars represent the mean ± SD of triplicates. (D) miR-27a expression was analyzed with qRT-PCR. GAPDH mRNA was used as the loading control. Results are presented as relative miR-27a levels. The asterisk indicates a significant difference compared with the vector transfectant (p < 0.05). The bars represent the mean ± SD of triplicates. (E) The cell extracts from the transfectants indicated were subjected to western blot analysis. GAPDH was used as the internal control. (F) The c-Myc promoter-driven luciferase reporter, together with the internal control TK reporter was transiently transfected into UMUC3(MEG3/Vector) and UMUC3(MEG3/miR-27a) cells. Twenty-four hours after transfection, the cells were extracted for luciferase assay. The asterisk indicates a significant increase compared with the vector transfectant (p < 0.05). The bars represent the mean ± SD of triplicates. (G) The PHLPP2 3′ UTR luciferase reporter P1 was transiently transfected into UMUC3(MEG3/Vector) and UMUC3(MEG3/miR-27a) cells, together with its internal control construct TK. (H) PHLPP2 3′-UTR luciferase reporter P1 and its point mutants (as indicated) were transiently transfected into T24T(Vector) and T24T(MEG3) cells, together with its internal control construct TK. Twenty-four hours after transfection, the cells were extracted for a luciferase assay. The asterisk indicates a significant inhibition compared with the vector transfectant (p < 0.05). The bars represent the mean ± SD of triplicates.
MEG3 Functions as a ceRNA of PHLPP2 through Competing for miR-27a in Bladder Cancer
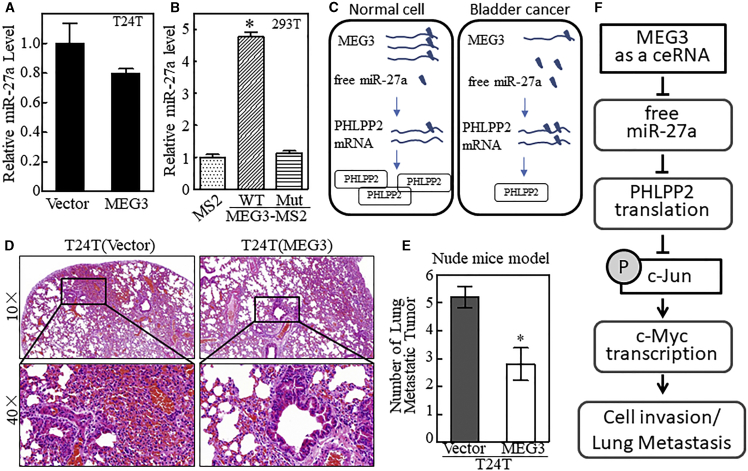
Given the above results showing that miR-27a mediates MEG3 upregulation of PHLPP2 protein translation and downregulation of c-Myc mRNA transcription, we anticipated that either miR-27a expression or miR-27a activity is regulated by MEG3 in human bladder cancer cells. To test this notion, we first analyzed the miR-27a expression in T24T(Vector) and T24T(MEG3) cells. As shown in Figure 7A and Table S7, miR-27a was slightly decreased in MEG3-overexpressing cells. Compared to the dramatic upregulation of PHLPP2 by MEG3, this slightly decreased miR-27a expression indicates a potential reduction of its activity, which may be involved in MEG3’s regulation of PHLPP2. We next determined the potential direct interaction between MEG3 and miR-27a at endogenous levels in the intact cells. To this end, we performed an RNA immunoprecipitation (RIP) analysis to pull down endogenous miRNAs associated with MEG3. As shown in Figure 7B and Table S8, WT MEG3 (WT-MEG3-MS2) can significantly enrich miR-27a, compared with empty vector (MS2) and MEG3, with mutation at the miR-27a binding site (Mut-MEG3-MS2), revealing that MEG3 is a good sponge for miR-27a. This result shows that MEG3 can function as a ceRNA and competitively binds to miR-27a, therefore reducing the free form of miR-27a and in turn releasing miR-27a’s inhibition of PHLPP2 translation, as diagrammed in Figure 7C, consequently attenuating c-Jun phosphorylation or activation as well as c-Myc transcription and bladder cancer invasion. To evaluate the effect of MEG3 on human bladder cancer metastasis in vivo, T24T(Vector) and T24T(MEG3) transfectants were injected into nude mice via an intravenous (i.v.) lateral tail vein injection. The nude mice were evaluated for the lung metastatic ability of both transfectants. As shown in Figures 7D and 7E and Table S9, ectopic overexpression of MEG3 significantly inhibited lung surface metastatic tumors in comparison with the mice injected with T24T(Vector) cells. Taken together, the results show that MEG3 can function as a ceRNA in bladder cancer and upregulate PHLPP2 translation by competitive binding with miR-27a, as diagrammed in Figure 7F.
Figure 7.
MEG3 Functions as a ceRNA with miR-27a to Regulate PHLPP2 Expression
(A) The miR-27a expression level was analyzed with qRT-PCR. U6 was used as the loading control. The results are presented as relative miR-27a levels. The bars represent the mean ± SD of triplicates. (B) The miR-27a expression level was analyzed with qRT-PCR. Input miR-27a was used as the loading control. The results are presented as relative miR-27a levels. The asterisk indicates a significant difference compared with the vector transfectant (p < 0.05). The bars represent the mean ± SD of triplicates. (C) A schematic diagram of the mechanisms underlying MEG3 functions as a ceRNA that regulates PHLPP2 expression in both normal cells and bladder cancer cells. (D and E) T24T(Vector) or T24T(MEG3) transfectants were subjected to lung metastatic assay in nude mice. Representative images of the histologic lung metastases were analyzed using H&E staining (D) and the number of lung metastatic tumors were graphed (E). The asterisk indicates a significant reduction of lung metastatic tumor by ectopic MEG3 expression, compared with the vector transfectant (p < 0.05). The bars represent the mean ± SD. (F) A schematic diagram of the mechanisms underlying MEG3’s function as a ceRNA that inhibits invasion and lung metastasis of bladder cancers.
Discussion
Muscle invasion and metastasis dramatically decreases the 5-year survival rate and results in a 100% death rate for bladder cancer patients.1 Thus, one of the needed therapeutic kernels for bladder cancer is to inhibit and block the invasiveness of bladder cancer cells. In the current study, we demonstrate that MEG3 acts as a ceRNA and markedly inhibits human bladder cancer cell invasion in vitro and lung metastasis in vivo. We found that MEG3 competes with miR-27a for PHLPP2, which leads to decreased activity of miR-27a, due to the reduced amount of free-form miR-27a and subsequent attenuation of the inhibitory effect of miR-27a on PHLPP2 protein translation. Consistent with our previous findings that PHLPP2 can directly bind to c-Jun protein and inhibit c-Jun activation,21 upregulated PHLPP2 inhibits c-Jun phosphorylation at Ser63/Ser73, in turn downregulating the c-Myc transcription and its mediation of bladder cancer cell invasion. This is the first demonstration that MEG3 acts as a ceRNA to inhibit the PHLPP2-c-Jun-c-Myc axis, consequently reducing invasion and lung metastasis of human bladder cancer cells.
It has been known that ceRNAs act as molecular sponges for microRNAs through their binding sites.16 MEG3 has been reported to be a ceRNA involved in regulation of several biological functions of gastric cancer cells.22 It has also been reported that MEG3 is dramatically decreased in liver metastatic colorectal tumors, compared with the level in primary colorectal tumors,11 suggesting a potential suppression of MEG3 in colorectal cancer metastasis. Our previous study showed that MEG3 downregulation could facilitate the transformation of normal bronchial epithelial cells by exposure to nickel.23 We demonstrated that downregulated MEG3 results in less interaction with c-Jun and therefore a failure to inhibit c-Jun activation, in turn increasing PHLPP1 transcription and finally leading to the transformation of normal bronchial epithelial cells. Our current study of human bladder cancer cells revealed that WT MEG3 can enrich miR-27a, whereas the miR-27a binding-site-mutated MEG3 loses its function to enrich the miR-27a, suggesting that MEG3 is an effective sponge of miR-27a. By such competitive binding with miR-27a, MEG3 reduces the free form of miR-27a, thereby decreasing miR-27a’s inhibition of PHLPP2 protein translation, in turn causing c-Jun phosphorylation or activation, c-Myc transcription, and regulation of bladder cancer invasion and lung metastasis. We noted that, although similar protein molecules were involved in the cell response to MEG3, biological consequences were quite different between human normal bronchial epithelial cells and human invasive bladder cancer cells. This difference could be due to either normal cells and cancer cells or the diversity of the two different cell types.
miR-27a has been reported to have oncogenic functions in various cancer cells. For example, miR-27a is reported to promote proliferation and progression in gastric and renal cancers.22, 24 miR-27a is also considered to be positively related to chemo-sensitivity in bladder cancer.25, 26 Although Xu et al.27 report that hypoxia elevates the expression of miR-27a and subsequently downregulates the angiogenic factor AGGF1 in bladder cancers, the biological functions of miR-27a in bladder cancers have not yet been explored. In the current study, miR-27a overexpression specifically inhibited PHLPP2 protein expression with marked induction of PHLPP2 mRNA 3′ UTR activity, while deletion of the miR-27a binding site in PHLPP2 mRNA 3′ UTR attenuated PHLPP2 mRNA 3′ UTR activity. These results suggest that miR-27a directly binds to the miR-27a binding sites of the PHLPP2 mRNA 3′ UTR, to inhibit PHLPP2 protein translation, further revealing a novel function of miR-27a in crosstalk with another tumor suppressor, PHLPP2, for suppression of bladder cancer invasion.
c-Myc is a very strong proto-oncogene and is frequently found to be overexpressed in many types of cancer. c-Myc activation has numerous biological effects, such as promotion of proliferation,28 enhancement of cancer cell invasion,19, 29 and induction of apoptosis.30 Our study indicates that c-Myc is an effector of MEG3 downstream and plays a crucial role in the regulation of bladder cancer invasion. We have discovered that MEG3 is downregulated in human highly invasive bladder cancers in comparison to non-invasive bladder cancers, whereas MEG3 overexpression attenuates c-Myc transcription through the inhibition of c-Jun phosphorylation in bladder cancer cells. The results of our investigation of the c-Myc promoter and its c-Jun binding site point mutation confirm that the c-Jun binding site is crucial for MEG3-mediated inhibition of c-Myc transcription.
In summary, the results obtained from the current study in both in vitro and in vivo experiments revealed the mechanisms of MEG3 downregulation in human bladder cancers and the role of such MEG3 downregulation in the promotion of bladder cancer’s invasive ability through the regulation of c-Myc transcription in a c-Jun-dependent manner. We also elucidated that MEG3 acts as a ceRNA, regulating the PHLPP2 translation via competing with miR-27a for binding to the PHLPP2 mRNA 3′ UTR. These findings not only provide novel insight into the mechanisms behind the MEG3 inhibition of bladder cancer cell invasion and metastasis, but also reveal a potential target of MEG3 for the management of invasive bladder cancer and for preventing non-muscle-invasive bladder cancer progression.
Materials and Methods
Cell Culture and Reagents
UMUC3 cells were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS and T24T cells were cultured in RPMI 1640/F12 medium (Invitrogen, Carlsbad, CA, USA) with 5% FBS. All cells were maintained in a humidified incubator at 37°C with a 5% CO2 atmosphere. Antibodies specific against c-Jun, c-Jun(D), Erk1/2, p-Erk1/2, P38, p-P38, and PP2A were purchased from Cell Signaling Technology (Beverly, MA, USA). Antibodies for c-Myc, MMP-2, Sp1, E2F1, and β-Actin were bought from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibodies specific for GAPDH and JNK1/2 were purchased from GeneTex, (Irvine, CA, USA), and antibodies for PHLPP1 and PHLPP2 were from Bethyl Laboratories (Montgomery, TX, USA). Act D was bought from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Plasmids and Cell Stable Transfection
The shRNAs specifically targeting human PHLPP1 and PHLPP2 were purchased from Open Biosystems (Lafayette, CO, USA). The c-Myc promoter luciferase reporter, HA-MS2-GFP overexpression vector and pSL-MS2-12X plasmids were obtained from Addgene (Cambridge, MA, USA). The TAM67 plasmid, a well-characterized dominant negative c-Jun mutant, was described in our previous study.31 WT PHLPP2 3′ UTR luciferase reporter plasmids were a kind gift from Professor Kai Fu (University of Nebraska Medical Center, Omaha, NE, USA).32 MEG3-overexpressing plasmid was bought form Genechem (Shanghai, China) and used in a prior study.33 The structure map is shown in Figure S9. The miR-27a binding site point mutations (Mut1 and Mut2) of PHLPP2 3′ UTR luciferase reporter were constructed by us, using the WT PHLPP2 3′ UTR luciferase reporter. The MEG3-MS2-overexpressing vector was constructed by us, based on the pSL-MS2-12X and MEG3 expression vectors. The miR-27a binding site point mutation of the MEG3-MS2 expressing vector was constructed by us. Cell transfections were performed with PolyJet DNA In Vitro Transfection Reagent (SignaGen Laboratories, Rockville, MD, USA) according to the manufacturer’s instructions. For stable transfection, transfected cells were subjected to G418 or puromycin selection based on the antibiotic resistance of the plasmids, and the surviving cells were pooled as stable mass transfectants.
RIP Assay
HEK293T cells were cultured in 10-cm dishes. When confluence reached 70–80%, the cells were transiently transfected with the HA-MS2-GFP plasmid or with MEG3-MS2 or its vector control construct. Forty-eight hours after transfection, the cells were extracted by using a polysome lysis buffer (10 mM HEPES [pH 7], 100 mM KCl, 5 mM MgCl2, 25 mM EDTA, 0.5% IGEPAL, 2 mM DTT, 50 U/mL RNaseOut and 50 U/mL SUPERase In, 0.2 mg/mL heparin, and complete proteinase inhibitor). The cell lysates were centrifuged at 14,000 × g for 10 min at 4°C. The same amount of cell extract from the HA-MS2-GFP transfectant was mixed with the cell extract from MEG3-MS2 or its vector transfectants. These mixtures were rotated in a cold room overnight. The anti-HA agarose A/G beads (purchased from Vector Laboratories, Burlingame, CA, USA) were added into the mixture and rotated for another 4 h at 4°C. The beads were washed three times with NET2 buffer (50 mM Tris-HCl [pH 7.4], 150 mM sodium chloride, 1 mM magnesium chloride, 0.05% IGEPAL, 50 U/mL RNaseOut, 50 U/mL SUPERase In, 1 mM dithiothreitol, and 30 mM EDTA), and resuspended in 100 μL NET2 and 100 μL SDS-TE (20 mM Tris-HCl [pH 7.5], 2 mM EDTA, and 2% sodium dodecyl sulfate [SDS]), then incubated at 55°C for 30 min with occasional mixing. The RNAs in the bead mixture were extracted using an miRNeasy Mini Kit (QIAGEN, Valencia, CA, USA), and real-time qPCR was performed to identify the miRNA presented in the immune complex.
Human Bladder Cancer Tissue Specimens
Twenty-seven primary human bladder cancer specimens and their paired adjacent non-tumorous bladder tissues were obtained from patients who underwent radical cystectomy at the Department of Urology of the Union Hospital of Tongji Medical College, as listed in Table S1. All specimens were obtained with approval by the Ethics Committee of Huazhong University of Science and Technology and with appropriate informed consent from the patients and were immediately snap frozen in liquid nitrogen after surgical removal. Histological and pathological diagnoses were confirmed by a pathologist based on the 2004 World Health Organization Consensus Classification and Staging System for bladder neoplasms.
Mouse Lung Metastasis Model
All animal studies were performed in the animal institute of the Tongji Medical College according to the protocols approved by the Laboratory Animal Ethics Committee of the Tongji Medical College & Laboratory Animal Centre of Huazhong University, Wuhan, China. Female BALB/c athymic nude mice (4–5 weeks old) were purchased from the Shanghai Silaike Experimental Animal Company (license no. SCXK, Shanghai, China). One week following purchase, 10 female nude mice were randomly divided into 2 groups (5 per group). The stable T24T(Vector) or T24T(MEG3) transfectants were injected into the nude mice via an i.v. lateral tail vein injection (1 × 107 in 100 μL PBS/mouse) to produce lung metastatic tumors. The mice were evaluated and weighed twice a week. None of the mice expired prior to the end of the experiment. Six weeks after injection, the mice were euthanized, and the lungs were removed by dissection. The lungs from the indicated groups were fixed in Bouin’s fixative solution (MilliporeSigma, Burlington, MA, USA) for 24 h and the number of superficial lung metastatic lesions were counted on each lobe of every specimen. The lungs in each group were fixed and embedded in paraffin for H&E staining and histopathological evaluation. The images were captured and analyzed, using a Pannoramic Digital Slide Scanner and Viewer (3DHISTECH, Budapest, Hungary).
Transwell Invasion Assay
The control (uncoated) and Matrigel inserts of BD Biocoat (BD Biosciences, Bedford, MA, USA) were used for a cell invasion assay. The invasion assay was performed, using a BD Falcon kit according to the manufacturer’s instructions (BD Biosciences, Bedford, MA, USA). Thirty thousand bladder cancer cells were seeded in the upper well with a 0.1% serum-containing medium and in the lower part with complete medium. After incubation in a humidified incubator with at 37°C and 5% CO2 atmosphere for 24 h, the cells on the inside and outside of the chamber were fixed with 3.7% formalin for 2 min, then washed twice with PBS, transferred to 100% methanol for 20 min, and washed twice with PBS. The cells were stained with Giemsa (1:20 diluted with PBS) at room temperature for 15 min in the dark, then washed again, twice. The non-invading cells were scraped off with a cotton swab (wet with PBS) four times. The number of the cells was quantified by the software Image J (NIH, Bethesda, MD, USA) and the images captured by a DP71 inverted microscope (Olympus, Center Valley, PA, USA), as described in our previous study.34
Western Blot Analysis
Whole-cell extracts were prepared with cell lysis buffer (10 mM Tris-HCl [pH 7.4], 1% SDS, and 1 mM Na3VO4), as described in our previous study.35 Fifty micrograms proteins resolved by SDS-PAGE were transferred to a PVDF membrane and probed with the indicated primary antibodies, together with the AP-conjugated secondary antibody. Protein signals were detected by the enhanced-chemiluminescence western blot system, as described in a previous report,36 and the images were acquired by scanning with a phosphorimager (Typhoon FLA 7000 imager, GE Healthcare, Pittsburgh, PA, USA).
Luciferase Reporter Assay
PHLPP2 3′ UTR luciferase reporters or c-Myc promoter-driven luciferase reporters were co-transfected into cultured bladder cancer cells with TK separately at a ratio of 10:1. Luciferase activity was determined by using the Luciferase Assay System kit (Promega, Madison, WI, USA). The results were normalized by internal TK signal as described in our previous study.23 All experiments were carried out in triplicate and the results expressed as the mean ± SE.
RT-PCR
Total RNA was extracted using TRIzol reagent, as described in the manufacturer’s instructions (Invitrogen, Grand Island, NY, USA). Five micrograms total RNA was used for first-strand cDNA synthesis with oligo(dT) primer, using the SuperScript IV First-Strand Synthesis system (Invitrogen, Grand Island, NY, USA). Specific primers were used for PCR amplification. The primers used in this study included: human c-Myc, forward: 5′-AAC ACA CAA CGT CTT GGA GC-3′, reverse: 5′-CCT TAC GCA CAA GAG TTC CG-3′, human phlpp2, forward: 5′-AGG TTC CTG AGC ATC TCT TC-3′, reverse: 5′-GTT CAG GCC CTT CAG TTG AG-3′, and human gapdh, forward: and 5′-AGA AGG CTG GGG CTC ATT TG-3′, reverse: 5′-AGG GGC CAT CCA CAG TCT TC-3′.
Real-Time qPCR
Cells were used for total RNA extraction with the miRNeasy Mini Kit (QIAGEN, Valencia, CA, USA). One microgram total RNA was used for RT. miR-27a was determined by the QuantStudio Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) with an miScript PCR kit (QIAGEN, Valencia, CA, USA). The primer for the microRNA assay was purchased from Invitrogen (Grand Island, NY, USA), and U6 was used for the internal control. The initial activation was performed at 95°C for 15 min, followed by 40 cycles (denaturation at 95°C for 15 s, annealing at 55°C for 30 s, and extension at 70°C for 30 s).
Statistical Analysis
The Student’s t test and Mann-Whitney U test were used to determine the significance of differences between the compared groups. Differences of p < 0.05 were considered significant.
Author Contributions
Chuanshu Huang and X.R.W. designed the study. Chao Huang and X.L. detected the cells’ biological function, conducted the real-time qPCR assays, carried out the western blot and luciferase reporter assays, and performed the statistical analysis. F.X. and F.Z. carried out the animal experiments. Chao Huang and Chuanshu Huang drafted the manuscript. X.L., H.J., J.L., C.Z., and G.J. helped to acquire the experimental data. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank Dr. Kai Fu from University of Nebraska Medical Center, Omaha, Nebraska, for the kind gift of PHLPP2 3′ UTR luciferase reporter. This work was partially supported by the following grants: NIH/National Cancer Institute CA165980, CA177665, CA229234, and CA217923; and NIH/NIEHS ES000260.
Footnotes
Supplemental information includes nine figures and nine tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2019.01.014.
Supplemental Information
References
- 1.Kamat A.M., Hahn N.M., Efstathiou J.A., Lerner S.P., Malmström P.U., Choi W., Guo C.C., Lotan Y., Kassouf W. Bladder cancer. Lancet. 2016;388:2796–2810. doi: 10.1016/S0140-6736(16)30512-8. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 3.Sullenger B.A., Nair S. From the RNA world to the clinic. Science. 2016;352:1417–1420. doi: 10.1126/science.aad8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitt A.M., Chang H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmitz S.U., Grote P., Herrmann B.G. Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 2016;73:2491–2509. doi: 10.1007/s00018-016-2174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Y., Zhang X., Klibanski A. MEG3 noncoding RNA: a tumor suppressor. J. Mol. Endocrinol. 2012;48:R45–R53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu J., Liu S., Ye F., Shen Y., Tie Y., Zhu J., Wei L., Jin Y., Fu H., Wu Y., Zheng X. Long Noncoding RNA MEG3 Interacts with p53 Protein and Regulates Partial p53 Target Genes in Hepatoma Cells. PLoS ONE. 2015;10:e0139790. doi: 10.1371/journal.pone.0139790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang C., Yan G., Zhang Y., Jia X., Bu P. Long non-coding RNA MEG3 suppresses migration and invasion of thyroid carcinoma by targeting of Rac1. Neoplasma. 2015;62:541–549. doi: 10.4149/neo_2015_065. [DOI] [PubMed] [Google Scholar]
- 9.Zhuang W., Ge X., Yang S., Huang M., Zhuang W., Chen P., Zhang X., Fu J., Qu J., Li B. Upregulation of lncRNA MEG3 Promotes Osteogenic Differentiation of Mesenchymal Stem Cells From Multiple Myeloma Patients By Targeting BMP4 Transcription. Stem Cells. 2015;33:1985–1997. doi: 10.1002/stem.1989. [DOI] [PubMed] [Google Scholar]
- 10.Cao X., Zhuang S., Hu Y., Xi L., Deng L., Sheng H., Shen W. Associations between polymorphisms of long non-coding RNA MEG3 and risk of colorectal cancer in Chinese. Oncotarget. 2016;7:19054–19059. doi: 10.18632/oncotarget.7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kong H., Wu Y., Zhu M., Zhai C., Qian J., Gao X., Wang S., Hou Y., Lu S., Zhu H. Long non-coding RNAs: novel prognostic biomarkers for liver metastases in patients with early stage colorectal cancer. Oncotarget. 2016;7:50428–50436. doi: 10.18632/oncotarget.10416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ying L., Huang Y., Chen H., Wang Y., Xia L., Chen Y., Liu Y., Qiu F. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol. Biosyst. 2013;9:407–411. doi: 10.1039/c2mb25386k. [DOI] [PubMed] [Google Scholar]
- 13.Duan W., Du L., Jiang X., Wang R., Yan S., Xie Y., Yan K., Wang Q., Wang L., Zhang X. Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget. 2016;7:78850–78858. doi: 10.18632/oncotarget.12880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karreth F.A., Pandolfi P.P. ceRNA cross-talk in cancer: when ce-bling rivalries go awry. Cancer Discov. 2013;3:1113–1121. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng L., Shi L., Lu Y.F., Wang B., Tang T., Fu W.M., He W., Li G., Zhang J.F. Linc-ROR Promotes Osteogenic Differentiation of Mesenchymal Stem Cells by Functioning as a Competing Endogenous RNA for miR-138 and miR-145. Mol. Ther. Nucleic Acids. 2018;11:345–353. doi: 10.1016/j.omtn.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun C., Li S., Zhang F., Xi Y., Wang L., Bi Y., Li D. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget. 2016;7:51784–51814. doi: 10.18632/oncotarget.10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin H., Xu J., Guo X., Huang H., Li J., Peng M., Zhu J., Tian Z., Wu X.R., Tang M.S., Huang C. XIAP RING domain mediates miR-4295 expression and subsequently inhibiting p63α protein translation and promoting transformation of bladder epithelial cells. Oncotarget. 2016;7:56540–56557. doi: 10.18632/oncotarget.10645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J., Zhao X., Shi J., Pan Y., Chen Q., Leng P., Wang Y. miR-451 suppresses bladder cancer cell migration and invasion via directly targeting c-Myc. Oncol. Rep. 2016;36:2049–2058. doi: 10.3892/or.2016.5040. [DOI] [PubMed] [Google Scholar]
- 20.Cargnello M., Roux P.P. Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 2011;75:50–83. doi: 10.1128/MMBR.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J., Zhang J., Huang H., Li J., Yu Y., Jin H., Li Y., Deng X., Gao J., Zhao Q., Huang C. Crucial role of c-Jun phosphorylation at Ser63/73 mediated by PHLPP protein degradation in the cheliensisin a inhibition of cell transformation. Cancer Prev. Res. (Phila.) 2014;7:1270–1281. doi: 10.1158/1940-6207.CAPR-14-0233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng H., Wang X., Zhang P., Sun T., Ren X., Xia Z. miR-27a promotes cell proliferation and metastasis in renal cell carcinoma. Int. J. Clin. Exp. Pathol. 2015;8:2259–2266. [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou C., Huang C., Wang J., Huang H., Li J., Xie Q., Liu Y., Zhu J., Li Y., Zhang D. LncRNA MEG3 downregulation mediated by DNMT3b contributes to nickel malignant transformation of human bronchial epithelial cells via modulating PHLPP1 transcription and HIF-1α translation. Oncogene. 2017;36:3878–3889. doi: 10.1038/onc.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S.H., Zhang W.J., Wu X.C., Weng M.Z., Zhang M.D., Cai Q., Zhou D., Wang J.D., Quan Z.W. The lncRNA MALAT1 functions as a competing endogenous RNA to regulate MCL-1 expression by sponging miR-363-3p in gallbladder cancer. J. Cell. Mol. Med. 2016;20:2299–2308. doi: 10.1111/jcmm.12920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nordentoft I., Birkenkamp-Demtroder K., Agerbæk M., Theodorescu D., Ostenfeld M.S., Hartmann A., Borre M., Ørntoft T.F., Dyrskjøt L. miRNAs associated with chemo-sensitivity in cell lines and in advanced bladder cancer. BMC Med. Genomics. 2012;5:40. doi: 10.1186/1755-8794-5-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deng Y., Bai H., Hu H. rs11671784 G/A variation in miR-27a decreases chemo-sensitivity of bladder cancer by decreasing miR-27a and increasing the target RUNX-1 expression. Biochem. Biophys. Res. Commun. 2015;458:321–327. doi: 10.1016/j.bbrc.2015.01.109. [DOI] [PubMed] [Google Scholar]
- 27.Xu Y., Zhou M., Wang J., Zhao Y., Li S., Zhou B., Su Z., Xu C., Xia Y., Qian H. Role of microRNA-27a in down-regulation of angiogenic factor AGGF1 under hypoxia associated with high-grade bladder urothelial carcinoma. Biochim. Biophys. Acta. 2014;1842:712–725. doi: 10.1016/j.bbadis.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 28.Seo H.K., Shin S.P., Jung N.R., Kwon W.A., Jeong K.C., Lee S.J. The establishment of a growth-controllable orthotopic bladder cancer model through the down-regulation of c-myc expression. Oncotarget. 2016;8:50500–50509. doi: 10.18632/oncotarget.10784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen H., Zhao L., Feng X., Xu C., Li C., Niu Y. Lin28A activates androgen receptor via regulation of c-myc and promotes malignancy of ER-/Her2+ breast cancer. Oncotarget. 2016 doi: 10.18632/oncotarget.11004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarveswaran S., Ghosh R., Parikh R., Ghosh J. Wedelolactone, an anti-inflammatory botanical, interrupts c-Myc oncogenic signaling and synergizes with enzalutamide to induce apoptosis in prostate cancer cells. Mol Cancer Ther. 2016 doi: 10.1158/1535-7163.MCT-15-0861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang D., Song L., Li J., Wu K., Huang C. Coordination of JNK1 and JNK2 is critical for GADD45alpha induction and its mediated cell apoptosis in arsenite responses. J. Biol. Chem. 2006;281:34113–34123. doi: 10.1074/jbc.M602821200. [DOI] [PubMed] [Google Scholar]
- 32.Rao E., Jiang C., Ji M., Huang X., Iqbal J., Lenz G., Wright G., Staudt L.M., Zhao Y., McKeithan T.W. The miRNA-17∼92 cluster mediates chemoresistance and enhances tumor growth in mantle cell lymphoma via PI3K/AKT pathway activation. Leukemia. 2012;26:1064–1072. doi: 10.1038/leu.2011.305. [DOI] [PubMed] [Google Scholar]
- 33.Luo G., Wang M., Wu X., Tao D., Xiao X., Wang L., Min F., Zeng F., Jiang G. Long Non-Coding RNA MEG3 Inhibits Cell Proliferation and Induces Apoptosis in Prostate Cancer. Cell. Physiol. Biochem. 2015;37:2209–2220. doi: 10.1159/000438577. [DOI] [PubMed] [Google Scholar]
- 34.Zuo Z., Che X., Wang Y., Li B., Li J., Dai W., Lin C.P., Huang C. High mobility group Box-1 inhibits cancer cell motility and metastasis by suppressing activation of transcription factor CREB and nWASP expression. Oncotarget. 2014;5:7458–7470. doi: 10.18632/oncotarget.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang C., Zeng X., Jiang G., Liao X., Liu C., Li J., Jin H., Zhu J., Sun H., Wu X.R., Huang C. XIAP BIR domain suppresses miR-200a expression and subsequently promotes EGFR protein translation and anchorage-independent growth of bladder cancer cell. J. Hematol. Oncol. 2017;10:6. doi: 10.1186/s13045-016-0376-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin H., Yu Y., Hu Y., Lu C., Li J., Gu J., Zhang L., Huang H., Zhang D., Wu X.R. Divergent behaviors and underlying mechanisms of cell migration and invasion in non-metastatic T24 and its metastatic derivative T24T bladder cancer cell lines. Oncotarget. 2015;6:522–536. doi: 10.18632/oncotarget.2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.