Abstract
Circular RNAs (circRNAs) are a specific form of non-coding RNAs, that serve a pivotal role in the development of human diseases, including Alzheimer's disease and cancer; however, only a few are known with respect to cancer. The present study identified a novel circRNA, circ lysophosphatidic acid receptor 1 (LPAR1) (hsa_circ_0087960), derived from two exons 226 base pairs in length, in muscle-invasive bladder cancer (MIBC) tissues. circLPAR1 was identified to be lowly expressed in MIBC tissues in a cohort of 125 cases, and predicted a poor disease-specific survival time, compared with patients with high circLPAR1 expression (52.4 vs. 56.0 months; P=0.001) by univariate and multivariate Cox regression analyses. Matrigel and wound healing assays also demonstrated that the invasion of 5637 and T24 bladder cancer cells were significantly enhanced following the knockdown of circLPAR1 by small interfering RNA (si-circLPAR1-1 in T24 cell line, P=0.01; si-circLPAR1-2 in 5637 cell line, P=0.003; si-circLPAR1-2 in T24 cell line, P=0.002; si-circLPAR1-2 in 5637 cell line, P=0.006). The bioinformatics analysis indicated that circLPAR1 may harbor specific microRNAs (miRNAs) according to the miRNAs seed sequence matching. A luciferase reporter assay revealed that miR-762 can inhibit the activity of the transfected luciferase gene when inserted in a circLPAR1 wild-type fragment, and this inhibition could be alleviated when the luciferase gene was inserted in a circLPAR1 fragment with the mutated miR-762 target site. In conclusion, the circLPAR1 may function as a potential novel and stable biomarker for the prognosis of MIBC and may be associated with the invasion and metastasis by miR-762.
Keywords: biomarker, bladder cancer, non-coding RNA, circular RNA, microRNA-762
Introduction
Bladder cancer was diagnosed in ~74,000 patients in the USA and in >430,000 patients worldwide, which makes it the fourth most common cancer in males and the 11th most common cancer in females in the USA during 2015 (1). Due to its invasive nature, muscle-invasive bladder cancer (MIBC) has a high rate of mortality with a poor quality of life and an overall 5-year survival rate of 35–60% (2). The burden of cancer in non-MIBC (NMIBC) patients is primarily caused by subsequent adjuvant therapy, including intravesical chemotherapy; and the progression risk and high recurrence rate ranged from 50–90% with a 5-year survival rate of 70–80% worldwide in 2018 (3). Owing to the high recurrence and heterogeneity of this disease, the characterization of the molecular profile of tumors and investigating putative biomarkers is imperative. A number of molecules, including microRNA (miRNA), long chain non-coding RNAs and other RNAs, have been demonstrated to be associated with the occurrence and development of bladder cancer, and the underlying molecular mechanisms and prognostic features have been analyzed extensively (4).
Circular RNAs (circRNAs) have been recently determined as a novel type of non-coding RNAs (5), and the enigmatic structure, origin, and classification have been elucidated (6). Previous studies also demonstrated that circRNAs are involved in the development of a number of types of diseases, including breast cancer and liver cancer (7,8). However, only a few articles have been reported, particularly with respect to bladder cancer (9,10).
In the present study, the differences in the expression of a valuable circRNA, circ lysophosphatidic acid receptor 1 (LPAR1) (hsa_circ_0087960), were detected, and its prognosis value in 125 patients with MIBC was evaluated. Additionally, its potential function and the underlying molecular mechanism were investigated.
Materials and methods
Patients and clinical specimens
A total of 125 bladder cancer tissues and 68 paired cancer tissue and adjacent non-tumorous tissues were obtained by radical cystectomy at Fudan University Cancer Center (Shanghai, China) between December 2006 and December 2015. Among them, the patients corresponding to the tissues used for disease-specific survival (DSS) analysis were treated with radical bladder radical resection between January 2007 and June 2012. All tissue specimens were preserved in RNA Later Stabilization Solution (Thermo Fisher Scientific, Inc., Waltham, MA, USA) at −80°C following surgical resection. Patients were excluded from the present study if they had received prior radiotherapy, systemic venous chemotherapy or immune therapy. The general clinical characteristics of the 125 participants were summarized in Table I.
Table I.
Comparison of patient clinicopathological characteristic for 125 patients with muscle-invasive bladder cancer.
| Characteristic | Low circLPAR1 level | High circLPAR1 level | P-valuea |
|---|---|---|---|
| Age, years | 0.126 | ||
| Median (range) | 62 (38–78) | 61 (25–74) | |
| Sex, n (%) | 0.465 | ||
| Male | 51 (48.1) | 55 (51.9) | |
| Female | 11 (57.9) | 8 (42.1) | |
| Pathological T stage, n (%) | 0.441 | ||
| T2 | 41 (47.1) | 46 (52.9) | |
| T3+T4 | 21 (55.3) | 17 (44.7) | |
| Pathological N stage, n (%) | 0.372 | ||
| N0 | 48 (47.5) | 53 (52.5) | |
| N1+N2 | 14 (58.3) | 10 (41.7) | |
| Pathological Grade, n (%) | 0.073 | ||
| Low | 10 (35.7) | 18 (64.3) | |
| High | 52 (53.6) | 45 (46.4) | |
| Maximum tumor diameter, cm | 0.318 | ||
| Median (range) | 4.0 (1.0–19.0) | 3.4 (0.2–18.0) | |
| Tumor number, n | 0.675 | ||
| Median (range) | 1.7 (1.0–5.0) | 1.5 (1.0–6.0) | |
| Recurrence after transurethral resection of bladder tumor, n (%) | 0.561 | ||
| Yes | 17 (44.7) | 21 (55.3) | |
| No | 45 (51.7) | 42 (48.3) | |
| Neoadjuvant chemotherapy prior to radical cystectomy, n (%) | 0.039b | ||
| Yes | 46 (56.8) | 35 (43.2) | |
| No | 16 (36.4) | 28 (63.6) | |
| Urinary diversion, n (%) | 0.086 | ||
| Continent reservoir | 40 (47.1) | 45 (52.9) | |
| Orthotopic neobladder | 15 (46.9) | 17 (53.1) | |
| Cutaneous ureterostomy | 7 (87.5) | 1 (12.5) |
P-value was calculated by Mann-Whitney U test for continuous variables and χ2 test for categorical variables.
P<0.05. circLPAR1, circular lysophosphatidic acid receptor 1.
The present study was conducted in accordance with the ethical standards of the Helsinki Declaration II and approved by the Ethics Committee of Fudan University Shanghai Cancer Center (approval no. 050432-4-1212B). Written informed consent was obtained from each patient prior to any study-specific experiments being performed.
RNA preparation
Total tissue RNA was isolated from the bladder cancer tissues and paired adjacent tissues using TRIzol® reagent (Thermo Fisher Scientific, Inc.) and was quantified by a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific, Inc.). The RNA integrity was assessed by 1.5% agarose gel electrophoresis. cDNA was synthesized using the PrimeScript RT Master mix (Takara Biotechnology Co., Ltd, Dalian, China) from 500 ng total RNA.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR was conducted and analyzed on a Mx3000P Quantitative PCR system Thermo Fisher Scientific, Inc.) using SYBR® Premix Ex Taq II (Takara Biotechnology, Co., Ltd.), according to the manufacturer's protocol. The divergent primers were designed to amplify specific circRNAs, based on previous literature (11). β-actin was used as an endogenous control. The primer sequences of the candidate circRNAs were listed as follows in the 5′ to 3′ direction: circLPAR1, forward (F), CGTGTTCACCACCTACAACCA, and reverse (R), ATGCTGTAGGTGTCAGTCCT; LPAR1 F, GCTGCCATCTCTACTTCCATC, and R, AAGCGGCGGTTGACATAGATT; and β-actin F, CATGTACGTTGCTATCCAGGC, and R, CTCCTTAATGTCACGCACGAT. The thermocycling conditions for RT-qPCR was as follows: 95°C for 1 min, 95°C for 5 sec, 40 cycles of 60°C for 30 sec, 95°C for 15 sec, 60°C for 1 min and 95°C 15 sec. Analysis of relative gene expression data was achieved using the 2−ΔΔCq method (11).
Sanger sequencing and RNase R digestion
The locations of the RT-qPCR amplified products were identified by 1.5% agarose gel electrophoresis at 150 V for 20 min. A total of 5 µl DNA sample was loaded per lane. Subsequently, these fragments were recovered from the gel and inserted into a pMD18 vector (Takara Biotechnology, Co., Ltd.). Following transformation and amplification, the plasmids were isolated and sequenced. Sanger sequencing was used to show the back-spliced events of candidate circRNAs. For RNase R digestion, 2 µg total RNA was incubated for 30 min at 37°C with 3 U/µg of RNase R (Epicenter Technologies, Madison, WI, USA), and the resulting product was subsequently purified using a RNeasy MinElute Cleaning kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer's protocol and previous literature (11). The visualization reagent gel loading dye purple was obtained from New England BioLabs, Inc., Ipswich, MA, USA.
Bioinformatics analysis
The predicted target miRNAs, including the conserved sites of the seed region binding to the circRNA, were analyzed by TargetScan 5.2 (http://www.targetscan.org/vert_72/) and miRanda 2.4 (http://www.microrna.org/) based on the miRBase (http:\www.mirbase.org) and The Cancer Genome Atlas network data of bladder cancer (11–13). Additionally, the Gene Ontology (GO; http:\www.geneontology.org) and Kyoto Encyclopedia of Genes and Genomes (KEGG; http:\www.genome.jpkegg) pathway annotation were also analyzed for circLPAR1 using the DAVID annotation tool (14), and these genes were categorized hierarchically. The Fisher's exact test was used to classify the enrichment of the pathway category. The false discovery rate algorithm was applied to adjust the P-values to the threshold of <0.05 (15). The National Centre for Biotechnology Information database (www.ncbi.nlm.nih.gov) was used to perform the literature review and TIGR (https://www.jcvi.org) was used for gene annotation.
Cell culture and treatments
The 5637 and T24 cells (American Type Culture Collection, Manassas, VA, USA)were cultured in RPMI-1640 (Hyclone; GE Healthcare Life Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone; GE Healthcare Life Sciences) and 1% penicillin/streptomycin (Hyclone; GE Healthcare Life Sciences) at 37°C and 5% CO2. For wound healing, Matrigel and Luciferase reporter assays 293 cells, obtained from the American Type Culture Collection (Manassas, VA, USA) were cultured in Dulbecco's modified Eagle's medium (Hyclone; GE Healthcare Life Sciences, with 10% FBS and 1% antibiotics. Transcription was blocked by the addition of 2 mg/ml Actinomycin D or dimethyl sulfoxide (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), which served as a control for the cell culture medium.
Vector construction
The PCR templates of circLPAR1 wide-type (WT) and mutation-type (MT) of the binding site with miR-762 was constructed by gene synthesis as follows: circLPAR1 WT (226 bp), 5′-CAGCAAACAAGAAAATTTGTCTCCCGTAGTTCTGGGGCGTGTTCACCACCTACAACCACAGAGCTGTCATGGCTGCCATCTCTACTTCCATCCCTGTAATTTCACAGCCCCAGGTGGACGTCTGATTTATGAAGCTCCCCATCCACCTATCTGAGTACCTGACTTCTCAGGACTGACACCTACAGCATCAGGTACACAGCTTCTCCTAGCATGACTTCGATCTGAT-3′; and circLPAR1 MT of the binding site with miR-762 (226 bp), 5′-CAGCAAACAAGAAAATTTGTCTCCCGTAGTTCTGGGGCGTGTTCACCACCTACAACCACAGAGCTGTCATGGCTGCCATCTCTACTTCCATCCCTGTAATTTCAGTCGGGCAGGTGGACGTCTGATTTATGAAGCTCCCCATCCACCTATCTGAGTACCTGACTTCTCAGGACTGACACCTACAGCATCAGGTACACAGCTTCTCCTAGCATGACTTCGATCTGAT-3′. The bold text represents the circLPAR1 mutation site, and the underlined text represents the back splice site.
The genomic region of circLPAR1-WT and MT was amplified by PrimerSTAR Max DNA Polymerase mix (Takara Biotechnology Co., Ltd.) and subcloned into a pcDNA3.0 vector, which was kindly provided by the Shenglin Huang lab (Fudan University Shanghai Cancer Center, Shanghai, China).
Oligonucleotide transfection
Oligonucleotides (si-circLPAR1, relative miRNA inhibitor and relative miRNA mimics) were synthesized by Guangzhou RiboBio Co., Ltd (Guangzhou, China). Cells at 70–80% confluency in 6-well plates were transfected with 3 µl oligonucleotides using 5 µl Lipofectamine® RNAiMax (Thermo Fisher Scientific, Inc.). The sequences of si-circLPAR1 are shown as follows (5′-3′): si-circPVT1-1, CAGGTGGACGTCTGATTTA; si-circPVT1-2, AGGTGGACGTCTGATTTAT; micrONTM hsa-miR-762 mimic, sense, GGGGCUGGGGCCGGGGCCGAGC, and antisense, GCUCGGCCCCGGCCCCAGCCCC; micrOFFTM hsa-miR-762 inhibitor, sense, GCUCGGCCCCGGCCCCAGCCCC; miRNA mimic negative control, sense, UUUGUACUACACAAAAGUACUG, and antisense, CAGUACUUUUGUGUAGUACAAA; miRNA inhibitor negative control, sense, CAGUACUUUUGUGUAGUACAAA. Subsequent experiments were performed 48 h after transfection.
Wound healing assay
The 5637 and T24 bladder cancer cells were seeded in 6-well culture plates at a density of 1×106 cells/well in RPMI-1640 (Hyclone; GE Healthcare Life Sciences) supplemented with 10% FBS (Hyclone; GE Healthcare Life Sciences) and 1% penicillin/streptomycin, and incubated for 12 h at 37°C. The cell monolayer was wounded by scratching a line with sterile 200 µl pipette tip. The detached cells were removed by washing the cell monolayer with PBS for 5 sec using a sterile rubber dropper. Subsequently, the mixtures of Lipofectamine RNAiMax and the si-circLPAR1 or negative control were added. The images were captured at 48 h using a digital camera.
Matrigel assay
si-circLPAR1-transfected 5637/T24 cells at a density of 2 ×105 cells/0.2 ml RPMI-1640 were seeded in the upper wells of Matrigel-precoated Transwell plates (Corning Costar Co., Lowell, CA, USA). Lower chambers comprised of 0.5 ml RPMI-1640 containing 10% FBS. At 48 h, the membranes were treated with 4% formaldehyde for 30 min at room temperature, followed by 2% crystal violet staining for 15 min at room temperature. Cells that invaded across the Transwell membrane were counted using a light microscope (magnification, ×40) in 10 randomly selected high-power fields.
Luciferase reporter assay
In the luciferase reporter assay, circLPAR1 was inserted into the region directly downstream of a cytomegalovirus promoter-driven firefly luciferase cassette in the pcDNA3.0 vector. Mutations in the miR-762 binding sites in the circLPAR1 sequence were inserted using a Mut Express II Fast Mutagenesis kit (Vazyme, Piscataway, NJ, USA) as described previously. The primers were listed as follows: Luc-cricLPAR1-F; AACGGCCGCCAGTGTGCTGGCAGCAAACAAGAAAATTTGTCTCCC and Luc-cricLPAR1-R; TAGAATAGGGCCCTCTAGAGATCAGATCGAAGTCATGCTAGGAG. 293 cells were seeded into 96-well plates at a density of 1×104 cells/well 24 h prior to transfection. The cells were then co-transfected using Lipofectamine® RNAiMax (Thermo Fisher Scientific, Inc.) with a mixture of 50 ng firefly luciferase reporter inserted in a circLPAR1 wide-type/mutation fragment, 5 ng Renilla luciferase reporter (from the Shenglin Huang lab, Fudan University Shanghai Cancer Center, Shanghai, China) and miRNA mimics/negative control (Guangzhou RiboBio Co., Ltd). At 48 h post-incubation, the firefly and Renilla luciferase activities were quantified using a dual-luciferase reporter assay (Promega Corporation, Madison, WI, USA). The relative luciferase activity was calculated for each miRNA relative to the negative control miRNA mimic.
Statistical analysis
In this retrospective study, Student's unpaired t-test for two groups or one-way analysis of variance followed by Tukey's post-hoc test for multiple comparisons were used to compare continuous variables in different groups. Data are presented as the mean ± standard error of the mean. Prognostic factors were assessed using univariate and multivariate Cox regression. The overall survival curve was plotted using the Kaplan-Meier method and a log-rank test. P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were performed using the SPSS software version 16.0 (SPSS, Inc., Chicago, IL, USA). Mann-Whitney U test was used for continuous variables and χ2 test was used for categorical variables.
Results
Identification and validation of circLPAR1 in bladder cancer tissue
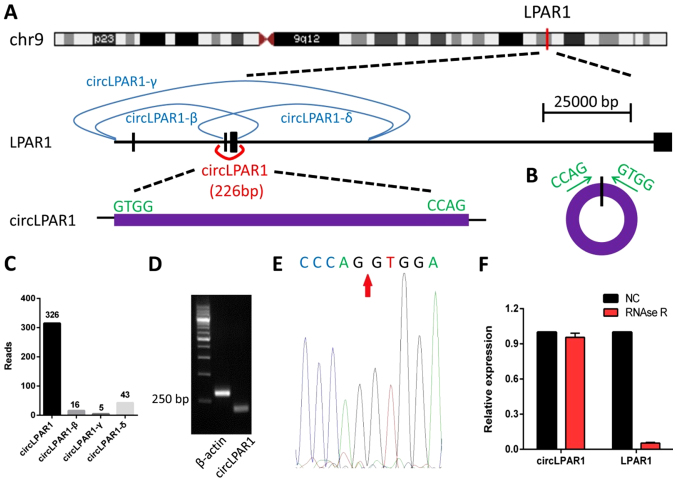
circLPAR1 (hsa_circ_0087960) is the product of gene LPAR1 formed during the transcription process, 226 bp in length, derived from exons 2 and 3 (Fig. 1A and B). The transcriptome sequencing results indicated that the LPAR1 gene encodes three other circRNAs, including LPAR1-β, LPAR1-γ and LPAR1-δ (Fig. 1C). The amplification products of circLAPR1 were assessed by RT-qPCR, and the divergent primers certified that the cyclization site was expressed in the bladder cancer samples (Fig. 1D). The Sanger sequencing results also indicated the occurrence of cyclization (Fig. 1E). The RNase R exonuclease digestion further confirmed that the RNA species was stable in circular form and resistant to digestion by RNase R (Fig. 1F).
Figure 1.
The location and structure characteristics of circLPAR1 and the proof of cyclization. (A) circLPAR1 is derived from the 2nd and 3rd exons from the LPAR1 gene which is located on chromosome 9. (B) Base sequence of the cyclization site. (C) Reads for four circRNAs from gene LPAR1 transcription by RNA-sequencing. (D) The amplification products with the divergent primers. (E) The Sanger sequencing results indicated the presence of cyclization. The red arrow indicates the splice site. (F) circLPAR1 was resistant to digestion with RNase R exonuclease. NC, negative control; circLPAR1, circular lysophosphatidic acid receptor 1.
circLPAR1 as a potential predictor of DSS for MIBC
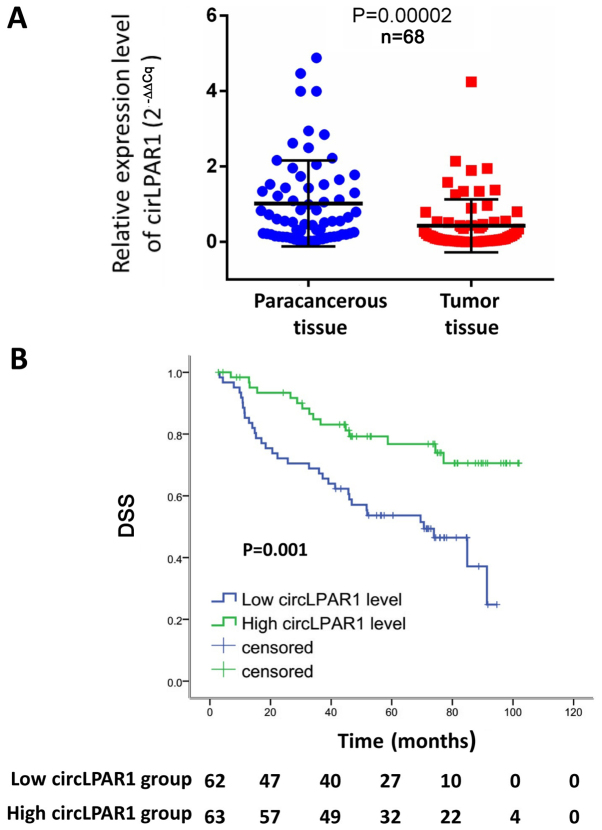
Firstly, the level of circLPAR1 expression was investigated by RT-qPCR in 68 MIBC tissues and paired adjacent non-tumorous tissues. The results demonstrated that the level of expression was significantly reduced in MIBC tissue, compared with para-carcinoma tissue (P=0.00002; Fig. 2A). The present study subsequently evaluated the potential prognostic value of the circRNA and identified that circLPAR1 expression was significantly associated with neoadjuvant chemotherapy prior to radical cystectomy (P=0.039; Table I). The mean DSS was 54.8±2.6 months (median, 53.2 months; 95% confidence interval CI, 49.7–60.0 months). In the univariate and multivariate analyses, a low circRNA expression level (2−∆∆Cq <0.0023) was significantly associated with poor DSS, compared with a high circRNA expression level (P=0.001; Table II). The median DSS was 52.4 months (95% CI, 42.9–57.3 months) and 56.0 months (95% CI, 52.0–66.6 months) for low and high circRNA expression groups, respectively (P=0.001), and the survival curve for DSS was illustrated in Fig. 2B.
Figure 2.
Expression level of circLPAR1 in MIBC tissues and survival curve of DSS for 125 patients. (A) Expression level of cirLPAR1 was significantly reduced in 68 MIBC tissues, compared with para-carcinoma tissues (P=0.00002). (B) There was significant difference in DSS between patients with a low circRNA expression level and a high circRNA expression level (52.4 vs. 56.0 months; P=0.001). MIBC, muscle-invasive bladder cancer; circRNA, circular RNA; LPAR1, lysophosphatidic acid receptor 1; DSS, disease specific survival.
Table II.
Univariate and multivariate Cox regression analyses for disease-specific survival in 125 patients with muscle-invasive bladder cancer.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Variables | P-value | HR (95% CI) | P-value | HR (95% CI) |
| Age (years) | ||||
| <61 vs. ≥61 | 0.910 | 0.968 (0.548–1.710) | ||
| Sex | ||||
| Male vs. female | 0.266 | 0.614 (0.260–1.451) | ||
| Pathological T stagea | ||||
| T2 vs. T3+T4 | 0.008 | 2.176 (1.228–3.857) | 0.027 | 1.885 (1.040–3.416) |
| Pathological N stagea | ||||
| N0 vs. N1+N2 | 0.070 | 1.780 (0.955–3.318) | ||
| Pathological Grade | ||||
| Low vs. high | 0.021 | 2.598 (1.154–5.851) | 0.042 | 2.355 (1.031–5.380) |
| Maximum tumor diameter (cm) | 0.089 | 1.682 (1.040–3.416) | ||
| <3.5 vs. ≥3.5 | 0.015 | 2.041 (1.149–3.626) | ||
| Tumor number (n) | ||||
| 1 vs. ≥2 | 0.403 | 0.762 (0.403–1.441) | ||
| circLPAR1 level | ||||
| Low vs. highb | 0.001 | 0.364 (0.197–0.673) | 0.011 | 0.444 (0.237–0.832) |
| Recurrence after TURBT | ||||
| Yes vs. no | 0.356 | 1.351 (0.713–2.558) | ||
| Chemotherapy before radical cystectomy | ||||
| Yes vs. no | 0.19 | 1.466 (0.828–2.596) | ||
According to the 2002 6th edition of the Union for International Cancer Control Tumor-Node-Metastasis staging system.
The low level of circLPAR1 expression was defined the level less than the median value (2−∆∆Cq=0.0023), and the high level was defined as the level equal or more than the median value. TURBT, transurethral resection of bladder tumor; DSS, disease-specific survival; circLPAR1, circular lysophosphatidic acid receptor 1; HR, hazard ratio; CI, confidence interval.
Altered invasion of bladder cancer cells by si-circLPAR1
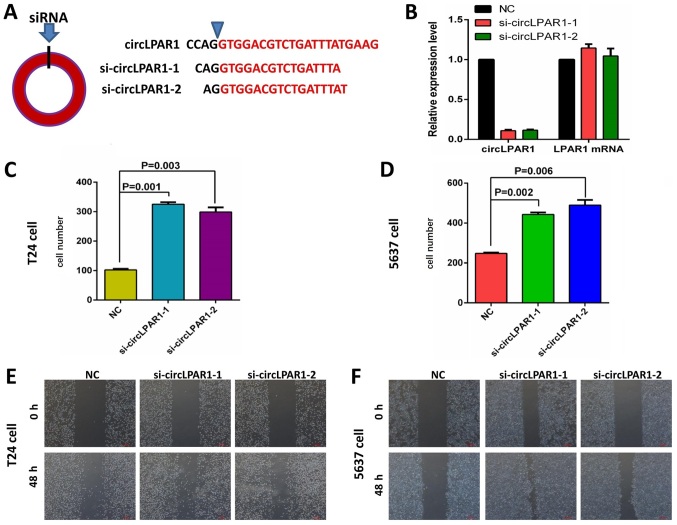
The present study designed and synthesized two siRNAs for circLPAR1 (Fig. 3A). The RT-qPCR results indicated that these siRNAs could successfully knockdown the expression of circLPAR1 without affecting the expression of the host gene, LPAR1 (Fig. 3B). Further cell function tests including Matrigel demonstrated that in T24 bladder cancer cells, the invasion was significantly enhanced following the knockdown of circLPAR1 (Fig. 3C). Similar results were also obtained for the 5637 bladder cancer cells (Fig. 3D). In addition, wound healing assays for these cell lines demonstrated similar results (Fig. 3E and F).
Figure 3.
The effect on invasion ability of bladder cancer cells with knockdown of circLPAR1 by siRNA. (A) The design of siRNAs against circLPAR1, in which removing joints can break the ring structure. (B) circLPAR1 and LPAR1 RNA expression following treatment with two siRNAs in 293 cells by reverse transcription-quantitative polymerase chain reaction analysis. The invasion ability of (C) T24 and (D) 5637 cells was significantly enhanced following the knockdown of circLPAR1. The invasion ability was more enhanced for (E) T24 and (F) 5637 cells by treatment with siRNA against circLPAR1, compared with the NC (magnification, ×40). NC, negative control; siRNA, small interfering RNA; circLPAR1, circular lysophosphatidic acid receptor 1.
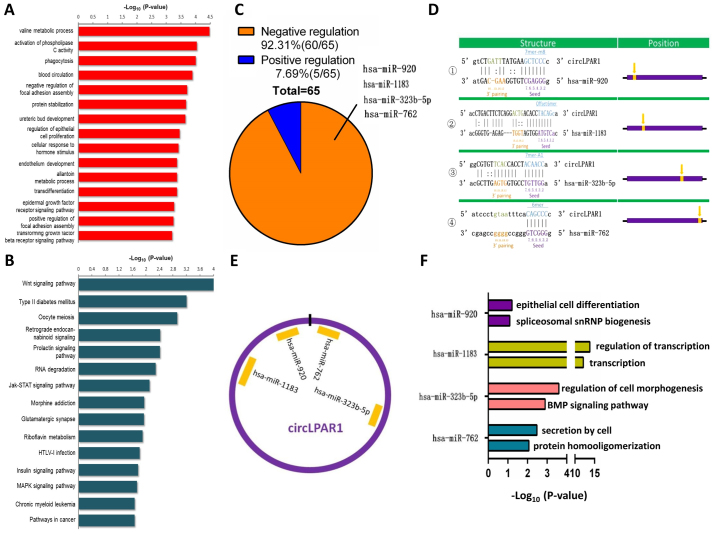
Bioinformatics analysis of circLPAR1 in bladder cancer
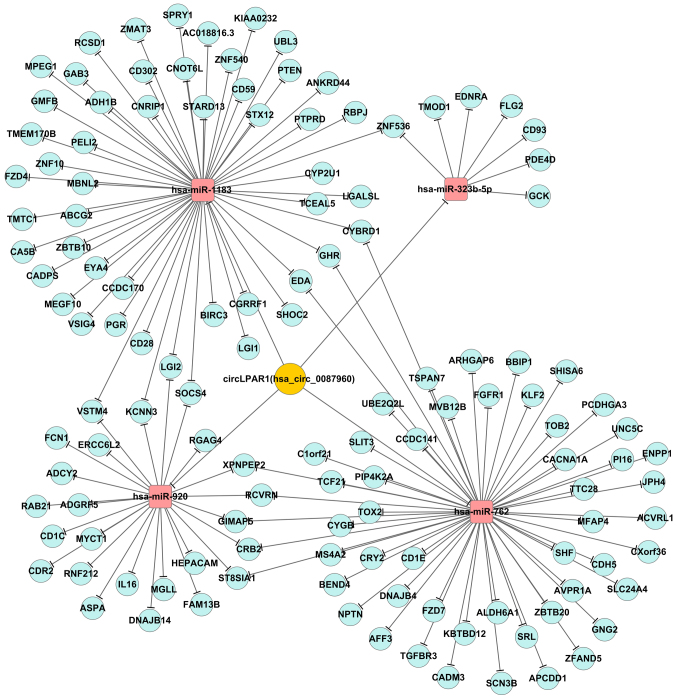
The present study identified that there were 56, 7, 52 and 25 associated mRNAs for these miRNAs (fold-change ≥2), respectively(data not shown). The bioinformatics analysis revealed that a total of 372 GO-Analysis-BP items were associated with circLPAR1, and that 22 KEGG pathways were associated with circLPAR1 (Fig. 4A and B). Additionally, there were 60 negatively regulated miRNAs and 5 positively regulated miRNAs for circLPAR1 (Fig. 4C). The data demonstrated that circLPAR1 harbors four miRNAs (hsa-miR-920, hsa-miR-1183, hsa-miR-323b-5p and hsa-miR-762) according to the matching seed sequences (Fig. 4D), and a schematic representation illustrated the putative binding sites of circLPAR1 with the four associated miRNAs (Fig. 4E). The present study also presented the two most markedly varying molecular function and biological process GO items for each of the four matched miRNAs, including miR-920, miR-1183, miR-323b-5p and miR-762 (Fig. 4F).
Figure 4.
Bioinformatics analysis for circLPAR1 in muscle-invasive bladder cancer. Top 15 (A) Gene Ontology-Analysis-BP items and (B) Kyoto Encyclopedia of Genes and Genomes pathways for circLPAR1. (C) The negative regulation between circLPAR1 and miRNAs. (D) circLPAR1 may harbor four miRNAs according to the miRNA seed sequence matching. (E) A schematic drawing illustrated the putative binding sites of the four miRNAs. (F) The two most markedly enriched GO-items for the four matched miRNAs. miRNAs, microRNAs; circLPAR1, circular lysophosphatidic acid receptor 1.
circLPAR1 binds to miR-762 and inhibits its activity
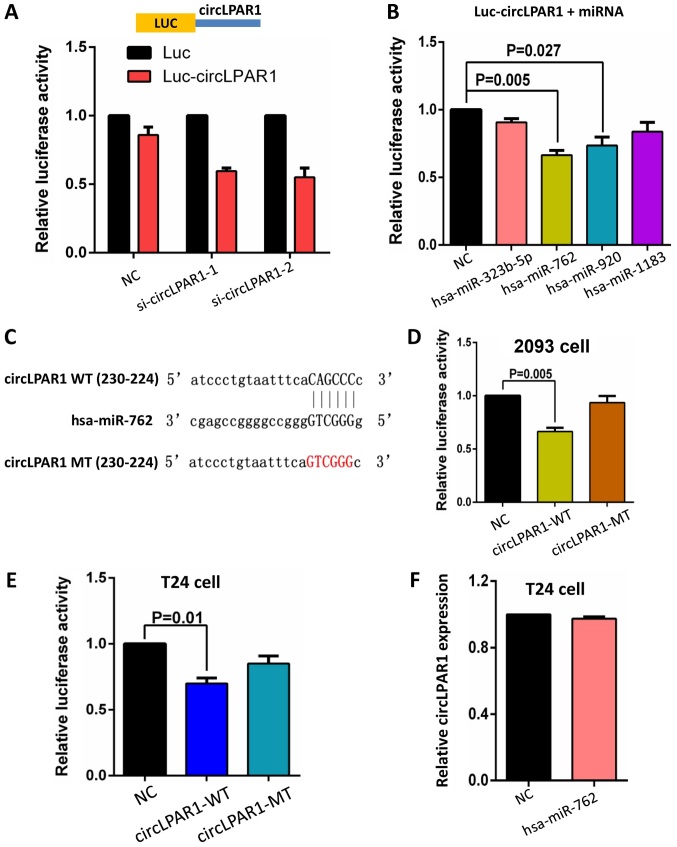
A circLPAR1 fragment was constructed and inserted downstream of the luciferase reporter gene. We hypothesized that miRNAs associated with circLPAR1 may potentially inhibit the luciferase activity by miRNA-mediated activation of deadenylation and the subsequent exonucleolytic degradation. The inclusion of circLPAR1 sequence in the 3′ untranslated region (3′UTR) was observed to cause the downregulation of luciferase activity and knockdown of the endogenous circLPAR1 further decreased the luciferase activity (Fig. 5A). Additionally, a luciferase reporter assay was performed using these four miRNAs. Each miRNA mimic was co-transfected with the luciferase reporters into 293 cells. Compared with the negative control RNA, miR-762 and miR-920 significantly reduced the luciferase reporter activity (P=0.005 and 0.027, respectively; Fig. 5B).
Figure 5.
circLPAR1 binds to miR-762 and inhibits its activity. (A) The entire circLPAR1 sequence was cloned into the downstream of the luciferase gene, as a Luc-circLPAR1 plasmid. Luciferase reporter assay for the activity of Luc-circLPAR1 in 293 cells co-transfected with siRNA against circLPAR1. (B) A luciferase reporter assay was used to detect the luciferase activity of Luc-circLPAR1 co-transfected with four miRNA mimics in 293 cells. Data are presented as the means ± standard deviation of three experiments. (C) The design of the circLPAR1 mutated the miR-762 binding site. (D) A luciferase reporter assay was used to detect the luciferase activity of Luc-circLPAR1 or the Luc-cPVT1-mutant co-transfected with miR-762 mimic in 2093 cells. (E) A luciferase reporter assay was used to detect the luciferase activity of Luc-circLPAR1 or the Luc-cPVT1-mutant co-transfected with miR-762 mimic in T24 cells. (F) Reverse transcription-quantitative polymerase chain reaction analysis demonstrated that miR-762 could not significantly change the expression level of circLPAR1. WT, wild type; MT, mutated; circLPAR1, circular lysophosphatidic acid receptor 1; miR, microRNA; NC, negative control.
The present study mutated the miR-762 target site by including the circLPAR1 sequence in the 3′UTR (Fig. 5C). The transfection of miR-762 and the luciferase reporter gene inserted circLPAR1 mutated fragment did not significantly affect the luciferase activity in 293 cells (Fig. 5D). Similar results were obtained in T24 cells (Fig. 5E). The transfection of miR-762 in T24 cells did not significantly decrease the level of circLPAR1, compared with the negative control, as assessed by RT-qPCR (Fig. 5F), which indicated that circLPAR1 may not be digested or suppressed by miR-762.
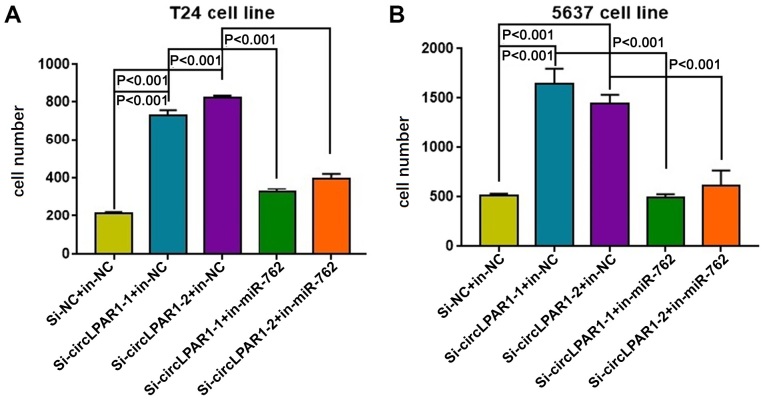
Matrigel assays demonstrated that in T24 bladder cancer cells, the invasion was significantly enhanced following the knockdown of circLPAR1 but suppressed by miR-762 (Fig. 6A). Similar results were also obtained for the 5637 bladder cancer cells (Fig. 6B). Additionally, the circRNA-miRNA-mRNA interaction network for circLPAR1 was also illustrated in Fig. 7.
Figure 6.
The effect on invasion ability of bladder cancer cells with knockdown of circLPAR1 by siRNA and it can return by inhibitor miR-762. The results demonstrated that the miR-762 inhibitor significantly attenuated the si-circLPAR1-induced increase in cell invasion in the (A) T24 and (B) 5637 cell lines. The number of the cells that pass through the Matrigel chamber are presented. NC, negative control; circLPAR1, circular lysophosphatidic acid receptor 1; miR, microRNA; si, small interfering.
Figure 7.
The circRNA-miRNA-mRNA network for circLPAR1. According to the result of bioinformatics analysis, there were four miRNAs negatively regulated by circLPAR1. In addition, certain mRNAs were also identified that may be negatively regulated by matched miRNAs. circLPAR1, circular lysophosphatidic acid receptor 1; miRNA, microRNA.
Discussion
Urothelial carcinoma is the most common type of bladder cancer (16). Certain drugs, such as PD-L1 inhibitors, propose novel treatment options and an improved patient prognosis (17); however, the heterogeneity of tumors necessitates the investigation of individualized treatments and prognostic biomarkers. Non-coding RNA is an intensive research topic in molecular biology and has been the focus of numerous studies (11–14); however, the oncologic value of circRNA, as a novel non-coding RNA, remains unclear in a clinical setting. Thus, the present study focused on the clinical application of circRNAs in bladder cancer and also investigated the underlying mechanism of action.
The majority of circRNAs are derived from exons and frequently exist in eukaryotic cells (18). A series of endogenous RNA molecules are speculated to serve a role in regulating gene and protein expression, and exhibit functions in several pathophysiological processes, including protein and miRNA binding (19–21). In the present study a closed ring structure is one of the most vital characteristics for circRNAs, and hence, circRNAs are evaluated for the occurrence of circular molecules. In our previous study, the parent gene LPAR1 in bladder cancer tissue could be formed from four circRNAs, of which one circRNA derived from exons 2 and 3, was highly abundant (11). In the present study, this circRNA was termed as circLPAR1, which constituted of 226 bp. The results of the present study confirmed that circLPAR1 was resistant to RNase R digestion. Divergent primers were used for amplification, and the RT-qPCR product also demonstrated that the fragment originated from across the back-splice site. Furthermore, the distinct band on agarose gel electrophoresis was confirmed as the ring structure by Sanger sequencing. These studies substantiated the circular structure of circLPAR1.
circRNAs are associated with numerous diseases, such as nervous system dysplasia, atherosclerosis and Alzheimer's disease (22–24). Additionally, circRNAs serve a major role in the occurrence and development of human tumors (8,25). Reportedly, the abundance of several circRNAs, including circHIPK3, differed significantly in the tumor and normal tissues (11,26). Some scholars indicated that the content of circular RNA (hsa_circ_002059) in gastric cancer was significantly reduced, compared with adjacent tissues, and may be a potential novel and stable biomarker for the diagnosis of gastric carcinoma (11,27). An additional study demonstrated that the level of the circular RNA hsa_circ_0001649 was not only decreased in the liver cancer tissue but was also associated with tumor size and lymph node metastasis (28). Zhong et al (29) elaborated the molecular mechanism of circTCF25 promoting the occurrence of bladder cancer by the inhibition of miR-103a-3p/miR-107.
In the present study, the expression of circLPAR1 was observed to be decreased in 68 bladder cancer tissues, compared with paired para-cancer tissues. The low level of circLPAR1 expression was defined the level less than the median value (2C∆∆q=0.0023), and the high level was defined as the level equal or more than the median value. Further survival analysis indicated that the DSS from patients with low a expression level of circLPAR1 was 4 months shorter than those with a high expression, and the risk of mortality in the low expression group was two times greater than that in the high expression group. A potential hypothesis was that circLPAR1 may activate or inhibit certain signaling pathways, thus resulting in disease progression (30). In order to investigate the effect of circLPAR1 on cell function, siRNA was employed to knockdown the expression of circLPAR1, and the invasion and metastasis ability of cells was observed to be significantly decreased. Collectively, these results indicate that that circLPAR1 may be associated with invasion and metastasis.
The most common mechanism of circRNA is speculated to involve the negative regulation of the expression and function of miRNAs (14,31). In the present study, the bioinformatics analysis indicated that four miRNAs may be the potential targets of circLPAR1. Based on the luciferase reporter assay, miR-762 and miR-920 were observed to reduce the luciferase activity, and miR-762 varies significantly. Additionally, the inhibition of luciferase activity could be alleviated in different cells after mutating the circLPAR1 sites that could potentially bind to miR-762. These results indicated that circLPAR1 could bind to miR-762 and inhibit its activity as a miRNA sponge. Previous studies also reported that miR-762 promoted the proliferation and invasion of breast cancer and the development of oral squamous cell carcinoma (32,33). Collectively, these results revealed a plausible explanation that circLPAR1 may serve a crucial role in the invasion and metastasis by miR-762.
However, the theory of circRNAs serving as the miRNA sponge remains controversial. Although the sponge-like effect of circRNAs has been proposed previously and confirmed experimentally (31), recent literature does not support this conclusion or indicate the presence of other interacting isoforms (7,20). Notably, a host gene can produce multiple circRNAs, one of which may exhibit abundant functions, including in cell proliferation and migration (11). Simultaneously, focus on the existing functional features of the host genes is required.
In conclusion, the present study demonstrated that circLPAR1, may be a potential novel and stable biomarker for the prognosis of bladder cancer that may associate with the invasion by miR-762. It indicated that the invasion of group si-circLPAR1 and inhibitor-NC is same as Fig. 3C and D. But for the group si-circLPAR1 and inhibitor-miR-762, the invasion returned to the level of si-NC and inhibitor-NC (Fig. 6).
circLPAR1 may be a potential novel and stable biomarker for the prognosis of MIBC and may be associated with the invasion and metastasis by miR-762.
Acknowledgements
The authors would like to thank Professor Sheng-Lin Huang of Fudan University Cancer Institute (Shanghai, China), who read this manuscript and provided some suggestions.
Funding
This study was supported by the Shanghai Health Bureau of Research Fund (20144Y0180) and Natural Science Foundation of China (grant nos. 81672544 and 81572531).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
GWL and HYS improved and refined the project details and performed the experiments; HYS and HYX provided important recommendations and all tissue specimens; GHS and DWY designed and supervised the study; QPZ made substantial contributions to conception and design; GWL, HY and YJS made substantial contributions to acquisition of clinicopathological data. All authors reviewed and approved the manuscript.
Ethics approval and consent to participate
The present study was conducted in accordance with the ethical standards of the Helsinki Declaration II and approved by the Ethics Committee of Fudan University Shanghai Cancer Center (approval no. 050432-4-1212B). Written informed consent was obtained from each patient prior to any study-specific experiments being performed.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.American Cancer Society; Atlanta, GA: 2015. American Cancer Society: Cancer facts and figures 2015. [Google Scholar]
- 2.Krogsbøll LT, Jørgensen KJ, Gøtzsche PC. Screening with urinary dipsticks for reducing morbidity and mortality. Cochrane Database Syst Rev. 2015;1:CD010007. doi: 10.1002/14651858.CD010007.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh R, Ansari JA, Maurya N, Mandhani A, Agrawal V, Garg M. Epithelial-to-Mesenchymal transition and its correlation with clinicopathologic features in patients with urothelial carcinoma of the bladder. Clin Genitourin Cancer. 2017;15:e187–e197. doi: 10.1016/j.clgc.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 4.Chen J, Miao Z, Xue B, Shan Y, Weng G, Shen B. Long Non-coding RNAs in urologic malignancies: Functional roles and clinical translation. J Cancer. 2016;7:1842–1855. doi: 10.7150/jca.15876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42:D92–D97. doi: 10.1093/nar/gkt1248. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73:5609–5612. doi: 10.1158/0008-5472.CAN-13-1568. [DOI] [PubMed] [Google Scholar]
- 9.Chi BJ, Zhao DM, Liu L, Yin XZ, Wang FF, Bi S, Gui SL, Zhou SB, Qin WB, Wu DM, Wang SQ. Downregulation of hsa_circ_0000285 serves as a prognostic biomarker for bladder cancer and is involved in cisplatin resistance. Neoplasma. 2018 Sep 16; doi: 10.4149/neo_2018_180318N185. (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Bi J, Dong W, Yang M, Shi J, Jiang N, Lin T, Huang J. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol Cancer. 2018;17:161. doi: 10.1186/s12943-018-0908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS. MicroRNA targets in drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pasquinelli AE. MicroRNAs and their targets: Recognition, regulation and an emerging reciprocal relationship. Nat Rev Genet. 2012;13:271–282. doi: 10.1038/nrg3162. [DOI] [PubMed] [Google Scholar]
- 14.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 15.Reiner A, Yekutieli D, Benjamini Y. Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics. 2003;19:368–375. doi: 10.1093/bioinformatics/btf877. [DOI] [PubMed] [Google Scholar]
- 16.Grivas PD, Melas M, Papavassiliou AG. The biological complexity of urothelial carcinoma: Insights into carcinogenesis, targets and biomarkers of response to therapeutic approaches. Semin Cancer Biol. 2015;35:125–132. doi: 10.1016/j.semcancer.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Aoun F, Kourie HR, Sideris S, Roumeguere T, van Velthoven R, Gil T. Checkpoint inhibitors in bladder and renal cancers: Results and perspectives. Immunotherapy. 2015;7:1259–1271. doi: 10.2217/imt.15.91. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 19.Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 21.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 22.Floris G, Zhang L, Follesa P, Sun T. Regulatory role of circular RNAs and neurological disorders. Mol Neurobiol. 2017;54:5156–5165. doi: 10.1007/s12035-016-0055-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6:e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukiw WJ. Circular RNA (circRNA) in Alzheimer's disease (AD) Front Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi: 10.1016/j.cca.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 28.Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2016;16:161–169. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 29.Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. doi: 10.1038/srep30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: From waste to treasure. Mol Cancer. 2017;16:58. doi: 10.1186/s12943-017-0630-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Huang R, Wang L, Hao J, Zhang Q, Ling R, Yun J. microRNA-762 promotes breast cancer cell proliferation and invasion by targeting IRF7 expression. Cell Prolif. 2015;48:643–649. doi: 10.1111/cpr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu T, Wang XY, Gong RG, Li A, Yang S, Cao YT, Wen YM, Wang CM, Yi XZ. The expression profile of microRNAs in a model of 7,12-dimethyl-benz[a]anthrance-induced oral carcinogenesis in Syrian hamster. J Exp Clin Cancer Res. 2009;28:64. doi: 10.1186/1756-9966-28-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.