Abstract
Gastric cancer (GC) is a common life-threatening cancer type worldwide, with an increasing prevalence and a high rate of mortality. Due to limitations in clinical treatment, surgery has become the most efficient strategy for the treatment of GC. It is urgent to identify novel biomarkers, which are useful for the diagnosis of GC and for improving the survival rate of patients with GC. HDACs are multi-functional proteins and are involved in regulating gene expression, cell proliferation and the epigenetic regulation. However, the precise role of HDACs in the progression of GC remains unknown. The present study demonstrated that HDAC1 is involved in the promotion of GC cell proliferation, possibly by upregulating the expression of the lncRNAs, BC01600 and AF116637, in the tissues of patients with GC. Abnormal expression profiles of lncRNAs were observed in the tissues of patients with GC. lncRNAs were analyzed in the GSE64951 and GSE19826 databases, and it was revealed that BC01600 and AF116637 were two typically upregulated lncRNAs. Furthermore, it was revealed that BC01600 and AF116637 are regulated by HDAC1, as evidenced by decreased expression of these two lncRNAs in HDAC1-knockout SC-M1 cell lines, and by reduced expression of HDAC1 in these two lncRNA-knockout SC-M1 cell lines. Silencing of HDAC1 decreased the proliferation and increased the apoptosis of SC-M1 cell lines, but had no effect on the migration of the SC-M1 cell lines. The present study provided evidence of the importance of HDAC1 in the progression of SC-M1, and the association between HDAC1 and the expression of lncRNAs. The results of the present study indicated that HDAC1 may be a promising target for the clinical treatment of GC.
Keywords: gastric cancer, BC01600, AF116637, HDACs, lincRNAs
Introduction
Gastric cancer (GC) is a common life-threatening cancer type worldwide, with an increasing prevalence and high rate of mortality, particularly in China (1). Etiological factors, and genetic and epigenetic factors are all involved in the progression of GC (2). At present, surgery is the most effective treatment for GC, and the survival rate primarily depends on the stage of GC (non-invasive detection of GC) (3). Therefore, identifying specific and effective biomarkers in the early stage of GC is important for the efficient treatment of patients with GC in order to increase the survival rate. Previous studies have demonstrated that genetic abnormalities, including aberrant genes, microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), participated in the initiation and progression of GC (competing endogenous RNA networks) (4).
Long non-coding RNAs are defined as transcripts >200 nucleotides, and are widely expressed in different tissues (5,6). lncRNAs are involved in a number of biological progresses, including the proliferation and differentiation of cells (7). lncRNA genes resemble protein-coding genes, and they have a variety of molecular functions across numerous cellular pathways and processes, including oncogenic signaling (8). Researchers have identified an increasing number of aberrantly expressed lncRNAs in GC, and have aimed to determine their role in the progression of GC (9–12).
Epigenetic modifications serve causative roles in the progression of GC (13). Histone deacetylases (HDACs), a family of 18 molecules divided into four sub-classes defined according to their structural similarities (14), serve functional roles, including modification of histone and non-histone proteins, regulation of chromatin structure, repression or activation of gene expression and regulation of cell metabolism (14). Previous studies have demonstrated that HDACs are crucial for the progression of several types of disease, including diabetes, melanoma, hepatocellular carcinoma and GC (15–18). However, the precise role of HDACs in the progression of GC remains unclear. HDACs have been demonstrated to serve roles in regulating the expression of microRNAs. However, little is known regarding whether HDACs may serve certain roles in regulating the expression of lncRNAs during the progression of GC.
The present study attempted to investigate the role of abnormal expression of HDACs in the progression of GC, and to determine the association between HDACs and lncRNAs. The present study provided evidence of the importance of HDACs in the progression of GC and identified them as promising targets for the clinical treatment of GC.
Patients and methods
Patient population
Gastric cancer tissues from 63 patients (mean age 61 years, range 45–73 years; 45 males and 18 females) with gastric cancer and normal tissues from 31 healthy individuals (mean age 58 years, range 40–75 years; 20 male and 11 female) were used in the present study, which was obtained from Jiangxi Province People's Hospital during the period from March 2015 to August 2016. Ethics approval for this study was approved by the Institutional Review Board and the Ethics Committee at Jiangxi Province People's Hospital (ID no. 2015083). Written informed consent was obtained from each patient. Ten cancer tissues and normal tissues were randomly selected for examining the expression profile of lncRNAs in the GSE64951 database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64951). Additionally, 9 cancer tissues and normal tissues were randomly selected for examining the expression profile of lncRNAs in the GSE19826 database (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi). The expression of lncRNAs in the tissues of patients with gastric cancer and healthy donors was analyzed by Affymetrix Human Genome U133 Plus 2.0 Array (platforms: GPL570, [HG-U133_Plus_2]; Affymetrix, Inc., California, USA).
Sample preparation for reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and gene chip analysis
The differential genes were verified using RT-qPCR. The procedure was as follows: Total RNA was isolated from the cultured SC-M1 cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), according to the manufacturer's protocol. Total RNA was used to synthesize cDNA using the PrimeScriptTM RT reagent kit (cat. no. RR037A; Takara Bio, Inc., Otsu, Japan), all processes were performed according to the manufacturer's protocol, then the cDNA was subjected to qPCR. The relative expression levels of lncRNAs were measured using a SYBR® Premix Ex TaqTM (cat. no. RR420A; Takara Bio, Inc.), according to the manufacturer's protocol, using GAPDH as an internal control. RT-qPCR primers were synthesized by Sangon Biotech Co., Ltd. (Shanghai, China). The 5′-3′ primer sequences used for RT-qPCR were as follows: GAPDH forward, GACTCATGACCACAGTCCATGC and reverse, AGAGGCAGGGATGATGTTCTGRT-PCR was performed on an ABI 7500 Sequence Detection system (ABI, Vernon, CA, USA). The thermocycling conditions were as follows: 95°C for 30 sec, 40 cycles of 95°C for 5 sec and 60°C for 31 sec. The dissociation stage was 95°C for 15 sec, 60°C for 1 min and 95°C for 15 sec. The expression of target genes was calculated based on the threshold cycle (CQ), and relative expression levels were calculated as 2−[(Cq of target genes)-(Cq of GAPDH)] (19) following normalization with reference to the quantification of GAPDH. Each experiment was repeated three times. The labeled target was fragmented and hybridized to probe arrays, using Encore Biotin Module from NuGEN Technologies, Inc. (San Carlos, CA, USA).
Stimulation of gastric cell line
The human gastric cancer SC-M1 cell line was obtained from American Type Culture Collection (ATCC; Manassas, VA, USA). The cells were cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific, Inc.), containing 10% fetal bovine serum (FBS; Hyclone; GE Healthcare Life Sciences, Logan, UT, USA), 100 U/ml penicillin and 100 mg/ml streptomycin. In certain experiments, cells (1×105 cells/ml) were cultured in 96-well plates for the examination of cell viability using an MTT assay (MTT Cell Proliferation kit; Sangon Biotech Co., Ltd., Shanghai, China). According to the manufacturer's protocol, formazan solubilization solution was used to dissolve the formazan crystals and measure the absorbance at 570 nm using a microplate reader (Thermo Fisher Scientific, Inc.).
For the cell migration assay by Transwell method (Transwell Permeable Supports, Corning, Cornstar, NY, USA), cells (1×105 cells/ml) were transfected with HDAC siRNAs and were cultured using Dulbecco's modified Eagle's medium (DMEM; Hyclone; GE Healthcare Life Sciences) in the upper chamber of 24-well plates, and 0.5 ml DMEM containing 10% FBS were added to the lower chambers. The total number of migrated cells were calculated 48 h later by inverted microscope (OLYMPUS IX51; Olympus Corporation, Tokyo, Japan. The magnification of the captured image was ×400.
RNA interference
The siRNA sequence targeting HDAC1s (NCBI; GenBank ID: NM_004964.2) and the scramble siRNA sequence were designed and synthesized by GenePharma (Shanghai, CHINA). The sequences for the si-HDAC1s were: siRNA1, 5′-gcaaagaaguccgaggc-3′; siRNA2, 5′-ucaaaggacacgccaagug-3′; and siRNA3, 5′-gagaagaaagaagucacc-3′. For in vitro experiments, siRNAs were transfected into three gastric cancer cell lines using Lipofectamine iMAX (Invitrogen; Thermo Fisher Scientific, Inc.), the terminal concentration of siRNA was 50 nM according to the manufacturer's protocol. At 48 h following transfection, the efficiency of RNA interference was determined by RT-qPCR that the method has been described in the upper.
Cell apoptosis assay
Gastric cancer cells were stained using a fluorescein isothiocyanate (FITC)-conjugated Annexin V and propidium iodide (PI) kit (BD Biosciences, Franklin Lakes, NJ, USA), according to the manufacturer's protocol. Cells (1×106 cells/ml) were cultured in 12-well plates, and following transfection with siRNAs for 24 h, the cells were harvested and washed twice with cold phosphate-buffered saline with gentle agitation. SC-M1 cells were then re-suspended and added to binding buffer (1X), and cell density was adjusted to 2–5×105/ml. In the dark, 5 µl Annexin V-FITC was added to the cell suspension volume of 195 µl and incubated for 10 min at room temperature prior to the addition of 190 µl binding buffer and 10 µl PI. A total of 10,000 events per sample were acquired using a FACS-scan flow cytometer (BD Biosciences, San Jose, CA, USA). The percentage of apoptotic cells was analyzed by Acri (BD Biosciences). The percentage of cells undergoing apoptosis was analyzed using BD CellQuestTM Pro Software Analysis Tutorial (BD Biosciences; version 5.1).
Western blot analysis
For western blotting, human gastric cancer SC-M1 cells were transfected with siRNAs in 12-well plates. The proteins were extracted from SC-M1 cells according to the standard protocol. Cells were lysed in cold lysis buffer (50 mM Tris, 150 mM NaCl, 10 mM ethylenediaminetetraacetic acid and 1% Triton X-100) containing protease and phosphatase inhibitors. The protein determination method was BCA (BCA Protein Assay kit, Sangon Biotech Co., Ltd. Shanghai, China). Briefly, the loading of protein sample was 40 µg each hole, the proteins were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels, and subsequently transferred to nitrocellulose membranes. The membrane was completely immersed in 5% skimmed milk powder for 60 min at room temperature. The following primary antibodies were used: Total p-21 (cat. no. PL0502562; dilution, 1:1,000; PLLABS, British Columbia, Canada; http://www.pllabs.com/total-p53 (cat. no. ab131442; dilution, 1:2,000: Epitomics; Abcam, Cambridge, MA, USA) and actin (cat. no. PA1-183; dilution, 1:2,000: Thermo Fisher Scientific, Inc.) antibodies. Incubate overnight at 4°C with primary antibodies, respectively. Membranes were subsequently incubated with horseradish peroxidase-conjugated secondary anti-rabbit antibodies (cat. no. 7074; Cell Signaling Technology, Inc., Danvers, MA, USA) at a dilution of 1:4,000 for 1 h at room temperature. Membranes were developed using the enhanced chemiluminescence detection system (Pierce; Thermo Fisher Scientific, Inc.).
Statistical analysis
Limma R package (1.10.1) was utilized to identify the DEGs in GSE64951 and GSE19826 database, with a padj ≤0.05. SPSS version 19.0 for Windows (IBM Corp., Armonk, NY, USA) was used for all analyses. All the data are expressed as the mean ± standard deviation. The correlation between HDAC1 and upregulated lncRNAs (BC01600 and AF116637 and uc004ebu.1) in gastric cancer were assessed using one-way analysis of variance followed by Tukey's post-hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
Abnormal expression of lncRNA is identified in gastric cancer
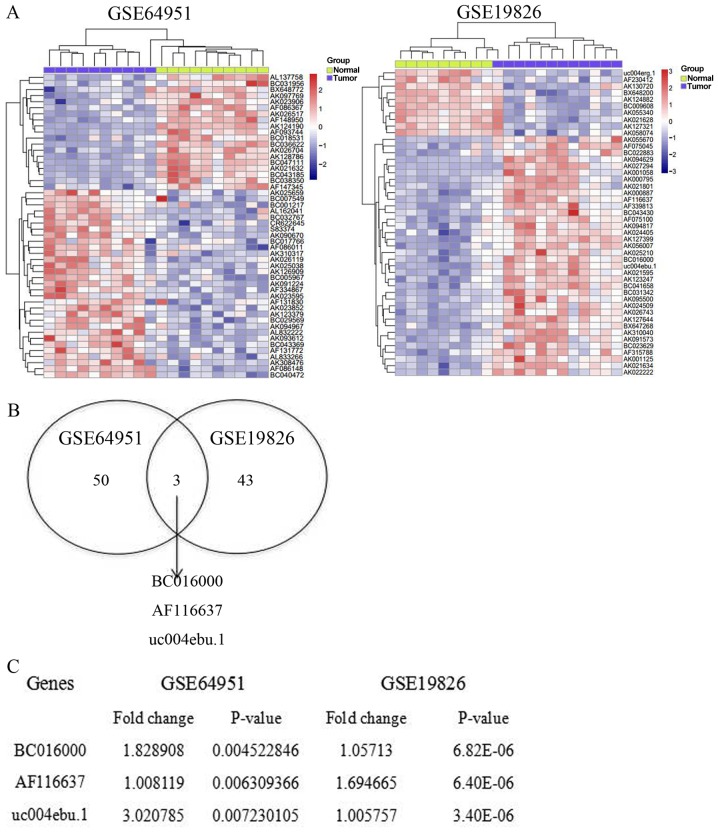
Accumulating evidence has demonstrated that the abnormal expression of lncRNAs is associated with the progression of certain diseases. In order to detect the expression profile of lncRNAs in gastric cancer tissues, the expression of lncRNAs in the tissues of patients with gastric cancer and healthy donors was analyzed by Affymetrix Human Genome U133 Plus 2.0 Array. Compared with normal controls, the expression profile of lncRNA is completely different in gastric cancer. A total of 5,621 lncRNAs were analyzed from the samples of patients with gastric cancer and healthy donors. In database no. GSE64951, 53 lncRNAs were significantly altered. Among these lncRNAs, 19 downregulated lncRNAs and 34 upregulated lncRNAs were identified from the samples of patients with gastric cancer, compared with matched samples from healthy donors (Fig. 1A). Meanwhile, in database no. GSE19826, 46 lncRNAs were abnormally expressed in the samples from patients with gastric cancer. Ten of these lncRNAs were upregulated, while 36 of these lncRNAs were downregulated (Fig. 1A). In order to further investigate the potential DE-lncRNAs, a Venn diagram was constructed using the upregulated lncRNAs in these two databases. The online software can be found at http://bioinfogp.cnb.csic.es/tools/venny/index.html. It was revealed that 3 lncRNAs overlapped between these two databases, GSE64951 and GSE19826 (Fig. 1B). The three significantly upregulated lncRNAs were BC016000, AF116637 and uc004edu.1, with a fold-change of 1.83, 1.01 and 3.02, respectively (Fig. 1C). Taken together, the results of the present study revealed that the expression profile is completely different in gastric cancer tissues compared with the normal tissues, indicating the abnormal expression profile of lncRNAs in the gastric cancer.
Figure 1.
Abnormal expression profile of lncRNAs in gastric cancer. (A) The expression profile of lncRNA in tumor and normal tissues was examined by heat-map analysis in GSE64951 and GSE19826 databases. Upregulated lncRNAs are shown in red and downregulated lncRNAs are shown in blue. (B) The overlapping lncRNAs of GSE64951 and GSE19826 are shown in a Venn diagram. (C) The expression of BC01600, AF116637 and uc004ebu.1 in gastric cancer. lncRNA, long non-coding RNA.
The correlation between HDAC1 and upregulated lncRNAs in gastric cancer
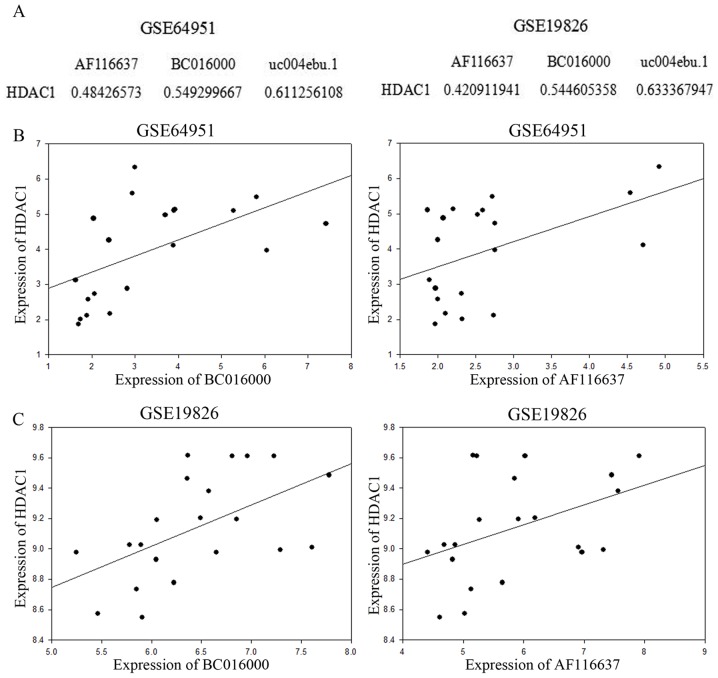
HDACs are a family of multi-functional proteins, which are involved in tumor suppression. HDAC1 is a very important member of the HDAC family. Therefore, the present study investigated the association between HDAC1 and the upregulated lncRNAs in gastric cancer. Correlation analysis of HDAC1 and the upregulated lncRNAs, BC016000 and AF116637, was conducted. The Pearson's correlation coefficient (PCC) between HDAC1 and BC016000 and AF116637 was calculated using the expression value. The absolute values of Correlation Coefficient were shown in Fig. 2A, for example: The absolute value of Correlation Coefficient between HDAC1 expression value and AF116637 expression value was 0.48426573 in the GSE64951 database. Correlation analysis was performed in the GSE64951 and GSE19826 databases. In the two aforementioned databases, lncRNAs BC016000 and AF116637 exhibited a moderate to strong positive correlation with HDAC1 with Correlation Coefficients of 0.545 and 0.421, respectively, in the GSE64951 database (Fig. 2A and B), and in the GSE19826 database, the Correlation Coefficients were 0.549 and 0.484, respectively (Fig. 2A and C).
Figure 2.
Correlation analyses of lncRNAs with HDAC1. (A) The association between abnormally expressed lncRNA and HDAC1. Scatter plots were drawn to demonstrate the associations between BC01600 or AF116637 and HDAC1 in two databases, (B) GSE64951 and (C) GSE19826, respectively; lncRNA, long non-coding RNA. lncRNAs, long non-coding RNAs.
Silencing of HDAC1 downregulates the expression of BC01600 and AF116637
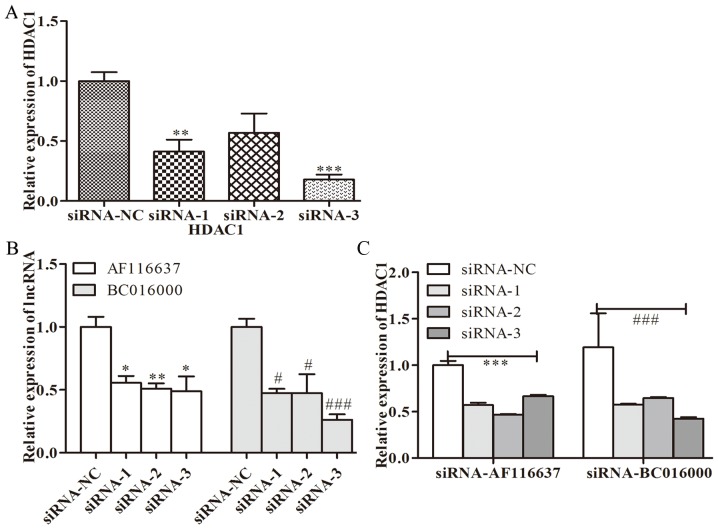
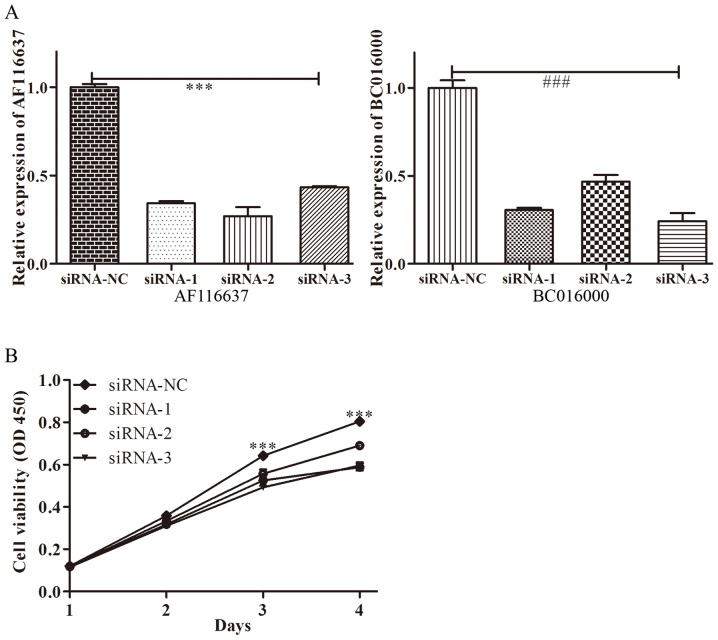
To further investigate whether BC01600 and AF116637 were correlated with HDAC1, the expression levels of BC01600 and AF116637 were evaluated following HDAC1-silencing with its specific siRNA. Three pairs of siRNAs targeting HDAC1 were designed. It was revealed that all these three siRNA sequences could efficiently inhibit the expression of HDAC1 (Fig. 3A). HDAC1 siRNA transfection with these three siRNAs significantly inhibited the expression of BC01600 and AF116637 (Fig. 3B). Additionally, three pairs of siRNAs targeting lncRNA, BC01600 and AF116637 were designed. Next, the expression of BC01600 and AF116637 was efficiently decreased by their respective siRNA sequences (Fig. 4). Subsequently, it was demonstrated that BC01600 and AF116637 lncRNA siRNA transfection with their respective siRNAs significantly inhibited the expression of HDAC1 (Fig. 3B). Furthermore, the expression of BC01600, AF116637 and HDAC1 was analyzed in gastric cancer tissues. It was revealed that these two lncRNAs and HDAC1 have an increased expression trend in gastric cancer tissue (Fig. 5). Taken together, these results indicated that HDAC1 could promote the expression of BC01600 and AF116637 in gastric cancer.
Figure 3.
Analysis of lncRNAs with HDCA1 using siRNA. (A) The expression of HDAC1 in gastric cancer cells following silencing of HDAC1 by siRNAs. **P<0.01 expression of HDAC1 in GC cells siRNA-1 group vs. siRNA-NC; ***P<0.001 expression of HDAC1 in GC cells siRNA-3 group vs. siRNA-NC. (B) The expression of BC01600 and AF116637 in gastric cancer cells following silencing of HDAC1. *P<0.05 AF116637 expression siRNA (−1/3) group vs. siRNA-NC; **P<0.01 AF116637 expression siRNA-2 group vs. siRNA-NC. #P<0.05 BC016000 expression siRNA (−1/2) group vs. siRNA-NC; ###P<0.001 BC016000 expression siRNA-3 group vs. siRNA-NC. (C) The expression of HDAC1 in gastric cancer cells after silencing of BC01600 and AF116637. ***P<0.001 HDAC1 expression siRNA-AF116637 (−1/2/3) group vs. siRNA-NC. ###P<0.001 HDAC1 expression siRNA-BC016000 (−1/2/3) group vs. siRNA-NC. siRNA, small interfering RNA; lncRNAs, long non-coding RNAs.
Figure 4.
LncRNA expression and the proliferation of GC cells by siRNA. (A) The respective expression of lncRNA, BC01600 and AF116637 in gastric cancer cells following silencing of these lncRNAs by siRNAs. ***P<0.001 AF116637 expression after silencing AF116637 vs. siRNA-NC. ###P<0.001 BC016000 expression after silencing BC016000 vs. siRNA-NC. (B) The cell viability of new GC cells at different time points following HDAC1 siRNA treatment. GC, gastric cancer. ***P<0.001 cell viability of GC between siRNA (−1/2/3) group and siRNA-NC at different time point. siRNA, small interfering RNA; lncRNA, long non-coding RNA.
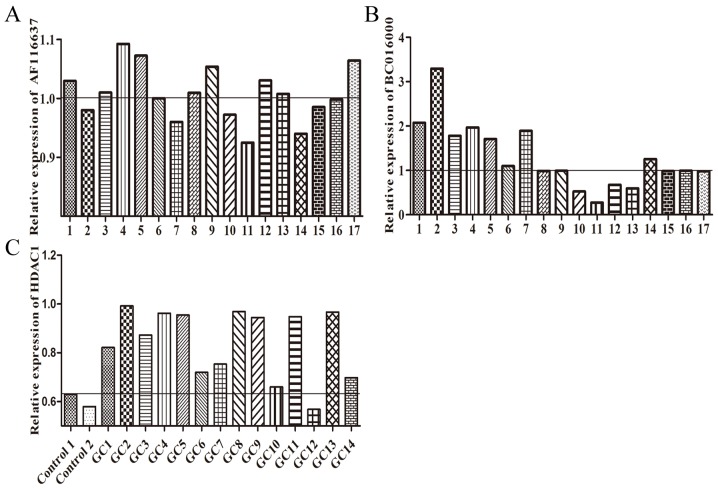
Figure 5.
Relative expression of (A) BC01600, (B) AF116637 and (C) HDAC1 in gastric cancer tissues.
HDAC1 promotes the proliferation of gastric cancer cells
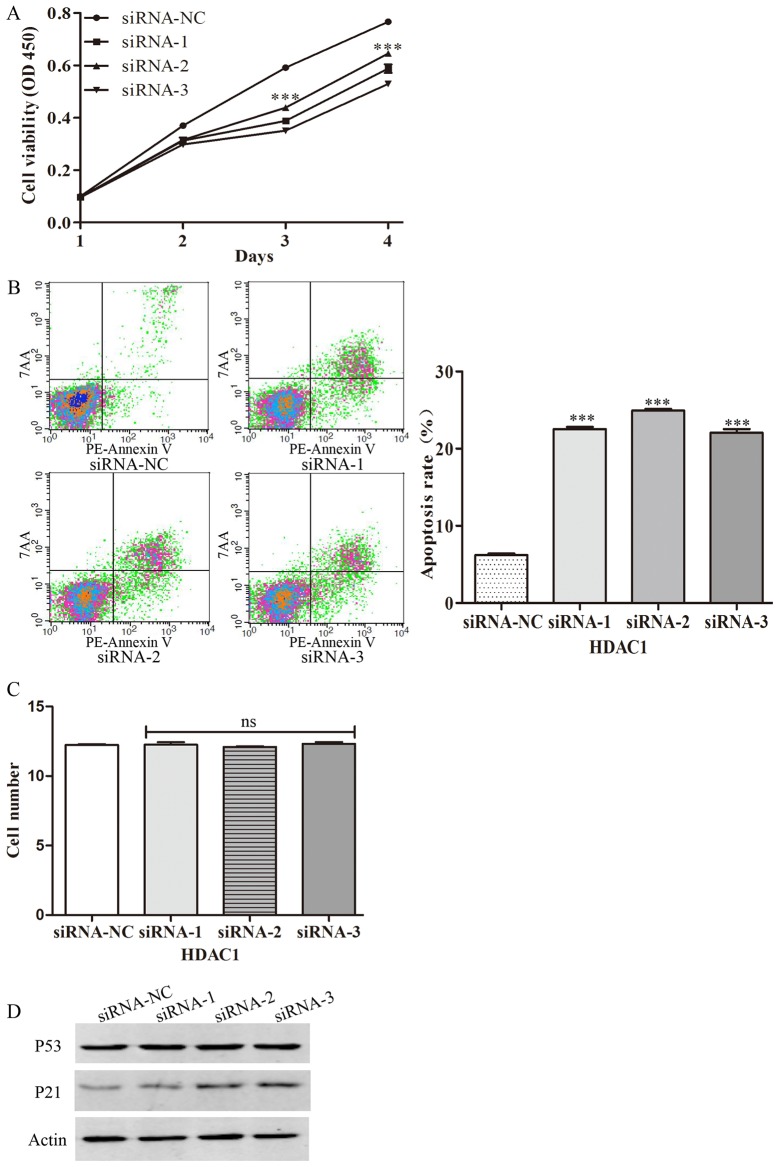
To further examine the role of HDACs in the progression of gastric cancer, HDAC1 was silenced by transfecting gastric cancer cells with specific siRNAs. It was revealed that silencing of HDAC1 decreased cell viability, indicating that HDAC1 can inhibit the proliferation of gastric cancer cells (Fig. 6A). Furthermore, the present study also examined the effect of HDAC1 on apoptosis by flow cytometry. It was demonstrated that silencing of HDAC1 increased the percentage of apoptotic cells, indicating that HDAC1 can decrease the apoptosis of gastric cancer cells (Fig. 6B). Following transfection of the gastric cancer cells with specific siRNAs, the present study also examined the migration of gastric cancer cells. The results demonstrated that silencing of HDAC1 with specific siRNAs has no effect on the migration of gastric cancer cells (Fig. 6C). Furthermore, the role of HDAC1 in the activation of tissue-associated genes, p53 and p21, was examined. It was demonstrated that transfection with HDAC1 siRNAs significantly increased the expression of total-p21, but had no effect on the expression of total-p53 (Fig. 6D). Taken together, the results of the present study demonstrated that HDACs could promote the proliferation of gastric cancer.
Figure 6.
HDAC1 was involved in the proliferation of gastric cancer cells. (A) The viability of gastric cancer cells at different time points following HDAC1 siRNA treatment. ***P<0.001 viability of GC cells siRNA (−1/2/3)group vs. siRNA-NC at different time points. (B) The apoptosis of gastric cancer cells was examined by flow cytometric analysis. ***P<0.001 apoptosis of GC cells siRNA (−1/2/3) group vs. siRNA-NC at different time points. (C) The migrated gastric cancer cells were counted. (D) The repression of P53 and P21 in gastric cancer was examined by western blot analysis. siRNA, small interfering RNA.
Discussion
The present study demonstrated that HDAC1 could promote the proliferation of GC cells, possibly by upregulating the expression of lncRNAs, BC01600 and AF116637. High expression of HDAC1 and abnormal expression of lncRNA was observed in the tissues of patients with GC. It was demonstrated that the expression of BC01600 and AF116637, two typically upregulated lncRNAs, is correlated with the expression of HDAC1, as evidenced by the decreased expression of these two lncRNAs in HDAC1-knockout GC cell lines, and by reduced expression of HDAC1 in lncRNA, BC01600 and AF116637, knockout GC cell lines, respectively. Silencing of HDACs increased the p21-dependent apoptosis of the GC cell lines.
Investigating novel treatments for GC, one of the most common types of cancer, is necessary. Limitations of current treatment options include the following: The patients in the later phase of GC experienced a lot of painful and a high mortality rate, even after surgery (20). It is useful to identify efficient biomarkers for the diagnosis of GC in the early phase of GC. Epigenetics has become a popular field of molecular biology recently and accumulating evidence has demonstrated that epigenetic mechanisms contribute toward differentiation, aging and disease development, particularly during the progression of cancer (13,21,22).
lncRNAs serve important roles in the progression of cancer and numerous hallmarks of cancer are controlled by lncRNAs (8-10,23-25). Various lncRNAs have been implicated in the regulation of individual genes via epigenetic regulation (26). Previous studies have demonstrated that overexpression of lncRNAs is implicated in the metastasis of human tumors (27,28). An increasing volume of evidence has indicated that specific lncRNA mutations are associated with the progression of cancer (8). Therefore, lncRNAs could be a promising candidate for the diagnosis and therapy of cancer (28,29). The present study demonstrated that the expression profile of lncRNAs is completely different in gastric cancer, compared with normal controls by using the bioinformatics methods according to the GSE64951 and GSE19826 databases. The following three lncRNAs were selected: BC016000, AF116637 and uc004edu.1, which were overlapped in these two databases, as determined by constructing a Venn diagram using the upregulated lncRNAs in these two databases. Therefore, the abnormal expression profile of lncRNAs may be involved in or be a factor in promoting the progression of GC. However, the way in which the expression of these lncRNAs is regulated remains unknown.
HDACs are multi-functional proteins, which serve critical roles in transcription regulation (30). Aberrant expression of HDACs is frequently observed in various types of human cancer. In the majority of cases, upregulation of HDACs is associated with advanced disease and poor outcomes in patients (15). HDACs have also been revealed to be broadly dysregulated in multiple myeloma (31). HDACs are also involved in the progression of several types of cancer, including melanoma and hepatocellular carcinoma (15,18). Participating in a range of cellular and molecular pathways, HDACs can either repress tumor suppressor gene expression or modify key molecules in oncogenic cell-signaling pathways. However, the precise role of HDACs has not yet been revealed. A previous study demonstrated that HDACs could upregulate the expression of microRNAs, indicating that HDACs may serve certain roles in regulating the expression of lncRNAs (32). HDAC1 is an important member of the HDACs family. In order to determine the role of HDAC1 in the expression of lncRNAs, correlation analysis of HDAC1 and the upregulated lncRNAs, BC016000 and AF116637, was conducted, and BC016000 and AF116637 lncRNAs exhibited a positive and strong correlation with HDAC1, indicating that HDAC1 serves roles in the expression of lncRNAs. The results of the present study demonstrated that HDACs may be involved in the progression of GC by upregulating the expression of lncRNAs.
Cancer cells are abnormally proliferated and differentiated cells. Proliferation, apoptosis and migration are important for the progression of cancer. The role of HDAC1 in the proliferation, apoptosis and migration of GC cancer cells was also examined. It was demonstrated that silencing of HDAC1 may decrease the proliferation and increase the apoptosis of gastric cancer cells. However, HDAC1 has no effect on the migration of gastric cancer cells. Therefore, HDAC1 is specifically involved in the proliferation of gastric cancer cells.
In summary, HDAC1 is critical for the progression of GC, by promoting the proliferation of cancer cells, which may be regulated by the HDAC1-mediated decreased or increased expression of lncRNAs. The present study demonstrated the correlation of HDACs and lncRNAs, and provided a possible mechanism by which HDACs regulate the progression of gastric cancer. The results of the present study provided a possible target for the clinical treatment of gastric cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the Jiangxi Province Natural Science Foundation of China (grant no. 2010BSA14300) and Science and Technology Department of Jiangxi Province, Key R&D Plan (grant no. 20171BBG700870).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
JC designed the research. ZhiY and JZ performed the research. TW and ZiY were responsible for clinical diagnosis and sample collection. HL contributed to data collection and statistical analysis. ZhiY, JZ and JC wrote the manuscript. All the authors read and approved the final manuscript.
Ethics statement and consent to participate
Human samples used in the present study were obtained from the patients with written informed consent. The present study was approved by the Ethics Committee of Jiangxi Province People's Hospital and was conducted according to The Declaration of Helsinki.
Patient consent for publication
Consent for publication was obtained from all patients.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Matsuda T, Saika K. Worldwide burden of cancer incidence in 2002 extrapolated from cancer incidence in five continents Vol.IX. Jpn J Clin Oncol. 2012;42:1111–1112. doi: 10.1093/jjco/hys176. [DOI] [PubMed] [Google Scholar]
- 2.Guo LL, Song CH, Wang P, Dai LP, Zhang JY, Wang KJ. Competing endogenous RNA networks and gastric cancer. World J Gastroenterol. 2015;21:11680–11687. doi: 10.3748/wjg.v21.i41.11680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu J, Huang CM, Zheng CH, Li P, Xie JW, Wang JB, Lin JX. Consideration of tumor size improves the accuracy of TNM predictions in patients with gastric cancer after curative gastrectomy. Surg Oncol. 2013;22:167–171. doi: 10.1016/j.suronc.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 4.Müller S, Raulefs S, Bruns P, Afonso-Grunz F, Plötner A, Thermann R, Jäger C, Schlitter AM, Kong B, Regel I, et al. Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol Cancer. 2015;14:94. doi: 10.1186/s12943-015-0401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Müller S, Raulefs S, Bruns P, Afonso-Grunz F, Plötner A, Thermann R, Jäger C, Schlitter AM, Kong B, Regel I, et al. Erratum to: Next-generation sequencing reveals novel differentially regulated mRNAs, lncRNAs, miRNAs, sdRNAs and a piRNA in pancreatic cancer. Mol Cancer. 2015;14:144. doi: 10.1186/s12943-015-0401-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Zhao T, Xu J, Liu L, Bai J, Xu C, Xiao Y, Li X, Zhang L. Identification of cancer-related lncRNAs through integrating genome, regulome and transcriptome features. Mol Biosyst. 2015;11:126–136. doi: 10.1039/C4MB00478G. [DOI] [PubMed] [Google Scholar]
- 8.Evans JR, Feng FY, Chinnaiyan AM. The bright side of dark matter: lncRNAs in cancer. J Clin Invest. 2016;126:2775–2782. doi: 10.1172/JCI84421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu Y, Chen T, Li G, Yu X, Lu Y, Wang H, Teng L. LncRNAs: Emerging biomarkers in gastric cancer. Future Oncol. 2015;11:2427–2441. doi: 10.2217/fon.15.175. [DOI] [PubMed] [Google Scholar]
- 10.Huang X, Zhi X, Gao Y, Ta N, Jiang H, Zheng J. LncRNAs in pancreatic cancer. Oncotarget. 2016;30:57379–57390. doi: 10.18632/oncotarget.10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jonsson P, Coarfa C, Mesmar F, Raz T, Rajapakshe K, Thompson JF, Gunaratne PH, Williams C. Single-molecule sequencing reveals estrogen-regulated clinically relevant lncRNAs in breast cancer. Mol Endocrinol. 2015;29:1634–1645. doi: 10.1210/me.2015-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng L, Yuan X, Jiang B, Tang Z, Li GC. LncRNAs: Key players and novel insights into cervical cancer. Tumour Biol. 2016;37:2779–2788. doi: 10.1007/s13277-015-4663-9. [DOI] [PubMed] [Google Scholar]
- 13.Ali Z, Deng Y, Tang Y, Zheng S, Ma N, He N. Epigenetic deregulations in gastric cancer. J Nanosci Nanotechnol. 2013;13:40–51. doi: 10.1166/jnn.2013.6705. [DOI] [PubMed] [Google Scholar]
- 14.Hildmann C, Riester D, Schwienhorst A. Histone deacetylases-an important class of cellular regulators with a variety of functions. Appl Microbiol Biotechnol. 2007;75:487–497. doi: 10.1007/s00253-007-0911-2. [DOI] [PubMed] [Google Scholar]
- 15.Mithraprabhu S, Kalff A, Chow A, Khong T, Spencer A. Dysregulated class I histone deacetylases are indicators of poor prognosis in multiple myeloma. Epigenetics. 2014;9:1511–1520. doi: 10.4161/15592294.2014.983367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christensen DP, Dahllöf M, Lundh M, Rasmussen DN, Nielsen MD, Billestrup N, Grunnet LG, Mandrup-Poulsen T. Histone deacetylase (HDAC) inhibition as a novel treatment for diabetes mellitus. Mol Med. 2011;17:378–390. doi: 10.2119/molmed.2011.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mutze K, Langer R, Becker K, Ott K, Novotny A, Luber B, Hapfelmeier A, Göttlicher M, Höfler H, Keller G. Histone deacetylase (HDAC) 1 and 2 expression and chemotherapy in gastric cancer. Ann Surg Oncol. 2010;17:3336–3343. doi: 10.1245/s10434-010-1182-1. [DOI] [PubMed] [Google Scholar]
- 18.Yang Z, Zhou L, Wu LM, Xie HY, Zhang F, Zheng SS. Combination of polymorphisms within the HDAC1 and HDAC3 gene predict tumor recurrence in hepatocellular carcinoma patients that have undergone transplant therapy. Clin Chem Lab Med. 2010;48:1785–1791. doi: 10.1515/CCLM.2010.353. [DOI] [PubMed] [Google Scholar]
- 19.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 20.Wan QS, Zhang KH. Noninvasive detection of gastric cancer. Tumour Biol. 2016;37:11633–11643. doi: 10.1007/s13277-016-5129-4. [DOI] [PubMed] [Google Scholar]
- 21.Gigek CO, Chen ES, Calcagno DQ, Wisnieski F, Burbano RR, Smith MA. Epigenetic mechanisms in gastric cancer. Epigenomics. 2012;4:279–294. doi: 10.2217/epi.12.22. [DOI] [PubMed] [Google Scholar]
- 22.Hagelkruys A, Sawicka A, Rennmayr M, Seiser C. The biology of HDAC in cancer: The nuclear and epigenetic components. Handb Exp Pharmacol. 2011;206:13–37. doi: 10.1007/978-3-642-21631-2_2. [DOI] [PubMed] [Google Scholar]
- 23.Chang YN, Zhang K, Hu ZM, Qi HX, Shi ZM, Han XH, Han YW, Hong W. Hypoxia-regulated lncRNAs in cancer. Gene. 2016;575:1–8. doi: 10.1016/j.gene.2015.08.049. [DOI] [PubMed] [Google Scholar]
- 24.Cao B, Song N, Zhang M, Di C, Yang Y, Lu Y, Chen R, Lu ZJ, Guo M. Systematic study of novel lncRNAs in different gastrointestinal cancer cells. Discov Med. 2016;21:159–171. [PubMed] [Google Scholar]
- 25.Cheng WS, Tao H, Hu EP, Liu S, Cai HR, Tao XL, Zhang L, Mao JJ, Yan DL. Both genes and lncRNAs can be used as biomarkers of prostate cancer by using high throughput sequencing data. Eur Rev Med Pharmacol Sci. 2014;18:3504–3510. [PubMed] [Google Scholar]
- 26.Cogill SB, Wang L. Co-expression network analysis of human lncRNAs and cancer genes. Cancer Inform. 2014;13(Suppl 5):S49–S59. doi: 10.4137/CIN.S14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han J, Rong LF, Shi CB, Dong XG, Wang J, Wang BL, Wen H, He ZY. Screening of lymph nodes metastasis associated lncRNAs in colorectal cancer patients. World J Gastroenterol. 2014;20:8139–8150. doi: 10.3748/wjg.v20.i25.8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang F, Zhang L, Zhang C. Long noncoding RNAs and tumorigenesis: Genetic associations, molecular mechanisms, and therapeutic strategies. Tumour Biol. 2016;37:163–175. doi: 10.1007/s13277-015-4445-4. [DOI] [PubMed] [Google Scholar]
- 29.Fayda M, Isin M, Tambas M, Guveli M, Meral R, Altun M, Sahin D, Ozkan G, Sanli Y, Isin H, et al. Do circulating long non-coding RNAs (lncRNAs) (LincRNA-p21, GAS 5, HOTAIR) predict the treatment response in patients with head and neck cancer treated with chemoradiotherapy? Tumour Biol. 2016;37:3969–3978. doi: 10.1007/s13277-015-4189-1. [DOI] [PubMed] [Google Scholar]
- 30.Seto E, Yoshida M. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6:a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terui Y. The epigenetic alteration and the effect of HDAC inhibitors in multiple myeloma. Nihon Rinsho. 2015;73:124–129. (In Japanese) [PubMed] [Google Scholar]
- 32.Srinivas C, Swathi V, Priyanka C, Anjana Devi T, Subba Reddy BV, Janaki Ramaiah M, Bhadra U, Bhadra MP. Novel SAHA analogues inhibit HDACs, induce apoptosis and modulate the expression of microRNAs in hepatocellular carcinoma. Apoptosis. 2016;21:1249–1264. doi: 10.1007/s10495-016-1278-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.