Abstract
This study evaluated the use of molecular breeding values (MBVs) for carcass traits to sort steers into quality grid and lean meat yield (LMY) groups. A discovery set of 2,609 animals with genotypes and carcass phenotypes was used to predict MBVs for LMY and marbling score (MBS) for 299 Angus, 181 Charolais, and 638 Kinsella Composite steers using genomic best linear unbiased prediction. Steers were sorted in silico into four MBV groups namely Quality (with MBVs greater than the mean for LMY and MBS), Lean (with MBVs greater than the mean for LMY but less than or equal to the mean for MBS), Marbling (with MBVs greater than the mean for MBS but less than or equal to the mean for LMY), and Other (with MBVs lower than the mean for LMY and MBS). Carcass phenotypes on the steers were then collected at slaughter and evaluated for consistency with the assigned MBV groups using descriptive statistics and least square analysis. Accuracy of MBV predictions was assessed by Pearson’s correlation between predicted MBV and adjusted phenotype divided by the square root of trait heritability. Genomic breed compositions were predicted for all steers to correct for possible population stratification and breed effects in the test model. The number of steers that met the expected carcass outcome was counted to produce actual percentages for each MBV group. Results showed that on average, Quality and Marbling groups had greater back-fat and more marbling across the three populations while Lean group had leaner carcasses. Carcass weights were similar across MBV groups. Within MBV groups, decreases in variability were observed for most traits suggesting improvement in carcass uniformity. Greater than 70% of the steers in Quality, Lean, and Marbling groups met the Quality Grid and Y1-LMY target for Angus and Charolais but not for Kinsella composite. The accuracy of MBV prediction ranged from 0.43 to 0.59 indicating that up to 35% of the observed carcass trait variability can be predicted, which suggests utility of MBV as a marker-assisted management tool to sort feeder cattle into uniform carcass endpoint groups under similar environmental and management conditions. Further investigation is warranted to evaluate the performance of feeder cattle sorted based on MBV and managed for different carcass endpoints as well as the cost–benefit implications for feedlot operations.
Keywords: beef carcass, genomics, marker-assisted management, steers
INTRODUCTION
The identification of feeder cattle that will produce greater carcass uniformity with acceptable yield and quality grade (QG) is a long-term goal of the beef cattle industry. In an effort to improve production efficiency, product uniformity, and carcass value, producers have applied visual inspection, individual animal weight, and ultrasound scan of back fat to sort incoming feedlot cattle into outcome groups (Basarab, et al., 1997, 1999; MacDonald et al., 2006). However, these strategies may be biased by preferential treatment and environmental variability especially when crossbreeding is used as a standard approach to produce commercial cattle (Nichols et al., 2014).
One possible solution to improve product uniformity and quality of beef is through genomic characterization (Rincker et al., 2006; DeNise et al., 2008; MacNeil et al., 2010). With the availability of affordable genotyping services for beef cattle, molecular breeding values (MBVs) of traits in the breeding objective can be accurately predicted for all possible candidates from dense SNP (Meuwissen et al., 2001). These MBVs can be used for preselection of individuals entering the feedlot station and in the decision about the optimum endpoint. This process is termed marker-assisted management which can possibly allow for earlier and more efficient sorting of cattle into uniform groups and towards a desired carcass outcome (Kononoff et al., 2015; Thompson et al., 2016). The objective of this study was to evaluate a genomic-enhanced sorting system that utilizes predicted MBVs of carcass traits to sort animals to quality grid and lean meat yield (LMY) groups.
MATERIALS AND METHODS
All management and procedures involving live animals conformed to the guidelines outlined in the Canadian Council on Animal Care (CCAC, 1993) and University of Alberta Care and Use Committee (AUP00000777).
Animals and Management
The animals used in this study were part of the Kinsella breeding project based at the University of Alberta Roy Berg Kinsella Research Ranch, Kinsella, AB, Canada. Three beef cattle herds existed at the ranch at the start of the project including Kinsella Composite (KC;n = 420), purebred Angus (AN; n = 200), and purebred Charolais (CH; n = 125) cows. The KC is heavily influenced by AN and CH breeds with infusion of Galloway and Hereford and other small breeds (Wang et al., 2005; Abo-Ismail et al., 2016). To produce the steers evaluated in this study, 20 KC bulls were used for breeding the KC cows through multi-sire group mating on pasture, and for the AN and CH herds one round of estrous synchronization and artificial insemination to industry registered sires was performed, followed by exposure to 10 AN and 5 CH bulls, respectively.
Calves were born between 2013 and 2016 from late-March until the beginning of June, with the purebred herds calving slightly earlier than the KC herds. All calves were raised by their dams until weaning which occurred from mid-October to the end of November. Each year, all calves received a backgrounding diet until they entered the GrowSafe feed intake measurement system (GrowSafe Systems Ltd., Airdrie, AB, Canada) at the Kinsella Ranch. Feed intake was measured using a silage-based diet supplemented with grain depending upon which production stage the cattle would spend a majority of their time. Typically, steers were in GrowSafe from April–June (KC) or July–September (AN and CH). All steers were managed similarly within their breed group (KC, AN, and CH) and received a single Revalor G (Merck Animal Health) growth implant at least 2 wk before entering the GrowSafe test and were fed ad libitum.
Data Collection
At birth, calves were tagged for identification using TypiFix ear tags (Life Technologies Inc., Burlington, ON, Canada), and birthdate, calving ease, calf vigor, and birth weight recorded. Ear tissue samples collected via the TypiFix tag were genotyped on the Illumina BovineSNP50 BeadChip (50K; Illumina Inc, San Diego, CA) at Delta Genomics, Edmonton, AB, Canada. Parentage determination was performed for animals with missing sire information using DNA genotypes. Each year, upon completion of the feed intake test, steers were slaughtered at an average age of 492 ± 26 d for AN, 517 ± 22 d for CH, and 499 ± 30 d for KC, when it was determined that a majority of the steers in a group of 50 had >8 mm back fat as predicted from ultrasound rib eye and back-fat measurement at the end of the GrowSafe test. Steers were slaughtered either at a commercial plant or at the Agriculture and Agri-Food Canada Lacombe abattoir. Each carcass was split and chilled for 48 h and hot carcass weight (HCW) in kg, grade fat thickness (FAT) in mm, ribeye area (REA) in cm2, percent LMY estimated with the formula: LMY = 57.96 – (0.027 × HCW) + (0.202 × REA) − (0.703 × FAT), marbling score (MBS) measured as >699 = abundant, 600 to 699 = modest, 500 to 599 = small, 400 to 499 = slight and 300 to 399 or less = traces, yield grade (YG) scored as Y1 = >59% LMY, Y2 = 54% to 58% LMY and Y3 = <53% LMY and QG scored as Prime = slightly abundant, AAA = small, AA = slight, and A = traces were recorded as well as video grading data tied to the individual animal. Grading of carcass was completed by graders certified under the Canadian Beef Grading Agency. A total of 1,118 steers consisting of 299 AN, 181 CH, and 638 KC slaughtered between 2014 and 2017 were evaluated in this study.
Prediction of MBVs
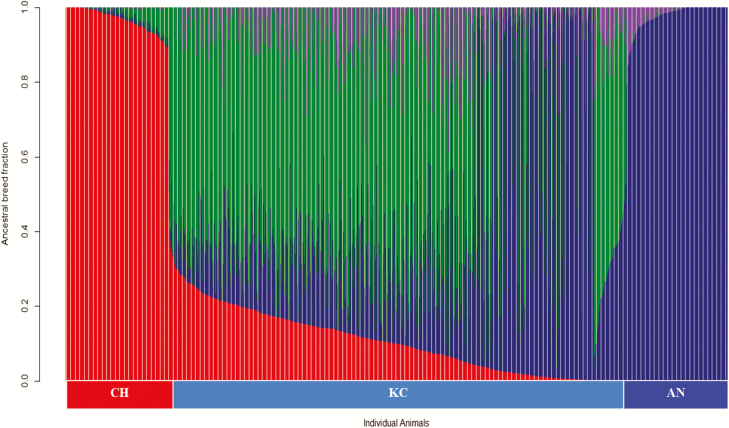
MBVs for the studied traits were generated for all slaughtered steers (n = 1,118) excluding their phenotypes and using existing records of purebred AN (n = 1,008), CH (n = 659), and KC (n = 942) animals accumulated from previous projects before 2013 (Akanno et al., 2014; Chen et al., 2015). This discovery population (n = 2,609) had 50K genotypes and matching phenotypes for HCW, FAT, REA, LMY, and MBS. Descriptive statistics and the genomic heritability of carcass traits in the discovery population are given in Table 1. To begin with, prediction of genomic breed composition was performed for the entire dataset using genotypes of all animals (n = 3,727). Here, all genotypes were initially filtered by removing SNPs with minor allele frequency <0.01 and call rate <0.90 leaving a total of 44,388 SNPs used in subsequent analysis. Genomic breed fractions were predicted for all individuals using a 10-fold cross-validation procedure available in the ADMIXTURE software (Alexander et al., 2009) to find the best possible K value with the smallest cross-validation error (Alexander et al., 2009), where K is the number of postulated ancestral populations. The resulting breed fractions for the target animals are shown in Figure 1.
Table 1.
The number of records (N), descriptive statistics, estimates of genomic heritability (h2 ± SE) for carcass traits of beef cattle in the discovery dataset
| Traits | N | Mean | SD | h 2 ± SE |
|---|---|---|---|---|
| Hot carcass weight, kg | 2,178 | 328.42 | 33.17 | 0.43 ± 0.05 |
| Average back fat, mm | 2,171 | 12.48 | 5.08 | 0.39 ± 0.05 |
| Rib eye area, cm2 | 2,174 | 83.81 | 11.06 | 0.45 ± 0.05 |
| Lean meat yield, % | 2,175 | 57.20 | 4.890 | 0.43 ± 0.05 |
| Marbling scores | 2,172 | 409.00 | 105.90 | 0.43 ± 0.05 |
Figure 1.
Distribution of breed fractions in the target animals (n = 1,118). Charolais is red, Angus is blue, Galloway is green, and Hereford is purple. AN = Purebred Angus, CH = Purebred Charolais, KC = Kinsella Composite
Thereafter, MBVs were predicted using the genomic best linear unbiased prediction (GBLUP) implemented in a bivariate animal model (1). A bivariate model was chosen for convenience considering that MBVs for LMY and MBS will be used jointly for the sorting program:
| (1) |
where and are vectors of observed trait 1 and trait 2, respectively; , are vectors of fixed effects for trait 1 and trait 2, respectively, including contemporary groups based on year of birth and management groups, covariates of genomic breed fractions including AN, CH, Galloway and Hereford breed fractions, and covariates of slaughter age; and are vectors of random genetic effects of trait 1 and trait 2, respectively, normally distributed as , where is the genetic variance–covariance matrix for trait 1 and trait 2. The matrix G is genomic additive relationship matrix constructed from genotype information according to the method described in detail by VanRaden (2008) as follows: G = WW′/2Σ (pj) (1 – pj). The matrix W is of the order n × m (i.e., the number of individuals by the number of SNPs). The elements in W are equal to −2pj, 1 − 2pj, and 2 − 2pj for genotypes AA, AB, and BB, while pj is the allele frequency of the B allele at the jth SNP; and are vectors of random residual effects of trait 1 and trait 2, respectively, that is normally distributed as , where and is the residual variance–covariance matrix of trait 1 and trait 2 and I is an identity matrix; , , , and are design matrices for fixed and random effects, respectively.
Sorting of Steers and Statistical Analyses
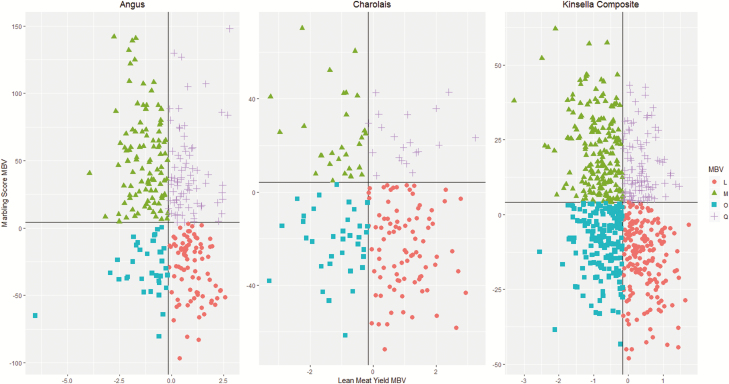
The goal of this study was to determine in silico how well MBV can be used to sort feeder cattle into uniform groups that meet the expected carcass outcome for 70:70 quality grid (70% AAA/Prime (MBS) and 70% Canada grade Y1/Y2), and a Y1–LMY program. This was designed to simulate how genomic information could be used in marker-assisted management strategies for the feedlot. The target MBVs of LMY and MBS were chosen as cattle producers are interested in a strategy that can improve carcass uniformity and revenue while maintaining beef quality and YGs (Basarab et al., 1999), and the choice of the two different end-point groups reflects desired and slightly opposing carcass outcomes for production within the Canadian beef cattle industry. In doing so, MBVs for LMY and MBS were used to sub-divide all the steers into four carcass endpoints. First, average MBVs of LMY and MBS were determined for all animals in the target populations. Subsequently, steers within each of the target populations were placed in silico into one of four groups: the quality (Q) group if their MBV values for LMY and MBS were greater than the average MBVs for LMY and MBS, respectively; the lean (L) group if their MBV values for LMY were greater than the average MBV for LMY but their MBV values of MBS were less than or equal to the average MBV of MBS, the marbling (M) group if their MBV values for MBS were greater than the average MBV for MBS but their MBV of LMY were less than or equal to the average MBV for LMY; and the other (O) group if their MBV values for LMY and MBS were less than the average MBVs for LMY and MBS, respectively (Figure 2).
Figure 2.
Quadrant distribution of MBV-based sorting using MBVs for lean meat yield and marbling score in Angus, Charolais, and Kinsella Composite beef steers (n = 1,118). The horizontal and vertical lines crosses at the mean MBVs for Marbling score and lean meat yield, respectively. Q = quality, L = lean, M = marbling, O = other
As carcass phenotypes became available for all in silico sorted steers in the target populations, they were evaluated and appraised for consistency with the assigned group. Least square analysis was performed according to model 2 for all carcass traits and MBV groups within each population
| (2) |
where yijk is the phenotypic observations for carcass trait under investigation for kth animal; µ is the overall mean; CGi is the fixed effects of ith contemporary group based on year of birth and management group, b1 to b4 is the regression coefficients for AN, CH, Galloway, and Hereford breed fractions, respectively, for kth animal; b5 is the regression coefficient for slaughter age of kth animal; SGj is the fixed effect of jth MBV group; and eijk is the random residual effect associated with kth animal. Also, descriptive statistics and the coefficient of variations were generated within each breed-group population for all carcass traits within MBV group and for all steers. Later, the number of animals that reached the target endpoints in each group was counted based on individual carcass grading for YG and QG, which was used to produce actual percentages for each MBV group.
Validation of Prediction Accuracy of Carcass Molecular Breeding Values
A scenario for evaluating the predictability of future carcass outcomes based on current carcass values was tested using phenotype and genotype of the slaughtered steers. Pearson’s correlation between predicted MBVs and adjusted phenotype of carcass traits divided by square root of heritability () was computed as the genomic prediction accuracy of MBVs, where was the adjusted phenotype for the steers generated using the following equation:
| (3) |
where yij is the phenotypic observations for carcass trait under investigation for jth animal; µ is the overall mean; CGi is the fixed effects of ith contemporary group based on year of birth and management group, b1 to b4 is the regression coefficients for AN, CH, Galloway, and Hereford breed fractions, respectively, for jth animal; b5 is the regression coefficient for slaughter age of jth animal; and eij is the random residual effect associated with jth animal. The slope of the regression of on MBV () was determined to measure the degree of bias due to genomic prediction. The estimates of close to 1 are indicative of predictions that are similar to that of corrected phenotype on scale. All analyses were conducted in R statistical software using default packages where applicable (Ihaka and Gentleman, 1996) and linked to ASReml software (Gilmour et al., 2015) for variance component estimation and genomic prediction.
RESULTS AND DISCUSSION
This study applied a genomic approach using MBVs of LMY and MBS to sort feeder cattle in silico into different carcass endpoint groups. The target MBVs of LMY and MBS were chosen as cattle producers are interested in a strategy that can improve carcass uniformity and revenue while maintaining beef quality and YGs (Basarab et al., 1999). Results of the least square analysis, means, standard deviations, and coefficient of variation for various carcass characteristics according to the carcass endpoint groups are summarized for AN, CH, and KC populations in Tables 2 to 4. Except for a few traits, the mean performance of the sorted group differed significantly (P < 0.05) among themselves. In general, Q and L groups had larger REA and LMY, and Q and M groups showed more marbling across the three populations. HCWs were consistently similar across MBV groups in CH and KC but not for AN (Tables 2 to 4).
Table 2.
Number of individuals in each MBV group (N), descriptive statistics, and percent CV for carcass characteristics in Angus steers
| Traits | Group | N | Mean1 | SD | CV (%) |
|---|---|---|---|---|---|
| Hot carcass weight, kg | Q—quality | 78 | 345.51ab | 36.22 | 10.48 |
| L—lean | 84 | 334.30a | 30.78 | 9.21 | |
| M—marbling | 101 | 342.84ab | 30.78 | 8.98 | |
| O—other | 36 | 351.38b | 33.53 | 9.54 | |
| Overall | 299 | 342.17 | 32.92 | 9.62 | |
| Average back fat, mm | Q—quality | 78 | 9.40a | 2.78 | 29.62 |
| L—lean | 84 | 8.97a | 2.64 | 29.39 | |
| M—marbling | 101 | 13.77b | 3.01 | 21.85 | |
| O—other | 36 | 12.68b | 3.21 | 25.31 | |
| Overall | 299 | 11.16 | 3.60 | 32.25 | |
| Rib eye area, cm2 | Q—quality | 78 | 81.09a | 10.06 | 12.40 |
| L—lean | 84 | 77.40a | 9.38 | 12.11 | |
| M—marbling | 101 | 71.72b | 8.02 | 11.18 | |
| O—other | 36 | 72.65b | 10.68 | 14.70 | |
| Overall | 299 | 75.87 | 10.02 | 13.21 | |
| Lean meat yield, % | Q—quality | 78 | 58.41a | 2.83 | 4.85 |
| L—lean | 84 | 58.34a | 2.65 | 4.54 | |
| M—marbling | 101 | 53.51b | 2.70 | 5.04 | |
| O—other | 36 | 53.40b | 4.39 | 8.22 | |
| Overall | 299 | 56.13 | 3.83 | 6.83 | |
| Marbling scores | Q—quality | 78 | 477.38a | 88.30 | 18.50 |
| L—lean | 84 | 378.50b | 55.89 | 14.77 | |
| M—marbling | 101 | 511.98a | 91.49 | 17.87 | |
| O—other | 36 | 381.17a | 54.13 | 14.20 | |
| Overall | 299 | 449.71 | 97.72 | 21.73 |
1Within columns for a given trait, means followed by the same letter are not significantly different according to Tukey (P < 0.05).
Table 4.
Number of individuals in each MBV group (N), descriptive statistics, and percent CV for carcass characteristics in Kinsella Composite steers
| Traits | Group | N | Mean | SD | CV (%) |
|---|---|---|---|---|---|
| Hot carcass weight, kg | Q—quality | 104 | 349.77ns | 30.54 | 8.73 |
| L—lean | 180 | 353.42ns | 32.02 | 9.06 | |
| M—marbling | 188 | 339.59ns | 32.39 | 9.54 | |
| O—other | 166 | 346.01ns | 31.57 | 9.12 | |
| Overall | 638 | 346.83 | 32.16 | 9.27 | |
| Average back fat, mm | Q—quality | 104 | 11.10a | 4.12 | 37.15 |
| L—lean | 180 | 10.30b | 4.46 | 43.26 | |
| M—marbling | 188 | 10.96b | 3.94 | 35.98 | |
| O—other | 166 | 11.96a | 4.59 | 38.39 | |
| Overall | 638 | 11.06 | 4.33 | 39.14 | |
| Rib eye area, cm2 | Q—quality | 104 | 80.41a | 10.39 | 12.92 |
| L—lean | 180 | 83.37a | 10.71 | 12.85 | |
| M—marbling | 188 | 78.25b | 9.44 | 12.07 | |
| O—other | 166 | 79.21b | 10.01 | 12.64 | |
| Overall | 638 | 80.32 | 10.30 | 12.83 | |
| Lean meat yield, % | Q—quality | 104 | 57.04a | 4.11 | 7.20 |
| L—lean | 180 | 58.02b | 4.33 | 7.47 | |
| M—marbling | 188 | 57.05a | 3.58 | 6.28 | |
| O—other | 166 | 56.40a | 4.28 | 7.59 | |
| Overall | 638 | 57.16 | 4.11 | 7.18 | |
| Marbling scores | Q—Quality | 104 | 406.36a | 81.50 | 20.06 |
| L—Lean | 180 | 380.47b | 77.45 | 20.36 | |
| M—Marbling | 188 | 417.41a | 77.16 | 18.49 | |
| O—Other | 166 | 379.78a | 86.13 | 22.68 | |
| Overall | 638 | 395.36 | 81.97 | 20.73 |
1Within columns for a given trait, means followed by the same letter are not significantly different according to Tukey (P < 0.05), ns = nonsignificant.
For AN steers, the genomic-enhanced sorting system numerically decreased variability in a majority of the carcass traits by 6.1% to 29.0% in Q, by 4.3% to 33.5% in L, by 6.7% to 32.3% in M and by 0.8% to 34.7% in O groups, on average, compared with the amount of variation in the overall population from which they were selected (Table 2). Similarly for CH steers, trait variability was decreased by 2.9% to 21.3% in Q, by 10.5% to 27.1% in L, by 8.8% to 17.5% in M, and by 0.1 to 27.5% in O groups across the studied traits, on average (Table 3). On the other hand, the KC steers showed several instances of increased variability for some of the MBV groups across the studied traits except for MBS where variability was slightly decreased (Table 4).
Table 3.
Number of individuals in each MBV group (N), descriptive statistics, and percent CV for carcass characteristics in Charolais steers
| Traits | Group | N | Mean | SD | CV (%) |
|---|---|---|---|---|---|
| Hot carcass weight, kg | Q—Quality | 22 | 379.45ns | 25.42 | 6.70 |
| L—Lean | 89 | 384.89ns | 29.63 | 7.70 | |
| M—Marbling | 30 | 385.73ns | 32.22 | 8.35 | |
| O—Other | 40 | 385.25ns | 28.25 | 7.33 | |
| Overall | 181 | 384.45 | 29.13 | 7.58 | |
| Average back fat, mm | Q—Quality | 22 | 5.93a | 2.46 | 41.56 |
| L—Lean | 89 | 5.65a | 2.17 | 38.31 | |
| M—Marbling | 30 | 8.72b | 3.23 | 37.04 | |
| O—Other | 40 | 8.48b | 2.92 | 34.51 | |
| Overall | 181 | 6.82 | 2.92 | 42.80 | |
| Rib eye area, cm2 | Q—Quality | 22 | 99.93a | 9.74 | 9.75 |
| L—Lean | 89 | 97.29a | 9.48 | 9.75 | |
| M—Marbling | 30 | 88.26b | 8.07 | 9.15 | |
| O—Other | 40 | 88.19b | 9.77 | 11.08 | |
| Overall | 181 | 94.10 | 10.43 | 11.09 | |
| Lean meat yield, % | Q—Quality | 22 | 63.73a | 2.67 | 4.18 |
| L—Lean | 89 | 63.25a | 2.34 | 3.69 | |
| M—Marbling | 30 | 59.48b | 2.68 | 4.50 | |
| O—Other | 40 | 59.42b | 2.62 | 4.42 | |
| Overall | 181 | 61.85 | 3.13 | 5.05 | |
| Marbling scores | Q—Quality | 22 | 432.09a | 55.09 | 12.75 |
| L—Lean | 89 | 353.45b | 41.74 | 11.81 | |
| M—Marbling | 30 | 432.47a | 63.93 | 14.78 | |
| O—Other | 40 | 340.13b | 39.93 | 11.74 | |
| Overall | 181 | 373.16 | 60.46 | 16.20 |
1Within columns for a given trait, means followed by the same letter are not significantly different according to Tukey (P < 0.05), ns = nonsignificant.
One of the main objectives of this study was to investigate a strategy for using MBVs of valued carcass traits to sort feeder cattle into uniform carcass endpoint groups prior to their entering the feedlot, or as they receive a second implant in the feedlot. Whereas concerted research efforts were made in the past evaluating different feeder cattle sorting techniques in order to improve carcass uniformity (Houghton and Turlington, 1992; Sainz and Oltjen, 1994; Basarab et al., 1997, 1999; Kononoff et al., 2015), a considerable amount of variation still exists for most carcass characteristics resulting in price discounts for finished cattle. Our in silico assessment of a genomic-enhanced sorting system resulted in lower coefficients of variation for most of the carcass traits than the average of the population from which they were selected, which suggests less trait variability and more uniform carcasses in the specific MBV groups.
The reduction in carcass variability for most of the assigned MBV groups as observed in purebred AN and CH steers is comparable with the findings of Basarab et al. (1997) using a video image and real-time ultrasound technique to sort feeder cattle. For these populations, efficiencies in animal management such as choice of hormone implant, energy concentration of feed, and especially days on feed could be planned for traits such as marbling that are difficult to visually inspect, and considering the animals that did not fit the breed norm, they could be removed from the pen group to achieve a higher premium for uniformity. Further, for commercial beef cattle production, where crossbreeding is a standard approach, the genomic-enhanced sorting system provides opportunity for sorting feeder cattle early in life such that they can be managed for different carcass endpoints. Current sorting practice in the beef cattle industry is the use of visual appraisal by one or several experienced animal sorters, with some measure of weights that can be used in the sorting process, which are recorded later in life of the animal. The lack of consistent reductions in trait variability observed for MBV groups in KC steers suggests that genomic prediction of carcass outcomes in crossbred steers or the genomic sorting process we have used needs improvement before consideration for use at the industry level.
The distributions of carcass quality and YGs for the various MBV groups and studied populations are shown in Table 5. The percentage of AN steers that achieved the AAA and prime grade carcasses were 95.2% for Q, 52.1% for L, 91% for M, and 52.0% for O. Within the AAA and prime grade classification, the distribution of Y1 YG was 40.5% (Q), 54.2% (L), 6.0% (M), and 16.0% (O). Similarly for CH, the percentage of carcasses classified as AAA and prime grade was 66.7% for Q, 10.9% for L, 56.0% for M, and 12.9% for O which also showed 100.0%, 96.9%, 72.0%, and 87.1% of Y1 YG, respectively. In KC steers, the percentage of carcasses that achieved AAA and Y1 grades were generally low ranging from 43.8% to 66.2% and from 33.1% to 46.0%, respectively, across the MBV groups.
Table 5.
Effect of genomic-enhanced sorting on the percentage of steers that reached the target endpoint of 70:70 quality grid (>70% AAA/Prime and Y1/Y2) and Y1-lean meat yield (>70% Y1) program
| Percentage (%) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Breed | Group1 | N1 | Prime/AAA | AA | A | B4 | Y1 | Y2 | Y3 | Prime/AAA | Y1 |
| Angus | Q—quality | 42 | 40 | 1 | 1 | 0 | 17 | 24 | 1 | 95.2 | 40.5 |
| L—lean | 48 | 25 | 21 | 1 | 1 | 26 | 22 | 0 | 52.1 | 54.2 | |
| M—marbling | 67 | 61 | 5 | 0 | 1 | 4 | 45 | 18 | 91.0 | 6.0 | |
| O—other | 25 | 13 | 12 | 0 | 0 | 4 | 17 | 4 | 52.0 | 16.0 | |
| Overall | 182 | 139 | 39 | 2 | 2 | 51 | 108 | 23 | 76.4 | 28.0 | |
| Charolais | Q—quality | 12 | 8 | 3 | 0 | 1 | 12 | 0 | 0 | 66.7 | 100.0 |
| L—lean | 64 | 7 | 48 | 7 | 2 | 62 | 2 | 0 | 10.9 | 96.9 | |
| M—marbling | 25 | 14 | 10 | 1 | 0 | 18 | 7 | 0 | 56.0 | 72.0 | |
| O—other | 31 | 4 | 24 | 3 | 0 | 27 | 4 | 0 | 12.9 | 87.1 | |
| Overall | 132 | 33 | 85 | 11 | 3 | 119 | 13 | 0 | 25.0 | 90.2 | |
| Kinsella Composite | Q—quality | 84 | 53 | 29 | 2 | 0 | 32 | 42 | 10 | 63.1 | 38.1 |
| L—lean | 137 | 60 | 70 | 5 | 2 | 63 | 58 | 16 | 43.8 | 46.0 | |
| M—marbling | 142 | 94 | 44 | 1 | 3 | 47 | 78 | 17 | 66.2 | 33.1 | |
| O—other | 121 | 55 | 56 | 9 | 1 | 40 | 60 | 21 | 45.5 | 33.1 | |
| Overall | 484 | 262 | 199 | 17 | 6 | 182 | 238 | 64 | 54.1 | 37.6 | |
1 N is the number of steers that were graded for quality and yield grade by graders certified under the Canadian Beef Grading Agency.
The high level of marbling achieved in Q (95.2%) and M (91.0%) for AN steers and the high amount of lean achieved in all MBV groups (72% to 100%) for CH steers are typical of these breeds and corroborates known differences between British and Continental cattle breeds. One criterion for judging the effectiveness of the feeder cattle sorting program was the percentage of carcasses that reached the expected endpoint target of 70:70 quality grid (>70% Y1/Y2 and AAA/Prime) and Y1-LMY (>70% Y1) program. In general, carcasses from the AN steers met the quality grid target in the Q and M groups while carcasses from the CH steers met the Y1-LMY target in all MBV groups. None of the carcasses from the KC steers met the desired carcass outcome in all MBV groups. This improvement in carcass quality and YG for sorted purebred AN and CH steers when using a genomic-enhanced sorting system warrants further investigation into the cost–benefit implications for feedlot operations.
This study also evaluated the accuracy of genomic prediction of future carcass outcomes based on MBV scores of slaughtered steers for each trait (Table 6). The results showed that genomic prediction accuracy obtained as Pearson’s correlation between predicted MBV and adjusted phenotype ranged from 0.53 for HCW to 0.59 for FAT, This accuracy of MBV predictions agrees with the results of earlier studies using a similar beef cattle population (Akanno et al., 2014, 2018; Chen et al., 2015). These moderately high genomic accuracies reflect the strong genetic ties between animals in the discovery set and the target population. For example, all animals in the target population are direct descendants of key ancestors in the discovery set for all three populations. However, the coefficients of regression of adjusted phenotypes on MBV ranged from 0.64 for FAT to 0.81 for MBS which are slightly lower than 1 across all traits (Table 6), indicating that the MBVs used in the sorting program were likely underestimated from their true breeding values.
Tables 6.
Number of records (N), genomic prediction accuracies (r), and regression coefficients (β) of adjusted phenotype on molecular breeding values for carcass traits of beef steers slaughtered between 2014 and 2017 in the Kinsella breeding project
| Traits | N | r | β |
|---|---|---|---|
| Hot carcass weight, kg | 1,118 | 0.53 | 0.73 |
| Average back fat, mm | 1,115 | 0.59 | 0.64 |
| Rib eye area, cm2 | 1,069 | 0.58 | 0.72 |
| Lean meat yield, % | 1,118 | 0.58 | 0.67 |
| Marbling scores | 1,117 | 0.58 | 0.81 |
Nevertheless, the accuracies of MBVs indicate that the MBV scores can predict up to 35% variance of the expected carcass endpoints. As shown in this study, the levels of MBV accuracy observed suggest utility of MBV as a marker-assisted management tool to sort feeder cattle into uniform carcass endpoint groups under similar environmental and management conditions. Thus, the MBVs generated by GBLUP using the existing reference dataset can potentially aid a marker-assisted management program for beef cattle.
CONCLUSIONS
This study utilized genomic tools to evaluate a strategy for sorting feeder cattle into uniform carcass endpoints and to ascertain the effectiveness of this technique for feedlot operations. In general, quality and lean groups had larger REA and LMY, while quality and marbling groups displayed higher marbling across the three populations. Decreases in trait variability were observed for the majority of the MBV groups within the AN and CH purebred populations, which suggest that more uniform carcasses resulted from the sorting program. For the KC crossbred population, the use of MBV to sort steers into carcass outcome groups needs to be refined before becoming an effective tool for the beef industry. Carcasses from AN steers met the expected quality grid target for the Quality and Marbling groups while carcasses from CH steers met the Y1-LMY target in all MBV groups which is typical of these breeds. The accuracy of genomic prediction showed that the MBVs could predict up to 35% of the observed carcass trait variability thus, can potentially aid a genome-based sorting program for feedlot operations under similar environmental and management conditions. Further investigation is warranted to evaluate the performance of feeder cattle sorted based on MBV and managed for different carcass endpoints as well as the cost–benefit implications for feedlot operations.
ACKNOWLEDGMENT
The authors acknowledge Lisa McKeown, Barry Irving, and Lynda Baker for their contributions to animal management and phenotypic data collection.
Conflict of interest statement. None declared.
Footnotes
This work was supported by the Kinsella breeding project (2013R027R, FDE.17.13). The management of Angus and Charolais herds and data collection were supported by Agriculture and Agri-Food Canada (AAFC) Livestock Genetics & Genomics Program funds and AAFC A-base peer-reviewed research projects (RBPI-1139, RBPI-1752).
LITERATURE CITED
- Abo-Ismail M. K., Akanno E. C., Khorshidi R., Crowley J., Chen L., Karisa B. K., Li X., Wang Z., Basarab J., Li C., et al. . 2016. 0310 Assessing genetic diversity in Canadian beef cattle populations using Illumina BovineSNP50 chip. J. Anim. Sci. 94 (Suppl. 5):148–149. doi: 10.2527/jam2016-0310 [DOI] [Google Scholar]
- Akanno E. C., Abo-Ismail M. K., Chen L., Crowley J. J., Wang Z., Li C., Basarab J. A., MacNeil M. D., and Plastow G. S.. 2018. Modeling heterotic effects in beef cattle using genome-wide SNP-marker genotypes. J. Anim. Sci. 96:830–845. doi: 10.1093/jas/skx002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akanno E. C., Plastow G., Woodward B. W., Bauck S., Okut H., Wu X. L., Sun C., Aalhus J. L., Moore S. S., Miller S. P., et al. . 2014. Reliability of molecular breeding values for warner-bratzler shear force and carcass traits of beef cattle – an independent validation study. J. Anim. Sci. 92:2896–2904. doi: 10.2527/jas.2013-7374 [DOI] [PubMed] [Google Scholar]
- Alexander D. H., Novembre J., and Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. doi: 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basarab J. A., Brethour J. R., ZoBell D. R., and Graham B.. 1999. Sorting feeder cattle with a system that integrates ultrasound backfat and marbling estimates with a model that maximizes feedlot profitability in value-based marketing. Can. J. Anim. Sci. 79: 327–334. [Google Scholar]
- Basarab J. A., Milligan D., McKinnon J. J., and Thorlakson B. E.. 1997. Use of video imaging and real-time ultrasound on incoming feeder steers to improve carcass uniformity. Can. J. Anim. Sci. 77: 385–392. [Google Scholar]
- Canadian Council on Animal Care. 1993. In: Olfert E. D., Cross B. M., and McWilliams A. A., editors, Guide to the care and use of experimental animals. Vol. 1 Canadian Council on Animal Care, Ottawa, ON. [Google Scholar]
- Chen L., Vinsky M., and Li C.. 2015. Accuracy of predicting genomic breeding values for carcass merit traits in angus and charolais beef cattle. Anim. Genet. 46:55–59. doi: 10.1111/age.12238 [DOI] [PubMed] [Google Scholar]
- DeNise S. K., Kerr R., Rosenfeld D., Holm T., Bates S., and Fantin D.. 2008. Methods and systems for inferring bovine traits. United States Patent No.: US 7.468, 248 B2, Date of Patent: December 23, 2008 [Google Scholar]
- Gilmour A. R., Gogel B. J., Cullis B. R., Welham S. J., and Thompson R.. 2015. ASReml user guide release 4.1. Hemel Hempstead, UK:VSN International Ltd; www.vsni.co.uk. [Google Scholar]
- Houghton P. L., and Turlington L. M.. 1992. Application of ultrasound for feeding and finishing animals: a review. J. Anim. Sci. 70:930–941. doi: 10.2527/1992.703930x [DOI] [PubMed] [Google Scholar]
- Ihaka R., and Gentleman R.. 1996. R: a language for data analysis and graphics. J. Comp. Graph. Stat. 5:299–314. doi: 10.1080/10618600.1996.10474713 [DOI] [Google Scholar]
- Kononoff P. J., Defoor P. J., Engler M. J., Swingle R. S., James S. Y., Deobald H. M., Deobald R. L., Woronuk G. N., and Marquess F. L.. 2015. Performance and carcass characteristics when sorting feedlot cattle on the basis of phenotype, and leptin genotype along with differential use of b-adrenergic agonists. Can. J. Anim. Sci. 95:455–463. doi: 10.4141/cjas-2014-052 [DOI] [Google Scholar]
- Macdonald J. C., Klopfenstein T. J., Erickson G. E., Macken C. N., Folmer J. D., and Blackford M. P.. 2006. Sorting strategies for long yearling cattle grown in an extensive forage utilization beef production system. The Prof. Anim. Sci. 22:225–235. http://digitalcommons.unl.edu/animalscifacpub/769 [Google Scholar]
- MacNeil M. D., Nkrumah J. D., Woodward B. W., and Northcutt S. L.. 2010. Genetic evaluation of angus cattle for carcass marbling using ultrasound and genomic indicators. J. Anim. Sci. 88:517–522. doi: 10.2527/jas.2009-2022 [DOI] [PubMed] [Google Scholar]
- Meuwissen T. H., Hayes B. J., and Goddard M. E.. 2001. Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829. ISSN: 0016-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols B. M., Reuter R. R., and Cook B. J.. 2014. Evaluation of using half-sibling beef cows to increase growth and carcass uniformity of calf crops. The Prof. Anim. Sci. 30:37–42. doi: 10.15232/S1080-7446(15)30080-2 [DOI] [Google Scholar]
- Rincker C. B., Pyatt N. A., Berger L. L., and Faulkner D. B.. 2006. Relationship among genestar marbling marker, intramuscular fat deposition, and expected progeny differences in early weaned simmental steers. J. Anim. Sci. 84:686–693. doi: 10.2527/2006.843686x [DOI] [PubMed] [Google Scholar]
- Sainz R. D., and Oltjen J. W.. 1994. Improving uniformity of feeder steers using ultrasound and computer modelling. Proc. Western Sec. Am. Soc. Anim. Sci. 45: 179–181. [Google Scholar]
- Thallman R. M., Hanford K. J., Quaas R. L., Kachman S. D., Tempelman R. J., Fernando R. L., Kuehn L. A., and Pollak E. J.. 2009. Estimation of the proportion of genetic variation accounted for by DNA Test. In: Proceedings of the Beef Improvement Federation 41st Annual Research Symposium; Sacramento, CA p. 184–209. [Google Scholar]
- Thompson N. M., DeVuyst E. A., Brorsen B. W., and Lusk J. L.. 2016. Using genetic testing to improve fed cattle marketing decisions. J. Agric. Resource Econ. 41: 286–306. http://www.waeaonline.org/UserFiles/file/JAREMay20167Thompson286-306.pdf [Google Scholar]
- VanRaden P. M. 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91:4414–4423. doi: 10.3168/jds.2007-0980 [DOI] [PubMed] [Google Scholar]
- Wang Z., Goonewardene L. A., Yang R. C., Price M. A., Makarechian M., Knapp J., Okine E. K., and Berg T. R.. 2005. Estimation of genetic parameters and trends in pre-weaning traits of beef lines subject to phenotypic selection. J. Anim. Vet. Adv. 4:202–209. [Google Scholar]