Abstract
Numerous studies have demonstrated that PABPC1 participates in the process of carcinogenesis and its function is inconsistent in different types of cancers. PABPC1-like (PABPC1L) is an important paralog of PABPC1 and few studies are available on the roles of PABPC1L in colorectal cancer (CRC) development. Hence, we explored the biological function and prognostic impact of PABPC1L in CRC. The mRNA expression of PABPC1L in CRC was determined based on the data obtained from The Cancer Genome Atlas (TCGA) database. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) was utilized to determine the PABPC1L mRNA expression level in CRC HT-29 and LS-174T cell lines. Kaplan-Meier method and Cox proportional-hazards model were utilized to conduct the survival and prognosis analyses. HT-29 cells with silenced PABPC1L were constructed to explore the effect of PABPC1L on cell proliferation, invasion and migration capacities using cell counting kit-8 (CCK-8), clone formation, wound-healing and Transwell assays, respectively. To uncover the potential mechanisms of how PABPC1L influences CRC proliferation and migration, we analyzed the expression of AKT, p-AKT, PI3K, and p-PI3K in HT-29 cells using western blotting. Our results revealed that PABPC1L was overexpressed in CRC tissues compared with normal tissues based on the data obtained from TCGA database. Similarly, the mRNA expression of PABPC1L was higher in HT-29 and LS-174T cells than that in CCD-18Co cells. The expression of PABPC1L in CRC was found to be significantly related to age, pathologic stage, pathologic-node, pathologic-metastasis, and death. In univariate and multivariate analyses, pathologic-tumor and pathologic-metastasis were identified as independent prognostic factors for CRC. After PABPC1L depletion, cell proliferation rate, colony numbers, and the invasive and migratory capacity of HT-29 cells were all reduced. Western blot analysis showed that reduction of PABPC1L significantly inhibited p-AKT, and p-PI3K expression level in HT-29 cells. Collectively, our results suggested that PABPC1L is a potential novel candidate oncogene in CRC, and targeting PABPC1L may provide clinical utility in CRC.
Keywords: colorectal cancer, cell proliferation, migration, PABPC1L
Introduction
Colorectal cancer (CRC) is regarded as the third leading culprit of cancer-related death (1). Globally, million new CRC cases are diagnosed per year, with ~21% proving fatal (2). Although there were advances in diagnostic and therapeutic techniques, the prognosis of CRC cases with metastases remains poor (3). Moreover, the molecular underpinning of CRC remains unclear. Therefore, an in-depth understanding of pathological mechanisms of CRC will be beneficial to development of novel therapeutic strategies.
Polyadenylate binding proteins (PABPs), the proteins interacting with single-stranded poly(A) by RNA recognition motif, are divided into PABPN1 in the nucleus and PABPC in the cytoplasm. PABPCs exert important functions in translation, mRNA decay, and controlling the rate of mRNA deadenylation (4). PABPC1, belonging to a family of PABPCs, is expressed in most eukaryotes (5). Numerous studies have reported that PABPC1 participates in the process of carcinogenesis. For example, PABPC1 was indispensable for translation beginning via interplay with EIF4G1, as a protein necessary for the formation of emboli in breast cancer (6). Another study revealed that PABPC1 acts as an oncogene in gastric cancer and promotes growth of gastric cancer cells (7). Eisermann et al (8) have demonstrated that PABPC1 is upregulated in prostate cancer tissues and this upregulation is associated with increased disease recurrence. However, downregulated PABPC1 was linked to tumor progression and worse prognosis in esophageal cancer (9). Accordingly, the role of PABPC1 in different types of cancers is inconsistent. PABPC1-like (PABPC1L) is an important paralog of PABPC1, which regulates and stabilizes the mRNA translation. Significantly, few studies have been done to investigate the roles of PABPC1L in CRC tissues and its relationship with the clinicopathological factors.
To explore the association between PABPC1L expression and the clinicopathological features, and prognosis of CRC patients, we conducted the corresponding analysis based on the The Cancer Genome Atlas (TCGA) data. To confirm our results in silico analysis, we utilized HT-29 cells to explore the influences of PABPC1L on CRC cell viability, invasion and migration in vitro. Moreover, we revealed the function mechanism of PABPC1L on modulating migration and proliferation in CRC. Our findings provide evidence that PABPC1L is upregulated in CRC and facilitates CRC proliferation and migration by regulating AKT signaling pathway.
Materials and methods
Data mining from the TCGA database
Based on the TCGA database (https://tcga-data.nci.nih.gov/tcga), PABPC1L mRNA expression level in normal tissues (n=41) and CRC samples (n=480) was analyzed using the limma package (10). The data were downloaded January 1, 2018.
Cell culture and small interfering RNA (siRNA)
Two human CRC cell lines (HT-29, and LS-174T) and a normal fibroblast cell line derived from human colon (CCD-18Co) were purchased from the Chinese Academy of Sciences Cell Bank (Shanghai, China) and cultivated at 37°C in 5% CO2 in RPMI-1640 medium containing 10% fetal bovine serum (FBS; HyClone, Logan, UT, USA), 100 U/ml penicillin, and 0.1 mg/ml streptomycin.
Cells were inoculated in 6-well plates (3×105/well) overnight, and siRNA against PABPC1L (si-PABPC1L) and one negative control (si-NC) were transfected into cells using Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The siRNA sequences were: 5′-CUUUGAGAUGCUCAAAGGC-3′ (sense) for siRNA PABPC1L 1# and 5′-CAUCUUCAUCAAGAACCUG-3′ (sense) for siRNA PABPC1L 2#. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and western blotting were utilized to measure the interfering efficiency 72 h following transfection, and silencing efficacy of >90% was selected for further assays.
RT-qPCR. HT-29, LS-174T and CCD-18Co cells were added in 6-well plates and total RNA from the cells was extracted by means of TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA was then reverse transcribed into cDNA using a HiFiScript cDNA Synthesis kit (CwBio, Beijing, China) on the basis of the manufacturer's description. Next, qPCR was implemented to quantify the level of PABPC1L and GAPDH. The primers used for qPCR were the following: PABPC1L forward, 5′-TCTCCAGAATCCCTCCATGC-3′ and reverse, 5′-AGTTAGAGTTTGCAGGGCCT-3′; GAPDH forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3′. The procedure for qPCR was 95°C for 30 sec, then for 40 cycles of 95°C for 5 sec, 60°C for 34 sec. All reactions were repeated three times. Relative gene expression of PABPC1L was determined through the 2−ΔΔCq method (11), and GAPDH was utilized as the reference gene.
Cell proliferation assay
The transfected cells were added to 96-well plates (2,000 cells/well). In line with the manufacturer's instructions of cell counting kit-8 (CCK-8), cell viability was tested at 24, 48, 72 and 96 h time-points. Subsequently, the absorbance at 450 nm were detected under a microplate reader (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The assays represent three independent experiments in triplicate.
Colony formation assay
Colony formation was conducted as described previously (12,13). Cells were placed in 6-well plate 48 h after transfection and incubated at 37°C, 5% CO2. On the 7th day, the medium was removed and the cells were washed twice using PBS. After that, colonies were fixed in 4% paraformaldehyde and stained using 0.1% crystal violet. Finally, colony numbers were analyzed using a light microscope (Olympus, Tokyo, Japan).
Wound-healing assay
Cells were added in 6-well plates. When the cells were confluent, the cultures were pre-treated with serum-free media for 12 h. Afterwards, the monolayers were scratched using a sterile micropipette tip size 10 µl. Baseline measurements were recorded at 0 h. At 24 h, the migration distance was measured. Three replicates were obtained from three independent experiments.
Cell migration and invasion assays
Cells after 24 h transfection were collected. In migration experiment, 5,000 cells were added to the upper chamber of Transwell inserts (Millipore, Bedford, MA, USA). Conditioned medium was added to the lower chamber. After incubating for 24 h, the migrated cells were fixed with 4% paraformaldehyde, stained using 0.1% crystal violet, and pictured using a light microscope. Cells were counted in five random fields.
For invasion, 10,000 cells were placed in the upper chamber with a Matrigel-coated membrane (BD Biosciences, Franklin Lakes, NJ, USA) in 500 µl serum-free RPMI-1640 accompanied by 500 µl 10% FBS/RPMI-1640 in the bottom chamber. The invasive ability was evaluated the same as in migration assay.
Western blot assay
Cells were lysed by RIPA lysate (plus protease inhibitor) (Beijing ComWin Biotech Co., Ltd., Beijing, China) to extract the total protein and protein concentration was determined by a BCA method. Cell lysates dissolved in SDS buffer were segregated using 10% SDS-PAGE (20 µg protein were loaded for each lane) and electro-transferred to a PVDF membrane. The membrane was blocked by 5% skimmed milk for 1 h and probed with specific primary antibodies [PABPC1L (dilution 1:1,000; cat. no. ab233280), PI3K (dilution 1:1,000; cat. no. ab232997) and p-PI3K (dilution 1:1,000, cat. no. ab182651) (all from Abcam, Cambridge, MA, USA), and AKT (dilution 1:1,000; cat. no. 4691), p-AKT (Ser473) (dilution 1:1,000; cat. no. 4060) and GAPDH (dilution 1:5,000; cat. no. 5174) (all from Cell Signaling Technology, Inc., Danvers, MA, USA)] overnight at 4°C. Then, the membrane was incubated with the HRP-conjugated goat anti-rabbit IgG mAb (dilution 1:5,000; PV-6001; ZSGB-BIO, Beijing, China) at room temperature for 1 h. Then, the signals were detected using an Immobilon™ western chemiluminescent HRP substrate (Millipore) and analyzed by a gel imaging analysis system (Bio-Rad Laboratories, Inc.). Detailed procedures were described previously (14).
Statistical analysis
In the present study, SPSS 22.0 software (IBM Corp., Armonk, NY, USA) was used to perform all statistical analyses. A Chi-square test was conducted to compare the difference between two groups, and one-way ANOVA model followed by a Dunnett's post hoc test was utilized to estimate the parameters of multi-groups. All the data were shown as mean ± standard deviation (SD). CRC samples were divided into high-expression or low-expression groups based on the median of PABPC1L mRNA expression level. Kaplan-Meier was applied to measure overall survival (OS), and log-rank test was conducted to determine the survival difference based on PABPC1L expression level. Cox proportional hazards regression analysis was conducted to estimate the prognostic features. Each variable with statistical difference in the univariate analysis was fitted into multivariate analysis to assess the independent prognostic values on CRC. P<0.05 was considered to indicate a statistically significant difference.
Results
PABPC1L is overexpressed predominantly in CRC tissues and cell lines
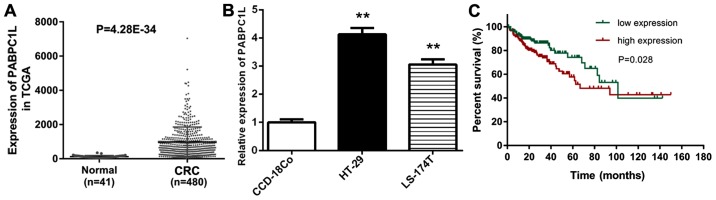
With an attempt to understand the relationship between PABPC1L and CRC, we analyzed the mRNA expression level of PABPC1L in CRC samples based on the data obtained from the TCGA database. The results demonstrated that PABPC1L was remarkably overexpressed in CRC tissues compared to normal samples (Fig. 1A, P<0.0001).
Figure 1.
PABPC1L level in CRC is remarkably increased, and high-expression of PABPC1L is remarkably related to worse prognosis. (A) PABPC1L expression level between the patients with CRC (n=480) and normal samples (n=41) retrieved from TCGA database. (B) Relative expression levels of PABPC1L in CRC cell lines (HT-29 and LS-174T) compared to a normal fibroblast cell line derived from human colon (CCD-18Co). The mRNA expression of PABPC1L in CRC cells was determined using RT-qPCR and normalized to GAPDH. Data are shown as the means ± SD. **P<0.001 vs. CCD-18Co group. (C) High PABPC1L expression level predicted poor survival based on Kaplan-Meier overall survival (OS) curves. PABPC1L, polyadenylate binding protein C1L; CRC, colorectal cancer.
Subsequently, we evaluated the expression level of PABPC1L in CCD-18Co, HT-29 and LS-174T cell lines (Fig. 1B). Both HT-29 and LS-174T cell lines were epithelial cells isolated from colorectal adenocarcinoma tissues. HT-29 cell line is positive for c-myc, K-ras, H-ras, N-ras, Myb, sis and fos. p53 antigen is overexpressed in HT-29 cell line. LS 174T cell line is positive for expression of c-myc, N-myc, H-ras, N-ras, Myb and fos. This cell line is negative for p53 antigen expression, but positive for mRNA expression. The culture property of these two cell lines was: adherent. Conforming to the condition in CRC tissues obtained from TCGA database, the expression level of PABPC1L in HT-29 and LS-174T cells was dramatically higher than that in the normal colon cells (P<0.001). Moreover, PABPC1L expression level was increased in HT-29 cells compared to LS-174T cells, thus, HT-29 cells were used for subsequent analysis in the present study.
Relationship of PABPC1L expression level with the clinicopathological parameters in CRC patients
Next, we classified the 380 CRC patients with integrated clinical data into two groups (high-expression or low-expression group) based on the median of PABPC1L expression level. Then, the clinical parameters were compared (age, sex, pathologic stage, pathologic-tumor, pathologic-node, pathologic-metastasis, and death) between the two groups (Table I). It was found that PABPC1L expression level was obviously associated with age (P=0.006), pathologic stage (P=0.002), pathologic-node (P=0.016), pathologic-metastasis (P=0.001), and death (P=0.025). Nevertheless, the PABPC1L expression level was not related to other clinical factors including sex (P=0.837), nor pathologic-tumor (P=0.796).
Table I.
Clinical association between PABPC1L expression and clinicopathological variables in CRC patients.
| Expression of PABPC1L | |||
|---|---|---|---|
| Characteristics | Low | High | P-value |
| Age (years) | 0.006a | ||
| <60 | 40 | 64 | |
| ≥60 | 150 | 126 | |
| Sex | 0.837 | ||
| Female | 90 | 88 | |
| Male | 100 | 102 | |
| Pathologic-stage | 0.002a | ||
| I+II | 123 | 93 | |
| III+IV | 67 | 97 | |
| Pathologic-T | 0.796 | ||
| T1+T2 | 36 | 38 | |
| T3+T4 | 154 | 152 | |
| Pathologic-N | 0.016a | ||
| N0 | 124 | 101 | |
| N1+N2 | 66 | 89 | |
| Pathologic-M | 0.001a | ||
| M0 | 171 | 148 | |
| M1 | 19 | 42 | |
| Death | 0.025a | ||
| Yes | 32 | 50 | |
| No | 158 | 140 | |
P<0.05. CRC, colorectal cancer; PABPC1L, polyadenylate binding protein C1L
PABPC1L predicts poor prognosis for CRC patients
We conducted Kaplan-Meier and log-rank test to compare the survival duration differences between the low- and high-expression groups. Fig. 1C shows that high PABPC1L expression was correlated with poor prognosis (P=0.028). PABPC1L was an important prognostic predictor of poor survival in human CRC.
Subsequently, univariate and multivariate model was used to prove the prognostic role of PABPC1L in CRC patients (Table II). In univariate analysis, PABPC1L expression (P=0.029), pathologic stage (P=0.001), pathologic-tumor (P=0.003), pathologic-node (P=0.001), as well as pathologic-metastasis (P=0.001) were significantly connected with shorter OS in CRC patients. In multivariate analysis, pathologic-tumor (P=0.029), and pathologic-metastasis (P=0.001) were predictive of OS. Multivariate analysis indicated that pathologic-tumor and pathologic-metastasis were independent prognostic factors for CRC.
Table II.
Univariate and multivariate analysis of clinical prognostic factors of CRC.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| Variables | P-value | HR | 95% CI | P-value | HR | 95% CI |
| PABPC1L expression (high/low) | 0.029a | 1.639 | 1.051–2.556 | 0.402 | 1.217 | 0.769–1.928 |
| Age (years) | 0.304 | 1.316 | 0.779–2.221 | |||
| Sex (female/male) | 0.960 | 0.989 | 0.640–1.527 | |||
| Pathologic-Stage (I+II/III+IV) | 0.001a | 3.271 | 2.053–5.212 | 0.076 | 2.768 | 0.898–8.530 |
| Pathologic-T (T1+T2/T3+T4) | 0.003a | 8.352 | 2.049–34.050 | 0.029a | 4.926 | 1.180–20.568 |
| Pathologic-M (M0/M1) | 0.001a | 4.699 | 2.979–7.410 | 0.001a | 2.577 | 1.469–4.522 |
| Pathologic-N (N0/N1+N2) | 0.001a | 2.627 | 1.680–4.107 | 0.334 | 0.620 | 0.235–1.636 |
P<0.05. PABPC1L, polyadenylate binding protein C1L; CRC, colorectal cancer
Knockdown of PABPC1L decreases cell proliferation
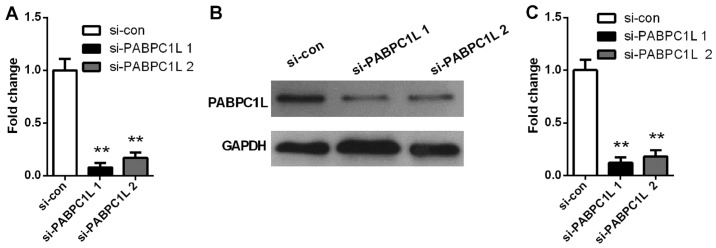
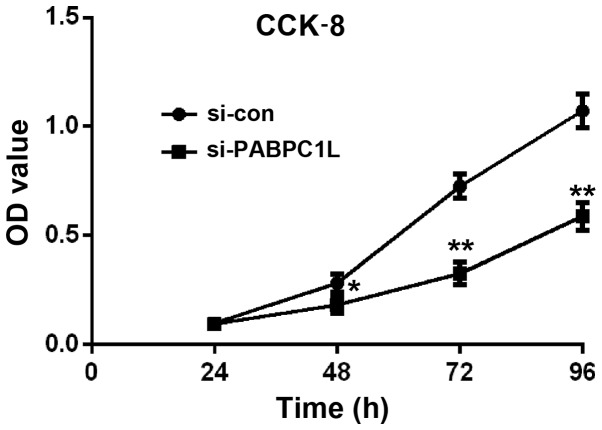
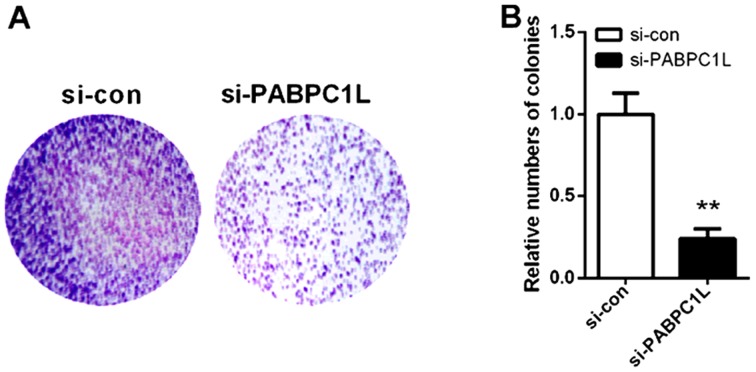
On the basis of the above findings, we inferred that PABPC1L may be involved in CRC development. To evaluate the role of PABPC1L in CRC growth, we firstly knocked down PABPC1L in HT-29 cells by siRNA. Fig. 2 shows that si-PABPC1L transfected cells had a significant decrease both in mRNA and protein level of PABPC1L, when comparing with those in non-transfected cells (P<0.001 both in RT-PCR and western blotting). Moreover, si-PABPC1L 1# transfected cells possessed higher silence efficiency relative to si-PABPC1L 2#, thus, we selected si-PABPC1L 1# for subsequent experiments. Then, the effects were investigated on cell proliferation using CCK-8 assay. The results revealed that PABPC1L silence suppressed cell growth in HT-29 cells relative to negative control (Fig. 3, P<0.05 for 48 h; P<0.001 for 72 and 96 h). Moreover, it was found that PABPC1L silencing HT-29 cells possessed reduced colony-formation capacity (Fig. 4, P<0.001). These results implicated that PABPC1L high-expression promoted cell growth.
Figure 2.
RT-qPCR and western blotting of PABPC1L expression levels in HT-29 cells infected with siRNA. (A) The expression of PABPC1L in HT-29 cells infected with siRNA detected by RT-qPCR. (B) Western blot analysis of the PABPC1L protein expression. (C) Quantification of PABPC1L protein expression detected by western blotting. Data are presented as the means ± SD. n=6, **P<0.001 vs. si-con group. PABPC1L, polyadenylate binding protein C1L.
Figure 3.
CCK-8 assay was performed to determine cell proliferation of HT-29 cells after 24, 48, 72 and 96 h infected with siRNA. Data are presented as the means ± SD. n=6, **P<0.001 and *P<0.05 vs. si-con group. CCK-8, cell counting kit-8.
Figure 4.
Colony formation assay implemented to measure the proliferation rate of HT-29 cells infected with siRNA. (A) Depletion of PABPC1L reduced colony formation of HT-29 cells. (B) The relative number of colonies was lower in si-PABPC1L group than that of si-con group. Colony formation data are presented as histograms. Data are presented as the means ± SD. n=6, **P<0.001 vs. si-con group. PABPC1L, polyadenylate binding protein C1L.
PABPC1L depletion inhibits the invasion and migration in HT-29 cells
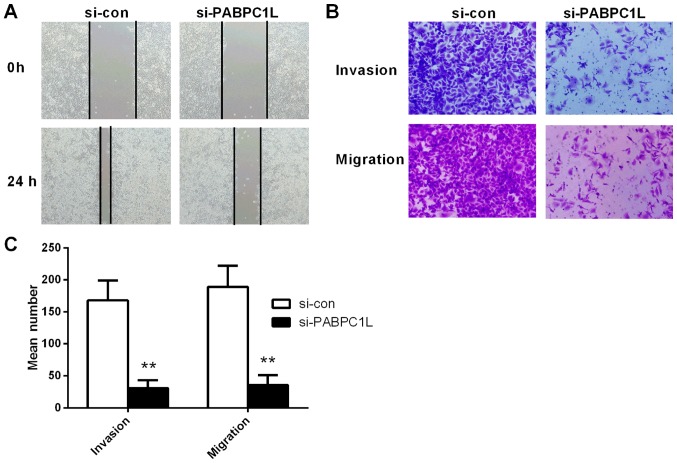
The wound-healing assays exhibited that PABPC1L depletion remarkably suppressed HT-29 cell migration (Fig. 5A), which was quantified based on the percentage of the wound area (Fig. 5A).
Figure 5.
PABPC1L silencing reduces the invasion and migration of HT-29 cells. (A) Migration ability of HT-29 cells with or without PABPC1L depletion determined using wound healing assay. (B and C) Transwell assay used to examine the invasion and migration capacity of HT-29 cells with or without PABPC1L knockdown. Data are presented as the means ± SD. n=6, **P<0.001 vs. si-con group. PABPC1L, polyadenylate binding protein C1L.
Results displayed that HT-29 cells after transfection had significantly smaller populations of invasive and migrant cells, relative to the negative controls (Fig. 5B and C). In summary, it was demonstrated that PABPC1L regulated HT-29 cell mobility in vitro.
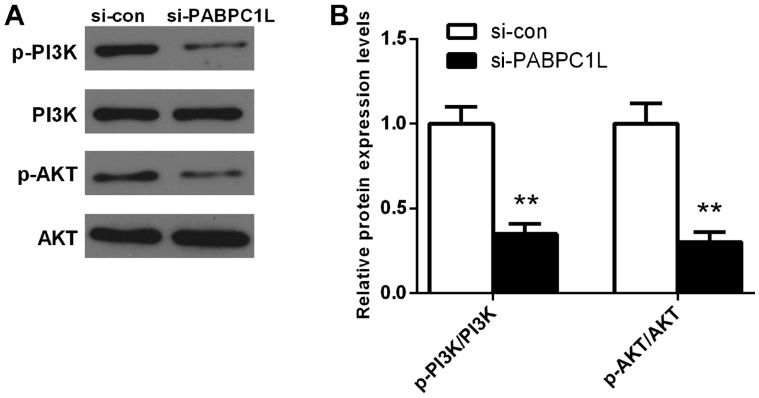
PABPC1L knockdown inhibits PI3K/AKT pathway activation in HT-29 cells
To uncover the potential mechanisms of how PABPC1L influences CRC proliferation and migration, we analyzed AKT, p-AKT, PI3K and p-PI3K protein expression levels in HT-29 cells using western blotting. Fig. 6 shows that PABPC1L knockdown significantly reduced p-AKT, and p-PI3K protein expression levels in HT-29 cells (P<0.001). However, no significant changes of AKT and PI3K protein levels were observed after silencing PABPC1L. Taken together, this result indicated that PABPC1L mediated CRC growth and migration via regulating the PI3K/AKT signaling pathway.
Figure 6.
The protein levels of AKT, p-AKT, PI3K and p-PI3K in CRC cell line (HT-29) after transfection determined using western blotting. (A) Western blot analysis of AKT, p-AKT, PI3K and p-PI3K expression in HT-29 cells. (B) Quantification of the protein expression ratios of p-PI3K/PI3K and p-AKT/AKT detected by western blotting. Data are presented as the means ± SD. n=6, **P<0.001 vs. si-con group. CRC, colorectal cancer.
Discussion
To date, few studies have investigated the role of PABPC1L in CRC. To study the expression of PABPC1L in CRC, in silico analysis was implemented using TCGA and the results implied that PABPC1L was overexpressed in CRC specimens compared with normal controls. We then utilized siRNA transfection to inhibit the expression of PABPC1L in HT-29 cells. We found that, PABPC1L mRNA and protein levels were significantly decreased after transfection. For the purpose of validating the effect of PABPC1L, CCK-8 and Transwell assays were applied to determine the proliferative, invasive and migrative abilities of HT-29 cells, wherein we observed that the proliferative, invasive and migrative abilities of HT-29 cells were significantly reduced after transfection. Our data demonstrated that PABPC1L may directly affect invasion and migration of cancer cells and is possibly a potential biomarker for early diagnosis of CRC.
PI3K/AKT plays important roles in the migratory and survival signaling pathway (15,16). Moreover, the activation of PI3K sparks a set of incidents resulting in the activation of AKT and mTOR (17), thereby inducing the expression of many target genes that mediate cell proliferation, differentiation as well as apoptosis (18,19). The hyperactivation of PI3K/AKT signaling pathway has been found in many kinds of tumors, including colon cancer (20–22). Additionally, the hyperactivation of this pathway was suggested to be correlated with a poor prognosis in colon cancer (23). Of note, blocking PI3K/AKT activity in colon cancer cells presented promising anti-cancer effects (24). In colon cancer, the mutations in PABPC1 have been found in minor tumor clones (25). The results of our study suggested that HT-29 cell migration and invasion were inhibited by PABPC1L silencing. These data demonstrate that PABPC1L exerts key functions in the progression of CRC cells. In addition, a positive relationship between PI3K/AKT expression and PABPC1L level was observed. Our findings verified that PABPC1L restrained CRC cell motility via downregulating the expression of PI3K/AKT.
Taken together, PABPC1L was upregulated in CRC, and dysrelated expression of PABPC1L could alter diverse biological processes of CRC cells, including proliferation, migration, and invasion, probably via regulating PI3K-AKT signaling pathway. Our data suggest that upregulation of PABPC1L was not only significantly connected with aggressive clinicopathological factors, but also was an independent poor prognostic indicator predicting worse OS in CRC patients. Thus, we infer that PABPC1L may be a diagnostic and prognostic biomaker in CRC and may be a novel target for developing a preventive agent for CRC prognosis. However, our results have not been veried using an animal model or an independent cohort of patients. Future studies are anticipated to explore the molecular mechanisms of the altered expression of PABPC1L in CRC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
YQW and RGW designed the study and were involved in data collection. YQW, CLJ and BJW carried out the experiments. YQW and CLJ performed the statistical analysis and prepared figures. YQW drafted the paper. RGW contributed substantially to its revision. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Ma J, Zou Z, Jemal A. Cancer Statistics, 2014. CA Cancer J Clin. 2014;64:10. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:11–30. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Weitz J, Koch M, Debus J, Höhler T, Galle PR, Büchler MW. Colorectal cancer. Lancet. 2005;365:153–165. doi: 10.1016/S0140-6736(05)17706-X. [DOI] [PubMed] [Google Scholar]
- 4.Villalba A, Coll O, Gebauer F. Cytoplasmic polyadenylation and translational control. Curr Opin Genet Dev. 2011;21:452–457. doi: 10.1016/j.gde.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 5.Grange T, de Sa CM, Oddos J, Pictet R. Human mRNA polyadenylate binding protein: Evolutionary conservation of a nucleic acid binding motif. Nucleic Acids Res. 1987;15:4771–4787. doi: 10.1093/nar/15.12.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silvera D, Arju R, Darvishian F, Levine PH, Zolfaghari L, Goldberg J, Hochman T, Formenti SC, Schneider RJ. Essential role for eIF4GI overexpression in the pathogenesis of inflammatory breast cancer. Nat Cell Biol. 2009;11:903–908. doi: 10.1038/ncb1900. [DOI] [PubMed] [Google Scholar]
- 7.Zhu J, Ding H, Wang X, Lu Q. PABPC1 exerts carcinogenesis in gastric carcinoma by targeting miR-34c. Int J Clin Exp Pathol. 2015;8:3794–3802. [PMC free article] [PubMed] [Google Scholar]
- 8.Eisermann K, Dar JA, Dong J, Wang D, Masoodi KZ, Wang Z. Poly (A) binding protein cytoplasmic 1 is a novel co-regulator of the androgen receptor. PLoS One. 2015;10:e0128495. doi: 10.1371/journal.pone.0128495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takashima N, Ishiguro H, Kuwabara Y, Kimura M, Haruki N, Ando T, Kurehara H, Sugito N, Mori R, Fujii Y. Expression and prognostic roles of PABPC1 in esophageal cancer: Correlation with tumor progression and postoperative survival. Oncol Rep. 2006;15:667–671. [PubMed] [Google Scholar]
- 10.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 12.Li L, Wang L, Song P, Geng X, Liang X, Zhou M, Wang Y, Chen C, Jia J, Zeng J. Critical role of histone demethylase RBP2 in human gastric cancer angiogenesis. Mol Cancer. 2014;13:81. doi: 10.1186/1476-4598-13-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang CG, Lv L, Liu FR, Wang ZN, Liu FN, Li YS, Wang CY, Zhang HY, Sun Z, Xu HM. Downregulation of connective tissue growth factor inhibits the growth and invasion of gastric cancer cells and attenuates peritoneal dissemination. Mol Cancer. 2011;10:122. doi: 10.1186/1476-4598-10-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tian Y, Pan Q, Shang Y, Zhu R, Ye J, Liu Y, Zhong X, Li S, He Y, Chen L, et al. MicroRNA-200 (miR-200) cluster regulation by achaete scute-like 2 (Ascl2): Impact on the epithelial-mesenchymal transition in colon cancer cells. J Biol Chem. 2014;289:36101–36115. doi: 10.1074/jbc.M114.598383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Assinder SJ, Dong Q, Kovacevic Z, Richardson DR. The TGF-beta, PI3K/Akt and PTEN pathways: Established and proposed biochemical integration in prostate cancer. Biochem J. 2009;417:411–421. doi: 10.1042/BJ20081610. [DOI] [PubMed] [Google Scholar]
- 16.Osaki M, Oshimura M, Ito H. PI3K-Akt pathway: Its functions and alterations in human cancer. Apoptosis. 2004;9:667–676. doi: 10.1023/B:APPT.0000045801.15585.dd. [DOI] [PubMed] [Google Scholar]
- 17.Kim SM, Park JH, Kim KD, Nam D, Shim BS, Kim SH, Ahn KS, Choi SH, Ahn KS. Brassinin induces apoptosis in PC-3 human prostate cancer cells through the suppression of PI3K/Akt/mTOR/S6K1 signaling cascades. Phytother Res. 2014;28:423–431. doi: 10.1002/ptr.5010. [DOI] [PubMed] [Google Scholar]
- 18.Liu YZ, Wu K, Huang J, Liu Y, Wang X, Meng ZJ, Yuan SX, Wang DX, Luo JY, Zuo GW, et al. The PTEN/PI3K/Akt and Wnt/β-catenin signaling pathways are involved in the inhibitory effect of resveratrol on human colon cancer cell proliferation. Int J Oncol. 2014;45:104–112. doi: 10.3892/ijo.2014.2392. [DOI] [PubMed] [Google Scholar]
- 19.Jiang QG, Li TY, Liu DN, Zhang HT. PI3K/Akt pathway involving into apoptosis and invasion in human colon cancer cells LoVo. Mol Biol Rep. 2014;41:3359–3367. doi: 10.1007/s11033-014-3198-2. [DOI] [PubMed] [Google Scholar]
- 20.Fruman DA, Rommel C. PI3K and cancer: Lessons, challenges and opportunities. Nat Rev Drug Discov. 2014;13:140–156. doi: 10.1038/nrd4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Josse C, Bouznad N, Geurts P, Irrthum A, Huynh-Thu VA, Servais L, Hego A, Delvenne P, Bours V, Oury C. Identification of a microRNA landscape targeting the PI3K/Akt signaling pathway in inflammation-induced colorectal carcinogenesis. Am J Physiol Gastrointest Liver Physiol. 2014;306:G229–G243. doi: 10.1152/ajpgi.00484.2012. [DOI] [PubMed] [Google Scholar]
- 22.Xiao ZM, Wang XY, Wang AM. Periostin induces chemoresistance in colon cancer cells through activation of the PI3K/Akt/survivin pathway. Biotechnol Appl Biochem. 2015;62:401–406. doi: 10.1002/bab.1193. [DOI] [PubMed] [Google Scholar]
- 23.Malinowsky K, Nitsche U, Janssen KP, Bader FG, Späth C, Drecoll E, Keller G, Höfler H, Slotta-Huspenina J, Becker KF. Activation of the PI3K/AKT pathway correlates with prognosis in stage II colon cancer. Br J Cancer. 2014;110:2081–2089. doi: 10.1038/bjc.2014.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nuvoli B, Santoro R, Catalani S, Battistelli S, Benedetti S, Canestrari F, Galati R. CELLFOOD™ induces apoptosis in human mesothelioma and colorectal cancer cells by modulating p53, c-myc and pAkt signaling pathways. J Exp Clin Cancer Res. 2014;33:24. doi: 10.1186/1756-9966-33-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu C, Yu J, Yao X, Wu WK, Lu Y, Tang S, Li X, Bao L, Li X, Hou Y, et al. Discovery of biclonal origin and a novel oncogene SLC12A5 in colon cancer by single-cell sequencing. Cell Res. 2014;24:701–712. doi: 10.1038/cr.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author on reasonable request.