Abstract
The objective of this study was to assess the impact of Saccharomyces cerevisiae in combination with Lactobacillus buchneri on the fermentation characteristics, aerobic stability, nutritive value, and microbial communities of corn silage. Whole crop corn (39% DM) was either uninoculated (Control) or inoculated with S. cerevisiae and L. buchneri at the following concentrations: S. cerevisiae 104 cfu/g fresh forage (S4), S. cerevisiae 105 cfu/g (S5), S. cerevisiae 104 cfu/g + L. buchneri 105 cfu/g (S4L5), and S. cerevisiae 105 cfu/g + L. buchneri 104 cfu/g (S5L4), and ensiled in mini silos for 118 d, followed by 7 d of aerobic exposure. Changes in fermentation characteristics and nutritive value were assessed in terminal silages. Saccharomyces cerevisiae, L. buchneri, and total yeast, fungal, and bacterial communities in silage were estimated using quantitative PCR. Composition of bacterial and fungal communities during ensiling and aerobic exposure was measured using 16S rDNA and ITS sequencing, respectively. In the first 7 d of ensiling, the concentration of lactic acid rapidly increased (P < 0.01) in all silages, with the pH declining to 4.0 (P < 0.001) and thereafter remaining stable (P = 0.23). Although S4L5 contained a higher (P = 0.03) concentration of acetic acid than Control, other fermentation characteristics were did not differ among terminal silages. Inoculation with S. cerevisiae had no detrimental effect on the aerobic stability of silage, whereas L. buchneri did not prevent spoilage as the pH across all silages averaged 8.0 after 7 d of aerobic exposure. Total yeast (P = 0.42), bacterial (P = 0.13), and fungal (P = 0.89) communities were not altered by the inoculants after ensiling or aerobic exposure. Sequencing identified temporal shifts of bacterial and fungal communities during ensiling and aerobic exposure. Concentrations of S. cerevisiae and L. buchneri in all inoculated silages remained greater (P < 0.01) than Control after ensiling, with numbers of S. cerevisiae increasing after 7 d of aerobic exposure. Bacterial communities in silages inoculated with S. cerevisiae and L. buchneri clustered separately from other silages, an observation that was not apparent for fungal communities. Our findings suggest that aerobic exposure could potentially increase the abundance of S. cerevisiae with probiotic properties in corn silage just prior to feeding.
Keywords: aerobic stability, corn silage, ensiling, inoculant, microbiome
INTRODUCTION
Various silage inoculants have been used to speed up the post-ensiling decline in pH, improve the aerobic stability, and enhance forage digestibility (Weinberg and Muck, 1996; Muck et al., 2018). The first-generation inoculants contained homolactic bacteria that accelerated the production of lactic acid and pH decline, enhancing the preservation of silage nutrients (McAllister et al., 1995). Subsequently, heterolactic bacteria, such as Lactobacillus buchneri, were introduced as second-generation inoculants as they produce residual amounts of acetic acid and 1,2-propanediol that improve aerobic stability through the inhibition of yeasts and molds (Reich and Kung, 2010). Apart from improving aerobic stability, some selected strains of L. buchneri were defined as third-generation inoculants as they possessed feruloyl esterases that may hydrolyze feruloylated polysaccharides within the forage plant cell wall and thus improve the digestibility of fiber in silage (Addah et al., 2012).
Directly fed microbials such as the yeast, Saccharomyces cerevisiae, have been reported to promote rumen health, improve feed efficiency, and mitigate methane emissions (Auclair, 2001; McAllister et al., 2011). Consequently, delivery within silage instead of as a separate additive to the diet may be a convenient means for livestock producers to deliver these beneficial microbes to ruminants. Such additives could be defined as fourth-generation silage inoculants where probiotic yeasts deliver direct benefits to the animal, whereas lactic acid bacteria (LAB) enhance the ensiling and digestibility of the forage. Only a few studies on fourth-generation inoculants have been undertaken, but a previous study in our laboratory found that inoculation of corn forage with 2 strains of S. cerevisiae or a single strain of Saccharomyces paradoxus had no detrimental effect on the aerobic stability of corn silage. Furthermore, S. cerevisiae and S. paradoxus populations increased during aerobic exposure, suggesting that the density of these beneficial yeast strains increases as the silage is exposed to oxygen prior to feeding (Dunière et al., 2015). Yeast strains used alone may not provide all the advantages of traditional silage inoculants. Consequently, further assessment of the effect of Saccharomyces combined with LAB bacteria on the fermentation and aerobic stability of silage is needed. The objectives of this study were to evaluate the effect of inoculation of corn forage with S. cerevisiae alone or in combination with L. buchneri on fermentation and aerobic stability of corn silage.
MATERIALS AND METHODS
Forages
Corn (Zea mays; 39T67; Pioneer Inc., Johnston, IA) was planted on 12 May 2014 at the Agriculture and Agri-Food Canada Lethbridge Research and Development Centre and harvested on 24 September at 2/3 milk line maturity (37 to 41% DM). Corn was planted at a depth of 4 cm, with 72-cm row spacing and a seed density of 79,000 seeds/ha. Crops were fertilized with 170 kg N/ha pre-seeding and 12.5 kg N/ha and 57 kg PO4/ha post-seeding and grown under a central pivot irrigation system. The harvested forage from the same plot and variety was chopped (9.5 mm average length) with a forage harvester (John Deere 6610; Moline, IL) equipped with a kernel processor and then used for the mini silo experiment.
Mini Silo Experiment
Samples of forages were divided into five 20-kg lots, spread on separate plastic sheets, and then sprayed with either water (i.e., 3 mL/kg; Control) or inoculated with S. cerevisiae NRRL Y-50736 and L. buchneri NRRL B-50733 in the same volume of water to achieve the following concentrations: S. cerevisiae 104 cfu/g fresh forage (S4), S. cerevisiae 105 cfu/g (S5), S. cerevisiae 104 cfu/g + L. buchneri 105 cfu/g (S4L5), and S. cerevisiae 105 cfu/g + L. buchneri 104 cfu/g (S5L4). The 4 corners of the sheet were drawn together with the forage being tumbled inside the sheet while inoculants were applied for approximately 3 min. Forage was hand mixed for an additional minute to ensure uniform inoculation. Forage (2.5 to 3 kg) was packed into mini PVC silos (10.4 cm diameter × 35.6 cm height) with a hydraulic press to a density of ~240 kg/m3. Prior to the hydraulic press, 2 temperature sensors (Thermochron iButtons; Embedded Data Systems, Lawrenceburg, KY) were embedded in the lower and middle layers of each silo, and an additional sensor was also placed in the silo storage room to record temperature every 2 h. Each silo was immediately weighed before and after filling and sealing. Triplicate silos for each treatment and sampling date were prepared and opened after 7, 30, and 61 d of ensiling, with 6 replicate silos opened at day 118 for the subsequent aerobic stability study. Prior to ensiling, triplicate samples of forage were collected for subsequent analysis. Silos were weighed prior to opening to estimate DM loss. The contents of each of the triplicate mini silos were mixed thoroughly after opening, and subsamples from each silo were collected for subsequent analysis.
Aerobic Stability
A total of 6 mini silos opened on day 118 for each treatment were pooled into 3 samples by bulking the content of 2 replicates into 3 samples. This resulted in 3 pooled silage samples for each treatment. Subsamples of silage (~1.2 kg) obtained from each silo of pooled silage samples at the end of ensiling day were placed into separate 4-L insulated containers, covered with 2 layers of cheesecloth and stored at 20 °C for 7 d. Two temperature sensors (Embedded Data Systems) were embedded in the lower and middle layers of each container and placed in the storage room to record temperature every 2 h. Contents of each container were thoroughly mixed and sampled after 3 and 7 d of aerobic exposure for subsequent analysis.
Chemical Analysis
For chemical analysis, 15 g of either fresh forage or silage samples was mixed with 135 mL of deionized water, blended for 30 s, and filtered through 2 layers of cheesecloth. The pH of the filtrate was immediately measured with a Symphony pH meter (VWR, Mississauga, ON, Canada). The filtrate was then divided into 2 portions. One portion was boiled for 10 min to halt fermentation and stored at −20 °C for subsequent analysis of water-soluble carbohydrates (WSC) by the Nelson–Somogyi method (Nelson, 1944) on a Dynatech MRX microplate reader (Dynatech Laboratories Inc., Chantilly, VA). The other portion of the filtrate was centrifuged for 15 min at 10,000 × g and collected for analyses of VFA, lactic acid, ammonia nitrogen (NH3-N), and ethanol as described by Addah et al. (2012). For starch determination, forage and silage samples were first freeze-dried and then ball-ground using a mixer mill (MM400, Retsch Inc., Newtown, PA). Starch was determined after hydrolysis to α-glucose polymers using amyloglucosidase (Megazyme Int. Ltd., Wicklow, UK) and 1,4 α-d-glucan glucano-hydrolase (Brennfag Canada Inc., Toronto, ON, Canada) as described by Herrera-Saldana et al. (1990). Total nitrogen (N) was determined by elemental analysis (Dumas Nitrogen) using a NA1500 Nitrogen/Carbon Analyzer (Carlo Erba Instruments, Milan, Italy). Crude protein was calculated as N × 6.25. The samples were analyzed for analytical DM by drying at 105 °C in a forced air oven for 24 h, for OM by ashing 1 g of dried sample in a muffle furnace at 550 °C for 5 h, and for NDF and ADF using an Ankom 200 system (Ankom Technology Corporation, Fairport, NY) with the addition of sodium sulfite and α-amylase in the NDF procedure. Acid detergent insoluble N was measured by combustion analysis as described previously.
Microbial Analysis
For microbial analyses, fresh forage or silage (10 g) was added to 90 mL of sterile 70 mM potassium phosphate buffer (pH 7.0) and agitated for 60 s at 260 rpm in a Stomacher 400 Circulator (Seward Stomacher, London, UK). The suspension was serially diluted and spread in triplicate onto semiselective lactobacilli media amended with 200 mg/mL of cycloheximide (MRS; Dalynn Biologicals, Calgary, AB, Canada) for enumeration of LAB, onto nutrient agar amended with 200 mg/mL of cycloheximide (NA; Dalynn Biologicals) for the enumeration of total bacteria (TB), and onto Sabouraud’s dextrose agar (SDA; Dalynn Biologicals) amended with 100 mg/mL of tetracycline and chloramphenicol for the enumeration of yeasts and other fungi. Lactobacilli MRS agar and NA plates were incubated aerobically at 37 °C for up to 48 h, whereas SDA plates were incubated at ambient temperature for 72 h. Colonies were counted from plates containing between 30 and 300 colonies. Numbers of yeasts and filamentous fungi (i.e., molds) were differentiated based on colony appearance and morphology.
DNA Extraction
For each sampling time, silage samples (30 g) from each mini silo were freeze-dried and ground through a 4-mm screen. Subsamples (5 g) were ball-ground for DNA extraction as described by Dunière et al. (2015), and yield and purity of extracted DNA were measured using a Nanodrop 3300 Spectrophotometer (Thermo Scientific, Waltham, MA) and stored at −20 °C prior to molecular analysis.
Quantitative PCR
Universal ribosomal DNA primer pairs as described in Dunière et al. (2015) were used to estimate TB, yeast, and fungi associated with fresh forage and silage during ensiling and aerobic exposure. Strain-specific primers for L. buchneri NRRL B-50733 were designed by alignment of its genomic sequence with known bacterial sequences in GenBank and selecting a region encoding a hypothetical protein, which did not match any other nucleotide sequences within the PubMed Nucleotide database (BLAST; Johnson et al., 2008). Similarly, primer sets for S. cerevisiae NRRL Y-50736 were designed by comparing its genomic sequence with 34 other yeast strains in the database. A total of 11 candidate primer sets were tested against 12 yeast strains within the DuPont Pioneer collection to ensure specificity and quantitative PCR (qPCR) efficiency. One of the candidate primer sets targeted a specific gene encoding for a C2 domain involved in delivering proteins to the cell membrane was selected. All PCR primers and annealing temperatures are summarized in Supplementary Table A1.
Standards for the quantification of TB, yeast, fungi, L. buchneri NRRL B-50733, and S. cerevisiae NRRL Y-50736 by qPCR were created as described in Dunière et al. (2015). Briefly, genomic DNA was extracted from L. buchneri NRRL B-50733 and S. cerevisiae NRRL Y-50736 using the protocols as described in Dunière et al. (2015) and amplified with primers (Supplementary Table A1). The PCR products were cloned into the pCR2.1 TOPO TA cloning vector (Life Technologies, Oakville, ON, Canada), and the plasmids were extracted using QIAprep Spin Miniprep Kit (Qiagen, Toronto, ON, Canada) and used as standards for qPCR. To confirm the standards encoded the appropriate gene fragments, plasmid inserts were commercially sequenced (Eurofins Genomics, Huntsville, AL) and analyzed by BLAST. All plasmid standard DNA was serially diluted from 102 to 107 copies/µL to generate a standard curve for qPCR. All reactions were conducted on an Applied Biosystems 7900HT Fast Real-Time PCR system (Life Technologies) and contained 1× iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA), 20 ng DNA, 0.2 µM primers, and 0.1 µg/µL bovine serum albumin (New England Biolabs, Pickering, ON, Canada) in a final volume of 25 µL. Conditions for PCR consisted of 1 cycle at 95 °C for 3 min, followed by 35 cycles of 15 s at 95 °C, 1 min at the respective annealing temperature for each primer pair, and 1 min at 72 °C. Melting curve analyses of the products were performed after the final extension step to ensure that the fluorescence signal originated from target amplification. Florescence normalization and data analysis were conducted using SDS software (Life Technologies).
Sequencing and Sequence Analysis
The V1 to V2 region of the 16S rDNA was PCR-amplified with primers 27F (5′-GTGCCAGCM GCCGCGGTAA-3′) and 341R (5′-CTGCWGCCN CCCGTAGG-3′) using Accuprime Pfx DNA polymerase (Life Technologies) in a reaction volume of 20 µL. A 12-bp barcode for sample identification was included in the forward primers. Amplification was achieved with the Applied Biosystems Veriti Thermocycler (Life Technologies) using an initial denaturation at 95 °C for 2 min, followed by 30 cycles of denaturation at 95 °C for 15 s, annealing at 50 °C for 30 s, and a final extension at 68ºC for 8 min. Polymerase chain reaction products were combined and assessed for integrity by agarose (2%) gel electrophoresis and quantified using the Qubit dsDNA BR Assay Kit in accordance with the manufacturer’s instructions (Life Technologies). Samples were pooled in equal proportions based on DNA concentrations. Pooled amplicons were then purified using a QIAquick Gel Extraction Kit (Qiagen). Paired-end reads of 300 bp were generated using the MiSeq v. 4 sequencing platform (Illumina Inc., San Diego, CA) from the purified amplicons. All paired ends were assembled, trimmed, quality filtered, and deconvoluted based on the 12-bp barcode sequence using Qiime 1.9.1’s split-libraries.fastq.py script-q 19. All other parameters used default values (Caporaso et al., 2010). Read pairs that could not be assembled and single reads were discarded. Chimeras were removed by script identify_chimeric_seqs.py with the usearch61 option (Edgar, 2010). Sequences that were shorter than 400 bp and longer than 550 bp with ambiguous bases or with homopolymers longer than 8 bases were removed and the remaining sequences were analyzed using QIIME (1.9.1) according to Caporaso et al. (2010). Operational–taxonomic units (OTUs) were picked at 97% identity threshold using the pick_de_novo_otus.py script. Representative OTUs were classified by Uclust (Edgar, 2010) against the GreenGenes reference database (release 13_08; DeSantis et al., 2006). Singletons were removed with the filter_otus_from_otu_table.py script (Bokulich et al., 2013) before further analyses. To allow for species-level classification, an in-house 16S rDNA database was used. The in-house 16S rDNA database consisted of a combination of 1) the bacterial 16S rDNA sequences from NCBI’s Bacterial 16S Ribosomal RNA RefSeq Targeted Loci Project (accession number: PRJNA33175), 2) 16S rDNA sequences extracted from genome sequences of L. buchneri PTA-2494 and NRRL B-50733, and 3) commonly known silage bacteria including Lactobacillus, Acetobacter, Weissella, Bacillus, Enterobacteriaceae, Clostridium, and Pediococcus. The representative sequences of each OTU were classified using the BLAST algorithm from the Qiime’s “Assign_taxonomy.py” script using the above-mentioned database. Most of OTUs classified to the species level.
The internal transcribed spacer region 1 (ITS1) of the fungal DNA was PCR amplified with primers ITSF (5′-AATGATACGGCGACCACCGAGATCTACACGGCTTGGTCATTTAGA-GGAAGTAA-3′) and ITS2 (5′-CAAGCAGAAGACGGCATACGAGATNNNNNNNNNNNN-CGGCTGCGTTCTTCATCGATGC-3′; Walters et al., 2016) using Accuprime Pfx DNA polymerase (Life Technologies) with a reaction volume of 20 µL. The sequence “NNNNNNNNNNNN” designates a 12-bp barcode for sample identification that was included in the reverse primers. Amplification was achieved with the Applied Biosystems Veriti Thermocycler (Life Technologies) using an initial denaturation at 95 °C for 2 min followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 50 °C for 30 s, and a final extension at 68 °C for 8 min. Polymerase chain reaction products were assessed, quantified, and purified using the same methods as described previously in the section of bacterial 16S rDNA sequencing. Reads of 300 bp were generated using the MiSeq v. 4 sequencing platform (Illumina Inc.) from the purified amplicons. The sequences were trimmed, quality filtered (≥Q20), and deconvoluted in the same manner as described previously for 16S rDNA sequences. Sequences that were shorter than 100 bp and longer than 250 bp with ambiguous bases or with homopolymers longer than 12 bases were removed. Operational–taxonomic units were picked at 97% identity threshold using the pick_open_reference_otus.py script with a UNITE reference database provided by “Unified system for the DNA based fungal species linked to the classification” Ver. 7.0 (Kõljalg et al., 2013). Representative OTUs were classified by BLAST against the same UNITE reference database using default parameters.
Statistical Analysis
The bacterial community structure (beta diversity) of each inoculant treatment and sampling time was evaluated using the weighted UniFrac distances (Lozupone and Knight, 2005) and visualized as principal coordinate analysis (PCoA) plots using Emperor (Vázquez-Baeza et al., 2013). The fungal community structure was assessed using Bray–Curtis distances and PCoA plots. Cultured microbial populations were estimated as cfu/g of forage or silage DM and then log transformed for statistical analysis. Copy numbers for each gene were log transformed before statistical analysis. Least square means of chemical and microbial parameters measured after ensiling that showed significant differences were separated by a pairwise Fisher’s LSD test at P > 0.05 using SAS (SAS Institute, Inc., Cary, NC). Changes in S. cerevisiae and L. buchneri as measured by qPCR over time were assessed using a repeated-measures analysis with each mini silo (n = 3) and each 4-L insulated container (n = 3) serving as random factors for ensiling and aerobic stability, respectively, whereas treatment was considered as a fixed factor. Main effects of sampling day, treatment, and their interactions were considered to be statistically significant at a probability level of <0.05.
RESULTS
Chemical and Microbial Analyses
The chemical characteristics of pre- and post-ensiled corn forages are shown in Table 1. Once ensiling started, the pH of all silages dropped to 4 before day 7. Temperature profiles of silages were similar during ensiling (Supplementary Fig. A1a), and all exhibited a temperature increase after 7 d of aerobic exposure (Supplementary Fig. A1b). An interaction between inoculant and sampling day (P < 0.001) affected pH during ensiling, but there was no treatment effect at day 118 (Table 1; Supplementary Fig. A2). Over 7 d of aerobic exposure, pH was affected by sampling time (P < 0.001) with an increase to an average of 8.0 at the end of the experiment (Supplementary Fig. A2). Corn DM, OM, and starch declined as a result of ensiling, whereas CP tended to increase, with no effect of treatment (Table 1). After ensiling, level of ADIN was higher (P = 0.03) in all inoculated silage than the control silage except S4. Corn NDF and ADF did not differ (P = 0.05; P = 0.14) before and after ensiling with no treatment effect after ensiling (P = 0.25; P = 0.38). Concentration of WSC declined (P < 0.001) after ensiling, but no treatment effect was observed (P = 0.26). Lactic and acetic acids were the major fermentation acids present in terminal silage, with propionic and butyric acids being below detectable limits. Acetic acid concentration in silage was influenced by L. buchneri in a dose-dependent manner with higher (P = 0.02) concentration in S4L5 than the control. Concentrations of lactic acid (P = 0.77), ethanol (P = 0.52), and NH3-N (P = 0.06) in silage did not differ among treatments (Table 1).
Table 1.
Effect of inoculants on chemical composition of whole-crop corn silage after 118 d of ensiling
| Day 1182 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Day 01 | Control | S4 | S5 | S4L5 | S5L4 | SEM3 | P-value4 | |
| pH | 5.70 | 3.89 | 3.92 | 3.91 | 3.94 | 3.92 | 0.018 | 0.51 |
| Nutrient composition, % DM | ||||||||
| DM | 39.06 | 37.10 | 37.66 | 38.59 | 39.10 | 39.04 | 0.902 | 0.48 |
| OM | 95.94 | 95.65 | 95.83 | 95.86 | 95.82 | 95.71 | 0.077 | 0.32 |
| CP | 8.70 | 9.54 | 9.50 | 9.88 | 9.47 | 9.41 | 0.333 | 0.86 |
| ADIN, % total N | 4.56 | 4.85b4 | 6.06ab | 6.87a | 6.98a | 6.34a | 0.409 | 0.03 |
| NDF | 48.04 | 48.81 | 53.92 | 54.39 | 53.58 | 53.77 | 1.833 | 0.25 |
| ADF | 23.03 | 23.81 | 25.65 | 25.70 | 25.52 | 25.30 | 0.731 | 0.38 |
| Starch | 28.07 | 28.15 | 26.13 | 27.01 | 26.63 | 27.58 | 1.262 | 0.81 |
| WSC,5 mg/g DM | 21.34 | 7.86 | 7.39 | 7.10 | 6.83 | 6.75 | 0.365 | 0.26 |
| Fermentation products, g/kg | ||||||||
| Lactic acid | 0.20 | 23.27 | 19.42 | 23.44 | 20.06 | 20.55 | 2.803 | 0.77 |
| Acetic acid | ND5 | 14.63b | 13.31b | 13.26b | 17.54a | 15.45ab | 0.790 | 0.02 |
| NH3-N | 0.14 | 0.92 | 0.86 | 0.84 | 0.85 | 0.80 | 0.024 | 0.06 |
| Ethanol | ND | 1.97 | 2.22 | 2.19 | 2.11 | 2.62 | 0.260 | 0.52 |
Data of fresh forage at day 0 were not included in statistical analysis.
Control = uninoculated; S4 = S. cerevisiae (1 × 104 cfu/g wet silage); S5 = S. cerevisiae (1 × 105 cfu/g wet silage); S4L5 = S. cerevisiae (1 × 104 cfu/g wet silage) and L. buchneri (1 × 105 cfu/g wet silage); S5L4 = S. cerevisiae (1 × 105 cfu/g wet silage) and L. buchneri (1 × 104 cfu/g wet silage).
Standard errors of least squares means (n = 3).
Within a row, means followed by different letters differ at P < 0.05.
WSC = water-soluble carbohydrates; ND = not determined.
Plate numbers of LAB increased (P < 0.0001) after 118 d with a trend toward higher numbers in S4L5 and S5L4 than control silage (Table 2). The viable counts of TB, yeasts, and fungi declined after ensiling with S4L5 containing higher (P = 0.01) numbers of bacteria than control silage after 118 of ensiling. Similar trends were confirmed by qPCR analysis with numerically higher copy numbers than cultured cfu numbers observed for bacterial (P = 0.35) and yeast (P = 0.64) populations, and statistically higher (P < 0.01) for the fungal population (Table 2). After ensiling, S5L4 silage contained higher (P = 0.04) copy numbers of yeasts and fungi than control, S4, and S5, but did not differ (P = 0.25) from S4L5 (Table 2).
Table 2.
Effect of inoculants on microbial populations of whole-crop corn silage as estimated by culturing and qPCR after 118 d of ensiling
| Day 1182 | ||||||||
|---|---|---|---|---|---|---|---|---|
| Day 01 | Control | S4 | S5 | S4L5 | S5L4 | SEM3 | P-value5 | |
| Culture, log10 cfu/g DM | ||||||||
| Lactic acid bacteria | 6.81 | 7.59 | 7.60 | 7.60 | 8.13 | 7.76 | 0.211 | 0.36 |
| Total bacteria | 8.38 | 7.90b4 | 7.87b | 7.85b | 8.44a | 8.01b | 0.100 | 0.01 |
| Total yeasts | 7.22 | 5.26 | 5.70 | 5.68 | 6.18 | 6.26 | 0.291 | 0.18 |
| Total fungi | 5.99 | 1.61 | 1.85 | 1.92 | 3.50 | 3.50 | 1.784 | 0.89 |
| qPCR, log10 copies/g DM | ||||||||
| Total bacteria | 10.24 | 9.93 | 9.88 | 9.63 | 9.92 | 9.87 | 0.114 | 0.42 |
| Total yeasts | 7.35 | 6.89 | 6.81 | 6.73 | 7.39 | 7.59 | 0.241 | 0.13 |
| Total fungi and yeasts | 10.55 | 9.77b | 9.47b | 9.29b | 10.17ab | 10.73a | 0.289 | 0.04 |
Data of fresh forage at day 0 were not included in statistical analysis.
Control = uninoculated; S4 = S. cerevisiae (1 × 104 cfu/g wet silage); S5 = S. cerevisiae (1 × 105 cfu/g wet silage); S4L5 = S. cerevisiae (1 × 104 cfu/g wet silage) and L. buchneri (1 × 10 cfu/g wet silage); S5L4 = S. cerevisiae (1 × 105 cfu/g wet silage) and L. buchneri (1 × 104 cfu/g wet silage).
Standard errors of least squares means (n = 3).
Within a row, means followed by different letters differ at P < 0.05.
Fate of S. cerevisiae and L. buchneri
An interaction between inoculant and sampling day affected copies of S. cerevisiae in silage during both ensiling (P = 0.004) and aerobic exposure (P = 0.03; Fig. 1a). Level of S. cerevisiae in all silages declined (P < 0.0001) after ensiling, reaching the lowest levels (P < 0.0001) on day 61. After 3 d of aerobic exposure, copy number of S. cerevisiae rapidly increased (P < 0.0001), with levels remaining constant (P = 0.26) after 7 d (Fig. 1a). Levels of S. cerevisiae were greater (P < 0.0001) in inoculated silage than control silage during ensiling and aerobic exposure. Silages inoculated with more S. cerevisiae (S5 and S5L4) exhibited higher (P < 0.0001) copies than S4 and S4L5 at day 30 (Fig. 1a). During aerobic exposure, all inoculated treatments had higher (P < 0.0001), levels of S. cerevisiae than the control, with copy numbers of S. cerevisiae also being greater (P = 0.02) in S5 than S5L4 after day 7 (Fig. 1a).
Figure 1.
Changes in the copy number (log10 copies/g DM) of (a) Saccharomyces cerevisiae and (b) Lactobacillus buchneri in whole-crop corn silage during both ensiling and aerobic exposure as quantified by qPCR. Control = uninoculated; S4 = S. cerevisiae (1 × 104 cfu/g wet silage); S5 = S. cerevisiae (1 × 105 cfu/g wet silage); S4L5 = S. cerevisiae (1 × 104 cfu/g wet silage) and Lactobacillus buchneri (1 × 105 cfu/g wet silage); S5L4 = S. cerevisiae (1 × 105 cfu/g wet silage) and L. buchneri (1 × 104 cfu/g wet silage). Treatments with different letters within each sampling day in (a) differ at P < 0.05. No letters in (b) indicate that L. buchneri did not exhibit a significant treatment × sampling day interaction.
During both ensiling and aerobic exposure, the level of L. buchneri was affected by inoculation (P < 0.001; Fig. 1b). Copies of L. buchneri in silage were positively correlated with dosage with the rank of S4L5 > S5L4 > control after 118 d of ensiling and 7 d of aerobic exposure. The level of L. buchneri was affected by sampling day during ensiling (P < 0.001), but not during aerobic exposure (P = 0.63) (Fig. 1b). Copy number of L. buchneri increased (P < 0.001) ~2 log10 copies/g DM after 7 d of ensiling, remained unchanged (P = 0.23) until day 61, and then increased (P < 0.01) ~4 log10 copies/g DM after 118 d of ensiling. Subsequently, the copy numbers of L. buchneri remained unchanged (P = 0.63) during aerobic exposure (Fig. 1b).
Sequencing of Bacterial Community
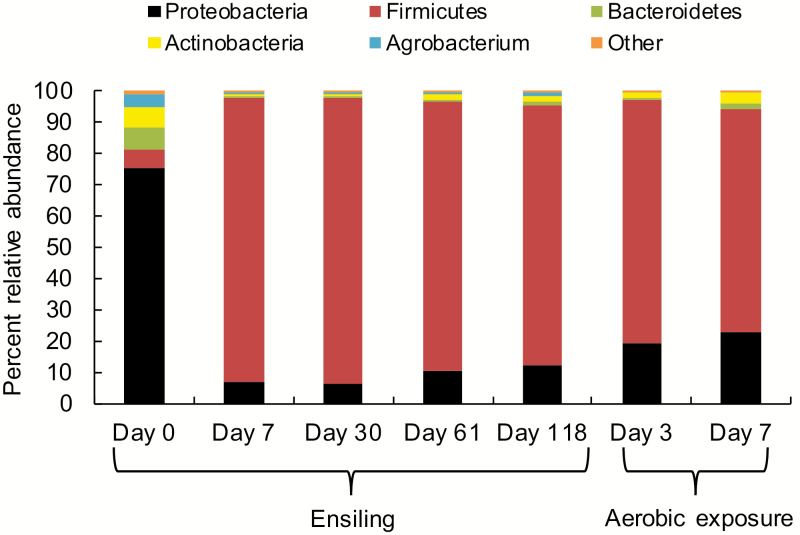
Taxonomic profile of the bacterial community varied among the sampling days during both ensiling and aerobic exposure (Supplementary Fig. A3a). Bacterial communities in silage were not altered by inoculants at day 7 but were altered by S4L5 at days 30 and 61 and by both S4L5 and S5L4 at day 118 (Supplementary Fig. A3b). A temporal shift in bacterial phyla was noted, with Proteobacteria (75.6%) being the most abundant in the epiphytic bacterial population and Firmicutes predominating during the ensiling period, accounting for 90.8% of the total population on day 7 and for 83.1% of the total population after 118 d of ensiling (Fig. 2). After aerobic exposure, Firmicutes still predominated in spoiled silage (77.9%), but continued to decline, primarily as a result of an increase in Proteobacteria and Actinobacteria (Fig. 2).
Figure 2.
Changes in the relative abundance (%) of bacterial phyla in whole-crop corn at harvest, during ensiling and aerobic exposure (n = 3).
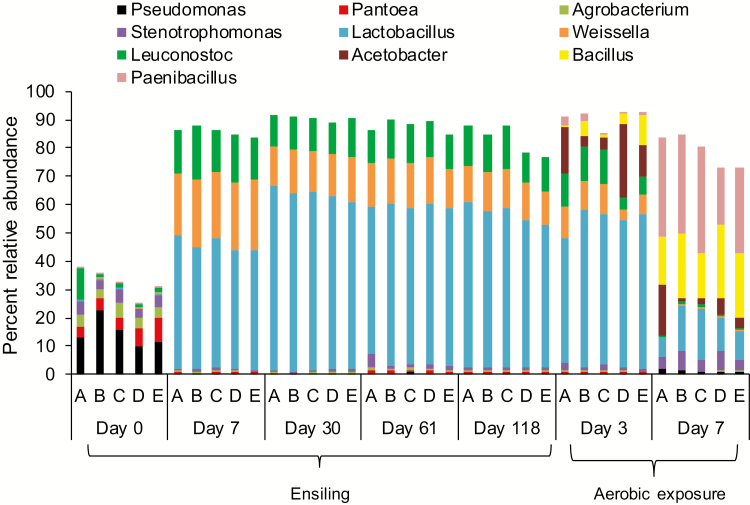
Pseudomonas, Pantoea, Agrobacterium, and Stenotrophomonas were identified in fresh forage, with Pseudomonas being most abundant ranging from 10.0 to 22.9% (Fig. 3). After 7 d of ensiling, Lactobacillus (44.5%), Weissella (23.3%), and Leuconostoc (16.5%) predominated in all silages, to the point that the original epiphytic populations only accounted for <1% of genera. After 30 d of ensiling, Lactobacillus peaked (63%), thereafter declining to 55.3% of genera in terminal silage (Fig. 3). The relative abundance of Weissella and Leuconostoc also declined after 7 d of ensiling. After 3 d of aerobic exposure, Lactobacillus, Weissella, and Leuconostoc remained relatively constant, but declined dramatically to 13.3, 0.3, and 0.7%, respectively, after 7 d of exposure (Fig. 3). Conversely, Pseudomonas, Stenotrophomonas, Bacillus, and Paenibacillus increased during aerobic exposure. After 7 d of exposure, Acetobacter was less evident in inoculated than control silage (Fig. 3).
Figure 3.
The 10 most relatively abundant bacterial genera in whole-crop corn at harvest, during ensiling and aerobic exposure (n = 3). (A) Control = uninoculated; (B) S4 = Saccharomyces cerevisiae (1 × 104 cfu/g wet silage); (C) S5 = S. cerevisiae (1 × 105 cfu/g wet silage); (D) S4L5 = S. cerevisiae (1 × 104 cfu/g wet silage) and Lactobacillus buchneri (1 × 105 cfu/g wet silage); (E) S5L4 = S. cerevisiae (1 × 105 cfu/g wet silage) and L. buchneri (1 × 104 cfu/g wet silage).
Sequencing of Fungal Community
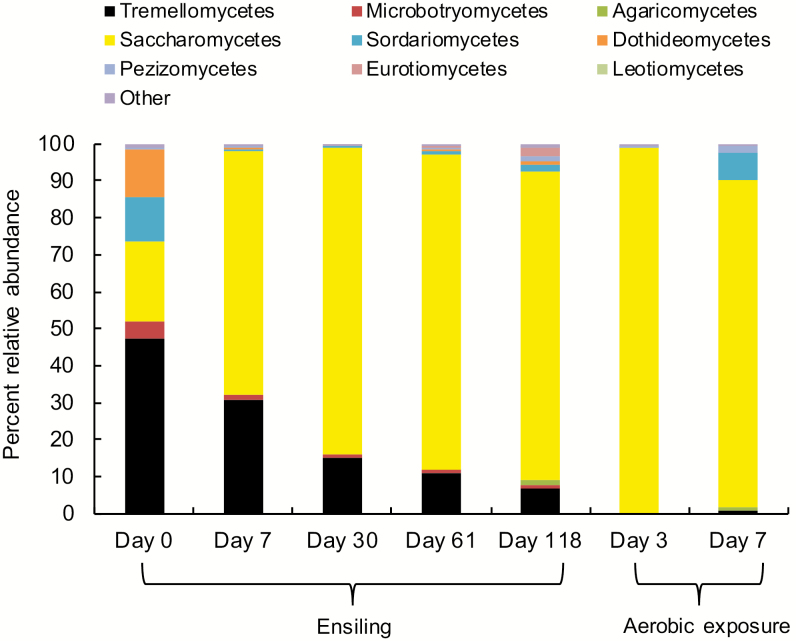
Similar to the bacterial community, the fungal communities differed with sampling time during both ensiling and aerobic exposure (Supplementary Fig. A4a), but were not altered by inoculation (Supplementary Fig. A4b). Tremellomycetes, Saccharomycetes, Dothideomycetes, Sordariomycetes, and Microbotryomycetes were identified as the most abundant fungi associated with fresh forage (Fig. 4). After 7 d of ensiling, Saccharomycetes was the most abundant class (65.8%), which continued to increase (83.6%) over the ensiling period. The abundance of other fungi, including Tremellomycetes, Dothideomycetes, and Microbotryomycetes, declined during ensiling. A slight increase in Sordariomycetes was noted after 118 d of ensiling, but the abundance was still far less than that associated with fresh forage. The abundance of other minor fungal classes including Agaricomycetes, Pezizomycetes, Eurotiomycetes, and Leotiomycetes also increased slightly at the end of ensiling (Fig. 4). Upon aerobic exposure, Saccharomycetes remained the most abundant class, accounting for a greater relative abundance than during ensiling. There was also a notable increase in the relative abundance of Sordariomycetes after 7 d of aerobic exposure (Fig. 4).
Figure 4.
Changes in the relative abundance (%) of fungal communities at the class level in whole-crop corn silage, averaged across treatments during both ensiling and aerobic exposure stages (n = 3).
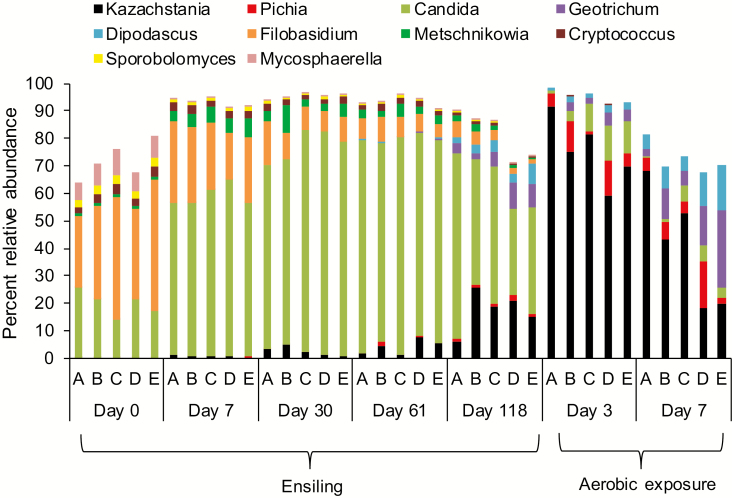
At the genus level, Filobasidium (26.2 to 47.6%), Candida (13.8 to 25.7%), and Mycosphaerella (6.5 to 9.4%) were the most abundant fungi identified in fresh corn forage (Fig. 5). After ensiling, Candida increased and was predominate in all silages, peaking at day 61 (75.3%), and thereafter declining by day 118 (46.7%). After 118 d of ensiling, Kazachstania, Pichia, Dipodascus, Metschnikowia, and Geotrichum increased, whereas Filobasidium, Cryptococcus, Sporobolomyces, and Mycosphaerella continued to decline. After aerobic exposure, Kazachstania was the most abundant genus with an increase at day 3 (75.4%), followed by a decline (40.4%) after 7 d. Although the abundance of Geotrichum and Dipodascus increased after 3 d of aerobic exposure, these genera had declined after 7 d (Fig. 5).
Figure 5.
The 10 most relatively abundant fungal genera in whole-crop corn silage during both ensiling and aerobic exposure (n = 3). (A) Control = uninoculated; (B) S4 = S. cerevisiae (1 × 104 cfu/g wet silage); (C) S5 = S. cerevisiae (1 × 105 cfu/g wet silage); (D) S4L5 = S. cerevisiae (1 × 104 cfu/g wet silage) and L. buchneri (1 × 105 cfu/g wet silage); (E) S5L4 = S. cerevisiae (1 × 105 cfu/g wet silage) and L. buchneri (1 × 104 cfu/g wet silage).
DISCUSSION
Silage Characteristics
As a component of a fourth-generation inoculant, yeast strains should not negatively affect the ensiling process and nutritional quality of silage (Dunière et al., 2015). In the present study, the pH of all yeast inoculated silages declined to below 4.0 and remained at this level after 118 d of ensiling, indicative of the successful ensiling of all silages. The temperature profiles also suggest that all treatment silages remained stable during 118 d of ensiling (Supplementary Fig. A1a). A previous study (Dunière et al., 2015) reported that inoculation with the same S. cerevisiae strain at 103 cfu/g of fresh forage did not alter the nutrient content or fermentation of corn silage after 90 d of ensiling. In this study, although the inoculate was increased to 104 and 105 cfu of S. cerevisiae/g of fresh forage, combined with or without L. buchneri, no changes in the nutrient profile or fermentation products in terminal corn silage were observed except for an increase in acetic acid and a minor increase in ADIN.
Lactobacillus buchneri can convert lactic acid to acetate and 1,2-propanediol during ensiling, which has been demonstrated to improve aerobic stability (Oude Elferink et al., 2001; Addah et al., 2012). Kleinschmit and Kung (2006) also reported inoculation with L. buchneri increased acetate content in a dose-dependent manner in corn silage. In the present study, despite an increase in the acetic acid concentration in silages inoculated with L. buchneri, all inoculated silages became unstable after 3 d of aerobic exposure as indicated by the temperature profiles (Supplementary Fig. A1b). Inoculation with a higher dose of L. buchneri resulted in a minor increase in acetic acid concentration (i.e., ~3 g/kg DM increase), which was insufficient to improve aerobic stability in ours and other studies (Kleinschmit and Kung, 2006). It was also found that a similar increase in the level of acetic acid in corn silage inoculated with L. buchneri and ensiled for 63 d did not improve aerobic stability (Addah et al., 2011).
Saccharomyces cerevisiae has a high ability to metabolize sugars (i.e., galactose, fructose, and glucose), but lacks the ability to metabolize lactic acid (Santos et al., 2016). In the present study, there was no evidence that S. cerevisiae dramatically altered the ensiling or aerobic stability of inoculated vs. control corn silage. Moreover, Kazachstania rather than Saccharomyces was identified as the primary genera associated with aerobically deteriorated corn silage (Fig. 5). Others have proposed that Saccharomyces do not play a significant role in the aerobic spoilage of silages (Rossi and Dellaglio, 2007). Consequently, if selected strains of S. cerevisiae are endowed with beneficial probiotic traits, silage may serve as a vehicle for their inclusion in the diets of ruminants.
Both S. cerevisiae and L. buchneri have strong ability to ferment sugars in silage (Oude Elferink et al., 2001). The present study showed that the WSC contents were lower in inoculated when compared with the control silage with this utilization ranking as Y5L4 > Y4L5 > Y5 > Y4. These findings suggest that the inoculated microbes survived and proliferated during the ensiling process. Compared with control silage, there was a minor increase in the ADIN content of inoculated silages, even though there was no evidence of an increase in the temperature profile of inoculated silages (Supplementary Fig. A1a). The increase in ADIN was only ~2% total N and would not be expected to seriously affect silage quality.
The current results showed that after 118 d of ensiling, compared with the control, neither S. cerevisiae alone nor in combination with L. buchneri altered the NDF, ADF, CP, or starch content of corn silage, an observation that agrees with previous studies where these inoculants were applied to corn silage individually (Kleinschmit and Kung, 2006; Addah et al., 2011; Dunière et al., 2015). In the absence of oxygen, Saccharomyces can convert WSC into CO2 and ethanol, an undesirable end product of ensiling (Pedroso et al., 2005), but as with previous research (Dunière et al., 2015), ethanol levels were not increased as result of inoculation with S. cerevisiae.
Fate of S. cerevisiae and L. buchneri
Quantitative PCR has been used to quantify abundance of whole microbial populations and the fate of specific species or even strains of bacteria and fungi during ensiling and feed out (McAllister et al., 2018). The results from both qPCR and direct plate counts showed similar changes of TB, yeasts, and fungi before and after ensiling, suggesting the suitability of this procedure to monitor the relative populations of microorganisms in silage. Copy numbers of TB, yeasts, and fungi in terminal corn silage estimated by qPCR were higher than those obtained through direct plating. This is probably because microbial numbers based on qPCR were overestimated as the copy numbers of 16S rDNA in bacteria and 18S rDNA, 28S rDNA in fungi and yeast varies among species (Case et al., 2007; Lindahl et al., 2013). Moreover, DNA from nonviable microorganisms can also be amplified and quantified, overestimating the metabolically active microbial population within silage (McAllister et al., 2018).
In the present study, qPCR results showed that S. cerevisiae applied at 104 or 105 cfu/g of fresh corn forage contained 102- to 106-fold higher copy numbers of S. cerevisiae than control corn forage. During ensiling, S. cerevisiae decreased, reflecting their intolerance for anaerobicity, high lactic acid concentrations and low pH. Upon aerobic exposure, S. cerevisiae numbers increased 103 to 105 times in inoculated silage, probably reflecting resuscitation of dormant yeast cells upon exposure to oxygen (Wang et al., 2018). These shifts in the S. cerevisiae population were also apparent in amplicon sequencing, particularly for S5 (Supplementary Fig. A5a). This suggests that this yeast strain would increase between the time that silage is removed from storage and the time it is fed to the animal. The qPCR results also showed that the copy numbers of S. cerevisiae after 7 d of aerobic exposure increased to pre-ensiling levels, suggesting that using S. cerevisiae as corn silage inoculant might offer similar numbers of cfu (i.e., 104 to 105 cfu/g of fresh forage) in terminal silage as that which is administered in direct fed microbials to ruminants.
Although qPCR and amplicon sequencing (Supplementary Fig. A5b) indicated that the numbers of L. buchneri increased during ensiling, particularly for S4L5, inoculation in combination with S. cerevisiae did not improve the aerobic stability of silage. The current results were similar to Dunière et al. (2015) who showed that L. buchneri increased during the first 7 d of aerobic exposure of corn silage but declined after 21 d. In the present study, increases in the concentration of acetic acid in corn silage were relatively minor and may have been insufficient to inhibit fungal populations. Further work is required to understand how the epiphytic microbial population influences the variation in the aerobic stability of silage inoculated with L. buchneri.
Bacterial Community in Ensiled and Aerobically Exposed Silage
Prior to ensiling, Pseudomonas, Pantoea, Agrobacterium, and Stenotrophomonas accounted for a significant portion of the epiphytic bacterial population. Pseudomonas have been shown to be associated with plant growth-promotion by suppressing pathogenic microorganisms (Preston, 2004). Pantoea, Agrobacterium, and Stenotrophomonas are also ubiquitous plant-associated proteobacteria associated with oats, corn, and potatoes (Rekosz-Burlaga and Garbolinska, 2006; Hayward et al., 2010; Wensing et al., 2010). These epiphytic genera were rapidly suppressed after ensiling, but Pseudomonas and Stenotrophomonas were once again apparent in aerobically exposed silage. Lactic acid bacteria constituted a small proportion of the epiphytic bacterial population in corn silage with the rank of Leuconostoc > Weissella = Lactobacillus. This is consistent with previous studies that observed a relatively low abundance of LAB in green grass, red clover, corn, and small grain forages (Mogodiniyai Kasmaei et al., 2016; Dunière et al., 2017).
After ensiling, LAB increased and dominated the ensiling process with Lactobacillus > Weissella > Leuconostoc. Both Weissella and Leuconostoc rapidly peaked at day 7, declining thereafter, with Lactobacillus peaking at day 30 and sustaining this level until day 118. Leuconostocs grow vigorously only in the early stage of the ensiling process. Although phylogenetic characterization of the bacterial community to the species level is tenuous using partial 16S rDNA sequencing, most of the reads detected in this study within Lactobacillus, Weissella, and Leuconostoc were associated with L. plantarum and L. brevis, W. cibaria and W. paramesenteroides, and Le. citreum and Le. lactis (data not shown). Ogunade et al. (2017) and Kraut-Cohen et al. (2016) also found Lactobacillus to be the predominant LAB in corn silage ensiled in mini silos for 120 d in the United States. However, Pang et al. (2011) identified Weissella as the dominant LAB in corn silage ensiled for 270 d on Chinese commercial farms. Moreover, sequencing results showed that L. buchneri altered the composition of the bacterial community after 118 d of ensiling (Supplementary Fig. A3b). After aerobic exposure, acetic acid bacteria such as Acetobacter became apparent. Acetobacter contributes to aerobic spoilage by primarily by oxidizing lactate and acetate (Dolci et al., 2011). In addition, butyric acid-producing bacteria including Bacillus and Paenibacillus predominated in aerobically exposed silage. These undesirable bacteria are believed to promote the growth of less acid-tolerant spoilage microorganisms, and their spores have been a source of contamination of milk and milk products (Vissers et al., 2007; Borreani et al., 2013).
Fungal Community in Ensiled and Aerobically Exposed Silage
Little information is available with regard to the epiphytic fungal community in silages, with most research focusing on the characterization of mycotoxigenic fungi (Dunière et al., 2017). Filobasidium, Mycosphaerella, and Candida were the main epiphytic fungi associated with fresh corn forage. However, other fungi including Sporobolomyces and Cryptococcus, previously shown to be associated with wheat forage (Karlsson et al., 2014), were also identified. Metschnikowia has been reported to be almost ubiquitous on flowers, fruits, and plants and has been used for wine fermentation (Oro et al., 2014). In this study, Metschnikowia increased after ensiling, but declined after aerobic exposure. Conversely, Geotrichum and Dipodascus were largely undetectable during early ensiling, but increased after 118 d and peaked after 7 d of aerobic exposure. Spadaro et al. (2015) also found that these fungal genera were readily apparent in corn silage after 7 d of aerobic exposure.
With the exception of Candida, most epiphytic fungi rapidly decreased after ensiling, reflecting the need for oxygen by the latter, and the ability of Candida to thrive under both aerobic and anaerobic conditions. Others have isolated Candida spp. from corn silage including C. holmii, C. krusei, and C. lambica (Middelhoven, 1998), offering further evidence that many species in this genera remain viable during ensiling. Residual levels of Candida were still observed during aerobic exposure, possibly as a result of their ability to utilize lactic acid, but Pichia and Kazachstania in particular were by far the dominate genera in aerobically exposed corn silage. Others have shown that Kazachstania and Pichia are associated with aerobically exposed corn silage (Dolci et al., 2011).
Sequencing results showed that Saccharomyces had a low relative abundance (<0.03%) with a trend of decline during the ensiling and increase after aerobic exposure (Supplementary Fig. A5a), which is in agreement with qPCR quantification of S. cerevisiae. However, the sequencing results did not reflect the treatment effect as seen with qPCR measurement, probably because fungal amplicon sequencing was not as effective at differentiating Saccharomyces to the species level.
In the present study, S. cerevisiae as a fourth-generation silage inoculant did not alter the fermentation of corn silage, but concentrations of acetic acid in silage were increased as a result of inoculation with L. buchneri. There was no evidence that the formation of this metabolite improved the aerobic stability of corn silage. A mixed inoculant of S. cerevisiae and L. buchneri did alter the composition of the bacterial population after ensiling, but not the fungal population. The qPCR findings showed that L. buchneri increased during the ensiling process and remained consistent after silo opening. Lactobacillus buchneri did not appear to alter S. cerevisiae populations, and this potential probiotic remained viable during ensiling and increased upon aerobic exposure. As a fourth-generation inoculant, S. cerevisiae combined with L. buchneri may confer silage as a vehicle to deliver beneficial yeast strains to ruminants.
Supplementary Material
Footnotes
Financial support for this work from DuPont Pioneer is gratefully acknowledged. We thank Z. Matic, C. Barkley, D. Vedres, B. Brent, and W. Smart for their invaluable technical assistance on this project.
LITERATURE CITED
- Addah W., Baah J., Groenewegen P., Okine E. K., and McAllister T. A.. 2011. Comparison of the fermentation characteristics, aerobic stability and nutritive value of barley and corn silages ensiled with or without a mixed bacterial inoculant. Can. J. Anim. Sci. 91:133–146. doi: 10.4141/CJAS10071 [DOI] [Google Scholar]
- Addah W., Baah J., Okine E. K., and McAllister T. A.. 2012. A third-generation esterase inoculant alters fermentation pattern and improves aerobic stability of barley silage and the efficiency of body weight gain of growing feedlot cattle. J. Anim. Sci. 90:1541–1552. doi: 10.2527/jas.2011-4085 [DOI] [PubMed] [Google Scholar]
- Auclair E. 2001. Yeast as an example of the mode of action of probiotics in monogastric and ruminant species. In: Brufau J., editor, Feed manufacturing in the Mediterranean region. Improving safety: From feed to food. Cahiers Options Méditerranéennes No. 54. Zaragoza, Spain: CIHEAM; p. 45–53. [Google Scholar]
- Bokulich N. A., Subramanian S., Faith J. J., Gevers D., Gordon J. I., Knight R., Mills D. A., and Caporaso J. G.. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10:57–59. doi: 10.1038/nmeth.2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borreani G., Dolci P., Tabacco E., and Cocolin L.. 2013. Aerobic deterioration stimulates outgrowth of spore-forming Paenibacillus in corn silage stored under oxygen-barrier or polyethylene films. J. Dairy Sci. 96:5206–5216. doi: 10.3168/jds.2013-6649 [DOI] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. doi: 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case R. J., Boucher Y., Dahllöf I., Holmström C., Doolittle W. F., and Kjelleberg S.. 2007. Use of 16S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl. Environ. Microbiol. 73:278–288. doi: 10.1128/AEM.01177-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., Huber T., Dalevi D., Hu P., and Andersen G. L.. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. doi: 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolci P., Tabacco E., Cocolin L., and Borreani G.. 2011. Microbial dynamics during aerobic exposure of corn silage stored under oxygen barrier or polyethylene films. Appl. Environ. Microbiol. 77:7499–7507. doi: 10.1128/AEM.05050-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunière L., Jin L., Smiley B., Qi M., Rutherford W., Wang Y., and McAllister T.. 2015. Impact of adding Saccharomyces strains on fermentation, aerobic stability, nutritive value, and select lactobacilli populations in corn silage. J. Anim. Sci. 93:2322–2335. doi: 10.2527/jas.2014-8287 [DOI] [PubMed] [Google Scholar]
- Dunière L., Xu S., Jin L., Elekwachi C., Wang Y., Turkington K., Forster R., and McAllister T.. 2017. Bacterial and fungal core microbiomes associated with small grain silages during ensiling and aerobic spoilage. BMC Microbiol. 17:50. doi: 10.1186/s12866-017-0947-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Hayward A. C., Fegan N., Fegan M., and Stirling G. R.. 2010. Stenotrophomonas and lysobacter: Ubiquitous plant-associated gamma-proteobacteria of developing significance in applied microbiology. J. Appl. Microbiol. 108:756–770. doi: 10.1111/j.1365-2672.2009.04471.x [DOI] [PubMed] [Google Scholar]
- Herrera-Saldana R. E., Huber J. T., and Poore M. H.. 1990. Dry matter, crude protein, and starch degradability of five cereal grains. J. Dairy Sci. 73:2386–2393. doi: 10.3168/jds.S0022-0302(90)78922-9 [DOI] [Google Scholar]
- Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., and Madden T. L.. 2008. NCBI BLAST: A better web interface. Nucleic Acids Res. 36:W5–W9. doi: 10.1093/nar/gkn201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson I., Friberg H., Steinberg C., and Persson P.. 2014. Fungicide effects on fungal community composition in the wheat phyllosphere. PLoS One 9:e111786. doi: 10.1371/journal.pone.0111786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschmit D. H., and Kung L. Jr.. 2006. A meta-analysis of the effects of Lactobacillus buchneri on the fermentation and aerobic stability of corn and grass and small-grain silages. J. Dairy Sci. 89:4005–4013. doi: 10.3168/jds.S0022-0302(06)72444-4 [DOI] [PubMed] [Google Scholar]
- Kõljalg U., Nilsson R. H., Abarenkov K., Tedersoo L., Taylor A. F. S., Bahram M., Bates S. T., Bruns T. D., Bengtsson-Palme J., Callaghan T. M., et al. 2013. Towards a unified paradigm of sequence-based identification of fungi. Mol. Ecol. 22:5271–5277. doi: 10.1111/mec.12481 [DOI] [PubMed] [Google Scholar]
- Kraut-Cohen J., Tripathi V., Chen Y., Gatica J., Volchinski V., Sela S., Weinberg Z., and Cytryn E.. 2016. Temporal and spatial assessment of microbial communities in commercial silages from bunker silos. Appl. Microbiol. Biotechnol. 100:6827–6835. doi: 10.1007/s00253-016-7512-x [DOI] [PubMed] [Google Scholar]
- Lindahl B. D., Nilsson R. H., Tedersoo L., Abarenkov K., Carlsen T., Kjøller R., Kõljalg U., Pennanen T., Rosendahl S., Stenlid J., et al. 2013. Fungal community analysis by high-throughput sequencing of amplified markers – A user’s guide. New Phytol. 199:288–299. doi: 10.1111/nph.12243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C., and Knight R.. 2005. Unifrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister T. A., Beauchemin K. A., Alazzeh A. Y., Baah J., Teather R. M., and Stanford K.. 2011. Review: The use of direct fed microbials to mitigate pathogens and enhance production in cattle. Can. J. Anim. Sci. 91:193–211. doi: 10.4141/cjas10047 [DOI] [Google Scholar]
- McAllister T. A., Dunière L., Drouin P., Xu S., Wang Y., Munns K., and Zaheer R.. 2018. Silage review: Using molecular approaches to define the microbial ecology of silage. J. Dairy Sci. 101:4060–4074. doi: 10.3168/jds.2017-13704 [DOI] [PubMed] [Google Scholar]
- McAllister T. A., Selinger L. B., McMahon L. R., Bae H. D., Lysyk T. J., Oosting S. J., and Cheng K.-J.. 1995. Intake, digestibility and aerobic stability of barley silage inoculated with mixtures of Lactobacillus plantarum and Enterococcus faecium. Can. J. Anim. Sci. 75:425–432. doi: 10.4141/cjas95-062 [DOI] [Google Scholar]
- Middelhoven W. J. 1998. The yeast flora of maize silage. Food Technol. Biotechnol. 36:7–11. [Google Scholar]
- Mogodiniyai Kasmaei K., Dicksved J., Spörndly R., and Udén P.. 2016. Separating the effects of forage source and field microbiota on silage fermentation quality and aerobic stability. Grass Forage Sci. 72:281–289. doi: 10.1111/gfs.12238 [DOI] [Google Scholar]
- Muck R. E., Nadeau E. M. G., McAllister T. A., Contreras-Govea F. E., Santos M. C., and Kung L. Jr. 2018. Silage review: Recent advances and future uses of silage additives. J. Dairy Sci. 101:3980–4000. doi: 10.3168/jds.2017-13839 [DOI] [PubMed] [Google Scholar]
- Nelson N. 1944. A photometric adaption of the Somogyi method for the determination of glucose. J. Biol. Chem. 153:375–380. [Google Scholar]
- Ogunade I. M., Jiang Y., Kim D. H., Cervantes A. A. P., Arriola K. G., Vyas D., Weinberg Z. G., Jeong K. C., and Adesogan A. T.. 2017. Fate of Escherichia coli O157:H7 and bacterial diversity in corn silage contaminated with the pathogen and treated with chemical or microbial additives. J. Dairy Sci. 100:1780–1794. doi: 10.3168/jds.2016-11745 [DOI] [PubMed] [Google Scholar]
- Oro L., Ciani M., and Comitini F.. 2014. Antimicrobial activity of Metschnikowia pulcherrima on wine yeasts. J. Appl. Microbiol. 116:1209–1217. doi: 10.1111/jam.12446 [DOI] [PubMed] [Google Scholar]
- Oude Elferink S. J., Krooneman J., Gottschal J. C., Spoelstra S. F., Faber F., and Driehuis F.. 2001. Anaerobic conversion of lactic acid to acetic acid and 1,2-propanediol by Lactobacillus buchneri. Appl. Environ. Microbiol. 67:125–132. doi: 10.1128/AEM.67.1.125-132.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang H., Qin G., Tan Z., Li Z., Wang Y., and Cai Y.. 2011. Natural populations of lactic acid bacteria associated with silage fermentation as determined by phenotype, 16S ribosomal RNA and reca gene analysis. Syst. Appl. Microbiol. 34:235–241. doi: 10.1016/j.syapm.2010.10.003 [DOI] [PubMed] [Google Scholar]
- Pedroso A. F., Nussio L. G., Paziani S. F., Loures D. R. S., Igarasi M. S., Coelho R. M., Packer I. H., Horii J., and Gomes L. H.. 2005. Fermentation and epiphytic microflora dynamics in sugar cane silage. Sci. Agric. 62:427–432. doi: 10.1590/S0103-90162005000500003 [DOI] [Google Scholar]
- Preston G. M. 2004. Plant perceptions of plant growth-promoting pseudomonas. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 359:907–918. doi: 10.1098/rstb.2003.1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich L. J., and Kung L. Jr. 2010. Effects of combining Lactobacillus buchneri 40788 with various lactic acid bacteria on the fermentation and aerobic stability of corn silage. Anim. Feed Sci. Technol. 159:105–109. doi: 10.1016/j.anifeedsci.2010.06.002 [DOI] [Google Scholar]
- Rekosz-Burlaga H., and Garbolinska M.. 2006. Characterization of selected groups of microorganisms occurring in soil rhizosphere and phyllosphere of oats. Pol. J. Microbiol. 55:227–235. [PubMed] [Google Scholar]
- Research Disclosure. 2015. Primers for identification of microbes. The industry standard disclosure publication service. Research Disclosure Database Number 612005. Questel Ireland Ltd., Killernan, Kilmaine, County Mayo, Ireland. [Google Scholar]
- Rossi F., and Dellaglio F.. 2007. Quality of silages from Italian farms as attested by number and identity of microbial indicators. J. Appl. Microbiol. 103:1707–1715. doi: 10.1111/j.1365-2672.2007.03416.x. [DOI] [PubMed] [Google Scholar]
- Santos A. O., Ávila C. L., Pinto J. C., Carvalho B. F., Dias D. R., and Schwan R. F.. 2016. Fermentative profile and bacterial diversity of corn silages inoculated with new tropical lactic acid bacteria. J. Appl. Microbiol. 120:266–279. doi: 10.1111/jam.12980. [DOI] [PubMed] [Google Scholar]
- Spadaro D., Bustos-Lopez M. P., Gullino M. L., Piano S., Tabacco E., and Borreani G.. 2015. Evolution of fungal populations in corn silage conserved under polyethylene or biodegradable films. J. Appl. Microbiol. 119:510–520. doi: 10.1111/jam.12852 [DOI] [PubMed] [Google Scholar]
- Vázquez-Baeza Y., Pirrung M., Gonzalez A., and Knight R.. 2013. Emperor: A tool for visualizing high-throughput microbial community data. Gigascience 2:16. doi: 10.1186/2047-217X-2-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers M. M., Driehuis F., Te Giffel M. C., De Jong P., and Lankveld J. M.. 2007. Concentrations of butyric acid bacteria spores in silage and relationships with aerobic deterioration. J. Dairy Sci. 90:928–936. doi: 10.3168/jds.S0022-0302(07)71576-X [DOI] [PubMed] [Google Scholar]
- Walters W., Hyde E. R., Berg-Lyons D., Ackermann G., Humphrey G., Parada A., Gilbert J. A., Jansson J. K., Caporaso J. G., Fuhrman J. A., et al. 2016. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009–e000015. doi: 10.1128/mSystems.00009-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Hao W., Ning T., Zheng M., and Xu C.. 2018. Characterization of culturable yeast species associating with whole crop corn and total mixed ration silage. Asian-Australas. J. Anim. Sci. 31:198–207. doi: 10.5713/ajas.17.0183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z. G., and Muck R. E.. 1996. New trends and opportunities in the development and use of inoculants for silage. FEMS Microbiol. Rev. 19:53–68. doi: 10.1016/0168-6445(96)00025-3 [DOI] [Google Scholar]
- Wensing A., Zimmermann S., and Geider K.. 2010. Identification of the corn pathogen Pantoea stewartii by mass spectrometry of whole-cell extracts and its detection with novel PCR primers. Appl. Environ. Microbiol. 76:6248–6256. doi: 10.1128/AEM.01032-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.