Key Points
We identify 2 new genetic loci associated with FVII activity and highlight REEP3 and JAZF1 as potential underlying causal genes.
We provide evidence for a causal effect of FVII activity on the risk of IS.
Abstract
Factor VII (FVII) is an important component of the coagulation cascade. Few genetic loci regulating FVII activity and/or levels have been discovered to date. We conducted a meta-analysis of 9 genome-wide association studies of plasma FVII levels (7 FVII activity and 2 FVII antigen) among 27 495 participants of European and African ancestry. Each study performed ancestry-specific association analyses. Inverse variance weighted meta-analysis was performed within each ancestry group and then combined for a trans-ancestry meta-analysis. Our primary analysis included the 7 studies that measured FVII activity, and a secondary analysis included all 9 studies. We provided functional genomic validation for newly identified significant loci by silencing candidate genes in a human liver cell line (HuH7) using small-interfering RNA and then measuring F7 messenger RNA and FVII protein expression. Lastly, we used meta-analysis results to perform Mendelian randomization analysis to estimate the causal effect of FVII activity on coronary artery disease, ischemic stroke (IS), and venous thromboembolism. We identified 2 novel (REEP3 and JAZF1-AS1) and 6 known loci associated with FVII activity, explaining 19.0% of the phenotypic variance. Adding FVII antigen data to the meta-analysis did not result in the discovery of further loci. Silencing REEP3 in HuH7 cells upregulated FVII, whereas silencing JAZF1 downregulated FVII. Mendelian randomization analyses suggest that FVII activity has a positive causal effect on the risk of IS. Variants at REEP3 and JAZF1 contribute to FVII activity by regulating F7 expression levels. FVII activity appears to contribute to the etiology of IS in the general population.
Visual Abstract
Introduction
As the initiator of the extrinsic coagulation pathway, coagulation factor VII (FVII) plays a central role in fibrin formation. Like many coagulation factors, FVII is produced as an inactivate zymogen, and is activated through proteolytic cleavage by other coagulation factors: mainly by FX, and also by thrombin, FIX, and FXII. Once activated FVII binds to tissue factor, its activity greatly increases. The complex of activated FVII and tissue factor activates FX and FIX, which ultimately leads to conversion of prothrombin to thrombin, which converts fibrinogen into fibrin. Plasma levels of FVII are associated with several clinical outcomes. For example, FVII deficiency is a rare bleeding disorder associated with hemorrhagic complications,1 whereas elevated levels of FVII have been associated in some studies with arterial thrombosis and venous thromboembolism (VTE).2-5
FVII activity and levels have a substantial heritable component, with estimates of the heritability of FVII activity ranging from 40% to 52%.6,7 The Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium previously conducted genome-wide association studies (GWASs) with data on over 2 million common single-nucleotide polymorphisms (SNPs) in 15 795 European-ancestry participants, identifying 4 new candidate genes for FVII in addition to the protein-coding locus, F7.8,9 The fact that lead variants at these known loci explain only 7.7% of the variance in FVII suggests that further heritability remains to be uncovered.8
To discover additional genetic variants associated with FVII, we performed an expanded GWAS with data on over 10 million common and low-frequency SNPs and insertion-deletions in 27 495 participants across 9 studies, including 3420 African American participants. Gene silencing in a human liver cell line was used to validate the genomic function of significantly associated loci. Lastly, we performed Mendelian randomization analyses to estimate the causal effects of FVII activity on atherosclerotic and thrombotic diseases, including coronary artery disease (CAD), ischemic stroke (IS), and VTE.
Methods
Study design and participating cohorts
This study was organized within the CHARGE Consortium Hemostasis Working Group.9 Nine studies were included: the Atherosclerosis Risk in Communities (ARIC) study,10 the Cardiovascular Health Study (CHS),11 the Coronary Artery Risk Development In young Adults (CARDIA) study,12 the Genetic Analysis for Idiopathic Thrombophilia 2 (GAIT2) study,13 the Framingham Heart Study (FHS),14 the LUdwigshafen RIsk and Cardiovascular Health (LURIC) study,15 the Multiple Environmental and Genetic Assessment of risk factors for venous thrombosis (MEGA) study,16 the Precocious Coronary Artery DISease (PROCARDIS) study,17 and the Rotterdam Study (RS).18 Descriptions and ancestry composition of participating cohorts are found in supplemental Methods and supplemental Table 1 (available on the Blood Web site). All studies were approved by appropriate research ethics committees and all respondents signed informed consent prior to participation. Seven studies (ARIC, CHS, CARDIA, GAIT2, LURIC, MEGA, RS) including 23 434 participants measured FVII activity (percentage or IU/mL × 100) and 2 studies (FHS, PROCARDIS) including 4061 participants measured FVII antigen (percentage or IU/mL × 100). CHS, FHS, RS, and a subset of ARIC were also included in the discovery analysis for the previous FVII GWAS,8 whereas PROCARDIS and a second subset of ARIC were included in the replication. CARDIA, GAIT2, LURIC, and MEGA were not included in the previous GWAS.
Genotyping and imputation
All participating cohorts performed genome-wide genotyping using commercial platforms available from Illumina or Affymetrix. Each study performed standard preimputation quality control checks and imputed autosomal and X-chromosome variants from the 1000 Genomes Project (1000G) phase 1 version 3 reference panel using available imputation methods.19-22 Genotyping, preimputation quality control, and imputation procedures are described in supplemental Table 2.
Cohort-specific association analyses
Natural log-transformed FVII was analyzed within each cohort. Participants with values 3 standard deviations (SDs) above or below the population mean were removed prior to cohort-level analysis and any individuals on anticoagulant therapy were also excluded. Ancestry-stratified, study-specific regression analyses using an additive genetic model were performed between genome-wide 1000G imputed variant dosages and phenotype levels, adjusted for age, sex, ancestry-informative principal components, and study-specific variables, such as center. Analyses of the X-chromosome were stratified by sex, with variants in males coded as 0/2. The covariates used in each of the studies are shown in supplemental Table 2. Quality control assessment of ancestry-specific result files from each study was conducted prior to meta-analysis using the EasyQC software package.23 Quality control procedures are further described in supplemental Methods.
Trans-ancestry meta-analysis
The discovery trans-ancestry meta-analysis was conducted in 2 steps. First, the METAL program was used to perform ancestry-specific inverse-variance weighted meta-analysis.24 We then used the same method to meta-analyze the ancestry-specific results. As suggested by Huang et al,25 we adopted a genome-wide significance threshold of P < 2.5 × 10−8. Compared with the traditional genome-wide significance threshold of 5 × 10−8, this stricter threshold additionally corrects for the low-frequency variants that were not included in the initial generation of GWASs.26 Finally, a locus was defined as ±1 Mb from the variant with the lowest P value.
To reduce heterogeneity, the primary trans-ancestry meta-analysis included the 7 studies that measured FVII activity, and not the 2 studies (FHS and PROCARDIS) that measured FVII antigen. In a secondary meta-analysis, we added results from the 2 studies that measured FVII antigen.
Postdiscovery analyses
Newly identified loci were validated and characterized by using small interfering RNA (siRNA) to silence candidate genes in human liver HuH7 cells, and measuring F7 messenger RNA (mRNA) levels and release of FVII protein levels. We based the design of these functional validation experiments on 2 assumptions: (1) that functional mechanisms underlying the genetic associations at new loci involve the candidate genes we selected and (2) that the functional mechanisms underlying the genetic associations involve FVII synthesis or release, as opposed to FVII activation or clearance. The methods used in these experiments are described in detail in supplemental Methods. As a positive control, we also silenced the F7 gene itself and measured the effect on F7 mRNA levels.
To identify additional independent signals at the associated loci, an approximate method implemented in the Genome-wide Complex Trait Analysis (GCTA) software was used for conditional and joint analysis using meta-analysis summary statistics from the trans-ancestry meta-analysis of FVII activity.27,28 Further details on the conditional analysis are provided in supplemental Methods.
Mendelian randomization is an approach that uses genetic variants associated with an exposure as an instrument to examine the causal effect of the exposure on an outcome. Although direct examination of the exposure-outcome association might be impeded by confounding or reverse causation, genetic instruments are less likely to be affected by these issues, allowing for causal inference. Mendelian randomization analyses were used to investigate the potential causal effect of FVII activity on CAD, IS, and VTE. We used 2-sample methods that rely on summary statistics (β coefficients with standard errors from GWASs).29 We used summary statistics from the trans-ancestry meta-analysis of FVII activity, and obtained summary statistics for CAD from the CARDIoGRAMplusC4D Consortium (http://www.cardiogramplusc4d.org/data-downloads/),30 summary statistics for IS from the MEGASTROKE Consortium,31 and summary statistics for VTE from the International Network against VENous Thrombosis (INVENT) Consortium.32 The methods used to perform Mendelian randomization can be found in supplemental Methods. In brief, we used 4 techniques to obtain causal-effect estimates based on the lead variants at the genome-wide significant loci: (1) inverse-variance weighted meta-analysis (primary analysis), (2) MR-Egger,33 (3) weighted median estimator,34 and (4) restriction of the analysis to the lead variant at the F7 locus. Given that the lead variant at the F7 locus is located in the gene that encodes the FVII protein, it is less likely to have clinical effects through pathways that are not directly mediated by FVII.35
Results
Baseline characteristics
In total, 20 014 European-ancestry and 3420 African-ancestry subjects from 7 studies were included in the meta-analysis of FVII activity and an additional 4061 European-ancestry subjects were included in the combined meta-analysis of FVII activity and antigen. Baseline characteristics of participants in each of the studies included in the GWAS are shown in supplemental Table 1. The mean age across the studies was 57.2 years, and 52.2% of the participants were women.
Trans-ancestry meta-analysis
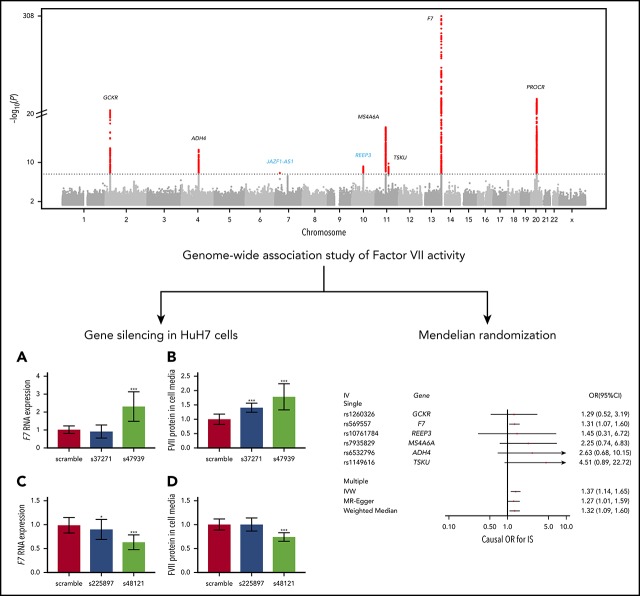
After quality control, 10 044 948 variants across the autosomal and X chromosomes were examined in the trans-ancestry meta-analysis of FVII activity. Of these variants, 9 316 598 were SNPs and 728 350 were insertions-deletions. The genomic inflation factors that were used to apply genomic control correction to each of the included studies were all <1.05 and are shown in supplemental Table 2. A QQ plot and Manhattan plot are shown in supplemental Figures 1 and 2, respectively.
Genome-wide significant results are presented in Table 1. Briefly, 1637 variants located in 8 loci exceeded the genome-wide significance level of P < 2.5 × 10−8. Among the associated regions, loci containing F7, PROCR, GCKR, MS4A6A, ADH4, and TSKU represented previously described loci (supplemental Figures 3),8 whereas 2 loci were novel: REEP3 and JAZF1-AS1. The most significant variant at the REEP3 locus was an intronic variant, rs10761784 (β = 0.013; P = 6.7 × 10−10) in REEP3 (supplemental Figure 4). At the second novel locus, the lead variant, rs498475, was located within the antisense RNA JAZF1-AS1 (β = 0.012; P = 1.5 × 10−8; supplemental Figure 4). Lead variants at PROCR and GCKR were identical to previously reported lead variants, whereas the lead variants at the remaining known loci differed from previously reported lead variants (supplemental Table 3). Forest plots showing study-specific results for known and novel loci are provided in supplemental Figure 5. In addition to these genome-wide significant findings, variants at a further 15 loci were suggestively associated (P < 2.5 × 10−6) with FVII activity (supplemental Table 4).
Table 1.
Lead variants at loci associated with FVII activity when excluding studies that measured FVII antigen from the trans-ancestry meta-analysis
| Variant rsID | Chr:Pos* | Alleles† | Freq‡ | Freq EA§ | Freq AA|| | β | SE | P | Variance explained, % | Closest gene | Annotation | Discovery status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs569557 | 13:113769917 | G/A | 0.88 | 0.88 | 0.88 | 0.157 | 0.003 | 6.4 × 10−600 | 13.9 | F7 | Intronic | Known |
| rs867186 | 20:33764554 | G/A | 0.10 | 0.11 | 0.09 | 0.057 | 0.003 | 3.3 × 10−64 | 1.6 | PROCR | Missense | Known |
| rs1260326 | 2:27730940 | T/C | 0.39 | 0.41 | 0.15 | 0.024 | 0.002 | 2.3 × 10−30 | 0.7 | GCKR | Missense | Known |
| rs7935829 | 11:59942815 | G/A | 0.39 | 0.40 | 0.21 | 0.018 | 0.002 | 6.3 × 10−18 | 0.4 | MS4A6A | Intronic | Known |
| rs6532796 | 4:100042242 | G/A | 0.70 | 0.69 | 0.83 | 0.016 | 0.002 | 2.6 × 10−13 | 0.3 | ADH4 | Downstream | Known |
| rs1149616 | 11:76498369 | T/C | 0.17 | 0.17 | 0.16 | 0.017 | 0.003 | 1.7 × 10−10 | 0.2 | TSKU | Intronic | Known |
| rs10761784 | 10:65308750 | A/T | 0.53 | 0.51 | 0.67 | 0.013 | 0.002 | 6.7 × 10−10 | 0.2 | REEP3 | Intronic | Novel |
| rs498475 | 7:28256240 | G/A | 0.42 | 0.38 | 0.74 | 0.012 | 0.002 | 1.5 × 10−8 | 0.2 | JAZF1-AS1 | ncRNA | Novel |
The variance explained shown in this table was calculated using the sample size weighted mean variance of log-transformed FVII and the betas and frequencies from the trans-ancestry meta-analysis summary statistics.
AA, African ancestry; Chr, chromosome; EA, European ancestry; Freq, frequency; ncRNA, noncoding RNA; Pos, position; rsID, reference SNP cluster ID; SE, standard error.
The Chr:Pos column shows the chromosome and position (build 37).
The Alleles column shows the FVII-increasing allele/FVII-decreasing allele.
The Freq column shows the frequency of the FVII-increasing allele.
The Freq EA column shows the frequency specifically in participants of European ancestry.
The Freq AA column shows the frequency specifically in participants of African ancestry.
Ancestry-specific results for FVII activity are presented in supplemental Table 5. The European-specific meta-analysis identified genome-wide significant associations at 7 of 8 loci (all but JAZF1-AS1). On the other hand, the African-specific meta-analysis only identified a genome-wide significant association at the F7 locus itself. Ancestry-specific Manhattan plots are shown in supplemental Figures 6 and 7, whereas ancestry-specific regional plots are shown in supplemental Figure 8. A comparison of the ancestry-specific betas (effect sizes) of the 8 genome-wide significant lead variants from the trans-ancestry analysis is shown in supplemental Figure 9. Overall, effect sizes were very similar across the 2 ancestry groups.
No additional genome-wide significant loci emerged when adding data from 2 additional studies in the combined analysis of FVII activity and antigen, but variants at the TSKU and JAZF1-AS1 loci were no longer genome-wide significant (supplemental Table 6). A QQ plot and Manhattan plot for the combined analysis of FVII activity and antigen are shown in supplemental Figures 10 and 11, respectively. The lead variants at TSKU and JAZF1-AS1 had opposing-effect directions on FVII activity and antigen, but lead variants at the remaining 6 loci had relatively similar effects on FVII activity and antigen (supplemental Figure 12). The variance in FVII activity explained by the lead variants at the 8 significant loci was 17.6%. The variance explained by each of the lead variants individually is shown in Table 1.
Conditional analysis
Conditional analysis identified 4 independent signals at the F7 locus as well as 2 independent signals at the PROCR locus. The conditional analysis of the trans-ancestry meta-analysis of FVII activity is shown in Table 2. Among the independently associated variants at the F7 locus was a low-frequency variant (minor allele frequency = 0.02) with the second largest effect size discovered in GWASs of FVII thus far (joint β = 0.08; joint P = 8.7 × 10−20). By considering these independent signals, the variance in FVII activity explained by the F7 locus increased from 13.9% to 15.2%, whereas the variance explained by the PROCR locus increased from 1.6% to 1.8%. The total variance explained therefore increased from 17.6% to 19.0%.
Table 2.
Conditional analysis results for FVII activity using the trans-ancestry meta-analysis results
| rsID | Chr:Pos* | Alleles† | Freq‡ | β§ | P§ | Joint β|| | Joint P|| | Variance explained, %¶ |
|---|---|---|---|---|---|---|---|---|
| F7 | ||||||||
| rs117989138 | 13:113697671 | A/G | 0.02 | 0.086 | 3.6 × 10−22 | 0.081 | 8.7 × 10−20 | 0.6 |
| rs36086577 | 13:113728498 | C/A | 0.87 | 0.035 | 2.2 × 10−19 | 0.031 | 7.5 × 10−15 | 0.6 |
| rs71446935 | 13:113734376 | A/G | 0.31 | 0.035 | 5.5 × 10−38 | 0.032 | 5.1 × 10−31 | 1.2 |
| rs1046205 | 13:113752057 | A/T | 0.79 | 0.121 | 3.9 × 10−573 | 0.121 | <1.0 × 10−320 | 12.1 |
| PROCR | ||||||||
| rs6119569 | 20:33672371 | G/A | 0.78 | 0.022 | 8.8 × 10−17 | 0.019 | 3.9 × 10−13 | 0.3 |
| rs867186 | 20:33764554 | G/A | 0.10 | 0.057 | 3.3 × 10−64 | 0.055 | 8.1 × 10−59 | 1.4 |
The variance explained shown in this table was calculated using the sample size weighted mean variance of log-transformed FVII and the betas and frequencies from the trans-ancestry meta-analysis summary statistics.
Abbreviations are explained in Table 1.
The Chr:Pos column shows the chromosome and position (build 37).
The Alleles column shows the FVII-increasing allele/FVII-decreasing allele.
The Freq column shows the frequency of the FVII-increasing allele.
The β and P columns are based on the association of each variant in isolation.
The Joint β and Joint P columns are based on the association of each variant test conditioned on the other variants.
The Variance Explained column is based on the joint analysis.
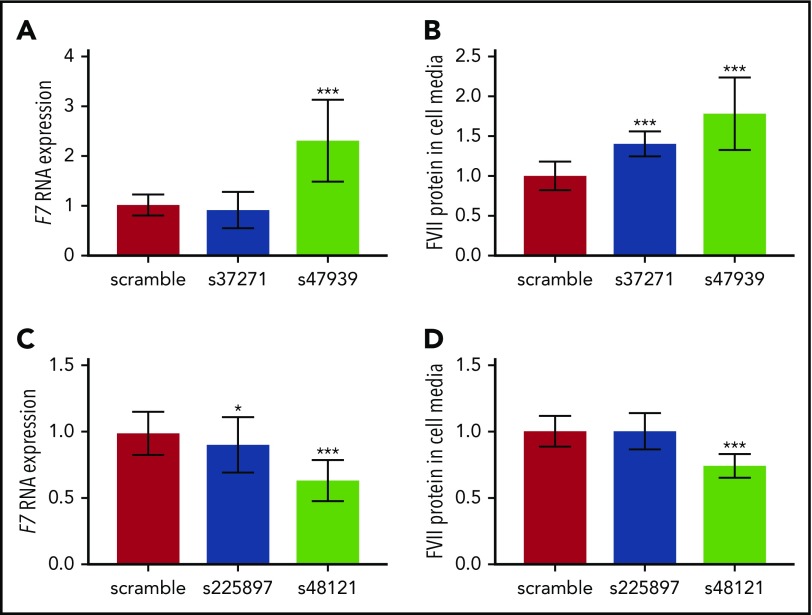
Functional validation of novel loci
The s47939 and s37271 silencing siRNAs both suppressed mRNA expression of their target gene, REEP3, by 88% compared with the scramble siRNA (negative control). Experiments were repeated 3 times with consistent results, showing that silencing of REEP3 resulted in upregulation of F7 mRNA (P = .0001 for s47939; P > .05 for s37271; Figure 1) and a corresponding increase in FVII protein levels (P = 9.1 × 10−5 for s47939; P = .0003 for s37271; Figure 1).
Figure 1.
Gene silencing in HuH7 cells. (A) F7 RNA expression after silencing REEP3, (B) FVII protein levels in cell media after silencing REEP3, (C) F7 RNA expression after silencing JAZF1, and (D) FVII protein levels in cell media after silencing JAZF1. *P = .01-.05; ***P < .001. Error bars indicate ±1 SD.
At the JAZF1-AS1 locus, we targeted the JAZF1 gene for silencing rather than the antisense RNA in which the lead variant was located. The s225897 silencer reduced mRNA expression of its target gene, JAZF1, by 68%, whereas the s48121 silencer reduced JAZF1 mRNA expression by 75%. As shown in Figure 1, silencing of JAZF1 resulted in modest downregulation of F7 mRNA (P = .02 for s225897; P < 2 × 10−6 for s48121) and a corresponding decrease in FVII protein expression: silencing experiments showed no effect on FVII protein in the media of cells silenced with s225897, but a significant decrease upon silencing with s48121 (P = 1.1 × 10−6).
As a positive control, we also silenced the F7 gene itself, which led to an 88% reduction in F7 mRNA levels (supplemental Figure 13). This demonstrates that our gene-silencing system was able to effectively assess differences in FVII expression.
Mendelian randomization
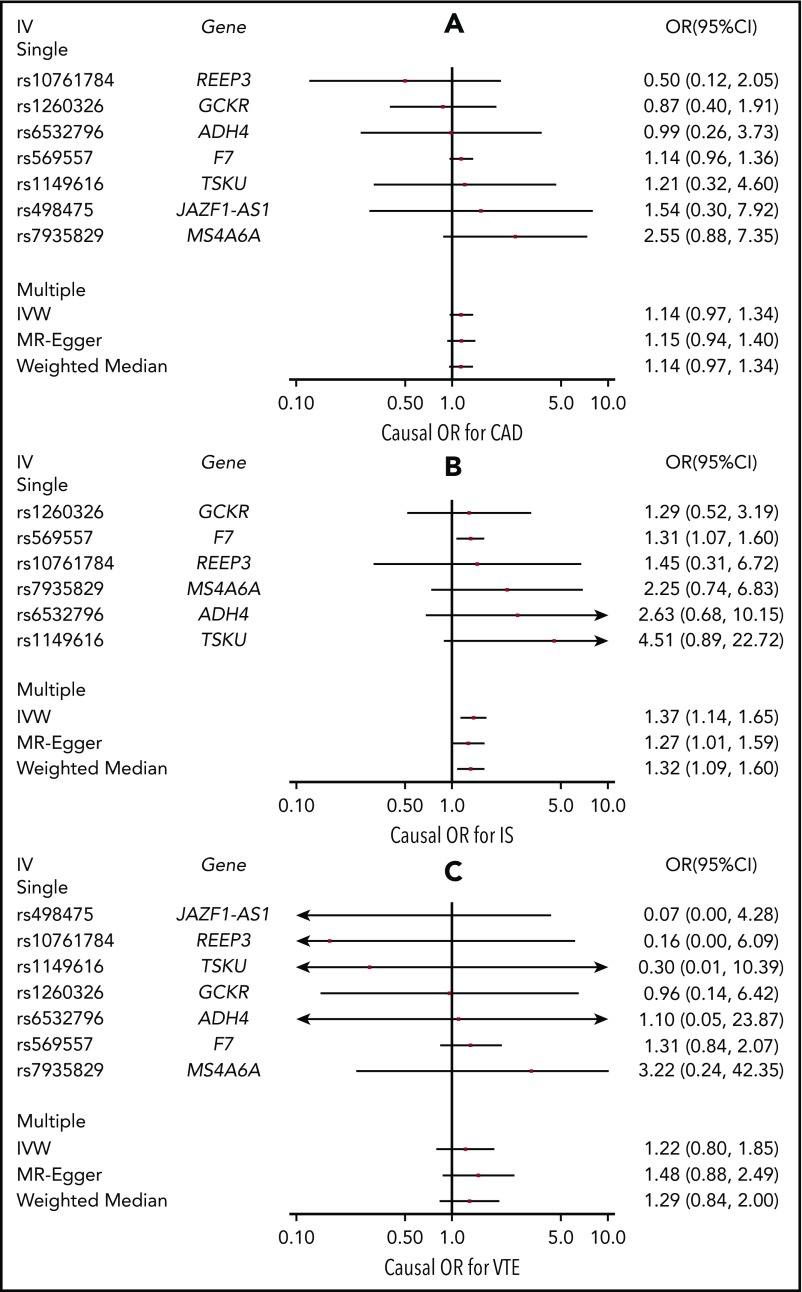
Figure 2 contains forest plots showing causal-effect estimates of FVII activity on (A) CAD, (B) IS, and (C) VTE. Causal effect estimates are given as odds ratios (ORs) per 1 unit higher natural log-transformed FVII activity (percentage or IU/mL × 100). The variant at the PROCR locus, rs867186, was removed from all analyses due to outlying causal -effect estimates for CAD, IS, and VTE. No heterogeneity in the single-variant causal-effect estimates was observed among the remaining variants for each outcome (Pheterogeneity > .05).
Figure 2.
Causal-effect estimates. Causal-effect estimates of FVII activity on CAD (A), IS (B), and VTE (C) using Mendelian randomization. Causal-effect estimates are shown as ORs and 95% CIs per 1 unit higher natural log-transformed FVII activity (percentage or IU/ml × 100) FVII activity. Causal estimates based on single variant (“Single” in plot title) instrumental variables (IVs) are shown, as well as causal estimates based on the combination of these variants (“Multiple” in plot title) using inverse variance weighted (IVW) meta-analysis, MR-Egger, and weighted median estimation.
All outcomes had positive point estimates for the causal effect across all main and sensitivity analyses. A significant causal effect of FVII activity on IS was detected using the inverse-variance weighted approach (OR = 1.37; 95% confidence interval [CI] = 1.14-1.65). Given that the SD of natural-log-transformed FVII activity ranged from 0.18 to 0.26 across our studies, the causal-effect estimate corresponds to an approximate OR of 1.06 to 1.09 per SD change in natural log-transformed FVII activity. Results were consistent across sensitivity analyses, including the use of MR-Egger, the weighted median estimator, and restriction of the analysis to the rs569557 variant at the F7 locus. These findings indicate that pleiotropy is unlikely to have biased the causal estimate. Causal-effect estimates for CAD (OR = 1.14; 95% CI = 0.97-1.34) and VTE (OR = 1.22; 95% CI = 0.80-1.85) using the inverse-variance weighted approach were more modest and had wider CIs. Nevertheless, the magnitude of these effect estimates was consistent across sensitivity analyses, including when the rs569557 variant at the F7 locus was examined in isolation (ORCAD = 1.14; 95% CI = 0.96-1.36; ORVTE = 1.31; 95% CI = 0.84-2.07).
Discussion
In this GWAS of circulating FVII levels, we identified the 6 previously known FVII loci as well as 2 new loci: REEP3 and JAZF1-AS1. In total, the 8 loci associated with FVII activity explained 19.0% of the variance. For each new discovery, we showed functional impact in vitro of candidate genes on F7 mRNA and FVII protein expression: REEP3 gene silencing in liver cells increased F7 mRNA and FVII protein expression, whereas JAZF1 gene silencing decreased F7 mRNA and FVII protein expression. Mendelian randomization analyses suggest that FVII activity has a positive causal effect on the risk of IS.
Annotation of associated loci
REEP3 encodes receptor accessory protein 3. Although this protein has not been widely studied, there is evidence that an absence of this protein leads to defects in mitosis and a proliferation of intranuclear membranes derived from the nuclear envelope.36 The locus containing REEP3 has been previously associated to several other coagulation phenotypes, namely circulating fibrinogen levels,37,38 mean platelet volume,39,40 and platelet aggregation,41 as well as to liver enzyme concentrations.42,43 For many of these phenotypes, the gene that was reported is not REEP3 but JMJD1C, with missense variants localized in the JMJD1C being associated with mean platelet volume.39 Functional studies in zebrafish indicate that JMJD1C has a major role in hematopoiesis.44 Although we did not examine the consequences of JMJD1C silencing on F7 expression and FVII release, our experiments implicate REEP3 as a causal gene for FVII. These results are consistent with tissue-specific pleiotropic effects at this locus, with JMJD1C being involved in hematopoiesis and REEP3 being of functional relevance in the liver, although further research is needed to confirm this hypothesis.
JAZF1-AS1 is a noncoding antisense RNA that may regulate the adjacent JAZF1 gene, which encodes a transcriptional repressor. We targeted JAZF1 in our gene-silencing experiments instead of JAZF1-AS1 because many antisense noncoding RNAs regulate the protein-coding gene that they are closest to. Silencing of JAZF1 in liver cells resulted in lower F7 mRNA and FVII protein expression, suggesting that the mechanism underlying the genetic association is likely to involve FVII levels. However, variants at the JAZF1-AS1 locus were associated with FVII activity, and their effect on FVII was attenuated when we included studies that measured FVII antigen. A possible explanation is that variants and this locus have an additional effect on FVII activity that is independent of FVII protein levels.
Apart from REEP3 and JAZF1-AS1, we identified 6 known loci: F7, PROCR, GCKR, MS4A6A, ADH4, and TSKU. The results of this study may aid in the identification of causal variants at these loci. For example, lead variants in PROCR and GCKR were both missense variants leading to amino acid substitutions (Ser219Gly in PROCR and Pro446Leu in GCKR). These variants were also the lead variants in their respective loci in the previous GWAS of FVII,8 and have been associated with other hemostatic phenotypes.45,46 In contrast, the lead variants that we identified at the F7, MS4A6A, ADH4, and TSKU loci differ from those published in the previous GWAS and may be in stronger linkage disequilibrium with the true causal variant.8
We also identified 15 additional loci that were suggestively associated with FVII activity (P < 2.5 × 10−6). These loci harbor several notable candidate genes, including MLXIPL, HNF4A, and XXYLT1. The first 2 genes have been previously found to be associated with FVII using a candidate gene design.47 XXYLT1 encodes xyloside xylosyltransferase 1, which elongates O-linked glycans by adding the second xylose to O-glucose-modified residues in the epidermal growth factor repeats of proteins. FVII has epidermal growth factor repeats that are known to be O-linked glycosylated.48
Mendelian randomization
Using the genetic-association results generated in this study, we performed Mendelian randomization analyses to estimate the causal effect of FVII activity on CAD, IS, and VTE. These analyses suggest that lifetime elevations in FVII activity influence the risk of IS in the general population. These results warrant further etiological research on the role of FVII in IS, as well as translational research on potential clinical applications involving FVII. Lifetime differences in FVII activity driven by genetic variants may have greater effects on outcomes than transient modifications made later in life. Therefore, even if a causal effect is confirmed, clinical research is needed to quantify the effect of specific interventions targeting FVII.49,50 Potential clinical applications that should be investigated include the reduction of FVII activity through lifestyle or pharmaceutical interventions for the prevention of IS. Our results also provide independent support for the restriction of off-label use of recombinant FVII, which trials have found to increase the risk of arterial thrombosis.51-53
For CAD and VTE, the point estimates of the causal effects were smaller than for IS and the CIs were wider. Although not statistically significant, these results do not exclude the possibility of additional causal effects of FVII activity on CAD and VTE. In fact, when using the rs569557 variant at the F7 locus in isolation as an instrumental variable, the point estimate of the causal effect on VTE was similar to, but less precise than, the estimate of the causal effect on IS. Lower statistical power for the Mendelian randomization analysis of VTE may explain the lack of a statistically significant causal effect of FVII activity on this outcome: the GWAS from which we obtained the effect of the variants on IS was composed of 60 341 cases and 454 450 controls,31 whereas the GWAS on VTE consisted of 7507 cases and 52 632 controls.32 Our results are thus consistent with positive and potentially clinically important causal effects of FVII activity on CAD and VTE. Further Mendelian randomization studies with increased sample sizes are warranted given the wide CIs.
Strengths and limitations
Our GWAS included 27 495 participants, a 74% increase in sample size as compared with the largest previous GWAS of FVII levels.8 Other strengths include insights into causal effects and disease etiology generated by functional validation of newly identified loci, through silencing candidate genes in human liver cell lines, and the use of Mendelian randomization. However, these approaches also have limitations. In the gene-silencing experiments, we silenced a single gene at each locus. Because an effect of FVII levels was observed in both cases, we did not pursue further experiments involving other genes at these loci. As such, we cannot exclude the possibility that other genes at these loci also influence FVII. However, our results provide a basis for the additional research that will be required to fully uncover the functional mechanisms linking these loci to FVII. Mendelian randomization analyses can be potentially biased by pleiotropic effects. To minimize the impact of pleiotropy on our results, we validated observed causal effects using MR-Egger, weighted median estimation, and restricting the Mendelian randomization analysis to the rs569557 variant at the F7 locus, each of which estimate causal effects using independent methodologies robust to bias from pleiotropy.33,34 The estimates of the causal effect of FVII activity on IS, CAD, and VTE were consistent across these sensitivity analyses. However, there is a degree of sample overlap between our GWAS of FVII and the GWAS of IS, CAD, and VTE, which may bias the effect estimates away from the null.54 At the same time, differences in the ancestral composition of the samples used to generate summary statistics for FVII activity, CAD, IS, and VTE may bias the effect estimates toward the null. Finally, we imputed genotypes using the 1000 Genomes Project reference panel. Compared with previous genetic-association studies on FVII, we therefore have improved coverage and more accurate determination of variants across the allele frequency spectrum.55 Nevertheless, the coverage of low-frequency and rare variants could be improved even further through the use of whole-genome sequencing instead of imputation genotypes. Thus, sequencing-based studies might identify further associations missed by our study, especially those involving rare variants.
In conclusion, this study identifies 2 novel loci associated with FVII activity, and functional studies suggest that REEP3 and JAZF1 are causal genes within these loci. Mendelian randomization analyses indicate that FVII activity is causally involved in the development of IS, while not excluding similar causal effects on CAD and VTE.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
The authors thank the staff and participants of each of the participating studies for their important contributions. The authors are grateful to Alexandra Bäcklund, Fariba Foroogh, and Angela Silveira for helpful advice in the cell culture and mesoscale experiments. The authors thank Martin Dichgans, Stéphanie Debette, and Rainer Malik for their assistance in coordinating access to data from the MEGASTROKE Consortium. The authors thank all colleagues from the French Centre National de Génotypage for the genotyping contribution.
This work was supported by American Heart Association grant #17POST33350042 (P.S.d.V.) and National Institutes of Health (NIH), National Heart, Lung, and Blood Institute grant R01HL134894 (N.L.S.). M.S.-L. was partially supported by a European Hematology Association (EHA)–International Society on Thrombosis and Haemostasis (ISTH) fellowship and by the Swedish Heart-Lung Foundation (20160290), and is a recipient of a Miguel Servet contract from the Spanish Ministry of Health (ISCIII CP17/00142). Infrastructure for the CHARGE Consortium was supported in part by National Heart, Lung, and Blood Institute grant R01HL105756. D.G. contributed to this work as part of a Wellcome Trust funded Clinical Research Training Fellowship. N.M.D. works at the Medical Research Council (MRC) and the University of Bristol supported the MRC Integrative Epidemiology Unit (MC_UU_12013/1, MC_UU_12013/9, MC_UU_00011/1). The Atherosclerosis Risk in Communities (ARIC) study was carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, and HHSN268201100012C), R01HL087641, R01HL59367, and R01HL086694; NIH, National Human Genome Research Institute contract U01HG004402; and NIH contract HHSN268200625226C. Infrastructure was partly supported by grant number UL1RR025005, a component of the NIH and NIH Roadmap for Medical Research. Funding support for “Building on GWAS for National Heart, Lung, and Blood Institute diseases: the US CHARGE Consortium” was provided by the NIH through the American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419). Cardiovascular Health Study (CHS) research was supported by National Heart, Lung, and Blood Institute contracts HHSN268201200036C, HHSN268200800007C, HHSN268200960009C, HHSN268201800001C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086; and National Heart, Lung, and Blood Institute grants U01HL080295, R01HL085251, R01HL087652, R01HL105756, R01HL103612, R01HL120393, and R01HL130114 with additional contribution from the National Institute of Neurological Disorders and Stroke (NINDS). Additional support was provided through R01AG023629 from the NIH, National Institute on Aging. A full list of principal CHS investigators and institutions can be found at CHS-NHLBI.org. The provision of genotyping data was supported in part by the NIH, National Center for Advancing Translational Sciences, Clinical and Translational Science Institute grant UL1TR000124, and the National Institute of Diabetes and Digestive and Kidney Disease Diabetes Research Center (DRC) grant DK063491 to the Southern California Diabetes Endocrinology Research Center. The Coronary Artery Risk Development in Young Adults Study (CARDIA) was conducted and supported by the National Heart, Lung, and Blood Institute in collaboration with the University of Alabama at Birmingham (HHSN268201300025C and HSN268201300026C), Northwestern University (HHSN268201300027C), University of Minnesota (HHSN268201300028C), Kaiser Foundation Research Institute (HHSN268201300029C), and Johns Hopkins University School of Medicine (HHSN268200900041C). CARDIA was also partially supported by the Intramural Research Program of the National Institute on Aging and an intra-agency agreement between the National Institute on Aging and the National Heart, Lung, and Blood Institute (AG0005). Genotyping was funded as part of the National Heart, Lung, and Blood Institute Candidate-gene Association Resource (N01-HC-65226) and the National Human Genome Research Institute Gene Environment Association Studies (GENEVA; U01-HG004729, U01-HG04424, and U01-HG004446). The Framingham Heart Study was partially supported by the National Heart, Lung, and Blood Institute’s Framingham Heart Study (contract no. N01-HC-25195) and its contract with Affymetrix, Inc for genotyping services (contract no. N02-HL-6-4278). A portion of this research used the Linux Cluster for Genetic Analysis (LinGA-II) funded by the Robert Dawson Evans Endowment of the Department of Medicine at Boston University School of Medicine and Boston Medical Center. The analyses reflect intellectual input and resource development from the Framingham Heart Study investigators participating in the SNP Health Association Resource (SHARe) project. J.E.H., C.S., M.-H.C., A.D.J., and C.J.O. were supported by National Heart, Lung and Blood Intramural Research Funds. The Genetic Analysis for Idiopathic Thrombophilia 2 (GAIT2) project was supported partially by grants PI-11/0184, PI-14/0582, and Red Investigación Cardiovascular RD12/0042/0032 from the Instituto Carlos III (Fondo de Investigación Sanitaria [FIS]), and 2014SGR-402 from the Consolidated Research Group of the Generalitat de Catalunya. The LUdwigshafen RIsk and Cardiovascular Health (LURIC) study was supported by the 7th Framework Program RiskyCAD (grant agreement number 305739) of the European Union. The work of W.M. and M.E.K. was supported as part of the Competence Cluster of Nutrition and Cardiovascular Health (nutriCARD), which was funded by the German Federal Ministry of Education and Research. The Multiple Environmental and Genetic Assessment of risk factors for venous thrombosis (MEGA) study was supported by the Netherlands Heart Foundation (NHS 98.113), the Dutch Cancer Foundation (RUL 99/1992), and the Netherlands Organisation for Scientific Research (912–03–033| 2003), and was partially supported by the GenMed LABEX (ANR-10-LABX-0013). The Precocious Coronary Artery DISease (PROCARDIS) study project was funded with 10 million Euros through the 6th Framework Program of the European Union (LSH-2005-2.1.1-1). It started in April 2007 and funding lasted until September 2011. Although the funding ended, PROCARDIS is still actively doing research in the field of coronary artery disease genetics. The generation and management of GWAS genotype data for the Rotterdam Study (RS) was supported by the Netherlands Organisation of Scientific Research NWO Investments (no. 175.010.2005.011, 911-03-012). This study was funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) project no. 050-060-810. The Rotterdam Study was funded by Erasmus Medical Center and Erasmus University, Rotterdam; Netherlands Organization for the Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly (RIDE); the Ministry of Education, Culture and Science; the Ministry for Health, Welfare and Sports; the European Commission (DG XII); and the Municipality of Rotterdam.
The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute, the National Institutes of Health, or the US Department of Health and Human Services. This manuscript does not necessarily represent US Environmental Protection Agency (EPA) policy. Mention of products or trade names does not constitute endorsement by the EPA.
This manuscript has been reviewed and approved by CARDIA for scientific content.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: P.S.d.V., M.S.-L., J.E.H., J.M., C.S., A.D.J., A.C.M., C.J.O., and N.L.S. contributed to the study design and conception, and to the drafting of the manuscript; M.S.-L., H.G.d.H., A.M.-P., M.P.M.d.M., M. Frånberg, M.E.K., F.R., J.M.S., W.T., G.H.T., A.G.U., A.v.H.V., S.S., E.B., A.-K.G., M.K.I., S.J.K., B.M.P., A.P.R., M.S., K.D.T., M. Fornage, A.H., W.M., F.R.R., J.C.S., A.D.J., A.C.M., and C.J.O. contributed to data collection and processing; P.S.d.V., M.S.-L., J.E.H., J.M., C.S., N.P., T.M.B., H.G.d.H., G.E.D., J.D.E., A.M.-P., C.K.W.-C., J.A.B., M.-H.C., D.G., M.E.K., N.M.D., B.M., A.D., and N.L.S. contributed to data analysis and interpretation; M.S.-L. conducted the gene-silencing experiments; and all authors critically reviewed and approved the manuscript.
Conflict-of-interest disclosure: W.M. reports grants and personal fees from AMGEN, BASF, Sanofi, Siemens Diagnostics, Aegerion Pharmaceuticals, Astrazeneca, Danone Research, Numares, Pfizer, and Hoffmann LaRoche; personal fees from MSD and Alexion; grants from Abbott Diagnostics, all outside of the submitted work; and is employed by Synlab Holding Deutschland GmbH. B.M.P. serves on the Data Safety Monitoring Board of a clinical trial funded by Zoll LifeCor and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson. N.M.D. reports a grant for research unrelated to this work from the Global Research Awards for Nicotine Dependence (GRAND), an independent grant-making body funded by Pfizer. The remaining authors declare no competing financial interests.
Complete lists of the members of the INVENT Consortium and the MEGASTROKE Consortium appear in the supplemental appendix.
Correspondence: Paul S. de Vries, Department of Epidemiology, Human Genetics, and Environmental Sciences, University of Texas Health Science Center at Houston, 1200 Pressler St, Suite E-429, Houston, TX 77030; e-mail: paul.s.devries@uth.tmc.edu; and Nicholas L. Smith, Department of Epidemiology, University of Washington, 1730 Minor Ave, Suite 1360, Seattle, WA 98101; e-mail: nlsmith@u.washington.edu.
REFERENCES
- 1.Lapecorella M, Mariani G; International Registry on Congenital Factor VII Deficiency. Factor VII deficiency: defining the clinical picture and optimizing therapeutic options. Haemophilia. 2008;14(6):1170-1175. [DOI] [PubMed] [Google Scholar]
- 2.Zakai NA, Lange L, Longstreth WT Jr, et al. Association of coagulation-related and inflammation-related genes and factor VIIc levels with stroke: the Cardiovascular Health Study. J Thromb Haemost. 2011;9(2):267-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith A, Patterson C, Yarnell J, Rumley A, Ben-Shlomo Y, Lowe G. Which hemostatic markers add to the predictive value of conventional risk factors for coronary heart disease and ischemic stroke? The Caerphilly Study. Circulation. 2005;112(20):3080-3087. [DOI] [PubMed] [Google Scholar]
- 4.Tsai AW, Cushman M, Rosamond WD, et al. Coagulation factors, inflammation markers, and venous thromboembolism: the longitudinal investigation of thromboembolism etiology (LITE). Am J Med. 2002;113(8):636-642. [DOI] [PubMed] [Google Scholar]
- 5.Folsom AR. Hemostatic risk factors for atherothrombotic disease: an epidemiologic view. Thromb Haemost. 2001;86(1):366-373. [PubMed] [Google Scholar]
- 6.Freeman MS, Mansfield MW, Barrett JH, Grant PJ. Genetic contribution to circulating levels of hemostatic factors in healthy families with effects of known genetic polymorphisms on heritability. Arterioscler Thromb Vasc Biol. 2002;22(3):506-510. [DOI] [PubMed] [Google Scholar]
- 7.Souto JC, Almasy L, Borrell M, et al. Genetic determinants of hemostasis phenotypes in Spanish families. Circulation. 2000;101(13):1546-1551. [DOI] [PubMed] [Google Scholar]
- 8.Smith NL, Chen MH, Dehghan A, et al. ; Wellcome Trust Case Control Consortium. Novel associations of multiple genetic loci with plasma levels of factor VII, factor VIII, and von Willebrand factor: the CHARGE (Cohorts for Heart and Aging Research in Genome Epidemiology) Consortium. Circulation. 2010;121(12):1382-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Psaty BM, O’Donnell CJ, Gudnason V, et al. ; CHARGE Consortium. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ Cardiovasc Genet. 2009;2(1):73-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The Atherosclerosis Risk in Communities (ARIC) study: design and objectives. The ARIC investigators. Am J Epidemiol. 1989;129(4):687-702. [PubMed] [Google Scholar]
- 11.Fried LP, Borhani NO, Enright P, et al. The Cardiovascular Health Study: design and rationale. Ann Epidemiol. 1991;1(3):263-276. [DOI] [PubMed] [Google Scholar]
- 12.Friedman GD, Cutter GR, Donahue RP, et al. CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol. 1988;41(11):1105-1116. [DOI] [PubMed] [Google Scholar]
- 13.Camacho M, Martinez-Perez A, Buil A, et al. Genetic determinants of 5-lipoxygenase pathway in a Spanish population and their relationship with cardiovascular risk. Atherosclerosis. 2012;224(1):129-135. [DOI] [PubMed] [Google Scholar]
- 14.Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham Offspring Study. Design and preliminary data. Prev Med. 1975;4(4):518-525. [DOI] [PubMed] [Google Scholar]
- 15.Winkelmann BR, März W, Boehm BO, et al. ; LURIC Study Group (LUdwigshafen RIsk and Cardiovascular Health). Rationale and design of the LURIC study--a resource for functional genomics, pharmacogenomics and long-term prognosis of cardiovascular disease. Pharmacogenomics. 2001;2(1 suppl 1):S1-S73. [DOI] [PubMed] [Google Scholar]
- 16.Blom JW, Doggen CJ, Osanto S, Rosendaal FR. Malignancies, prothrombotic mutations, and the risk of venous thrombosis. JAMA. 2005;293(6):715-722. [DOI] [PubMed] [Google Scholar]
- 17.Barlera S, Chiodini BD, Franzosi MG, Tognoni G. PROCARDIS: a current approach to the study of the genetics of myocardial infarct [in Italian]. Ital Heart J Suppl. 2001;2(9):997-1004. [PubMed] [Google Scholar]
- 18.Hofman A, Brusselle GG, Darwish Murad S, et al. The Rotterdam Study: 2016 objectives and design update. Eur J Epidemiol. 2015;30(8):661-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44(8):955-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34(8):816-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winkler TW, Day FR, Croteau-Chonka DC, et al. ; Genetic Investigation of Anthropometric Traits (GIANT) Consortium. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 2014;9(5):1192-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J, Ellinghaus D, Franke A, Howie B, Li Y. 1000 Genomes-based imputation identifies novel and refined associations for the Wellcome Trust Case Control Consortium phase 1 data. Eur J Hum Genet. 2012;20(7):801-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fadista J, Manning AK, Florez JC, Groop L. The (in)famous GWAS P-value threshold revisited and updated for low-frequency variants. Eur J Hum Genet. 2016;24(8):1202-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Ferreira T, Morris AP, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44(4):369-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88(1):76-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikpay M, Goel A, Won HH, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47(10):1121-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malik R, Chauhan G, Traylor M, et al. ; MEGASTROKE Consortium. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat Genet. 2018;50(4):524-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Germain M, Chasman DI, de Haan H, et al. ; Cardiogenics Consortium. Meta-analysis of 65,734 individuals identifies TSPAN15 and SLC44A2 as two susceptibility loci for venous thromboembolism. Am J Hum Genet. 2015;96(4):532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swerdlow DI, Kuchenbaecker KB, Shah S, et al. Selecting instruments for Mendelian randomization in the wake of genome-wide association studies. Int J Epidemiol. 2016;45(5):1600-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlaitz AL, Thompson J, Wong CC, Yates JR III, Heald R. REEP3/4 ensure endoplasmic reticulum clearance from metaphase chromatin and proper nuclear envelope architecture. Dev Cell. 2013;26(3):315-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Vries PS, Chasman DI, Sabater-Lleal M, et al. A meta-analysis of 120 246 individuals identifies 18 new loci for fibrinogen concentration. Hum Mol Genet. 2016;25(2):358-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sabater-Lleal M, Huang J, Chasman D, et al. ; CARDIoGRAM Consortium. Multiethnic meta-analysis of genome-wide association studies in >100 000 subjects identifies 23 fibrinogen-associated Loci but no strong evidence of a causal association between circulating fibrinogen and cardiovascular disease. Circulation. 2013;128(12):1310-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eicher JD, Chami N, Kacprowski T, et al. ; Myocardial Infarction Genetics Consortium. Platelet-related variants identified by Exomechip meta-analysis in 157,293 individuals. Am J Hum Genet. 2016;99(1):40-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soranzo N, Spector TD, Mangino M, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009;41(11):1182-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson AD, Yanek LR, Chen MH, et al. Genome-wide meta-analyses identifies seven loci associated with platelet aggregation in response to agonists. Nat Genet. 2010;42(7):608-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chambers JC, Zhang W, Sehmi J, et al. ; Meta-analyses of Glucose and Insulin-Related Traits Consortium (MAGIC). Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43(11):1131-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.D’Cruz OJ, Haas GG Jr, de La Rocha R, Lambert H. Occurrence of serum antisperm antibodies in patients with cystic fibrosis. Fertil Steril. 1991;56(3):519-527. [PubMed] [Google Scholar]
- 44.Gieger C, Radhakrishnan A, Cvejic A, et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480(7376):201-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sennblad B, Basu S, Mazur J, et al. Genome-wide association study with additional genetic and post-transcriptional analyses reveals novel regulators of plasma factor XI levels. Hum Mol Genet. 2017;26(3):637-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang W, Basu S, Kong X, et al. Genome-wide association study identifies novel loci for plasma levels of protein C: the ARIC study. Blood. 2010;116(23):5032-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor KC, Lange LA, Zabaneh D, et al. A gene-centric association scan for coagulation factor VII levels in European and African Americans: the Candidate Gene Association Resource (CARe) Consortium. Hum Mol Genet. 2011;20(17):3525-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sethi MK, Buettner FF, Krylov VB, et al. Identification of glycosyltransferase 8 family members as xylosyltransferases acting on O-glucosylated notch epidermal growth factor repeats. J Biol Chem. 2010;285(3):1582-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burgess S, Butterworth A, Malarstig A, Thompson SG. Use of Mendelian randomisation to assess potential benefit of clinical intervention. BMJ. 2012;345:e7325. [DOI] [PubMed] [Google Scholar]
- 50.Davey Smith G, Paternoster L, Relton C. When will Mendelian randomization become relevant for clinical practice and public health? JAMA. 2017;317(6):589-591. [DOI] [PubMed] [Google Scholar]
- 51.Logan AC, Yank V, Stafford RS. Off-label use of recombinant factor VIIa in U.S. hospitals: analysis of hospital records. Ann Intern Med. 2011;154(8):516-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yank V, Tuohy CV, Logan AC, et al. Systematic review: benefits and harms of in-hospital use of recombinant factor VIIa for off-label indications. Ann Intern Med. 2011;154(8):529-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Levi M, Levy JH, Andersen HF, Truloff D. Safety of recombinant activated factor VII in randomized clinical trials. N Engl J Med. 2010;363(19):1791-1800. [DOI] [PubMed] [Google Scholar]
- 54.Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol. 2016;40(7):597-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Vries PS, Sabater-Lleal M, Chasman DI, et al. Comparison of HapMap and 1000 Genomes reference panels in a large-scale genome-wide association study. PLoS One. 2017;12(1):e0167742. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.