Abstract
Zinc finger protein 91 (ZFP91) gene has been recently acknowledged to possess oncogenic properties. To date, its expression has been examined only in a handful of human organs and cancer types. The aim of the present study was to characterize, for the first time, the ZFP91 expression pattern in a range of human tissues and cancer types. ZFP91 mRNA expression was examined using Cancer Survey cDNA sets. Utilized cDNA samples represented 15 human organs and 17 cancer types. ZFP91 mRNA expression was the highest in the testes and lymph nodes. It was downregulated in testis cancer, lymphoma and thyroid cancer, and upregulated in prostate cancer. Among the analyzed cancer types, ZFP91 expression was markedly elevated in sarcomas and melanoma. On a protein level, a large-scale reverse phase protein array was employed providing samples from 11 organ types and from cancers derived from these organs. ZFP91 protein expression was revealed to be generally stable across the tested samples and was only moderately elevated in breast, ovarian and pancreatic cancers. To the best of our knowledge, this is the first study to thoroughly analyze the ZFP91 expression pattern in human tissues and cancers. The obtained results provide the foundation for further work aiming to reveal its full biological significance.
Keywords: zinc finger protein 91, nuclear factor-κB, hypoxia inducible factor-1α, cancer pathogenesis
Introduction
ZFP91 zinc finger protein (ZFP91) is a little-studied gene discovered in mouse in 1995 by Saotome et al (1). In 2003 Unoki et al (2) found, in a screening-type study, ZFP91 overexpression in leukemic cells and neoplastic blood cell lines. These authors were the first to detect ZFP91 expression (using northern blot method) in several human tissues and confirm ZFP91 protein presence in human cells-in cultured colon and endometrial cancer cell lines. They were also the first to characterize ZFP91 protein structure and potential properties. It is a protein with a molecular mass of 63.4 kDa, composed of 570 amino acid residues. It contains five zinc-finger motives, a leucine zipper, a coiled-coil structure and nuclear localization sequences. In mammals, it is highly conserved among species. Based on its structure, ZFP91 was predicted to be localized in nucleus and to act as a transcription factor (2).
ZFP91 expression is positively regulated by NF-κB signaling pathway through NF-κB complex binding with ZFP91 gene's 5′upstream promotor region (3). ZFP91 overexpression, on the other hand, leads to increased NF-κB signaling pathway activation in a manner dependent on NF-κB inducing kinase (NIK) presence (4). This kinase regulates the activity of NF-κB non-canonical (alternative) signaling pathway (5). ZFP91 acts as an atypical E3 ubiquitin-protein ligase in NIK ubiquitinization which results in NIK stabilization and activation of the non-canonical NF-κB signaling pathway and its target genes expression (4,6). NIK activity and its overexpression has been connected to cancer pathogenesis in e.g., melanoma, pancreatic-, breast- and lung cancer (7). The potential role of ZFP91 in above mentioned NIK relationships remains to be elucidated.
Another important intracellular signaling pathway besides NF-κB has been recently discovered to be dependent on ZFP91 expression-the hypoxia inducible factor (HIF-1) signaling pathway (8). ZFP91 expression was found to be increased in colon cancer and positively associated with HIF-1α expression. ZFP91 via interaction with NF-κB/p65 protein binds to HIF-1α promoter region and upregulates its expression. It was proven that ZFP91 has the potential to promote proliferation of colon cancer cells in vitro and tumor growth in vivo via HIF-1α (8). HIF-1 is a key transcription factor responsible for cellular response to hypoxia and plays a crucial role in adaptive responses of cancer cells to the hypoxic microenvironment (9,10). HIF-1α together with NF-κB are two transcription factors involved on many levels in tumors growth, progression and resistance to chemotherapy. Novel therapy strategies based on molecular targets within these factors' pathways are being investigated (11–13).
Oncogenic properties of ZFP91 were revealed also in experiments were its expression was inhibited using RNA interference method. Unoki et al (2) found that ZFP91 inhibition in colon cancer and endometrial cancer cell lines resulted in increased apoptotic rate. Lee et al (3), found similarly increased apoptosis in cultured breast and stomach cancer cell lines. What is more, cells overexpressing ZFP91 as a result of transfection exhibited increased growth rate and metastatic potential (3).
To date, ZFP91 expression in human cancers has been studied almost exclusively in cancer cell lines. On a protein level, using immunohistochemistry, ZFP91 upregulation in colon cancer specimens was noted (8). On a mRNA level, using in situ hybridization technique, increased ZFP91 mRNA staining was observed in liver-, prostate- and stomach cancer specimens (3). As reported in our earlier work, ZFP91 mRNA overexpression was revealed in benign prostate hyperplasia (BPH) specimens, however without concomitant protein overexpression. In prostate cancer cell lines ZFP91 protein was markedly upregulated compared to normal prostate epithelial cells (14). In most recent work ZFP91 mRNA was found to be significantly overexpressed in prostate cancer specimens (15).
To the best of our knowledge, besides above mentioned colon cancer and prostate studies, expression of ZFP91 was not analyzed in human tissues and cancers specimens on a protein level. Similarly, on a mRNA level only qualitative data regarding ZFP91 expression in several normal human tissues and few cancer types exist. Taking into consideration ZFP91 important oncogenic properties, studies are lacking analyzing this gene expression across a variety of human normal tissues and cancer types. Presented study is the first aimed at this gap in our knowledge about ZFP91 biology and addressing it both on mRNA and protein level.
Materials and methods
Prostate cancer cDNA samples
Cancer Survey cDNA Arrays I, II and III (CSRT101, CSRT103, CSRT104) from OriGene Technologies (Rockville, USA) were utilized providing 573 cDNA samples from a variety of human normal tissues and cancer types. Description of every sample includes relevant clinical information, full pathology report and RNA quality data (data available at www.origene.com/qPCR/Tissue-qPCR-Arrays.aspx). The range of analyzed data was narrowed to organs and cancer types with a sufficient number of independent samples. Cancer types with insufficient number of samples e.g. adrenal cancer were excluded. Moreover, across all three cDNA arrays a fraction of samples occurred more than once and therefore were treated as technical repetitions and not independent samples. Final analysis included 397 cDNA samples containing 86 normal tissues and 311 cancer samples (Table I).
Table I.
Analyzed types of normal tissue and cancerous cDNA samples (from the Cancer Survey sets I, II and III-OriGene).
| Organ type | Total n | Cancer type | Total n |
|---|---|---|---|
| Breast | 5 | Breast cancer | 30 |
| Colon | 11 | Colon cancer | 16 |
| Endometrium | 5 | Endometrial cancer | 21 |
| Esophagus | 4 | Esophageal cancer (adenocarcinoma) | 14 |
| Kidney | 8 | Kidney cancer | 19 |
| Liver | 6 | Liver cancer | 15 |
| Lung | 6 | Lung (non-small cell carcinoma) | 21 |
| Lymph node | 4 | Lymphoma (non-Hodgkin) | 33 |
| Ovary | 5 | Ovarian cancer | 27 |
| Pancreas | 5 | Pancreatic cancer (adenocarcinoma) | 6 |
| Prostate | 6 | Prostate cancer | 20 |
| Stomach | 5 | Stomach cancer | 8 |
| Testis | 6 | Testicular cancer (7 seminomas, 10 non-seminomas) | 17 |
| Thyroid | 6 | Thyroid cancer (papillary and follicular types) | 20 |
| Urothelium | 4 | Urothelial cancer | 23 |
| – | – | Melanoma | 11 |
| – | – | Sarcoma | 10 |
In order to choose most suitable reference genes to test a wide range of tissue types, in a preliminary study, in 96 cDNA samples from the Cancer Survey arrays the expression of five reference genes were tested: Tubulin alpha 1b (TUBA1B), aminolevulinate, delta-, synthase (ALAS1), β2-microglobulin (B2 M), actin beta (ACTB) and hypoxanthine phosphoribosyltransferase 1 (HPRT1). Using NormFinder algorithm (16) the expression of TUBA1B and ALAS1 was found to be the most stable and this pair of genes was used as a reference for all examined samples.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) analysis
Analyses were performed as described earlier (14,17–19). Briefly, primers were designed by Primer 3 software (Whitehead Institute for Biomedical Research, Cambridge, USA) and purchased from the Laboratory of DNA Sequencing and Oligonucleotide Synthesis (Institute of Biochemistry and Biophysics, Polish Academy of Sciences, Warsaw, Poland). Primers sequences are listed in the Table II. RT-qPCR was carried out in a LightCycler 2.0 thermocycler (Roche Diagnostics, Basel, Switzerland) with software version 4.05. SYBR Green detection system was used based on LightCycler FastStart DNA Master SYBR Green I mix (Roche Diagnostics). PCR reactions were carried out in 20 µl mixtures, containing 4 µl template cDNA, 0.2 µM of each gene specific primer and 3.5 mM of Mg2+ ions. The RT-qPCR program included a 10 min denaturation step to activate the Taq DNA polymerase, followed by a three-step amplification program: Denaturation at 95.0°C for 9 sec, annealing at 58.0°C for 5 sec, and extension at 72.0°C for 5 sec. Specificity of the reaction products was routinely checked by determination of melting points (0.1°C/s transition rate) and random sample separation in a 2.5% ethidium bromide/agarose gel. PCR efficiency was assessed by a serial dilution method. Briefly, products of PCR reactions were separated in a 2.5% agarose gel and specific bands were extracted using a DNA gel extraction kit (EMD Millipore, Billerica, MA, USA). The amount of extracted DNA was estimated spectrophotometrically. Extracted DNA was diluted (10-fold serial dilutions) in order to generate a standard curve for efficiency calculation (LightCycler software v.4.05).
Table II.
Oligonucleotide sequences of sense and antisense primers.
| cDNA | Genbank accession no. | Primer | Primer sequence (5′-3′) | Position | PCR product size (bp) |
|---|---|---|---|---|---|
| ZFP91 | NM_053023 | S | TGTCCTTGCCCATCCTCGCTA | 1,128–1,148 | 190 |
| A | ACTCTTGAAGGCCCGAGCAC | 1,298–1,317 | |||
| TUBA1B | NM_006082 | S | TGGAACCCACAGTCATTGATGA | 430–451 | 135 |
| A | TGATCTCCTTGCCAATGGTGTA | 543–564 | |||
| ALAS1 | NM_000688 | S | AGACATAACATCTACGTGCAA | 2,031–2,051 | 167 |
| A | GAATGAGGCTTCAGTTCCA | 2,179–2,197 | |||
| B2M | NM_004048 | S | CAGCCCAAGATAGTTAAGTG | 385–404 | 262 |
| A | CCCTCCTAGAGCTACCTGT | 628–646 | |||
| ACTB | NM_001101 | S | CAGCCATGTACGTTGCTATCCAG | 473–496 | 151 |
| A | GAGGTCCAGACGCAGGATGGCATG | 601–623 | |||
| HPRT1 | NM_000194 | S | CTCCTCTGCTCCGCCACCG | 103–121 | 218 |
| A | TCGAGCAAGACGTTCAGTCC | 301–320 |
S, sense; A, antisense; ZFP91, zinc finger protein 91; TUBA1B, tubulin α1b; ALAS1, aminolevulinate, δ-synthase; B2M, β2-microglobulin; ACTB, β-actin; HPRT1, hypoxanthine phosphoribosyltransferase 1; PCR, polymerase chain reaction.
Cancer protein lysate array samples
ZFP91 protein expression studies were performed using a large-scale reverse phase protein array (RPPA)-ProteoScan Cancer Lysate Array 2.0 from OriGene. It contains 431 protein lysates of normal and cancer specimens from 11 different tissue types. The specimens come from accredited academic and medical institutions in the USA and were collected according to proper bioethical standards. All samples are provided with detailed pathology reports and basic clinical information regarding patient age, gender and disease staging. Array layout, sample location and associated clinical data for each biospecimen can be found at the OriGene website (http://www.origene.com).
As for protein extraction, all tissues were processed within 30 min of ischemia, frozen in OCT embedding agent at −80°C. The samples were homogenized and extracted in modified RIPA buffer containing protease inhibitors. Protein content was determined by BCA assay. All lysates were adjusted to 1 mg/ml using modified RIPA buffer and diluted to 500, 250, 125 and 62.5 µg/ml in RIPA buffer. The arrays were printed on Grace-Bio lab SuperNova nitrocellulose slides using non-contact, inkjet printing technologies. The total amount deposited for each spot was approximately 300 pl. The quality of each print was verified by Syproruby protein staining. Each sample was spotted on a slide at five different protein concentrations and these were spotted in triplicate on three different subarrays.
Each organ and its associated cancer type are represented by approximately 40 samples-25 cancer samples and 15 from normal tissues. In some cases, the number of samples analyzed was narrowed in order to include only specific cancer-type group e.g. in case of pancreatic adenocarcinoma. All tissue types and group sizes are listed in Table III.
Table III.
Analyzed types of normal tissue and cancerous protein samples (from Proteoscan Cancer Lysate Array-OriGene).
| Organ type | Total n | Cancer type | Total n |
|---|---|---|---|
| Breast | 15 | Breast cancer | 24 |
| Colon | 15 | Colon cancer | 25 |
| Kidney | 14 | Kidney cancer | 24 |
| Liver | 15 | Liver cancer | 25 |
| Lung | 15 | Lung cancer (non-small cell carcinoma) | 21 |
| Lymph node | 15 | Lymphoma (non-Hodgkin) | 24 |
| Skin | 11 | Melanoma | 19 |
| Ovary | 15 | Ovarian cancer | 24 |
| Pancreas | 9 | Pancreatic cancer (adenocarcinoma) | 7 |
| Prostate | 13 | Prostate cancer | 22 |
| Stomach | 12 | Stomach cancer | 23 |
Immunoblotting
The array immunoblotting procedure was optimized using ProteoScan Assay Optimization Array. After optimization, following procedure was utilized. The array was hydrated with 10 ml of ultrapure water for 30 min. Afterwards it was equilibrated with TBST washing buffer for 5 min, followed by 20 min incubation in background reducing buffer (0.5% polyvinyl alcohol; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). The membrane was quickly washed in washing buffer and blocked with 5 ml of array blocking buffer for 30 min (StartingBlock T20 (TBS) Blocking Buffer; Thermo Fisher Scientific, Inc) and again rinsed with washing buffer. Membrane was then incubated at 4°C with rabbit anti-ZFP91 at 1:50 (sc-102172; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) overnight. Afterwards, membrane was thoroughly washed and incubated with an anti-rabbit HRP-linked antibody at 1:500 (cat. no. 7074; Cell Signaling Technology, Inc., Danvers, MA, USA) for 1 h at room temperature. After washing, membrane was incubated with Biotinyl-tyramide solution (Tyramide Signal Amplification (TSA™) from PerkinElmer, Inc., Waltham, MA, USA) for 25 min and washed again. Streptavidin-Texas red conjugate diluted 1:100 in blocking buffer was added for 30 min at room temperature. The membrane was thoroughly washed and finally rinsed for 10 min in ultrapure water. Afterwards the array was air-dried and scanned using Genepix microarray scanner. Signal intensities from samples spotted at maximum protein concentration (1 mg/ml) were found to be falling best within dynamic range of the detection method for all tested samples and were utilized for comparative analysis.
Statistical analysis
GraphPad Prism v.5.00 (GraphPad Software, Inc., San Diego, CA, USA) was used to perform statistical analyses. P<0.05 was considered to indicate a statistically significant difference. Depending on the number of groups being statistically compared either a Mann-Whitney or Kruskal-Wallis test was performed with Dunn's test. The results are presented as the median and the interquartile range, or as box and whisker plots with the whiskers representing the 5 and 95th percentile.
Results
ZFP91 mRNA expression pattern in normal human tissues and cancers
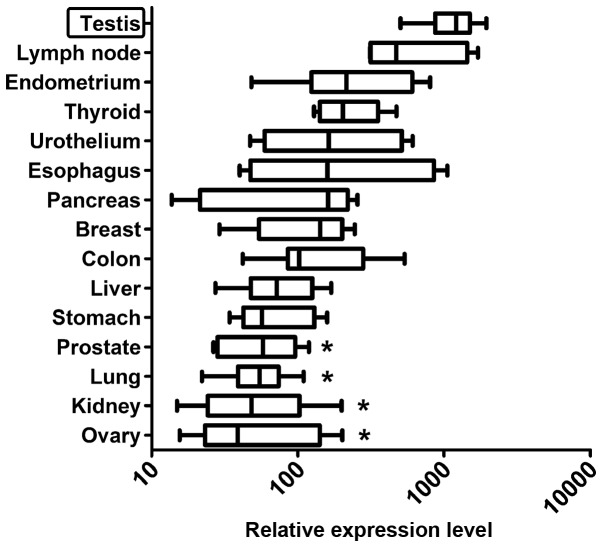
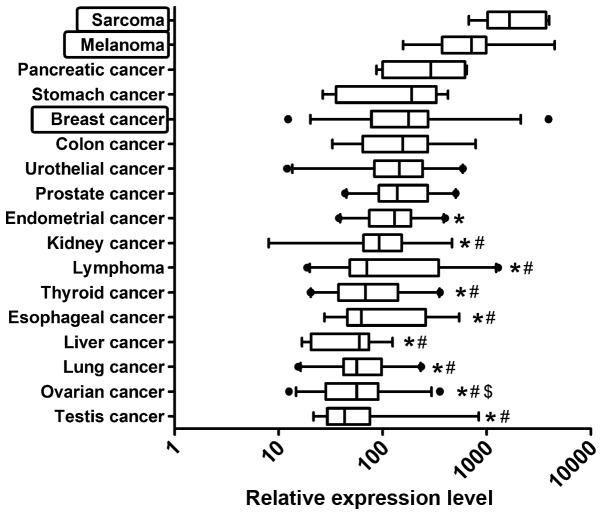
Firstly, cDNA samples from 15 normal human tissues were analyzed. The highest ZFP91 expression was found in testis (Fig. 1). It was significantly higher than in prostate, lung, kidney and ovary. The difference between highest expression-in testis and lowest-in ovary was more than 10-fold. Among the 17 cancer types analyzed, ZFP91 expression was the highest in sarcomas and melanoma (Fig. 2), while the lowest in testis and ovarian cancer.
Figure 1.
Reverse transcription-quantitative polymerase chain reaction analysis of Zinc Finger Protein 91 mRNA levels relative to the reference genes in 15 human normal organs. Median expression in all data was assigned a value of 100. Results are presented as box-and-whiskers plots with the whiskers showing the 5 and 95th percentile. Statistical comparisons were conducted using the Kruskal-Wallis test followed by Dunn's test. *P<0.05 vs. Testis.
Figure 2.
Reverse transcription-quantitative polymerase chain reaction analysis of Zinc Finger Protein 91 mRNA levels relative to the reference genes in 17 cancer types. Median expression in all data was assigned a value of 100. Results are presented as box-and-whiskers plots with the whiskers showing the 5 and 95th percentile. Statistical comparisons were conducted using the Kruskal-Wallis test followed by Dunn's test. *P<0.05 vs. Sarcoma; #P<0.05 vs. Melanoma; $P<0.05 vs. Breast cancer.
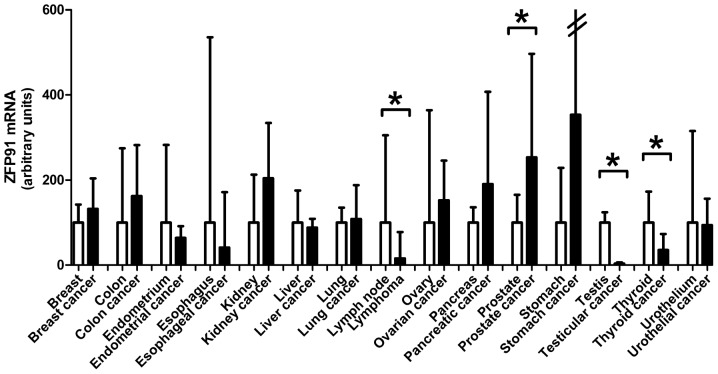
Finally, studied gene expression was compared between 15 normal tissue types and cancers derived from them (Fig. 3). Testis cancer deriving from organ with the highest ZFP91 expression-testis, had its expression significantly lower. No difference with this regard was noted between seminomas and non-seminomas. Second organ with highest ZFP91 expression-lymph nodes, also differed significantly from analyzed cases of non-Hodgkin lymphomas. In the case of thyroid cancer, similarly a downregulation of ZFP91 expression compared to normal thyroid was noted. The only cancer type with elevated ZFP91 mRNA expression compared to normal tissue was prostate cancer where the results are in accordance with our earlier reports of ZFP91 upregulation in this disease. In majority of the analyzed cancer types no significant differences in ZFP91 mRNA levels were observed between normal tissue and cancer.
Figure 3.
Comparison of ZFP91 expression between 15 human normal tissues and cancer types. Bars represent the mRNA levels relative to the reference genes. Median expression in each organ was independently assigned a value of 100 and the expression in the corresponding cancer type was normalized accordingly. Results are presented as the medians with the interquartile range. Statistical comparisons were conducted using the Mann-Whitney test. *P<0.05, as indicated. ZFP91, Zinc Finger Protein 91.
ZFP91 protein levels in 11 human organs and cancer types
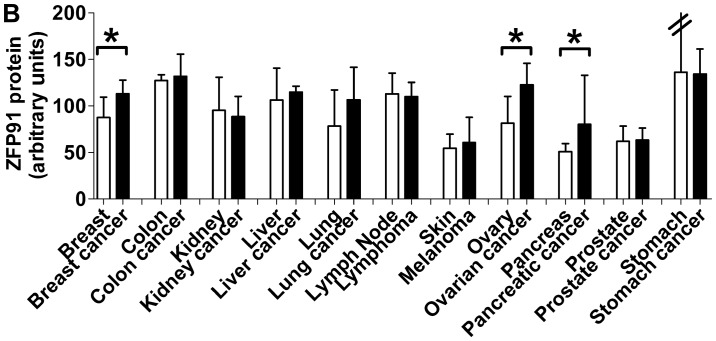
A large scale reverse phase protein array allowed for analysis of 431 protein lysates from normal and cancer specimens spanning 11 different tissues (Fig. 4A). ZFP91 protein expression was found to be ubiquitous and markedly less diverse than ZFP91 mRNA levels (Fig. 4B). Analyzed tissues and cancers presented similar ZFP91 protein levels. In most cases its expression did not differ between normal tissue and cancer derived from it. What is interesting, neither melanoma with observed exceptionally high ZFP91 mRNA levels nor prostate cancer with significant ZFP91 mRNA overexpression were found to present similar changes on a protein level. On the other hand, in the case of breast cancer, ovarian cancer and pancreatic cancer protein analysis revealed significant upregulation of ZFP91 levels, despite no differences noted in mRNA expression analysis.
Figure 4.
ZFP91 expression. (A) ZFP91 protein expression in a large-scale reverse-phase protein array (ProteoScan Cancer Lysate Array-OriGene) containing 431 protein lysates of normal and cancer specimens derived from 11 tissues and cancer types. (B) Comparison of ZFP91 protein levels between 11 human normal tissues and cancer types. Bars represent protein levels relative to total protein. Median expression in all data was assigned a value of 100. Results are presented as the median with the interquartile range. Statistical comparison was conducted using the Mann-Whitney test. *P<0.05, as indicated. ZFP91, Zinc Finger Protein 91.
Discussion
Earlier findings regarding ZFP91 expression in normal human tissues showed ZFP91 mRNA presence in all studied organs, with a particularly high expression in testis (2). The present study confirmed ZFP91 expression in tissues not to date examined: lymph node, endometrium, thyroid gland, urothelium, esophagus, breast and stomach. Particularly high ZFP91 expression in testis was also observed in our material. It is difficult to assess the value of this observation. Studies have proved that some of the mRNA transcripts found in testis are not present in other tissues and no evidence of their translation and function can be found (20–22). What is interesting, in mouse ZFP91 is one of the genes potentially causing male infertility (23). Unfortunately, this issue has not been further studied.
Another tissue with relatively high ZFP91 expression is lymph node and leukocytes it contains. This might be connected with NF-κB signaling pathway activity in these cells. NF-κB signaling pathway is present in its inducible form in virtually all cell types (24). However, in most cells of the immune system it is constitutionally activated (25–27). The constitutive activation of NF-κB signaling pathway is necessary to block apoptosis in mature lymphocytes in phase G0 of cell cycle (28). This suggests that higher ZFP91 expression in lymph node may be a result of higher NF-κB signaling pathway activity.
Among the 17 cancer types examined, two have shown markedly increased ZFP91 expression-sarcomas and melanoma. Overactivity of the NF-κB signaling pathway observed in these cancer types (29,30) seems not to be a sole cause of this phenomenon since it is also present in other types of aggressive cancers (31). The highest expression of ZFP91 in analyzed cases of sarcomas may reflect unique features associated with this type of cancer-aggressive, non-epithelial neoplasm of mesenchymal origin. Only sarcomas among studied cancer types originate from transformed mesenchymal stem cells (32). ZFP91 expression was at the second-highest level in melanoma, a cancer type possessing some distinct similarities to sarcomas. This cancer type is also an aggressive neoplasm but of neuroectodermal origin. A transformation towards mesenchymal phenotype plays, however, an important role in melanoma's development (33). In the embryogenesis, melanocytes migrate from the neural crest and this process is facilitated by transient mesenchymal phenotype. Mature melanocytes present some of the epithelial cells markers, but their neoplastic transformation into melanoma cells and melanoma's progression are related to their change in phenotype towards mesenchymal cells (34). They become more mobile, acquire the capacity to interact with extracellular matrix, to disseminate and metastasize (35). A question arises whether ZFP91 overexpression in sarcomas and melanoma is connected to their mesenchymal properties or presents just a concomitant phenomenon.
Comparative analysis of ZFP91 expression in human organs and respective cancer types revealed significant differences in a few cases. In testis cancer, non-Hodgkin lymphoma and thyroid cancer, ZFP91 expression was markedly lower in cancer samples. This may be connected to the fact that testis, lymph node and thyroid were all organs with the highest ZFP91 expression compared to other organs. Decreased ZFP91 expression in the cancer types stemming from them seems to be secondary to the changes in cellular phenotype. Further studies are required to elucidate full physiological role of ZFP91 in human organs. In the case of lymphoma cells a decrease in ZFP91 expression is particularly interesting, taking into consideration data showing its overexpression in leukemia cells (2). ZFP91 overexpression is therefore not a common feature of all cancers originating from immune system cells.
Among studied cancer types only in the case of prostate cancer ZFP91 expression was significantly elevated in comparison to the unchanged prostatic tissue. Similarly as in our previous study on this subject (15), the increased (even 10-fold) ZFP91 mRNA quantity was present only in the fraction of cancer cases. ZFP91 overexpression on mRNA level was observed by Lee et al (3) in cancers of prostate, stomach and liver. In our material, no differences were noted in cases of stomach and liver cancer. Based on the obtained results, it can be stated that ZFP91 mRNA regulation is not a phenomenon typical for oncogenic transformation.
Results of the reverse phase protein array analysis are the first to our knowledge to survey ZFP91 expression over a wide range of tissues and cancer types. Changes in ZFP91 mRNA levels observed in corresponding tissues were not reflected on a protein level. A discrepancy between mRNA and protein levels is often encountered (36–39) and in the case of ZFP91 has been described in our previous works (14,15) as well as observed by the team of Lee et al (3). At this point of knowledge it seems that no clear correlation between ZFP91 mRNA and protein levels exist. Based on previous experiments one can hypothesize that ZFP91 is subjected to posttranscriptional regulation within the cells. Further experiments in the field should establish whether this regulation involves changes in ZFP91 protein structure affecting its stability, particularly in neoplastic cells. Neither ZFP91 overexpression in melanoma and prostate cancer nor underexpression in lymphoma was observed on a protein level. On the other hand, in the case of breast cancer, ovarian cancer and pancreatic cancer a significant, although limited, protein upregulation was observed in our material. On a mRNA level these three types of cancer showed also limited upregulation although observed changes did not reach statistical significance. What is important, in the case of colon cancer no ZFP91 protein upregulation was noted, contrary to the results of the study performed by Ma et al (8). It can be hypothesized, that differences in the methodology and in the material may be the factors responsible for it. Furthermore, the character of RPPA analysis performed in this study makes it suitable for survey purposes only and may have limited sensitivity to detect more subtle changes in protein expression.
Several limitations of the presented study can be brought up and should be addressed in future experiments. Firstly, an immunohistochemical staining of ZFP91 protein in cancer tissue and adjacent healthy tissue would provide an interesting comparison as well as analysis of paired samples. This was not done in this study as provided sample sets consisted mostly of unpaired samples. Another interesting issue would be to analyze ZFP91 expression with regard to cancer sub-types and grading, however limited sample size of each group prevented us from further subdivisions. As for relation between ZFP91 expression and patients' prognosis and survival this could not be addressed in this study due to lack of such data. It would be interesting to analyze particular cancer types highlighted by this study e.g. melanoma, with the use of cell lines and patient samples including evaluation of patients' prognosis and treatment results.
Presented study is the first to examine ZFP91 expression pattern over a wide range of human tissues and cancer types. Obtained results indicate that ZFP91 is a ubiquitously expressed gene with overall stable expression levels. Revealed differences in studied gene expression, especially high mRNA levels in melanoma and sarcomas and elevated protein levels in cancers of breast, ovary and pancreas, are particularly interesting with regard to ZFP91 oncogenic potential. Further studies are, however, required to assess biological significance of the observed changes.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from Ministry of Science and Higher Education of the Republic of Poland (grant no. DI2011 0219 41).
Availability of data and materials
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.
Authors' contributions
LP, LKM and MR designed the study. LP, KJ, MS, MT and MR performed the experiments and analyzed the data. LP wrote the manuscript. LKM and MR revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Saotome Y, Winter CG, Hirsh D. A widely expressed novel C2H2 zinc-finger protein with multiple consensus phosphorylation sites is conserved in mouse and man. Gene. 1995;152:233–238. doi: 10.1016/0378-1119(94)00717-7. [DOI] [PubMed] [Google Scholar]
- 2.Unoki M, Okutsu J, Nakamura Y. Identification of a novel human gene, ZFP91, involved in acute myelogenous leukemia. Int J Oncol. 2003;22:1217–1223. [PubMed] [Google Scholar]
- 3.Lee JJ, Lee JH, Lee K, Hong Ys, Jin X. Therapeutic agent for cancer, inflammation and auto-immune disease containing inhibitor of Zinc Finger Protein 91. US Patent 20: 080,248,024. 2008
- 4.Jin X, Jin HR, Jung HS, Lee SJ, Lee JH, Lee JJ. An atypical E3 ligase zinc finger protein 91 stabilizes and activates NF-kappaB-inducing kinase via Lys63-linked ubiquitination. J Biol Chem. 2010;285:30539–30547. doi: 10.1074/jbc.M110.129551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun SC. Non-canonical NF-κB signaling pathway. Cell Res. 2011;21:71–85. doi: 10.1038/cr.2010.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin HR, Jin X, Lee JJ. Zinc-finger protein 91 plays a key role in LIGHT-induced activation of non-canonical NF-κB pathway. Biochem Biophys Res Commun. 2010;400:581–586. doi: 10.1016/j.bbrc.2010.08.107. [DOI] [PubMed] [Google Scholar]
- 7.Xiao G, Fu J. NF-κB and cancer: A paradigm of Yin-Yang. Am J Cancer Res. 2011;1:192–221. [PMC free article] [PubMed] [Google Scholar]
- 8.Ma J, Mi C, Wang KS, Lee JJ, Jin X. Zinc finger protein 91 (ZFP91) activates HIF-1α via NF-κB/p65 to promote proliferation and tumorigenesis of colon cancer. Oncotarget. 2016;7:36551–36562. doi: 10.18632/oncotarget.9070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mabjeesh NJ, Amir S. Hypoxia-inducible factor (HIF) in human tumorigenesis. Histol Histopathol. 2007;22:559–572. doi: 10.14670/HH-22.559. [DOI] [PubMed] [Google Scholar]
- 10.Masoud GN, Li W. HIF-1α pathway: Role, regulation and intervention for cancer therapy. Acta Pharm Sin B. 2015;5:378–389. doi: 10.1016/j.apsb.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tafani M, Pucci B, Russo A, Schito L, Pellefrini L, Perrone AG, Villanova L, Salvatori L, Ravenna L, Petrangeli E, Russo MA. Modulators of HIF1α and NFkB in cancer treatment: Is it a rational approach for controlling malignant progression? Front Pharmacol. 2013;4(13) doi: 10.3389/fphar.2013.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoesel B, Schmid JA. The complexity of NF-κB signaling in inflammation and cancer. Mol Cancer. 2013;12:86. doi: 10.1186/1476-4598-12-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu Y, Liu J, Huang H. Recent agents targeting HIF-1α for cancer therapy. J Cell Biochem. 2013;114:498–509. doi: 10.1002/jcb.24390. [DOI] [PubMed] [Google Scholar]
- 14.Paschke L, Rucinski M, Ziolkowska A, Zemleduch T, Malendowicz W, Kwias Z, Malendowicz LK. ZFP91-a newly described gene potentially involved in prostate pathology. Pathol Oncol Res. 2014;20:453–459. doi: 10.1007/s12253-013-9716-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paschke L, Jopek K, Szyszka M, Tyczewska M, Ziolkowska A, Rucinski M, Malendowicz LK. ZFP91: A noncanonical NF-κB signaling pathway regulator with oncogenic properties is overexpressed in prostate cancer. Biomed Res Int. 2016;2016:6963582. doi: 10.1155/2016/6963582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 17.Paschke L, Zemleduch T, Rucinski M, Ziolkowska A, Szyszka M, Malendowicz LK. Adiponectin and adiponectin receptor system in the rat adrenal gland: Ontogenetic and physiologic regulation, and its involvement in regulating adrenocortical growth and steroidogenesis. Peptides. 2010;31:1715–1724. doi: 10.1016/j.peptides.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Szyszka M, Paschke L, Tyczewska M, Rucinski M, Grabowska P, Malendowicz LK. Lack of expression of preproorexin and orexin receptors genes in human normal and prostate cancer cell lines. Folia Histochem Cytobiol. 2015;53:333–341. doi: 10.5603/fhc.a2015.0035. [DOI] [PubMed] [Google Scholar]
- 19.Jopek K, Celichowski P, Szyszka M, Tyczewska M, Milecka P, Malendowicz LK, Rucinski M. Transcriptome profile of rat adrenal evoked by gonadectomy and testosterone or estradiol replacement. Front Endocrinol (Lausanne) 2017;8:26. doi: 10.3389/fendo.2017.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobner PR, Kislauskis E, Wentworth BM, Villa-Komaroff L. Alternative 5′exons either provide or deny an initiator methionine codon to the same alpha-tubulin coding region. Nucleic Acids Res. 1987;15:199–218. doi: 10.1093/nar/15.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrett JE, Collard MW, Douglass JO. Translational control of germ cell-expressed mRNA imposed by alternative splicing: Opioid peptide gene expression in rat testis. Mol Cell Biol. 1989;9:4381–4389. doi: 10.1128/MCB.9.10.4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stallard BJ, Collard MW, Griswold MD. A transferrinlike (hemiferrin) mRNA is expressed in the germ cells of rat testis. Mol Cell Biol. 1991;11:1448–1453. doi: 10.1128/MCB.11.3.1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trachtulec Z, Mnuková-Fajdelová M, Hamvas RM, Gregorová S, Mayer WE, Lehrach HR, Vincek V, Forejt J, Klein J. Isolation of candidate hybrid sterility 1 genes by cDNA selection in a 1.1 megabase pair region on mouse chromosome 17. Mamm Genome. 1997;8:312–316. doi: 10.1007/s003359900430. [DOI] [PubMed] [Google Scholar]
- 24.Lenardo MJ, Baltimore D. NF-kappa B: A pleiotropic mediator of inducible and tissue-specific gene control. Cell. 1989;58:227–229. doi: 10.1016/0092-8674(89)90833-7. [DOI] [PubMed] [Google Scholar]
- 25.Kolenko V, Bloom T, Rayman P, Bukowski R, Hsi E, Finke J. Inhibition of NF-kappa B activity in human T lymphocytes induces caspase-dependent apoptosis without detectable activation of caspase-1 and −3. J Immunol. 1999;163:590–598. [PubMed] [Google Scholar]
- 26.Ward C, Chilvers ER, Lawson MF, Pryde JG, Fujihara S, Farrow SN, Haslett C, Rossi AG. NF-kappaB activation is a critical regulator of human granulocyte apoptosis in vitro. J Biol Chem. 1999;274:4309–4318. doi: 10.1074/jbc.274.7.4309. [DOI] [PubMed] [Google Scholar]
- 27.Griffin GE, Leung K, Folks TM, Kunkel S, Nabel GJ. Activation of HIV gene expression during monocyte differentiation by induction of NF-kappa B. Nature. 1989;339:70–73. doi: 10.1038/339070a0. [DOI] [PubMed] [Google Scholar]
- 28.Bureau F, Vanderplasschen A, Jaspar F, Minner F, Pastoret PP, Merville MP, Bours V, Lekeux P. Constitutive nuclear factor-kappaB activity preserves homeostasis of quiescent mature lymphocytes and granulocytes by controlling the expression of distinct Bcl-2 family proteins. Blood. 2002;99:3683–3691. doi: 10.1182/blood.V99.10.3683. [DOI] [PubMed] [Google Scholar]
- 29.Horiuchi K, Morioka H, Nishimoto K, Suzuki Y, Susa M, Nakayama R, Kawai A, Sonobe H, Takaishi H, Ozaki T, et al. Growth suppression and apoptosis induction in synovial sarcoma cell lines by a novel NF-kappaB inhibitor, dehydroxymethylepoxyquinomicin (DHMEQ) Cancer Lett. 2008;272:336–344. doi: 10.1016/j.canlet.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 30.Madonna G, Ullman CD, Gentilcore G, Palmieri G, Ascierto PA. NF-κB as potential target in the treatment of melanoma. J Transl Med. 2012;10:53. doi: 10.1186/1479-5876-10-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin Y, Bai L, Chen W, Xu S. The NF-kappaB activation pathways, emerging molecular targets for cancer prevention and therapy. Expert Opin Ther Targets. 2010;14:45–55. doi: 10.1517/14728220903431069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao W, Mohseny AB, Hogendoorn PC, Cleton-Jansen AM. Mesenchymal stem cell transformation and sarcoma genesis. Clin Sarcoma Res. 2013;3:10. doi: 10.1186/2045-3329-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikesh LM, Kumar M, Erdag G, Hogan KT, Molhoek KR, Mayo MW, Slingluff CL., Jr Evaluation of molecular markers of mesenchymal phenotype in melanoma. Melanoma Res. 2010;20:485–495. doi: 10.1097/CMR.0b013e32833fafb4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alonso SR, Tracey L, Ortiz P, Pérez-Gómez B, Palacios J, Pollán M, Linares J, Serrano S, Sáez-Castillo AI, Sánchez L, et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007;67:3450–3460. doi: 10.1158/0008-5472.CAN-06-3481. [DOI] [PubMed] [Google Scholar]
- 35.Grunert S, Jechlinger M, Beug H. Diverse cellular and molecular mechanisms contribute to epithelial plasticity and metastasis. Nat Rev Mol Cell Biol. 2003;4:657–665. doi: 10.1038/nrm1175. [DOI] [PubMed] [Google Scholar]
- 36.Maier T, Güell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 37.Maier T, Schmidt A, Güell M, Kühner S, Gavin AC, Aebersold R, Serrano L. Quantification of mRNA and protein and integration with protein turnover in a bacterium. Mol Syst Biol. 2011;7:511. doi: 10.1038/msb.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 2010;329:533–538. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analyzed during the present study are available from the corresponding author on reasonable request.