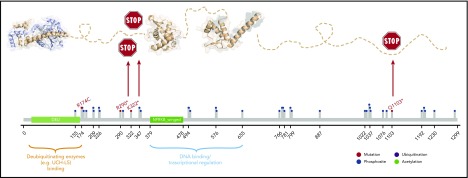
Figure 2.
Modeling of NFRKB mutations. NFRKB mutations (red lollipops) annotated on protein primary sequence with additional information regarding posttranslational modifications (PTMs) and domain composition. Three-dimensional structural model showing the deubiquitinating enzyme-binding domain (Protein Data Bank identifier [PDB ID]: 4UF5), the winged domain (PDB ID: 3U21), and the transcriptional regulation domain (by homology; PDB ID: 3BY6). The 2 stop-gain mutations R290* and K322* are predicted to delete the protein region responsible for mediating interactions with transcriptional regulators and/or DNA whereas leaving the N-terminal region deputed to the interaction with deubiquitnating enzymes. The R174C mutation is predicted to perturb adjacent phosphosites, for example, S176, and is located in a motif predicted by the Eukaryotic Linear Motif (ELM; elm.eu.org) to be either the phosphorylation site of GSK3 and MAPK or the docking site for USP7 or the Pin1 WW domain. The C-terminal deletion by Q1103* is predicted to have detrimental consequences, as it is rich in phosphorylation sites (eg, CK1 and GSK3) or recognition sites for FHA and WDR5.