Abstract
Lynch syndrome is the most common form of hereditary colorectal carcinoma. However, establishing the diagnosis of Lynch syndrome is challenging, and ancillary studies that distinguish between sporadic DNA mismatch repair (MMR) protein deficiency and Lynch syndrome are needed, particularly when germline mutation studies are inconclusive. The aim of this study was to determine if MMR protein deficient non-neoplastic intestinal crypts can help distinguish between patients with and without Lynch syndrome. We evaluated the expression of MMR proteins in non-neoplastic intestinal mucosa obtained from colorectal surgical resection specimens from patients with Lynch syndrome-associated colorectal carcinoma (n=52) and patients with colorectal carcinoma without evidence of Lynch syndrome (n=70), including sporadic MMR protein deficient colorectal carcinoma (n=30), MMR protein proficient colorectal carcinoma (n=30), and “Lynch-like” syndrome (n=10). MMR protein deficient non-neoplastic colonic crypts were identified in 19 of 122 (16%) patients. MMR protein deficient colonic crypts were identified in 18 of 52 (35%) patients with Lynch syndrome compared to only 1 of 70 (1%) patients without Lynch syndrome (p<0.001). This one patient had “Lynch-like” syndrome and harbored two MSH2 deficient non-neoplastic colonic crypts. MMR protein deficient non-neoplastic colonic crypts were not identified in patients with sporadic MMR protein deficient or MMR protein proficient colorectal carcinoma. Our findings suggest that MMR protein deficient colonic crypts are a novel indicator of Lynch syndrome, and evaluation for MMR protein deficient crypts may be a helpful addition to Lynch syndrome diagnostics.
Keywords: Lynch syndrome, Lynch-like syndrome, suspected Lynch syndrome, mismatch repair protein, MSI, colorectal carcinoma, immunohistochemistry
INTRODUCTION
Roughly one of every 35 patients with colorectal carcinoma has Lynch syndrome, making Lynch syndrome the most common hereditary form of colorectal carcinoma. Lynch syndrome is genetically heterogeneous with most patients harboring pathogenic variants in one of four DNA mismatch repair (MMR) genes, MLH1, MSH2, MSH6, or PMS2, or germline deletions of the 3’ end of EPCAM that lead to epigenetic silencing of the MSH2 gene (1, 2). Identification of individuals who have Lynch syndrome is essential as these individuals and their relatives benefit from genetic counseling and increased surveillance that can result in early cancer detection and decreased mortality. Universal screening for Lynch syndrome among all newly diagnosed colorectal carcinomas has been advocated by most major professional organizations, including the Evaluation of Genomic Applications in Practice and Prevention (EGAPP, a working group sponsored by the Centers for Disease Control) (1), the US Multi-Society Task Force for colorectal carcinoma (3), the American College of Gastroenterology (ACG) (4), the National Comprehensive Cancer Network (NCCN), American Society of Clinical Oncology (ASCO) (5), and European Society of Medical Oncology (ESMO) (6). Increasing universal screening efforts in colorectal carcinoma have led to the identification of patients with abnormal MMR protein expression within their tumor concerning for Lynch syndrome. However, some of these patients will have no evidence of a pathogenic germline variant in MMR genes or EPCAM and have been provisionally labeled as having “Lynch-like” syndrome (7–9) or “suspected Lynch syndrome” (8). The management of these patients represents a major challenge in Lynch syndrome diagnostics and ancillary studies that can help distinguish between sporadic MMR protein deficiency and Lynch syndrome are still needed.
Two recent publications from the same institution have demonstrated that histologically normal intestinal crypts in patients with Lynch syndrome can exhibit loss of MMR protein expression (10, 11). However, this finding has not been independently verified by other groups. In this study, we evaluated the expression of MMR proteins in non-neoplastic intestinal mucosa from patients with Lynch syndrome-associated colorectal carcinoma, “Lynch-like” syndrome, sporadic MMR protein deficient colorectal carcinoma, and MMR protein proficient colorectal carcinoma. In so doing, we demonstrate that MMR protein deficient non-neoplastic colonic crypts can be identified in patients with Lynch syndrome and “Lynch-like” syndrome but are not seen in patients with sporadic MMR protein deficiency or MMR protein proficient colorectal carcinoma. Our findings suggest that evaluation for MMR protein deficient intestinal crypts may be a helpful ancillary study in Lynch syndrome diagnostics and has the potential to identify patients with Lynch syndrome when germline mutation studies are inconclusive.
METHODS
Case Selection
The study group consisted of 52 patients with Lynch syndrome identified between 1994 and 2017 from the pathology archives of: University of Pittsburgh Medical Center, Mayo Clinic, and the Australasian Colorectal Cancer Family Registry (ACCFR). Patients were included in the study group with Lynch syndrome if they had 1) a confirmed MMR gene or EPCAM germline mutation classified as a pathogenic variant and 2) a surgically resected colorectal carcinoma with non-neoplastic intestinal tissue available for immunohistochemical analysis. The study group of patients with Lynch syndrome was compared to a control group of 70 patients with colorectal carcinoma without evidence of Lynch syndrome. The control group included three categories of patients: 1) 10 patients with “Lynch-like” syndrome defined as abnormal MMR protein expression by immunohistochemistry suggestive of Lynch syndrome but with no germline MMR gene or EPCAM pathogenic variant identified and no evidence of somatic MLH1 promoter hypermethylation, as previously described (9); 2) a consecutive series of 30 patients with colorectal carcinoma with sporadic MMR protein deficiency characterized by loss of MLH1 and PMS2 expression by immunohistochemistry and concurrent BRAF V600E mutation or MLH1 promoter hypermethylation within the tumor; and 3) a consecutive series of 30 patients with colorectal carcinoma with proficient MMR protein expression by immunohistochemistry. Clinicopathologic and demographic information were obtained from medical records under the guidelines of the University of Pittsburgh Institutional Review Board (IRB# PR016040136), Mayo Clinic Institutional Review Board (IRB16–008881), and University of Melbourne (Ethics ID 1339757).
MMR Protein Immunohistochemistry, BRAF Mutation, and MLH1 Promoter Hypermethylation
For the cases analyzed at Mayo Clinic and the University of Pittsburgh, MMR protein immunohistochemistry was performed using primary monoclonal antibodies against MLH1 (clone M1, Ventana, Tucson, AZ), MSH2 (clone G219–1129, Ventana), MSH6 (clone 44, Ventana), and PMS2 (clone EPR3947, Cell Marque, Rocklin, CA). For the ACCFR cases, the primary monoclonal antibodies were MLH1, clone G168–15 (BD PharMingen); MSH2, clone G219–1129 (BD PharMingen); MSH6, clone 44 (BD Transduction Labs); PMS2, clone A16–4 (BD PharMingen) (12). Normal preserved expression was defined as nuclear staining, using infiltrating lymphocytes and stromal cells as a positive internal control. Loss of protein expression in colorectal carcinoma was defined as complete absence of nuclear staining within tumor cells with concurrent positive labeling in internal non-neoplastic tissues. Colorectal carcinomas with loss of MLH1 and PMS2 protein expression were analyzed for BRAF mutations using fluorescent allele-specific PCR assays as previously described at the University of Pittsburgh (13) and for ACCFR participants (14). For University of Pittsburgh participants, if the tumor was BRAF wild-type with loss of MLH1 and PMS2 expression, further analysis for MLH1 promoter hypermethylation was performed at Mayo Clinic Laboratories (Test ID MLH1M). For the ACCFR participants, tumors demonstrating loss of the MLH1 and PMS2 by immunohistochemistry were tested for methylation of the MLH1 promoter region regardless of BRAF status using a MethyLight assay as previously described (15, 16).
For both the study group and control group, non-neoplastic intestinal mucosa from the resection specimens taken as part of routine pathologic evaluation were evaluated by MMR protein immunohistochemistry. The non-neoplastic intestinal mucosa analyzed included sections taken from the grossly normal surgical resection specimen margins and uninvolved grossly normal mucosa away from the tumor. For patients with Lynch syndrome, MMR protein immunohistochemistry was performed for the gene in which a germline pathogenic variant had been identified. For patients with “Lynch-like” syndrome, MMR protein immunohistochemistry was performed based on the abnormal MMR protein immunohistochemistry results in the patient’s colorectal carcinoma. For patients with sporadic MMR protein deficiency, MLH1 immunohistochemistry was performed. For patients with MMR protein proficient colorectal carcinoma, MMR protein immunohistochemistry for all four proteins (MLH1, MSH2, MSH6, and PMS2) was performed. Loss of protein expression in non-neoplastic intestinal mucosa was defined as complete absence of nuclear staining within the crypt epithelial cells in the base of the intestinal crypts with concurrent nuclear expression in adjacent stromal cells and lymphocytes. The number of crypt bases with loss of protein expression, if any, was recorded. MMR protein deficient non-neoplastic crypts were further categorized based on the number of crypts exhibiting loss of protein expression. Single, isolated crypts with loss of MMR protein expression were classified as solitary MMR protein deficient crypts. Two or more adjacent crypts with loss of MMR protein expression were classified as grouped MMR protein deficient crypts.
The linear length in millimeters of non-neoplastic intestinal mucosa examined by immunohistochemistry was measured microscopically on the histologic sections taken from the grossly normal mucosa of the surgical resection specimens. The non-neoplastic mucosa was sectioned along the long axis of the colonic crypts perpendicular to the muscularis mucosae. The number of intestinal crypts in the linear length of non-neoplastic intestinal mucosa analyzed was estimated by dividing the linear length by mean crypt diameter of 90 μm for the colon and 70 μm for the small intestine.
Germline Mutation Analysis
Direct germline MMR gene sequencing and rearrangement analysis were performed at various clinical laboratories, including commercial laboratories (17, 18). After obtaining patient consent, a peripheral blood sample from patients with tumors demonstrating loss of MMR protein expression concerning for Lynch syndrome were analyzed for germline MMR and EPCAM gene mutations based on the pattern of abnormal MMR protein expression within the patient’s tumor. For the ACCFR participants, germline MMR gene sequencing and rearrangement testing was performed as previously described (19).
Based on the germline sequencing analysis, patients were stratified into two groups. Those patients with a germline MMR gene pathogenic variant or EPCAM deletion were classified as having Lynch syndrome. Patients with abnormal MMR protein immunohistochemical expression in their colorectal carcinoma but without a germline MMR gene pathogenic variant or EPCAM deletion were classified as having “Lynch-like” syndrome. Individuals with negative germline mutation studies and whose tumors demonstrated loss of MLH1 and PMS2 expression were only considered to have “Lynch-like” syndrome if their tumor had no evidence of a BRAF V600E mutation and no evidence of MLH1 promoter hypermethylation. Patients with MMR gene variants of uncertain significance (VUS) detected by germline mutation analysis were not included in the study.
Statistical Analysis
Chi-square or Fisher exact tests were used to characterize the relationship between categorical variables, as appropriate. Kruskal-Wallis tests were used to characterize the relationship between continuous variables. All statistics were assessed using two-sided tests with p-values <0.05 considered statistically significant. Statistical analyses were performed using SPSS (for Windows 23, IBM, Armonk, NY).
RESULTS
MMR Protein Deficient Intestinal Crypts in Lynch Syndrome and “Lynch-like” Syndrome
A total of 52 patients with Lynch syndrome comprise the study group. All patients with Lynch syndrome presented with surgically resected colorectal carcinoma and were identified during Lynch syndrome screening efforts at three institutions. For each patient, germline mutation testing confirmed the diagnosis of Lynch syndrome. Most patients (48%) harbored a germline pathogenic variant in MSH2, followed by MLH1 (25%), MSH6 (15%), and PMS2 (12%) (Table 1). For 51 patients, the colorectal carcinoma exhibited the expected pattern of abnormal MMR protein expression as predicted by the germline mutation studies. One patient harbored a germline MSH2 pathogenic variant; however, the colorectal carcinoma identified in this patient exhibited proficient MMR protein expression.
Table 1.
Clinical and Pathologic Features of Patients with and without Lynch Syndrome
| Clinical and Pathologic Features | Confirmed Lynch Syndrome N (%) | Non-Lynch Syndrome |
p-value*** | |||
|---|---|---|---|---|---|---|
| Overall N (%) | Lynch-like Syndrome** N (%) | Sporadic MLH1 Protein Deficiency N (%) | MMR Proficient N (%) | |||
| No. of cases | 52 | 70 | 10 | 30 | 30 | NA |
| Tumor location | ||||||
| Right colon | 32 (62) | 49 (70) | 6 (60) | 26 (87) | 17 (57) | 0.3 |
| Left colon / rectum | 20 (40) | 21 (30) | 4 (40) | 4 (13) | 13 (43) | |
| Mean age in years (range) | 52 (27–80) | 70 (37–93) | 60 (37–76) | 76 (57–93) | 68 (46–92) | <0.001 |
| Gender, Male/Female | 25 (48) / 27 (52) | 31 (56) / 39 (44) | 7 (70) / 3 (30) | 9 (30) / 21 (70) | 15 (50) / 15 (50) | 0.7 |
| Germline mutation analysis | ||||||
| MLH1 pathogenic variant present | 13 (25) | NA | 0 (0) | NA | NA | NA |
| MSH2 pathogenic variant present | 25 (48) | 0 (0) | ||||
| MSH6 pathogenic variant present | 8 (15) | 0 (0) | ||||
| PMS2 pathogenic variant present | 6 (12) | 0 (0) | ||||
| MMR protein expression in tumor | ||||||
| Intact expression of all 4 proteins | 1 (2) | 30 (43) | 0 (0) | 0 (0) | 30 (100) | <0.001 |
| MLH1 and PMS2 Loss | 13 (25) | 35 (50) | 5 (50) | 30 (100) | 0 (0) | |
| MSH2 and MSH6 Loss | 24 (46) | 2 (3) | 2 (20) | 0 (0) | 0 (0) | |
| Isolated MSH6 Loss | 8 (15) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Isolated PMS2 Loss | 6 (12) | 3 (4) | 3 (30) | 0 (0) | 0 (0) | |
| MMR protein expression of affected | ||||||
| MMR gene in normal intestinal crypts* | ||||||
| Intact expression | 34 (65) | 69 (99) | 9 (90) | 30 (100) | 30 (100) | <0.001 |
| Loss of expression | 18 (35) | 1 (1) | 1 (10) | 0 (0) | 0 (0) | |
| MLH1 | 4 | 0 | 0 | |||
| MSH2 | 7 | 1 | 1 | |||
| MSH6 | 4 | 0 | 0 | |||
| PMS2 | 3 | 0 | 0 | |||
| Mean length of non-neoplastic mucosa evaluated by IHC in millimeters (range) | 105 (5–336) | 112 (10–399) | 187 (88–290) | 138 (41–399) | 61 (10–179) | 0.6 |
| Mean estimated number of non-neoplastic intestinal crypts evaluated by IHC (range) | 1163 (56–3733) | 1330 (111–5424) | 2195 (1089–3533) | 1659 (567–5424) | 712 (111–2268) | 0.3 |
MMR, mismatch repair; IHC, immunohistochemistry; Loss, loss of expression
Affected MMR gene is that which has germline MMR gene pathogenic variant or obligate partner of the MMR protein pair with a tumor showing concurrent loss of MMR protein expression.
Defined as loss of expression of one or more MMR proteins but with no explanation found (no germline variants or MLH1 promoter hypermethylation).
p-value is patients with Lynch syndrome versus patients without Lynch syndrome
The control group of 70 patients with colorectal carcinoma included three categories of patients: patients with “Lynch-like” syndrome (n=10), patients with sporadic MLH1/PMS2 protein deficient colorectal carcinoma (n=30), and patients with MMR protein proficient colorectal carcinoma (n=30) (Table 1). Of the “Lynch-like” syndrome patients, five had colorectal carcinomas with loss of MLH1 and PMS2 immunohistochemical expression, concurrent wild-type BRAF, negative MLH1 promoter hypermethylation, and negative germline MMR gene mutation studies. Two patients had colorectal carcinoma with concurrent loss of MSH2 and MSH6 expression, and three patients had colorectal carcinoma with isolated loss of PMS2; all five of which had negative germline mutation studies. Patients with Lynch syndrome presented with colorectal carcinoma at a younger age than the control group of patients without Lynch syndrome (mean age 52 years vs. 70 years, p<0.001) (Table 1).
Table 1 compares the frequency of MMR protein deficient intestinal crypts in each patient group. A total of 13,305 millimeters (mm) of non-neoplastic intestinal mucosa were evaluated, including 11,494 mm of non-neoplastic colonic mucosa and 1,811 mm of non-neoplastic small intestinal mucosa from the terminal ileum. Overall, an estimated 127,718 non-neoplastic colonic crypts and 25,871 non-neoplastic small intestinal crypts were evaluated. There was no significant difference in number of non-neoplastic intestinal crypts evaluated in patients with Lynch syndrome and patients without Lynch syndrome (mean number of crypts 1163 vs. 1330, p>0.05). MMR protein deficient non-neoplastic intestinal crypts were identified in 19 of 122 (16%) patients (Table 2, Figure 1). Eight patients had MSH2 deficient crypts followed by 4 patients with MLH1 deficient crypts, 4 patients with MSH6 deficient crypts, and 3 patients with PMS2 deficient crypts (Tables 1 and 2). MMR protein deficient colonic crypts were identified in 18 of 52 (35%) patients with Lynch syndrome compared to only 1 of 70 (1%) of patients in the control group (p<0.001). No patients with colorectal carcinoma with either sporadic MMR protein deficiency or MMR protein proficiency harbored MMR deficient non-neoplastic intestinal crypts. The sensitivity and specificity estimates for MMR deficient crypts in Lynch syndrome are 0.35 (95% CI 0.22–0.49) and 0.99 (95% CI 0.92–1.00) respectively, although the sensitivity estimate is likely influenced by the amount of crypts evaluated.
Table 2.
Patients with DNA Mismatch Repair (MMR) Protein Deficient Non-Neoplastic Colonic Crypts
| Case Number | Tumor MMR Protein Expression Pattern by IHC | Germline MMR Gene Affected | Age at Diagnosis (years) | Gender | Linear Length of Non-Neoplastic Colonic Mucosa Evaluated (mm) | Estimated Number of Non-Neoplastic Colonic Crypts Evaluated | MMR Protein Evaluated in Non-neoplastic Mucosa by IHC | Number of MMR Deficient Non-Neoplastic Colonic Crypts by IHC | Location of MMR Deficient Non-Neoplastic Colonic Crypts | Pattern of MMR Deficient Non-Neoplastic Colonic Crypts (Solitary vs. Grouped) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | MSH2/MSH6 loss | MSH2 | 65 | Female | 145 | 1611 | MSH2 | 13 | Center colon | Solitary and Grouped |
| 2 | MSH2/MSH6 loss | MSH2 | 55 | Male | 131 | 1456 | MSH2 | 1 | Right colon | Solitary |
| 3 | Isolated MSH6 loss | MSH6 | 66 | Male | 180 | 2000 | MSH6 | 49 | Center colon | Grouped |
| 4 | Isolated MSH6 loss | MSH6 | 48 | Male | 102 | 1133 | MSH6 | 1 | Right colon | Solitary |
| 5 | Isolated PMS2 loss | PMS2 | 52 | Male | 158 | 1756 | PMS2 | 1 | Center colon | Solitary |
| 6 | MLH1/PMS2 loss | MLH1 | 49 | Male | 61 | 678 | MLH1 | 1 | Center colon | Solitary |
| 7 | Isolated PMS2 loss | PMS2 | 79 | Male | 167 | 1856 | PMS2 | 5 | Right colon | Grouped |
| 8 | Isolated PMS2 loss | PMS2 | 71 | Female | 118 | 1311 | PMS2 | 1 | Right colon | Solitary |
| 9 | MSH2/MSH6 loss | MSH2 | 34 | Female | 171 | 1900 | MSH2 | 4 | Right colon | Grouped |
| 10 | Isolated MSH6 loss | MSH6 | 65 | Female | 199 | 2211 | MSH6 | 2 | Right colon | Solitary |
| 11 | MSH2/MSH6 loss | MSH2 | 27 | Female | 90 | 1000 | MSH2 | 6 | Right colon | Grouped |
| 12 | MLH1/PMS2 loss | MLH1 | 60 | Female | 21 | 233 | MLH1 | 8 | Right colon | Grouped |
| 13 | MSH2/MSH6 loss | MSH2 | 53 | Female | 62 | 688 | MSH2 | 1 | Right colon | Solitary |
| 14 | MLH1/PMS2 loss | MLH1 | 64 | Male | 101 | 1117 | MLH1 | 2 | Right colon | Solitary |
| 15 | MSH2/MSH6 loss | MSH2 | 47 | Female | 56 | 624 | MSH2 | 3 | Right colon | Solitary |
| 16 | Isolated MSH6 loss | MSH6 | 73 | Male | 269 | 2989 | MSH6 | 2 | Right colon | Solitary |
| 17 | MSH2/MSH6 loss | MSH2 | 40 | Female | 336 | 3733 | MSH2 | 4 | Right colon | Grouped |
| 18 | MLH1/PMS2 loss | MLH1 | 69 | Female | 76 | 844 | MLH1 | 1 | Right colon | Solitary |
| 19 | MSH2/MSH6 loss | None | 47 | Male | 80 | 889 | MSH2 | 2 | Right colon | Solitary |
MMR, mismatch repair; IHC, immunohistochemistry
Figure 1.
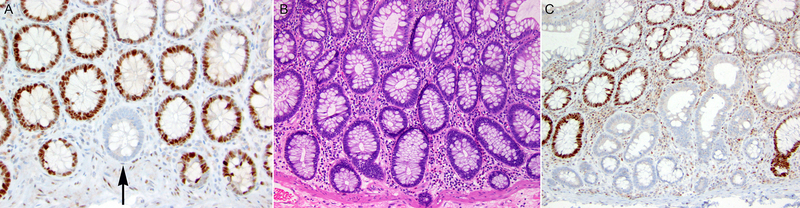
A. Solitary non-neoplastic colonic crypt with loss of MSH6 protein expression by immunohistochemistry (black arrow) in a patient with Lynch syndrome due to a pathogenic germline variant in MSH6 (MSH6 immunohistochemistry, 200x magnification) (Table 2, case 4). B. Non-neoplastic colonic mucosa in another patient with Lynch syndrome due to a pathogenic germline variant in MSH6 (Table 2, case 3) (H&E stain, 100x magnification). C. The non-neoplastic colonic mucosa depicted in B (Table 2, case 3) harbored a group of colonic crypts demonstrating loss of MSH6 protein expression by immunohistochemistry (MSH6 immunohistochemistry, 100x magnification).
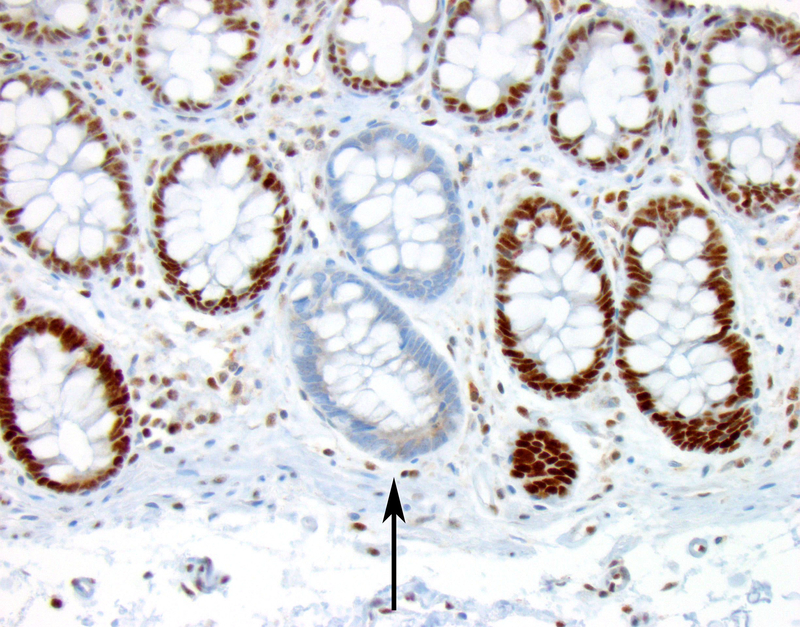
Only a single patient in the control group, from the “Lynch-like” subgroup, was found to have MMR protein deficient intestinal crypts (Table 2, case 19) (Figure 2), demonstrating two MSH2 deficient colonic crypts. This 47 year old patient with “Lynch-like” syndrome had a personal history of two primary colon cancers and a family history of early-onset Lynch syndrome-related cancers. Despite the personal and family cancer history and loss of MSH2/MSH6 protein expression in the tumor all suggestive of Lynch syndrome, germline mutation studies for MSH2, MSH6, and EPCAM identified no pathogenic mutation.
Figure 2.
This patient (Table 2, case 19) had a MSH2 and MSH6-deficient colorectal carcinoma but had negative germline MSH2, MSH6, and EPCAM mutation studies fulfilling criteria for “Lynch-like” syndrome. The patient had two solitary MSH2-deficient intestinal crypts, one of which is depicted (black arrow), that suggest Lynch syndrome despite the negative germline mutation studies (MSH2 immunohistochemistry, 100x magnification).
Overall, a total of 104 MMR protein deficient non-neoplastic intestinal crypts were identified in 19 patients. All MMR protein deficient non-neoplastic intestinal crypts were identified in the colon with none observed in the small intestinal mucosa analyzed. The pattern and number of MMR protein deficient non-neoplastic colonic crypts varied. For most patients (12 of 19, 63%), only solitary MMR protein deficient colonic crypts were identified. For seven patients, MMR protein deficient non-neoplastic colonic crypts were identified in groups ranging in number from two to 22 crypts. One patient (Table 2, case 3) with a germline MSH6 gene pathogenic variant had multiple foci of grouped MMR protein deficient colonic crypts (Figure 1). None of the cases with MMR deficient non-neoplastic colonic crypts displayed any histologic features of dysplasia.
Factors Affecting Detection of MMR Protein Deficient Colonic Crypts in Lynch Syndrome
Table 3 compares the features of patients with Lynch syndrome stratified by the presence of MMR deficient non-neoplastic colonic crypts. The number of non-neoplastic colonic crypts evaluated significantly affected the detection of MMR protein deficient non-neoplastic colonic crypts. Patients with Lynch syndrome with MMR protein deficient non-neoplastic colonic crypts had more non-neoplastic crypts evaluated compared to those who did not harbor MMR protein deficient crypts (mean number of crypts 1508 vs. 981, p=0.03). In addition, a trend to increasing frequency of MMR protein deficient crypts identified in the right colon (14/32, 44%) versus the left colon (4/20, 20%) was identified, although this did not reach statistical significance (p=0.08).
Table 3.
Patients with Lynch Syndrome Stratified by the Presence of DNA Mismatch Repair (MMR) Protein Deficient Colonic Crypts
| Clinicopathologic Features | Lynch Syndrome with MMR Deficient Non-Neoplastic Colonic Crypt N (%) | Lynch Syndrome without MMR Deficient Non-Neoplastic Colonic Crypt N (%) | p-value |
|---|---|---|---|
| No. of Cases | 18 | 34 | NA |
| Mean Age in years (range) | 57 (27–79) | 51 (28–80) | 0.1 |
| Gender, Male/Female | 10 (56) / 8 (44) | 17 (50) / 17 (50) | 0.9 |
| Location | |||
| Right Colon | 14 (78) | 18 (53) | 0.08 |
| Left Colon / Rectum | 4 (22) | 16 (47) | |
| MMR IHC Pattern in Carcinoma | |||
| Intact expression of all 4 proteins | 0 (0) | 1 (3) | 0.7 |
| MLH1 and PMS2 Loss | 4 (22) | 9 (26) | |
| MSH2 and MSH6 Loss | 7 (39) | 17 (50) | |
| Isolated MSH6 Loss | 4 (22) | 4 (12) | |
| Isolated PMS2 Loss | 3 (17) | 3 (9) | |
| Germline Mutation Analysis | |||
| MLH1 pathogenic variant present | 4 (22) | 9 (27) | 0.6 |
| MSH2 pathogenic variant present | 7 (39) | 18 (53) | |
| MSH6 pathogenic variant present | 4 (22) | 4 (12) | |
| PMS2 pathogenic variant present | 3 (17) | 3 (17) | |
| Mean length of colonic mucosa evaluated by IHC in millimeters (range) | 136 (21–336) | 88 (5–244) | 0.03 |
| Mean estimated number of colonic crypts evaluated by IHC (range) | 1508 (233–3733) | 981 (56–2711) | 0.03 |
MMR, mismatch repair; IHC, immunohistochemistry
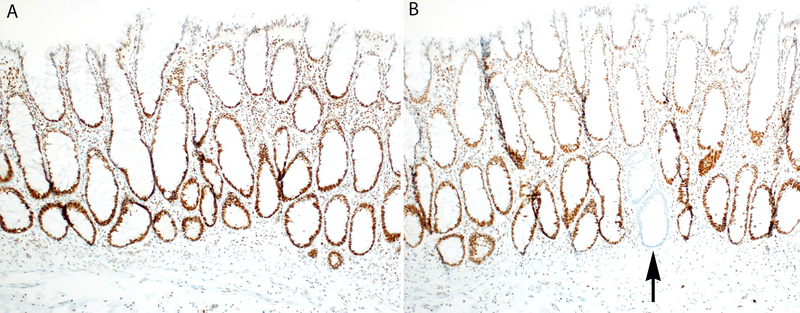
To determine if examination of additional non-neoplastic colonic mucosa could increase detection of MMR protein deficient colonic crypts, seven patients with Lynch syndrome who originally had no MMR protein deficient crypts seen on initial sections were selected for additional immunohistochemical analysis. For each patient, one paraffin block containing non-neoplastic colonic mucosa was serially sectioned at 80 to 90 μm intervals and an additional six sections were evaluated by MMR protein immunohistochemistry. These level sections resulted in between 633 to 2800 additional non-neoplastic colonic crypts for MMR protein immunohistochemical analysis. Of the seven patients with additional level sections analyzed, three displayed MMR protein deficient non-neoplastic colonic crypts (Table 2, cases 16, 17, and 18) (Figure 3).
Figure 3.
A. Original immunohistochemical stain for MLH1 demonstrated preserved expression in all non-neoplastic colonic crypts (Table 2, case 18) in a Lynch syndrome patient with a pathogenic germline variant in MLH1 (MLH1 immunohistochemistry, 40x magnification). B. A deeper section (80 μm deeper) reveals a single crypt with loss of MLH1 expression by immunohistochemistry (black arrow) (MLH1 immunohistochemistry, 40x magnification).
The frequency of MMR protein deficient crypts was estimated using the data from Table 2 but excluding case 3 (a relative outlier with 49 MMR protein deficient crypts identified) and case 19 (the patient with “Lynch-like” syndrome). Approximately 1 MMR protein deficient crypt can be identified per 1063 colonic crypts corresponding to 1 MMR protein deficient crypt per 8.6 mm2 of colonic mucosa. Using binomial distribution with a 0.00094 probability of occurrence, we estimate that immunohistochemical evaluation of all colonic crypts in 26.3 mm2 of colonic mucosa (~3250 crypts) would yield a 95% probability of detecting at least one MMR protein deficient crypt in a patient with Lynch syndrome. Practically, this could be accomplished by immunohistochemical evaluation of 10 sections cut at 90 μm intervals from 30 mm of normal colonic mucosa obtained from surgical resection specimens.
DISCUSSION
We demonstrate that MMR protein deficient non-neoplastic colonic crypts can be identified in 35% of patients with Lynch syndrome and are not seen in patients with colorectal carcinoma with sporadic MMR protein deficiency or MMR protein proficiency. One patient with “Lynch-like” syndrome also harbored MSH2 deficient non-neoplastic colonic crypts suggesting that the patient may have an undetected germline MMR gene or EPCAM pathogenic variant. Our findings indicate that identification of MMR protein deficient non-neoplastic colonic crypts is a specific indicator of Lynch syndrome and inclusion of MMR protein immunohistochemical evaluation of non-neoplastic colonic mucosa in the Lynch syndrome diagnostic workflow may be worthwhile, particularly in patients where germline MMR gene and EPCAM testing is inconclusive.
Two recent publications from the same institution have demonstrated that histologically normal crypts in patients with Lynch syndrome can exhibit loss of expression of MMR proteins (10, 11). Kloor et al. evaluated intestinal mucosa in 40 patients with Lynch syndrome and identified 10 (25%) patients with MMR protein deficient intestinal crypts by immunohistochemistry, a lower proportion than observed in our series (10). In a follow-up study from the same institution, Staffa et al. identified MMR protein deficient intestinal crypts in 11 of 34 (32%) patients with Lynch syndrome, similar to the 35% of patients with Lynch syndrome in our study with MMR protein deficient crypts (11). Kloor et al. also found that the incidence of MMR deficient crypts was higher in colonic compared with small intestinal mucosa and estimated that 1.6 times more MMR protein deficient crypts are present in colonic mucosa compared to small intestinal mucosa of patients with Lynch syndrome. This may help to explain why we only observed MMR protein deficient intestinal crypts in colonic mucosa and not small intestinal mucosa. However, there are a number of differences between our study and those of Kloor et al. and Staffa et al. First, our study included patients with Lynch syndrome due to germline pathogenic variants in all 4 MMR genes. In contrast, Kloor et al. and Staffa et al. analyzed patients with Lynch syndrome primarily due to MLH1 and MSH2 germline pathogenic variants with only one patient harboring a MSH6 germline pathogenic variant and none with a PMS2 germline pathogenic variant. Second, Staffa et al. identified that the frequency of MMR protein deficient crypts increases with patient age and that MMR protein deficient crypts are more often seen in the left colon. In contrast, we did not identify an association between patient age and the identification of MMR protein deficient crypts. In addition, in our analysis, MMR protein deficient crypts were more often identified in the right colon, although this did not reach statistical significance. Third, we observed a pattern of solitary crypts with loss of MMR protein expression in most (12 of 19, 63%) of the patients with Lynch syndrome with MMR protein deficient crypts. In contrast, Kloor et al. identified groups of MMR protein deficient crypts in most cases (9 of 10, 90%). Finally, 13 of the 27 MMR protein deficient crypt foci identified by Kloor et al. displayed “unusual morphological changes” including “nuclear enlargement of cells at the crypt bottom”. In addition, seven of the 27 MMR protein deficient crypt foci identified by Staffa et al. are described as harboring “dysplastic changes”. In contrast, the MMR protein deficient colonic crypts in our series lacked any features of dysplasia and appear histologically to be non-neoplastic.
Universal screening for Lynch syndrome in colorectal cancer is routinely performed in most pathology practices; however, the identification of abnormal MMR protein expression within a patient’s tumor should not be considered diagnostic of Lynch syndrome. Between 25% to 40% of patients will have no evidence of a germline alteration in MMR genes or EPCAM despite harboring tumors with MMR protein deficiency concerning for Lynch syndrome (9, 19). Such patients have been labeled with provisional terms such as “Lynch-like” syndrome or “suspected Lynch syndrome” (7–9, 20, 21). Recent literature indicates that up to 70% of patients with “Lynch-like” syndrome acquire biallelic (somatic) tumor mutations in MMR genes that explain the MMR deficiency within the tumor in the presence of negative germline MMR gene and EPCAM mutation studies (22–25). These somatic MMR gene mutations are likely sporadic events, suggesting that such tumors are most likely cancers with sporadic MMR protein deficiency. However, the possibility that these somatic biallelic mutations are secondary to other germline gene defects, including within MUTYH and POLE/POLD1 still remains (26–28). Still other patients who undergo germline mutation analysis will be identified as having germline MMR gene variants of unknown significance (VUS). Identification of an MMR gene VUS does not establish the diagnosis of Lynch syndrome. Inconclusive germline mutation studies represent a major challenge in Lynch syndrome diagnostics. With the adoption of universal screening efforts for Lynch syndrome, identification of patients with tumors harboring abnormal MMR protein expression concerning for Lynch syndrome but with inconclusive germline mutation testing is more frequently encountered (8, 22). In a recent series at one of the contributing institutions, negative germline MMR gene mutation studies were identified in 32% of patients harboring abnormal MMR protein expression concerning for Lynch syndrome (9). It is unclear if patients with “Lynch-like” syndrome or MMR gene VUS should undergo the same intensive lifelong screening protocol used for patients with confirmed Lynch syndrome. In addition, the MMR gene VUS cannot be used with certainty in any of the patient’s asymptomatic relatives to evaluate if they have Lynch syndrome.
Evaluation for MMR protein deficient non-neoplastic colonic crypts may be a helpful complementary test to the germline and somatic MMR gene mutation analyses currently used to establish a diagnosis of Lynch syndrome, although further studies are needed assess its clinical utility. This is highlighted by the patient in our series with “Lynch-like” syndrome with a personal and family history and loss of MSH2/MSH6 protein expression in the tumor collectively suggestive of Lynch syndrome (Table 2, case 19). The presence of multiple MMR protein deficient colonic crypts in this patient adds further support for clinical management consistent with a diagnosis Lynch syndrome despite not being able to identify a germline mutation with the diagnostic testing approaches currently used. In addition, in individuals who are found to have a germline VUS in the gene that is implicated by the abnormal MMR protein immunohistochemistry pattern, we predict that observing MMR protein deficient crypts in normal colon would provide evidence that the VUS is pathogenic. However, additional study is needed to assess the clinical utility of detecting MMR protein deficient colonic crypts in patients with germline MMR gene VUS.
In daily practice, if a patient is found to have a colorectal carcinoma with MMR protein immunohistochemistry that is suggestive of Lynch syndrome, testing normal colonic mucosa for MMR protein deficient intestinal crypts may be useful. Although we only identified MMR protein deficient colonic crypts in 35% of patients with known Lynch syndrome, this is likely due to the amount of mucosa analyzed. This is supported by the association between the number of crypts analyzed and the presence of MMR protein deficient crypts as well as the demonstration that analysis of deeper levels was able to identify MMR protein deficient non-neoplastic colonic crypts in three of seven cases that initially lacked these crypts. Thus, in the setting of a colorectal carcinoma with MMR protein immunohistochemistry that is suggestive of Lynch syndrome, it may be prudent for pathologists to submit additional normal colorectal mucosa for analysis. This would include patients with colorectal carcinomas demonstrating isolated loss of MSH6 expression, isolated loss of PMS2 expression, concurrent loss of MSH2/MSH6 expression, and concurrent loss of MLH1/PMS2 expression with negative MLH1 promoter hypermethylation studies. Combined, these scenarios would account for less than 5% of all patients with colorectal carcinoma. In these patients, if germline and somatic MMR gene mutation testing is inconclusive, additional sections of normal colonic mucosa obtained from the surgical resection specimen can be examined by immunohistochemistry for MMR protein deficient colonic crypts. Based on our estimated frequency of MMR protein deficient crypts, analysis of all colonic crypts in 26.3 mm2 of mucosa would have a 95% probability of identifying at least one MMR protein deficient crypt. This could be accomplished by immunohistochemical evaluation of 10 sections cut at 90 μm intervals from 30 mm of mucosa obtained from surgical resection specimens. Identification of MMR protein deficient colonic crypts would support the diagnosis of Lynch syndrome and indicate that the patient would benefit from intensive lifelong screening similar to that offered to patients with Lynch syndrome confirmed by germline mutation testing. However, the absence of MMR protein deficient intestinal crypts does not exclude the possibility of Lynch syndrome.
There are many pitfalls in MMR protein immunohistochemistry interpretation particularly with regards to aberrant staining patterns and variability in staining intensity that could affect evaluation for MMR protein deficient non-neoplastic colonic crypts (29–31). Variation in staining patterns has resulted in a less than perfect interobserver agreement in the interpretation of MMR protein immunohistochemical stains (29, 30). Equivocal staining patterns include punctate/speckled nuclear staining, nucleolar staining (31), and nuclear membrane staining. Weak nuclear staining can also be a diagnostic challenge. Comparison with the internal control is often helpful to point to technical issues. In normal colonic mucosa, MMR protein deficiency should only be considered present if the entire base of a colonic crypt lacks nuclear MMR protein expression. A normal colonic crypt with equivocal nuclear staining of MMR proteins by immunohistochemistry should not be considered as evidence supporting a diagnosis of Lynch syndrome.
The significance and etiology of the MMR protein deficient crypts needs further study. MMR protein deficient colonic crypts may represent the earliest precursor to neoplasia in patients with Lynch syndrome. However, most patients with Lynch syndrome do not develop polyposis (32). Thus, it is unlikely that MMR protein deficient colonic crypts often progress to dysplasia and carcinoma without the accumulation of other genetic mutations. MMR protein deficient colonic crypts are likely the result of a clonal biallelic inactivation of the affected MMR gene as the MMR protein loss is seen within the entire crypt base and can colonize adjacent intestinal crypts to form groups of MMR protein deficient crypts. Microsatellite instability has been identified by PCR analysis in microdissected MMR protein deficient crypt foci (10, 11). In total, Kloor et al. and Staffa et al. evaluated 30 MMR protein deficient non-neoplastic crypt foci, including grouped and solitary MMR protein deficient crypts, and identified microsatellite instability by PCR in at least one of the tested markers in 27 crypt foci (90%) (10, 11). Additional study is needed to determine the sequence of molecular alterations in MMR protein deficient colonic crypts that can lead to the development of neoplasia, including evaluation for mutations in MMR genes and other genes involved in DNA replication and repair such as MUTYH and POLE/POLD1.
Our analysis has limitations, including the retrospective design and the size of the study cohort. Although the number of patients with Lynch syndrome in our study is relatively small by some standards, our analysis is the largest to date evaluating MMR protein expression in non-neoplastic intestinal mucosa. Another limitation of this study is that germline mutation analysis was performed at multiple laboratories over a 20-year period. At the time, most of these laboratories did not evaluate for an inversion of MSH2 (exons 1–7) which has been found to explain a subset of cases of “Lynch-like” syndrome in which no germline MSH2 mutation is identified by conventional analyses (33). For patients with “Lynch-like” syndrome, we also did not have data on somatic MMR gene mutations. We also did not include evaluation of other anatomic sites, such as the endometrium, in which Lynch syndrome-associated carcinomas frequently arise. Lastly, we did not evaluate for MMR protein deficient crypts in unaffected and asymptomatic Lynch syndrome mutation carriers, and additional studies to evaluate for MMR deficient non-neoplastic crypts in unaffected Lynch syndrome mutation carriers may also be of interest.
In conclusion, we demonstrate that MMR protein deficient colonic crypts are a novel indicator of Lynch syndrome and are not seen in the colonic mucosa of patients with sporadic MMR protein deficient colorectal carcinoma resulting from somatic MLH1 promoter methylation or in patients with MMR protein proficient colorectal carcinoma. MMR protein deficient crypts are identified in colonic mucosa in more than one-third of patients with Lynch syndrome and can either be solitary crypts or small groups of crypts. The detection of MMR protein deficient colonic crypts largely depends on the amount of normal mucosa evaluated. Thus, additional sampling of non-neoplastic colorectal mucosa, including evaluation of deeper levels, in patients with colorectal carcinoma harboring MMR protein deficiency concerning for Lynch syndrome may increase the detection of MMR protein deficient colonic crypts. Evaluation for MMR protein deficient colonic crypts may be a helpful addition to Lynch syndrome diagnostics, particularly in the setting of “Lynch-like” syndrome and patients with germline MMR gene VUS.
Acknowledgments
Financial support: This work was supported by grant UM1 CA167551 from the National Cancer Institute and through cooperative agreements with Australasian Colorectal Cancer Family Registry (U01 CA074778, U01/U24 CA097735, and U01/U24 CA074800) and was conducted under Colon-CFR approval C-AU-1014–01. Christophe Rosty is the Jass Pathology Fellow. Daniel D. Buchanan is a University of Melbourne Research at Melbourne Accelerator Program (R@MAP) Senior Research Fellow and National Health and Medical Research Council of Australia (NHMRC) R.D. Wright Career Development Fellow. Mark A. Jenkins is a NHMRC Senior Research Fellow.
Footnotes
Disclosures: The authors have no conflict of interest.
References
- 1.Evaluation of Genomic Applications in Practice, Prevention Working Group. Recommendations from the EGAPP Working Group: genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives.Genet Med. 2009;11:35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ligtenberg MJ, Kuiper RP, Geurts van Kessel A, et al. EPCAM deletion carriers constitute a unique subgroup of Lynch syndrome patients. Fam Cancer. 2013;12:169–174. [DOI] [PubMed] [Google Scholar]
- 3.Giardiello FM, Allen JI, Axilbund JE, et al. Guidelines on genetic evaluation and management of Lynch syndrome: a consensus statement by the US Multi-Society Task Force on colorectal cancer. Gastroenterology. 2014;147:502–526. [DOI] [PubMed] [Google Scholar]
- 4.Syngal S, Brand RE, Church JM, et al. ACG clinical guideline: Genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol. 2015;110:223–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoffel EM, Mangu PB, Gruber SB, et al. Hereditary colorectal cancer syndromes: American Society of Clinical Oncology Clinical Practice Guideline endorsement of the familial risk-colorectal cancer: European Society for Medical Oncology Clinical Practice Guidelines. J Clin Oncol. 2015;33:209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balmana J, Balaguer F, Cervantes A, et al. Familial risk-colorectal cancer: ESMO Clinical Practice Guidelines. Ann Oncol. 2013;24 Suppl 6:vi73–80. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez-Soler M, Perez-Carbonell L, Guarinos C, et al. Risk of Cancer in Cases of Suspected Lynch Syndrome Without Germline Mutation. Gastroenterology. 2013;144:926–932. [DOI] [PubMed] [Google Scholar]
- 8.Buchanan DD, Rosty C, Clendenning M, et al. Clinical problems of colorectal cancer and endometrial cancer cases with unknown cause of tumor mismatch repair deficiency (suspected Lynch syndrome). Appl Clin Genet. 2014;7:183–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mas-Moya J, Dudley B, Brand RE, et al. Clinicopathological comparison of colorectal and endometrial carcinomas in patients with Lynch-like syndrome versus patients with Lynch syndrome. Hum Pathol. 2015;46:1616–1625. [DOI] [PubMed] [Google Scholar]
- 10.Kloor M, Huth C, Voigt AY, et al. Prevalence of mismatch repair-deficient crypt foci in Lynch syndrome: a pathological study. Lancet Oncol. 2012;13:598–606. [DOI] [PubMed] [Google Scholar]
- 11.Staffa L, Echterdiek F, Nelius N, et al. Mismatch repair-deficient crypt foci in Lynch syndrome--molecular alterations and association with clinical parameters. PloS one. 2015;10:e0121980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walsh MD, Cummings MC, Buchanan DD, et al. Molecular, pathologic, and clinical features of early-onset endometrial cancer: identifying presumptive Lynch syndrome patients. Clin Cancer Res. 2008;14:1692–1700. [DOI] [PubMed] [Google Scholar]
- 13.Yousem SA, Nikiforova M, Nikiforov Y. The histopathology of BRAF-V600E-mutated lung adenocarcinoma. Am J Surg Pathol. 2008;32:1317–1321. [DOI] [PubMed] [Google Scholar]
- 14.Buchanan DD, Sweet K, Drini M, et al. Risk factors for colorectal cancer in patients with multiple serrated polyps: a cross-sectional case series from genetics clinics. PloS one. 2010;5:e11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchanan DD, Tan YY, Walsh MD, et al. Tumor mismatch repair immunohistochemistry and DNA MLH1 methylation testing of patients with endometrial cancer diagnosed at age younger than 60 years optimizes triage for population-level germline mismatch repair gene mutation testing. J Clin Oncol. 2014;32:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poynter JN, Siegmund KD, Weisenberger DJ, et al. Molecular characterization of MSI-H colorectal cancer by MLHI promoter methylation, immunohistochemistry, and mismatch repair germline mutation screening. Cancer Epidemiol Biomarkers Prev. 2008;17:3208–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myriad Genetics. Lynch syndrome. 2015. [cited 2015 June 1, 2015]. Available from: https://www.myriadpro.com/.
- 18.Ambry Genetics. Lynch syndrome. [cited 2015 June 6, 2015]. Available from: http://www.ambrygen.com/tests/lynch-syndrome.
- 19.Buchanan DD, Clendenning M, Rosty C, et al. Tumor testing to identify lynch syndrome in two Australian colorectal cancer cohorts. J Gastroenterol Hepatol. 2017;32:427–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Win AK, Buchanan DD, Rosty C, et al. Role of tumour molecular and pathology features to estimate colorectal cancer risk for first-degree relatives. Gut. 2015;64:101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hampel H, Frankel WL, Martin E, et al. Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med. 2005;352:1851–1860. [DOI] [PubMed] [Google Scholar]
- 22.Haraldsdottir S, Hampel H, Tomsic J, et al. Colon and endometrial cancers with mismatch repair deficiency can arise from somatic, rather than germline, mutations. Gastroenterology. 2014;147:1308–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mensenkamp AR, Vogelaar IP, van Zelst-Stams WA, et al. Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology. 2014;146:643–646. [DOI] [PubMed] [Google Scholar]
- 24.Geurts-Giele WR, Leenen CH, Dubbink HJ, et al. Somatic aberrations of mismatch repair genes as a cause of microsatellite-unstable cancers. J Pathol. 2014;234:548–559. [DOI] [PubMed] [Google Scholar]
- 25.Sourrouille I, Coulet F, Lefevre JH, et al. Somatic mosaicism and double somatic hits can lead to MSI colorectal tumors. Fam Cancer. 2013;12:27–33. [DOI] [PubMed] [Google Scholar]
- 26.Castillejo A, Vargas G, Castillejo MI, et al. Prevalence of germline MUTYH mutations among Lynch-like syndrome patients. Eur J Cancer. 2014;50:2241–2250. [DOI] [PubMed] [Google Scholar]
- 27.Jansen AM, van Wezel T, van den Akker BE, et al. Combined mismatch repair and POLE/POLD1 defects explain unresolved suspected Lynch syndrome cancers. Eur J Hum Genet. 2016;24:1089–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Elsayed FA, Kets CM, Ruano D, et al. Germline variants in POLE are associated with early onset mismatch repair deficient colorectal cancer. Eur J Hum Genet. 2015;23:1080–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Jong AE, van Puijenbroek M, Hendriks Y, et al. Microsatellite instability, immunohistochemistry, and additional PMS2 staining in suspected hereditary nonpolyposis colorectal cancer. Clin Cancer Res. 2004;10:972–980. [DOI] [PubMed] [Google Scholar]
- 30.Overbeek LI, Ligtenberg MJ, Willems RW, et al. Interpretation of immunohistochemistry for mismatch repair proteins is only reliable in a specialized setting. Am J Surg Pathol. 2008;32:1246–1251. [DOI] [PubMed] [Google Scholar]
- 31.Radu OM, Nikiforova MN, Farkas LM, et al. Challenging cases encountered in colorectal cancer screening for Lynch syndrome reveal novel findings: nucleolar MSH6 staining and impact of prior chemoradiation therapy. Hum Pathol. 2011;42:1247–1258. [DOI] [PubMed] [Google Scholar]
- 32.Kalady MF, Kravochuck SE, Heald B, et al. Defining the adenoma burden in lynch syndrome. Dis Colon Rectum. 2015;58:388–392. [DOI] [PubMed] [Google Scholar]
- 33.Rhees J, Arnold M, Boland CR. Inversion of exons 1–7 of the MSH2 gene is a frequent cause of unexplained Lynch syndrome in one local population. Fam Cancer. 2014;13:219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]