Abstract
One of the most commonly employed bioorthogonal reactions with azides is copper-catalyzed azide–alkyne [3+2] cycloaddition (CuAAC, a ‘click’ reaction). More recently, the strain-promoted azide–alkyne [3+2] cycloaddition (SPAAC, a copper-free ‘click’ reaction) was developed, in which an alkyne is sufficiently strained to promote rapid cycloaddition with an azide to form a stable triazole conjugate. In this report, we show that an internal alkyne in a strained ring system with two electron-withdrawing fluorine atoms adjacent to the carbon–carbon triple bond reacts to yield covalent adducts not only with azide moieties but also reacts with free sulfhydryl groups abundant in the cytosol. We have identified conditions that allow the enhanced reactivity to be tolerated when using such conformationally strained reagents to enhance reaction rates and selectivity for bioorthogonal applications such as O-GlcNAc detection.
Keywords: Copper-catalyzed azide–alkyne, cycloaddition, Strain-promoted azide–alkyne cycloaddition, O-GlcNAc, Glycoconjugates
1. Introduction
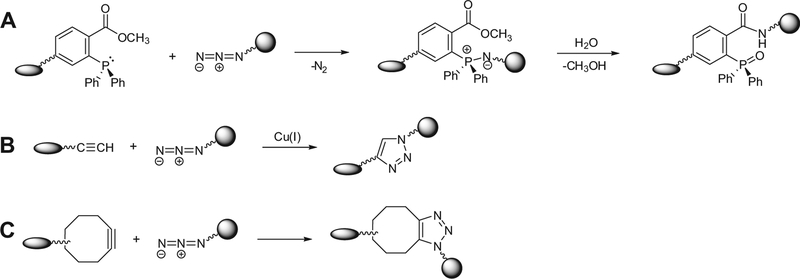
Detection and isolation of metabolites and posttranslationally modified biomolecules such as glycoproteins and glycolipids can be achieved by tagging target biomolecules with a bioorthogonal functional group, a ‘chemical handle’, and using highly selective chemical reactions (chemoselective reactions) between the chemical handle and an appropriate reagent probe. The chemical handle should be unreactive with native biological functional groups (i.e., bioorthogonal), and it should be incorporated into the target bio-molecules using either the cell’s biosynthetic machinery1–6 or added to macromolecules by the action of enzymes in vitro.7 Once incorporated into bioconjugates, chemical handles can be covalently ligated to either a visualization tag or an enrichment tag through a chemoselective reaction.8,9 Among the chemical handles that have been used, the azide is valuable as it is not typically found in biological systems and does not interact with any biological functionality. In addition, the azide has unique reactivity with phosphine- and alkyne-based probes. The most commonly employed reactions involving azido-functionality include the modified-Staudinger ligation with a triaryl phosphine probe3,10,11 (Fig. 1A) and the copper(I)-catalyzed cycloaddition with a terminal alkyne probe, which is called CuAAC12,13 a ‘click’ reaction14,15 (Fig. 1B).
Figure 1.
Chemoselective reactions with azide. Molecules containing azide react via the Staudinger ligation (A), Cu-catalyzed ‘click’ chemistry (B), or strain-promoted cycloaddition (C) to produce ligated products.
Compared to the Staudinger ligation, Cu(I)-catalyzed ‘click’ chemistry is faster and more sensitive, making this method attractive for bioconjugate chemistry.16–18 However, applying CuAAC to bioconjugate chemistry introduces several challenges, specifically in biological settings. First, the more stable Cu(II) ion is initially introduced as a Cu(II) salt and the Cu(I) catalyst is generated in situ by reduction of Cu(II) using a reducing agent such as sodium ascorbate. The Cu(I)-catalyzed ‘click’ reaction requires optimization of several factors such as metal, ligand, and auxiliary reductant concentrations. Furthermore, the copper ion(II) used in many CuAAC is known to oxidize amino acid side chains19 and is attributed to DNA cleavage.20,21 More recently, strain-promoted [3+2] azide–alkyne cycloaddition (SPAAC), also known as copper-free ‘click’ chemistry was developed22–24 in order to eliminate the use of a cytotoxic copper catalyst. Copper-free ‘click’ reactions use a cyclooctyne structure in which a triple bond in a ring is sufficiently strained to promote rapid cycloaddition with an azide in order to form a stable triazole conjugate (Fig. 1C). Unfortunately, the chemical reaction of cyclooctyne and azide-functionality generally proceeds at a slower rate than that of CuAAC.25 Therefore, several groups have made considerable effort toward improving the reaction rate of strain-promoted copper-free 1,3-dipolar cycloaddition between azide-moi6eties and cyclooctynes. These efforts have produced a number of strained cycloalkynes with enhanced reactivity22,26–28 making SPAAC more practical and effective for application in biological and physiological studies.26,29–32 Foremost among the new generation of reagents are difluorinated cyclooctyne (so-called DIFO)-based reagents.26,27 Ess et al.33 explained the accelerated rate effect of introduction of an electron-withdrawing group (fluorine) adjacent to the alkyne on the SPAAC reaction by calculating the energy of the transition states of 1,3-dipolar cycloadditions involving azide and different alkynes using density functional theory (B3LYP). Calculations of the transition states of different azide–alkyne combinations disclosed that difluoro substitution lowers the activation energy by 2.0 kcal/mol compared to that of non-fluorinated cyclooctyne, likely due to the increase in the stabilizing interaction caused by fluorination. Consistent with this result, calculations on the frontier orbitals of the cyclooctyne and difluorinated cyclooctyne revealed that difluoro substitution lowers the LUMO energy and increases the HOMO energy thereby narrowing the HOMO–LUMO gap and enhancing the reactivity.33
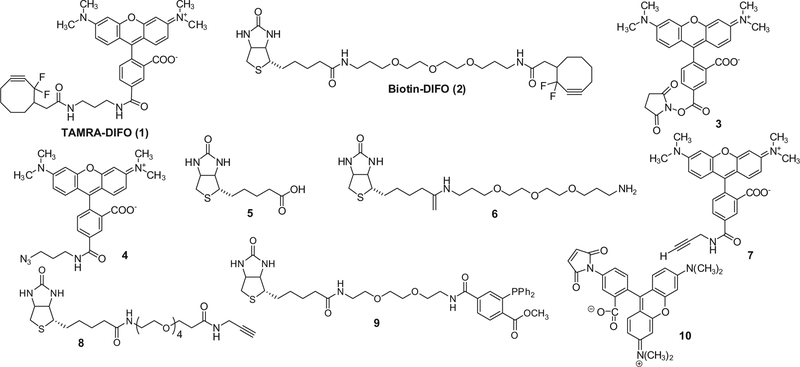
Herein, we report that difluorinated cyclooctyne core structures conjugated with either a rhodamine-based fluorophore (TAMRA) or a biotin tag (1 and 2, see Fig. 2) exhibit reactivity that causes these reagents to form an adduct with not only azide-functionality but also reactive sulfhydryls, which are abundant in many biological systems. Since reactive sulfhydryls are maintained by a reducing intracellular environment this is particularly problematic when examining intracellular glycosylation events. We describe a strategy that greatly improves the specificity of the DIFO reactivity under these conditions.
Figure 2.
Structures of DIFO-based reagents (1, 2), synthetic intermediates (3–6), terminal alkyne reagents (7, 8), triaryl phosphine reagent (9), and TAMRA–Mal (10).
2. Results and discussion
2.1. Syntheses of DIFO-based reagents
DIFO-based reagents (TAMRA–DIFO 1 and Biotin–DIFO 2) used in copper-free strain-promoted cycloadditions were prepared according to previously published procedures27 with minor modifications. Syntheses of these reagents are presented in detail in the Section 3 and the structures of DIFO-based reagents and synthetic intermediates are found in Figure 2. Briefly, commercially available 5/6-carboxy-tetramethylrhodamine succinimidyl ester 3 was reacted with 3-azidopropylamine to produce mixed isomers of azide-tagged tetramethylrhodamine. Azide-tagged tetramethyl-rhodamine single isomer 4 was obtained by flash column chromatography from mixed isomers. Compound 4 was subjected to hydrogenation and subsequent coupling reaction with 2-(2,2-difluorocyclooct-3-yn-1-yl)acetic acid to produce DIFO–TAMRA 1 in 96% yield. Biotin–DIFO 2 was prepared by direct amide coupling of biotinyl-4,7,10-trioxatridecanediamine 6 and 2-(2,2-difluorocyclooct-3-yn-1-yl)acetic acid using a HATU coupling reagent in 35% yield.
2.2. DIFO-based reagents are reactive toward both azide-labeled and unlabeled proteins
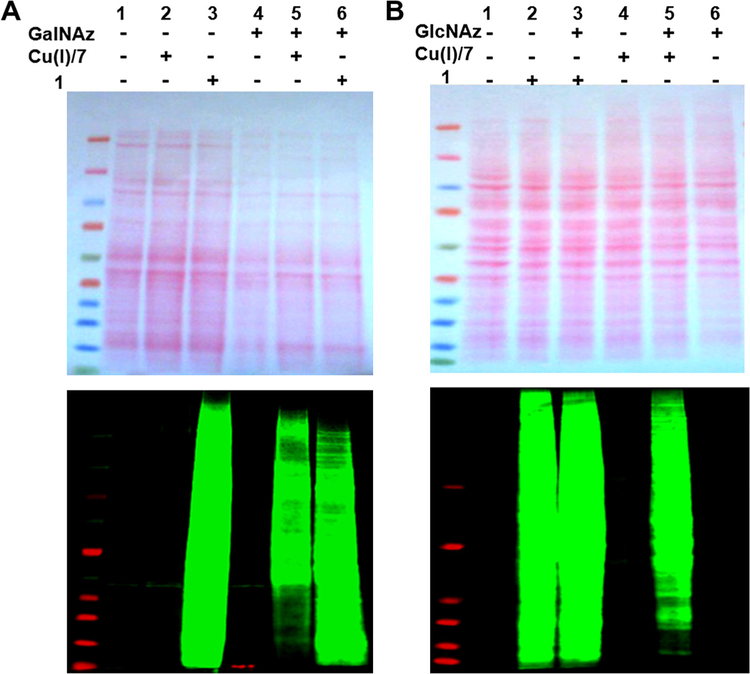
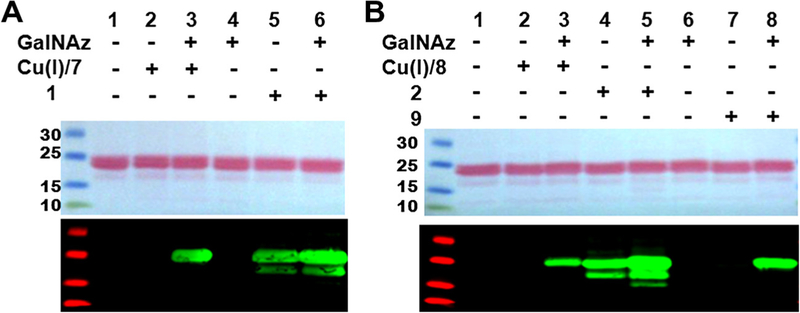
Additional reactivity of DIFO-based reagents was observed in unlabeled cell lysates (Fig. 3). In this study, azide groups were installed on proteins in two ways: in vitro enzymatic transfer and metabolic labeling. In in vitro enzymatic transfer, proteins in Caenorhabditis elegans’ lysates were tagged with the azide-functionality using a mutant Gal-T1 enzyme that is capable of transferring an unnatural N-azidoacetylgalactosamine (GalNAz) sugar from UDPGalNAz onto proteins that have a terminal N-acetylglucosamine (O-GlcNAc). Alternatively, HeLa cell proteins were metabolically labeled by culturing the cells in a medium containing 100 μM peracetylated N-azidoacetylglucosamine ((Ac)4GlcNAz). In order to examine reaction specificity, both azide-labeled and unlabeled cell lysates were treated with either a DIFO-based probe (1) or terminal alkyne (TAMRA–Alkyne 7) at room temperature for 20 min. Results of each reaction were monitored by gel electrophoresis and the immunoblot analysis. As necessary, blots were normalized to the intensity of the Ponceau S stained gels quantified by Image J. As expected, copper-catalyzed ‘click’ reactions of azide-labeled C. elegans lysate (Fig. 3A, lane 5) and HeLa cell lysate (Fig. 3B, lane 5) with TAMRA–alkyne were specific for azide-moiety compared with those reactions of unlabeled lysates (Fig. 3A, lane 2; Fig. 3B, lane 4). However, strain-promoted copper-free ‘click’ reactions with TAMRA–DIFO did not show specificity for azide-functionality, since treatment of unlabeled lysates with TAMRA–DIFO also generated normalized fluorescent signals as strong as those generated from the treatment of azide-labeled lysates (Fig. 3A, lane 3; Fig. 3B, lane 2). Previous results from Boons et al.23 indicated low background interactions of cell lysates with cyclooctyne reagents and provided a possible explanation about the source of background staining. By careful experimentation, they excluded the possibility of unwanted side reaction of the cyclooctyne reagents with proteins and suggested the background was generated from potential noncovalent interactions of the FITC-conjugated avidin agent used for capturing biomolecules. Although dyes are known to interact with lysates in a non-specific manner, the TAMRA dye itself is unlikely to have generated the numerous non-specific bands detected in the samples lacking azide-functionality depicted in Figure 3. As demonstrated in Figure 3A (lane 2) and B (lane 4), although TAMRA–Alkyne (7) possesses the same dye component as found in TAMRA–DIFO, it did not give distinctive signals arising from non-specific interactions with unlabeled lysates. Only one band found in the lanes 1, 2, and 4 of Figure 3A most likely represents a noncovalent interaction with C. elegans lysates. Likewise, treatment of a different tag-containing DIFO reagent, Biotin–DIFO 2, with azide-labeled and unlabeled cell lysates yielded the same observation as obtained with TAMRA–DIFO (Fig. 4).
Figure 3.
Ponceau S-stained nitrocellulose membrane (upper) and immunoblots (lower) of C. elegans (A) and HeLa cell (B) extracts treated with and without 1 or 7. Immunoblots were probed with rabbit anti-TAMRA antibody followed by IRDye®800CW-conjugated goat anti-rabbit antibody. Quantitation of Ponceau S-stained gels was performed using Image J and used to generate normalized values referred to in the text. In (A), Ponceau S staining of lanes 4–6 was 62% of the intensity of lanes 1–3 and was used to normalize the fluorescence intensity values for comparison.
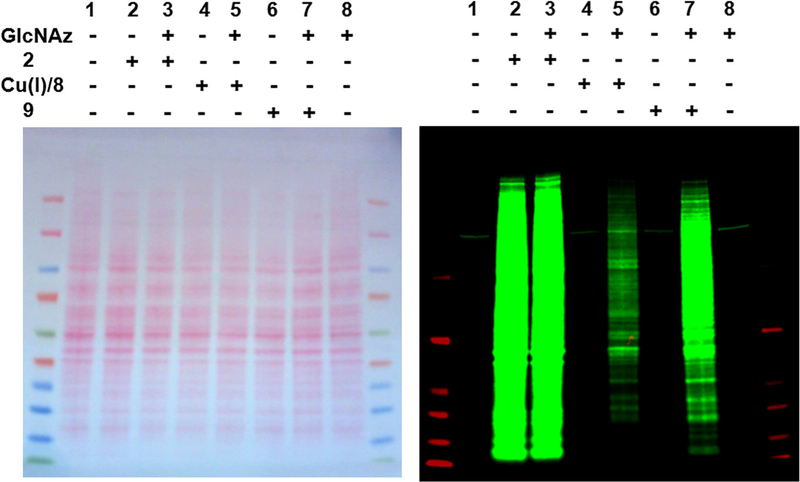
Figure 4.
Ponceau S.-stained nitrocellulose membrane (left) and immunoblot (right) of HeLa cell extracts reacted with and without 2, 8, or 9. The immunoblot was probed with IRDye800CW-Streptavidin.
It is important to note that copper-catalyzed ‘click’ reaction of terminal alkyne and azide-containing molecules does not occur in the commercial T-PER or M-PER lysis buffer (Thermo Scientific Pierce). We speculate that the copper catalyst is no longer available for alkyne–azide cycloaddition due to chelation of copper ions by these commercial buffers.
As expected, copper-catalyzed ‘click’ reaction and Staudinger ligation using biotinylated terminal alkyne (Biotin–Alkyne, 8) in combination with Cu(I) or triaryl phosphine (Biotin–Phosphine, 9), exhibited high specificity for azide-functionality. Immuno-reactive bands were detected only in the azide-labeled lysates (Fig. 4, lanes 5 and 6, respectively) and not in lanes containing unlabeled lysates (lanes 4 and 6). In contrast, Biotin–DIFO (2) produced numerous strong signals in both the absence and presence of azide-labeled lysates (Fig. 4, lanes 2 and 3, respectively).
We used purified α-crystallin to further test the specificity of the DIFO-based probes. α-crystallin is a protein modified by several mapped O-GlcNAc posttranslational modifications and has a mass range of 15–25 kDa. Azide-labeling of this protein was performed via an in vitro enzymatic transfer strategy using UDP-Gal-NAz and the Gal-Tl enzyme. Consistent with the observations in Figure 4, treatment of unlabeled or azido-labeled α-crystallin with either DIFO-based probes (1 or 2) produced strong immuno-reactive bands, indicating the formation of adducts between DIFO-based reagents and α-crystallin, regardless of the presence of an azide-functional group, as shown in Figure 5 (Fig. 5A, lanes 5 and 6; Fig. 5B, lanes 4 and 5). In contrast, only azide-labeled a-crystallin treated with either terminal alkyne reagent (7 or 8) or Biotin–Phosphine (9) showed strong immunoreactivity, indicating the high specificity of these reagents for azide-functionality (Fig. 5A, lanes 2 and 3 for reaction with 7; Fig. 5B, lanes 2 and 3 for reaction with 8; Fig. 5B, lanes 7 and 8 for reaction with 9).
Figure 5.
Ponceau S.-stained nitrocellulose membranes (upper) and corresponding immunoblots (lower) of α-crystallin reacted with various azide-reactive reagents: α-crystallin is treated with TAMRA-tagged reagents (Cu+/terminal alkyne 7 or DIFO-based reagent 1) (A) and with Biotin-tagged reagents (Cu+/terminal alkyne 8, DIFO-based reagent 2, or triaryl Phosphine reagent 9) (B). Immuoblots were analyzed using rabbit anti-TAMRA antibody followed by IRDye®800CW-conjugated goat anti-rabbit antibody for A and IRDye800 CW-conjugated Streptavidin for B.
According to a previous report from Agard et al., non-specific covalent adduct formation between proteins lacking azido-moieties and cyclooctyne-based probes can occur when the reaction temperature is elevated to boiling.25 Our experiments were performed either at, or lower than, room temperature, ruling out elevated temperature as the cause for the non-specific adduct formation. Additionally, noncovalent interactions due to Biotin and TAMRA do not appear to play a major role in adduct formation since reactions of unlabeled proteins with the other Biotin- or TAMRA–containing reagents (7, 8, and 9) gave little to no reactivity.
2.3. Sulfhydryls are highly reactive toward DIFO-based reagents
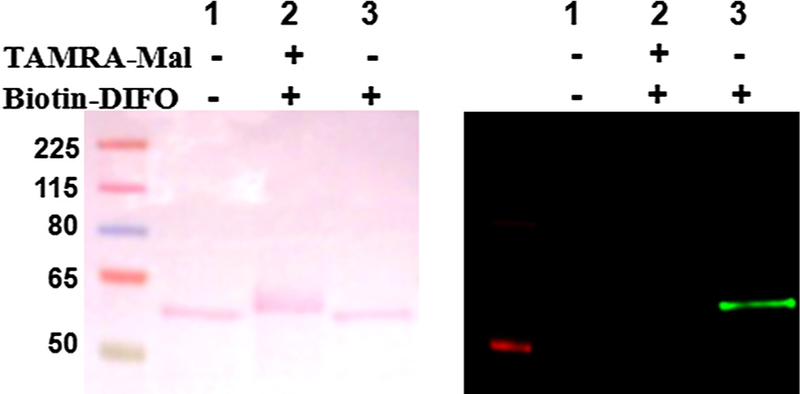
A study by van Geel et al. reported the azide-independent reactions of three strained cyclooctynes, dibenzocyclooctyne (DIBO), azadibenzocyclooctyne (DIBAC), and bicyclo[6.1.0]nonyne (BCN) and suggested that a free-thiol moiety could serve as a reactive functional group for strained cyclooctynes.34 To determine if the reactivity we observed was due to a free thiol, we used purified recombinant human nuclear pore protein Nup62, which is heavily modified with O-GlcNAc and contains three cysteine residues with at least one cysteine that is not part of a disulfide-bridge. We confirmed the presence of a free thiol moiety in Nup62 using the fluorophore-labeled sulfhydryl alkylating tetramethylrhodamine–5-maleimide (TAMRA–Mal, 10; data not shown). Exposing unlabeled Nup62 to Biotin–DIFO (2) produced a distinct signal (Fig. 6, lane 3). Prior treatment of Nup62 with TAMRA–Mal to block free cysteine’s thiol(s) resulted in an almost complete abolishment of the adduct formation between the DIFO-based probe and Nup62 (Fig. 6, lane 2).
Figure 6.
Ponceau S.-stained nitrocellulose membranes (left) and corresponding immunoblots (right) of Nup62: lane 1 is Nup62 as a negative control, lane 2 is Nup62 sequentially treated with TAMRA–Mal (10) and then with Biotin–DIFO (2), and lane 3 is Nup62 treated with only DIFO–Biotin. Western blots were analyzed using IRDye800 CW-conjugated Streptavidin.
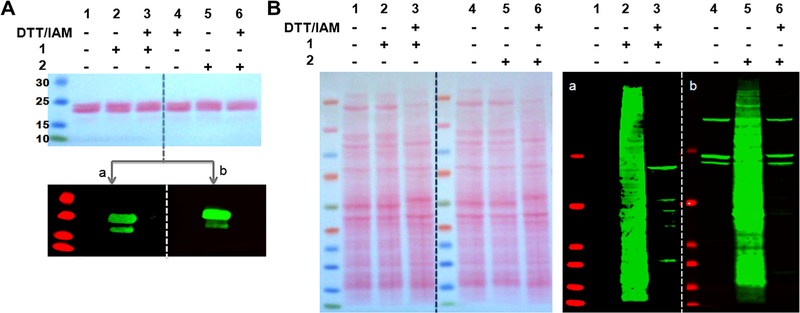
Likewise, prior treatment of α-crystallin with 1,4-dithio-d-threitol (DTT) to generate free cysteines and blocking the moieties using another sulfhydryl-alkylating reagent, 2-iodoacetamide (IAM), followed by its exposure to DIFO-based probes completely abolished the adduct formation (Fig. 7A, lanes 2 and 3 for the reaction with TAMRA–DIFO, 1; lanes 5 and 6 for the reaction with Biotin–DIFO, 2).
Figure 7.
Ponceau S.-stained nitrocellulose membrane (upper in A and left in B) and immunoblots (bottom in A and right in B) of α-crystallin (A) and C. elegans lysate (B) treated with DTT/IAM, followed by reactions with and without either DIFO–TAMRA (1) or DIFO–Biotin (2). Each immunoblot was cut into two pieces and probed with rabbit anti-TAMRA antibody and IRDye800CW-conjugated goat anti-rabbit antibody (a) or with IRDye800CW-Streptavidin (b). Three strong signals in the negative control of C. elegans (Fig. 6B, lane 4) represent non-specific interactions with IRDye800CW-Streptavidin.
Additionally, prior treatment of unlabeled C. elegans lysate with DTT/IAM resulted in a marked reduction of non-specific reactions arising from the formation of adducts between unlabeled lysate and DIFO-based probes (Fig. 7B, lane 2 and lane 3 for the reaction with TAMRA–DIFO and lane 5 and lane 6 for the reaction with Biotin–DIFO). In the C. elegans negative control, three strong bands appear representing non-specific binding between IR-dye conjugated to Streptavidin and C. elegans proteins supporting that these three bands are not due to covalent labeling reactions of the reagent. Unlike the pure proteins, DTT/IAM-treatment of unlabeled C. elegans extract did not completely remove the background signals shown in the lane 3 of Figure 7B. Since azide-lacking C. elegans lysates treated with TAMRA–Alkyne reagent (7) produced the same background signal (Fig. 3A, lane 2) as negative controls of C. elegans (Fig. 3A, lane 1 and lane 4), we can exclude the possibility that higher background signals shown in DTT/IAM-treated unlabeled C. elegans exposed to TAMRA–DIFO are simply due to non-covalent TAMRA–dye interaction with unlabeled proteins. Faint signals were also seen in the unlabeled lysate that was sequentially reacted with DTT/IAM and Biotin–DIFO.
It is interesting that not all sulfhydryl-containing molecules are reactive toward DIFO-based reagents: when sulfhydryl-containing small molecules such as DTT, l-cysteine, and glutathione were reacted with DIFO-based reagents at the same molar ratio at room temperature for 1 h, corresponding adduct products were not detected at all. However, when 2-mercaptoethanol was reacted with Biotin–DIFO, a small amount of the adduct product was observed by MS at m/z 709.3 and the percentage of adduct product increased as the concentration of 2-mercaptoethanol was increased (data not shown). Thus, we speculate that sulfhydryls with increased reactivity due to their surrounding context within a protein could potentially add to DIFO-based probes. These results are different from those obtained with other cyclooctynes.34 Previously, DIBO, DIBAC, and BCN were reported to produce their adducts with a nonpeptidyl thiol such as DTT, 2-mercaptoethanol, and glutathione.34 It implies that DIFO-based probes are less prone to thiolyne addition than three other cyclooctynes.
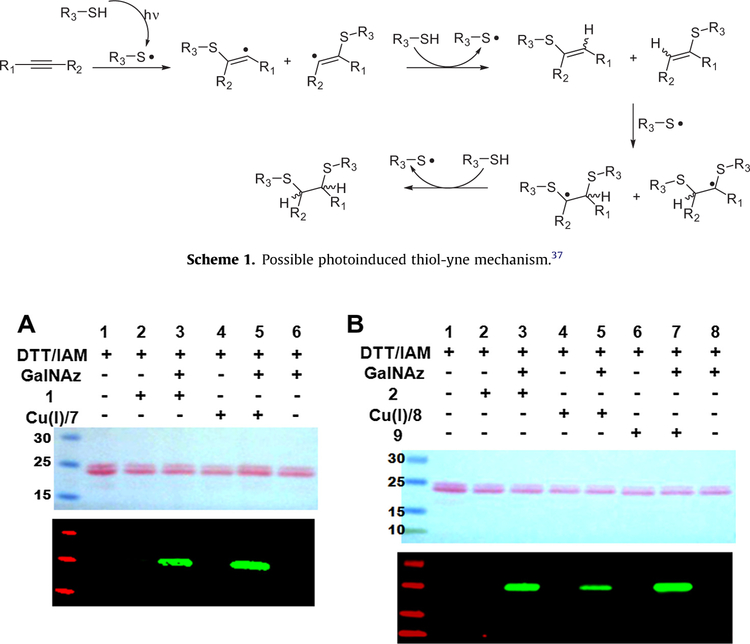
Although the addition of thiols to alkynes was described as early as the 1930s,35 photoinitiated radical addition of thiols to various alkynes including cyclooctyne was only recently investigated.36–38 Thiol-yne coupling is generally known to proceed by a radical mechanism as shown in Scheme 1. Briefly, a thiyl radical generated under the photoinduced reaction condition adds to a triple bond to give a vinyl sulfide radical which abstracts hydrogen from the thiol to afford the thiyl radical and a regioisomeric mixture of vinyl sulfide. Subsequently, the regenerated thiyl radical adds to the double bond of the vinyl sulfide eventually yielding a dithioether through a dithioether radical. The second addition of thiol to the vinyl sulfide is known to depend on the initial yne structure.37 In addition to the general photoinitiated thiol-yne reaction, spontaneous thiol addition to cyclooctyne under the ‘dark’ reaction condition (absence of light or a photoinitiator) was also reported by Fairbanks et al.37 Although, the precise mechanism of the ‘dark’ reaction of thiols with cyclooctyne has not been elucidated, this reaction is thought to proceed through a radical mechanism probably initiated by some reactive oxygen species introduced by the diffusion of molecular oxygen into the reaction mixture.37 This may be of particular importance in the cytoplasm of the cell and cell extracts where free radical generation is thought to occur under tightly controlled conditions.
2.4. Free thiol-blocking is necessary prior to reaction with DIFO-based probes
Exhaustive alkylation of all sulfhydryl groups of unlabeled α-crystallin was conducted by protein treatment with DTT to first break all disulfide bonds followed by subsequent treatment with IAM. Treated α-crystallin was then completely inactive toward DIFO-based probes as demonstrated before (Fig. 8A and B, lane 2). However, azide-labeling of DTT/IAM and Gal-T/UDP-GalNAz-treated α-crystallin followed by the reaction with DIFO-based reagents restored a strong signal which represents the adduct formation between the azide-labeled protein and the DIFO-based probe (Fig. 8A, lane 3 and B, lane 3). In the parallel experiment, copper-catalyzed ‘click’ reactions using TAMRA–Alkyne (7) and Biotin–Alkyne (8) and Staudinger ligation reaction using Biotin–Phosphine (9) were carried out as positive controls for the chemoselective cycloaddition of the azide-labeled α-crystallin (Fig. 8A, lane5 and B, lane 5 and lane 7).
Figure 8.
Ponceau S-stained nitrocellulose membranes (upper) and immunoblots (lower) of sequentially reacted α-crystallin: α-crystallin is first subjected to reduction of disulfide bond and then alkylation of all sulfhydryls generated. Next, the protein was subjected to GalNAz labeling followed by reaction with TAMRA-tagged reagents (Cu+/terminal alkyne 7 or TAMRA–DIFO 1) (A) or with Biotin-tagged reagents (Cu+/terminal alkyne 8 or Biotin–DIFO 2, or Biotin–Phosphine 9) (B). Immunoblots were analyzed using rabbit anti-TAMRA antibody and IRDye800CW-conjugated goat anti-rabbit antibody for A and IRDye800CW-Streptavidin for B.
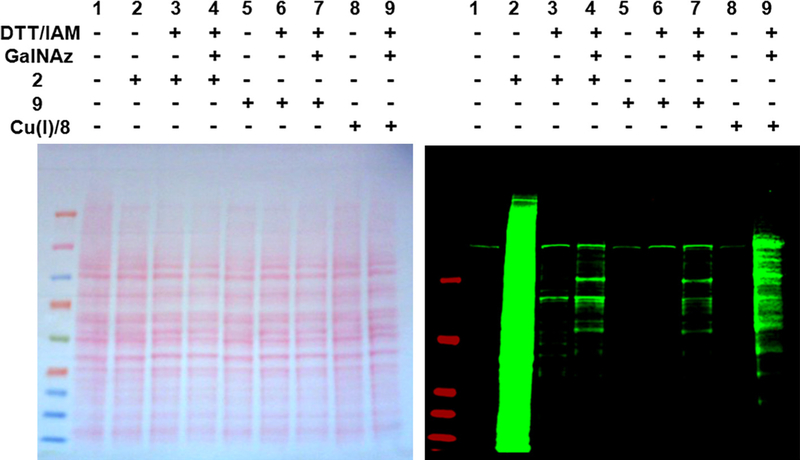
Treatment of GalNAz-labeled HeLa cell extract with DTT/IAM followed by exposure to the Biotin–DIFO (2) generated many signals as shown in Fig. 9 (lane 4). The reaction of GalNAz-labeled HeLa cell extracts with Biotin–Alkyne 8 in combination with Cu(I) (Fig. 9, lane 9) and the reaction with Biotin–Phosphine 9 were performed (Fig. 9, lane 7) as positive controls. The two background signals depicted in the immunoblot in Fig. 9 (lanes 1, 5, and 8) are due to the non-specific interaction with IR-dye conjugated Streptavidin. Although DTT/IAM-treatment affected the blocking of reactive functionalities toward DIFO-based probes, there are still DIFO-reactive species remaining (Fig. 9, lane 3). It is currently unclear whether these remaining weak signals are caused from failing to complete sulfhydryls’ alkylation or from the existence of another functional group that can also react with DIFO-based probes. However, based on the reaction results obtained with the pure proteins such as Nup62 and α-crystallin where DTT/IAM-treatment of the proteins completely blocked the non-specific, covalent additions with DIFO-based probes, it is more likely that the background observed in Figure 9, lane 3 is from the incomplete alkylation of free thiols.
Figure 9.
Ponceau S-stained nitrocellulose membrane (left) and immunoblot (right) of unlabeled HeLa cell extracts sequentially treated with or without DTT/IAM, GalNAz labeling reagents, and azide-reactive reagent 2, 8, or 9. Immunoblot was analyzed using IRDye800CW-Streptavidin.
More recently, thiol-yne reactions using more biologically-relevant biomolecules were explored by Conte et al.34,39 Three previous reports34,37,39 examining thiol-yne reactions, expressed concern about orthogonal reactions with the reactive cyclooctyne probes. Our in vitro experimental results also show that not only azide-moieties but also reactive sulfhydryl groups in protein lysates can rapidly react with highly reactive strained alkynes such as DIFO-based reagents to give adduct products. The observation that two DIFO-based reagents examined here do not form detectable adducts with nonpeptidyl thiols suggests they are less prone to thiol-yne addition than other cyclooctynes such as DIBO, DIBAC, and BCN. In addition, in order to use DIFO-based reagents to tag azide-labeled biomolecules in biological contexts, the bioconjugate thiol moieties can be blocked first to diminish the potential unwanted side-reaction with DIFO-based reagents. Although this step is probably less important for cell surface applications where free thiols are less likely to exist,6 this thiol-blocking step is essential when applying DIFO-based labeling to intracellular glycans.
2.5. Conclusion
Difluorinated cyclooctynes (DIFO) conjugated with either a rhodamine-based fluorophore (TAMRA) or a biotin tag are useful dipolarphiles developed for rapid Strain-Promoted Azide Alkyne Cycloaddition (SPAAC) protein labeling and have been widely applied in several outstanding studies. Nevertheless, the bioorthogonality of these probes is not absolute and minimizing non-specific labeling (background labeling) is still a major concern in these chemical biology approaches. Previous reports illustrate the potential non-specific labeling with strained cyclooctynes such as DIBO, DIBAC, and BCN.34 Here, we extend these studies suggesting that two widely used DIFO-based probes also show the same concern for bioorthogonality. In this study, we demonstrated that reactive thiols present in biomolecules rapidly add to DIFO-based probes to form the covalent adducts and thus potentially limit the bioorthogonality of the strain-promoted copper-free azide–alkyne cycloadditions. However, we demonstrate that thorough blocking of the free-thiols using exhaustive alkylation prior to copper-free ‘click’ reactions restores the bioorthogonal nature of these reactions and this may be particularly important when examining intracellular glycans such as the widespread O-GlcNAc modification.
3. Experimental
3.1. Material
All chemicals were purchased from Sigma–Aldrich (St. Louis, MO, USA) unless otherwise indicated. All solutions were prepared using ultrapure deionized water. 5-(and 6)-carboxytetramethylrhodamine, succinimidyl ester (3, Invitrogen, Carlsbad, CA, USA), 3-azidopropylamine, d-Biotin, 4,7,10-trioxa-1,13-tridecanediamine, 2-iodoacetamide (Pierce Biotechnology, Rockford, IL, USA), DTT, β-mercaptoethanol, and l-cysteine were used as received. UDP-GalNAz, Gal-T1 enzyme, and α-crystallin are contained in the Click-iT™ O-GlcNAc Enzymatic Labeling Kit (Invitrogen, Carlsbad, CA, USA). TAMRA–Alkyne and Biotin–Alkyne with copper(II)-salt and reducing agent are contained in the Click-iT™ Protein Analysis Detection Kits (Invitrogen). DMEM, FBS, PBS, NuPAGE pre-cast gels, 4 × NuPAGE®LDS sample buffer, and rabbit anti-TAMRA antibody were purchased from Invitrogen (Carlsbad, CA, USA). Complete, mini, EDTA-free protease inhibitor cocktail tablets were purchased from Roche Applied Science (Indianapolis, IN, USA). M-PER and T-PER lysis buffers, and BCA protein assay reagent were purchased from Thermo Scientific Pierce (Rockford, IL, USA). IRDye®800CW-conjugated goat anti-rabbit antibody was purchased from LI-COR Biosciences (Lincoln, NE, USA). Recombinant human nuclear pore glycoprotein, p62 (Nup62) was purchased from Bioclone (San Diego, CA, USA). Synthetic intermediates, and target compounds (TAMRA–DIFO and Biotin–DIFO) were confirmed by high-resolution ESI MS analysis. High-resolution mass measurements were performed on a Micromass/Waters LCT Premier Electrospray Time of Flight (TOF) mass spectrometer coupled with a Waters HPLC system. The immunoblots were imaged according to the manufacturer’s instructions using the Odyssey Infrared Imaging System (LI-COR Biosciences).
3.2. Syntheses of DIFO-based reagents
Difluorinated cyclooctyne (DIFO) core structure [2-(2,2-difluoro-cyclooct-3-yn-1-yl)acetic acid] was prepared according to the procedure reported by Codelli et al.27
3.2.1. 5-(3-Azidopropylcarbamoyl)-2-(6-dimethylamino-3-dimethylimino-3H-xanthen-9-yl) benzoate (4)
A solution of 2-(6-dimethylamino-3-dimethyliminio-3H-xan-then-9-yl)-5(and 6)-[(2,5-dioxopyrrolidin-1-yloxy)carbonyl]benzoate 3 (9.8 mg, 0.019 mmol, Invitrogen) and triethylamine (10.1 mg, 0.0998 mmol) in methanol (1 mL) was treated with 3-azidopropylamine (11.6 mg, 0.116 mmol) and then stirred for 16 h at ambient temperature. Methanol was removed by evaporation and the residue was separated by flash column chromatography eluting with CH2Cl2:MeOH = 10:1–4:1. The product fractions were collected, concentrated, and lyophilized to afford the compound 4 (5.7 mg, 60%). HRMS (ESI): [M+H]+ calculated for C28H29N6O4: m/z = 513.2250; found m/z = 513.2230.
3.2.2. 5-{3-[2-(2,2-Difluorocyclooct-3-yn-1-yl)acetamido]propylcarbamoyl}−2-(6-dimethyl-amino-3-dimethyliminio-3H-xanthen-9-yl)benzoate (1)
To a solution of 4 (2.0 mg, 0.0039 mmol) in ethanol (4 mL) was added 5% Pd on carbon (2 mg) and hydrogenated for 24 h. Reaction progress was monitored by MS. When the azide compound 4 disappeared and amine compound existed alone, the catalyst was removed by filtration. The filtrate was concentrated and lyophilized. The amine compound was dissolved in DMF (1 mL) and 2-(2,2-difluorocyclooct-3-yn-1-yl)acetic acid (1.5 mg, 0.0074 mmol), HATU (3.2 mg, 0.0082 mmol), and diisopropyl-ethylamine (5.0 mg, 0.039 mmol) were added and stirred for 2 h at ambient temperature. The reaction mixture was lyophilized to remove DMF. The residue was separated by flash column chromatography eluting with CH2Cl2:MeOH = 10:1–4:1. The product fractions were collected, concentrated, and lyophilized to afford the titled compound 1 (2.5 mg, 96%). HRMS (ESI): [M+H]+ calculated for C38H41N4O5F2: m/z = 671.3045; found m/z = 671.3054.
3.2.3. N-(3-{2-[2-(3-Aminopropoxy)ethoxy]ethoxy}propyl)-5-[(4S)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide (6)
To a solution of biotin 5 (109.5 mg, 0.444 mmol) in 1,4-dioxane (3 mL) were added 4,7,10-trioxa-1,13-tridecanediamine (102 mg, 0.449 mmol), EDC (132 mg, 0.689 mmol), HOBT (95 mg, 0.703 mmol), and triethylamine (145 mg, 1.43 mmol). The reaction mixture was stirred for 16 h at ambient temperature and then 1,4-dioxane was removed. The residue was separated by flash column chromatography eluting with CH2Cl2:MeOH:AcOH = 100:10:2–30:10:2. The product fractions were collected, and concentrated to afford the titled compound 6 (56.1 mg, 28%). HRMS (ESI): [M+H]+ calculated for C20H39N4O5S: m/z = 447.2641; found m/z = 447.2659.
3.2.4. N-[1-(2,2-Difluorocyclooct-3-yn-1-yl)-2-oxo-7,10,13-trioxa-3-azahexadecan-16-yl]-5-[(4S)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide (2)
To a solution of 6 (4.4 mg, 0.00985 mmol) in DMF (0.5 mL) was added 2-(2,2-difluorocyclooct-3-yn-1-yl)acetic acid (1.8 mg, 0.00890 mmol), HATU (4.7 mg, 0.0120 mmol), and diisopropylethylamine (2.0 mg, 0.0155 mmol). The reaction mixture was stirred for 16 h at room temperature and then lyophilized to remove DMF. The residue was separated by flash column chromatography eluting with CH2Cl2:MeOH = 20:1–5:1. The product fractions were collected, and concentrated to afford the titled compound 2 (2.2 mg, 35%). HRMS (ESI): [M+H]+ calculated for C30H49N4O6F2S: m/z = 631.3341; found m/z = 631.3359.
3.3. Biological assays
3.3.1. Preparation of azide-labeled C. elegans and HeLa cell lysates
20–50 μL of worms was lysed in 100 μL T-PER lysis buffer with protease inhibitors (complete, mini, EDTA-free protease inhibitor cocktail and PMSF). Worm lysate was sonicated in an ice bath in Misonic sonicator for 10 min followed by centrifugation at 20,000 × g for 10 min at 4 °C. Supernatant was transferred into a fresh tube and protein concentration was determined by BCA assay and samples were stored at −80 °C until use. 500 lg of wild-type C. elegans proteins prepared in T-PER lysis buffer was buffer-exchanged into 100 μL of 20 mM HEPES buffer containing 0.1% SDS using a 10 K-cutoff Amicon Ultra-0.5 Centrifugal filter device (EMD Millipore, Billerica, MA, USA) according to the manufacturer’s protocol. Then, 500 μg of C. elegans proteins in the 20 mM HEPES buffer was subject to N-azidoacetylgalactosamine (Gal-NAz)-labeling using a Click-iT™ O-GlcNAc Enzymatic Labeling Kit. Briefly, 500 μg of C. elegans proteins in 100 μL of 20 mM HEPES buffer containing 0.1% SDS was placed into a 1.5 mL microcentrifuge tube and 122.5 μL of DPEC-treated water, 200 μL of labeling buffer, 27.5 μL of 100 mM MnCl2, 25 μL of 0.5 mM of UDP-GalNAz, and 20 μL of Gal-T1 were added. The reaction mixture was incubated at 4 °C overnight. Finally, the buffer was exchanged into a buffer appropriate for the following chemoselective reaction using a 10 K-cutoff Amicon Ultra-0.5 Centrifugal filter device. Metabolic azide-labeling of HeLa cells was achieved by culturing the cells in a 10 mL DMEM supplemented with 10% FBS in the presence of 100 μM of peracetylated N-azidoacetylglucosamine (Ac4GlcNAz) in a humidified 5% CO2 incubator for 2 days. Cultured cells were washed with PBS at pH 7.2, transferred into a 15 mL-felcon tube and then centrifuged at 2500 × g for 10 min at 4 °C. The pellets were lysed in 0.6 mL of M-PER protein extraction buffer containing an EDTA-free protease inhibitor cocktail, according to the manufacturer’s protocol. The supernatant was obtained after centrifugation at 20,000 × g for 15 min at 4 °C and stored in aliquots at −80 °C until use. GalNAz-labeling of unlabeled HeLa cell extracts was performed using a Click-iT™ O-GlcNAc Enzymatic Labeling Kit as described above.
3.3.2. Preparation of azide-labeled α-crystallin and Nup62
GalNAz-labeling of the pure proteins such as α-crystallin and Nup62 was achieved by in vitro enzymatic labeling strategy using a Click-iT™ O-GlcNAc Enzymatic Labeling Kit. Briefly, 50 μg of either α-crystallin or Nup62 was reconstituted in 10 μL of 20 mM HEPES buffer containing 0.1% SDS. Sequentially, 8.2 μL of DPEC-treated water, 16 μL of labeling buffer, 2.2 μL of 100 mM MnCl2, 2 μL of 0.5 mM of UDP-GalNAz, and 1.6 μL of Gal-T1 were added and the reaction mixture was incubated at 4 °C overnight. Then, reaction buffer was exchanged into an appropriate buffer for copper-catalyzed or copper-free ‘click’ reaction or Staudinger ligation using a 10 K-cutoff Amicon Ultra-0.5 Centrifugal filter device.
3.3.3. Reactions of azide-labeled and unlabeled proteins with various azide-specific reagents
Before carrying out the reaction of cell lysates or pure proteins with various azide-specific reagents, buffer-exchange into an appropriate buffer was performed. For copper-catalyzed or copper-free ‘click’ reactions, 50 mM Tris–HCl, pH 7.4 buffer containing 0.1% SDS was used as reaction buffer. For Staudinger ligation reaction, PBS was used.
3.3.4. Reactions of azide-labeled and unlabeled proteins with DIFO-based probes (copper-free ‘click’ reaction)
To 10 μg of GalNAz-labeled and unlabeled (without any incorporated azides) α-crystallin in 39.8 μL of 50 mM Tris–HCl buffer containing 0.1% SDS was added 0.2 μL of 10 mM TAMRA–DIFO (or Biotin–DIFO). Reaction mixtures were agitated at room temperature for 20 min. In the parallel experiments, same reaction mixtures were carried out in the dark at room temperature for 20 min. The reactions were quenched by removing remaining small molecules including excess DIFO-TAMRA reagent using a 10 K-cutoff Amicon Ultra-0.5 Centrifugal filter device. Therefore, the whole reaction mixture was transferred into a 10 K-cutoff Amicon filter device and centrifuged at 14,000 × g at 4 °C for 15 min. Washes and buffer exchange were performed by placing 250 μL of 50 mM Tris–HCl buffer containing 0.1% SDS into the centrifuged filter device and by re-centrifuging at 14,000 × g at 4 °C for 30 min. This buffer exchange process was repeated three times. In order to recover the residues remaining in the filter device, 30 μL of 50 mM Tris–HCl buffer containing 0.1% SDS was added into the filter to give a total volume of 30 μL and the filter device was placed upside down in a clean microcentrifuge tube and was centrifuged at 1000 × g at 4 °C for 2 min. The reaction results were evaluated by gel electrophoresis followed by Western blot analysis. Samples were prepared for gel electrophoresis by adding 5 μL of distilled water and 15 μL of 4 NuPAGE® LDS sample buffer to give a total volume of 60 μL. After incubation at 80 °C for 5 min, 20 μL of each reaction sample was loaded onto a SDS–PAGE gel (10% or 4–12% NuPAGE® Bis–Tris gel) and run with 1 × NuPAGE®MOPS SDS Running Buffer for 50 min at 200 V. Then, the proteins were electrophoretically transferred onto the nitrocellulose membrane for Western blot analysis. The membrane was probed either with rabbit anti-TAMRA antibody at 1:1000 dilution, followed by IRDye®800CW-conjugated goat anti-rabbit secondary antibody at 1:10,000 dilution for the evaluation of reactions with TAMRA-tagged reagent, or with IRDye®800CW-Streptavidin at 1:5000 dilution for the evaluation of reactions with Biotin-tagged reagent. The blot was imaged according to the manufacturer’s instructions by using the Odyssey Infrared Imaging System.
To 150 μg of GalNAz-labeled and unlabeled C. elegans lysates (or HeLa cell lysates) in 30 μL of 50 mM Tris–HCl buffer containing 0.1% SDS was added 1 μL of 10 mM TAMRA–DIFO (or Biotin–DIFO). Reaction mixtures were agitated at room temperature for 20 min. In the parallel experiments, same reaction mixtures were carried out in the dark at room temperature for 20 min. Then, reaction buffer was exchanged into 80 μL of 50 mM Tris–HCl buffer containing 0.1% SDS to remove unreacted TAMRA–DIFO (or Biotin–DIFO), using a 10 K-cutoff Amicon Ultra-0.5 Centrifugal filter device. For gel electrophoresis and subsequent Western blot analysis, 10 μL of distilled water and 30 μL of 4 × NuPAGE® LDS sample buffer were added to each reacted sample. Protocols for gel electrophoresis and Western blot analysis were identical as described above.
3.3.5. Reactions of azide-labeled and unlabeled proteins with terminal alkyne and copper catalyst (copper-catalyzed ‘click’ reaction)
Reactions of azide-labeled and unlabeled α-crystallin with TAMRA–Alkyne (7) (or Biotin–Alkyne, 8) were carried out using a Click-iT™ Protein Analysis Detection Kit according to the manufacturer’s protocol. Briefly, to 10 μg of GalNAz-labeled and unlabeled α-crystallin in 2 μL of 50 mM Tris–HCl buffer containing 0.1% SDS were added 10 μL of distilled water and 20 μL of 2 Click-iT™ reaction buffer containing TAMRA–Alkyne (or Biotin–Alkyne). Subsequently, 2 μL of 40 mM CuSO4 solution and 2 μL of Click-iT™ reaction buffer additive 1 were added. After 2–3 min, 4 μL of Click-iT™ reaction buffer additive 2 was added and the reaction tube was covered with foil to minimize light exposure and agitated at room temperature for 20 min. Reaction was quenched by removing excess DIFO-based reagent by performing buffer-exchange into a total volume of 40 μL of 50 mM Tris–HCl buffer containing 0.1% SDS, using a 10 K-cutoff Amicon Ultra-0.5 Centrifugal filter device. The reaction results were determined by gel electrophoresis followed by Western blot analysis. Reactions of azide-labeled and unlabeled C. elegans lysates (or HeLa cell lysates) with TAMRA–Alkyne (7) and Biotin–Alkyne (8) were carried out using a Click-iT™ Protein Analysis Detection Kit, according to the manufacturer’s protocol. Briefly, 150 μg of GalNAz-labeled and unlabeled C. elegans lysates (or HeLa cell lysates) in 30 μL of 50 mM Tris–HCl buffer containing 0.1% SDS were added 50 μL of 2 Click-iT™ reaction buffer containing TAMRA–Alkyne reagent (or Biotin–Alkyne). Subsequently, 5 μL of 40 mM CuSO4 solution, and 5 μL of Click-iT™ reaction buffer additive 1 were added. After 2–3 min, 10 μL of Click-iT™ reaction buffer additive 2 was added. The reaction tube was covered with foil to minimize light exposure and agitated at room temperature for 20 min. Reaction was quenched by removing excess DIFO-based reagent by performing buffer-exchange into a total 80 μL volume of 50 mM Tris–HCl buffer containing 0.1% SDS, using a 10 K-cutoff Amicon Ultra-0.5 Centrifugal filter device. The reaction results were determined by gel electrophoresis followed by Western blot analysis.
3.3.6. Reactions of azide-labeled and unlabeled proteins with Biotin–Phosphine (Staudinger ligation)
Staudinger ligation reactions of azide-labeled and unlabeled α-crystallin were performed using Biotin–Phosphine (9). To 2 μL of 0.1% SDS, 50 mM Tris–HCl buffer containing either 10 μg of Gal-NAz-labeled or unlabeled α-crystallin was added 36 μL of PBS buffer. Subsequently, 2 μL of 10 mM Biotin–Phosphine (9) was added and the reaction mixture was agitated at 37 °C for 90 min. Reaction was quenched by removing excess Biotin–Phosphine reagent by performing buffer-exchange into a total 40 μL volume of 50 mM Tris–HCl buffer containing 0.1% SDS, using a 10 K-cutoff Amicon Ultra-0.5 Centrifugal filter device. The reaction results were determined by gel electrophoresis followed by Western blot analysis. Staudinger ligation reactions of azide-labeled and unlabeled HeLa cell lysates were performed using Biotin–Phosphine (9). To 20 μL of 0.1% SDS, 50 mM Tris–HCl buffer containing either 100 μg of GalNAz-labeled or unlabeled HeLa cell lysate were added 52 μL of PBS and 8 μL of 10 mM Biotin–Phospine (9). The reaction mixture was agitated at 37 °C for 90 min. Then, buffer-exchange into a total 60 μL volume of 50 mM Tris–HCl buffer containing 0.1% SDS was performed to remove the excess, unreacted reagent using a 10 K-cutoff Amicon Ultra-0.5 Centrifugal filter device. The reaction results were determined by gel electrophoresis followed by Western blot analysis.
3.3.7. Treatment of unlabeled Nup62 with TAMRA–Mal and reaction of prior TAMRA–Mal-treated Nup62
1 μg of Nup62 in 49 μL of PBS was treated with and without 1 μL of 10 mM tetramethylrhodamine–5-maleimide (TAMRA–Mal) solution at room temperature in the dark for 1 h. The reaction was quenched by removing excess, unreacted TAMRA–Mal reagent by performing buffer-exchange into a total volume of 49 μL of 50 mM Tris–HCl buffer containing 0.1% SDS, using a 10 K-cutoff Amicon Ultra-0.5 Centrifugal filter device. Then, resulting Nup62 was reacted with 1 μL of 10 mM DIFO–biotin solution at room temperature for 30 min. Reaction results were determined by gel electrophoresis and Western blot analysis as described before.
3.3.8. Treatment of GalNAz-labeled or unlabeled α-crystallin (or Nup62) with DTT and IAM
To 20 μg of either GalNAz-labeled or unlabeled α-crystallin reconstituted in 99 μL of 0.1% SDS, 50 mM Tris–HCl buffer was added 1 μL of 1 M DTT. The reaction was heated at 80 °C on a heating block for 15 min and then cooled at room temperature for 15 min. Immediately before use, sulfhydryl alkylating agent was prepared by dissolving 14.4 mg of iodoacetamide in 0.1% SDS, 50 mM Tris–HCl, pH 7.5, to give a total volume of 1.4 mL to make 55.6 mM iodoacetamide solution. Then, 100 μL of the 55.6 mM iodoacetamide (IAM) was added to the DTT-treated α-crystallin and the reaction mixture was protected from light and agitated at room temperature for 40 min. The reaction was quenched by removing excess, unreacted IAM reagent by performing buffer-exchange into a total volume of 40 μL of 50 mM Tris–HCl buffer containing 0.1% SDS, using a 10 K-cutoff Amicon Ultra-0.5 Centrifugal filter device. GalNAz-labeling of prior DTT/IAM-treated α-crystallin (or Nup62) was carried out using a Click-iT™ O-GlcNAc Enzymatic Labeling Kit as described previously. GalNAz-labeled or unlabeled α-crystallin (or Nup62) prior treated with DTT/IAM was then reacted with various azide-reactive reagents as described above. Reaction results were determined by gel electrophoresis and Western blot analysis as described before.
3.3.9. Treatment of unlabeled C. elegans lysate (or HeLa cell lysate) with DTT and IAM
To 200 μg of either GalNAz-labeled or unlabeled C. elegans lysate (or HeLa cell lysate) constituted in a 198 μL volume of 0.1% SDS, 50 mM Tris–HCl buffer was added 2 μL of 1 M DTT. Reaction was heated at 80 °C on a heating block for 20 min and then cooled at room temperature for 15 min. Immediately before use, sulfhydryl alkylating agent was prepared by dissolving 14.4 mg of iodoacetamide in 0.1% SDS, 50 mM Tris–HCl, pH 7.5, to give a total volume of 1.4 mL to make 55.6 mM iodoacetamide solution. 200 μL of the 55.6 mM iodoacetamide (IAM) was added to the DTT-treated lysate and the reaction mixture was protected from light and agitated for 40 min at room temperature. Reaction was quenched by removing excess, unreacted IAM reagent by performing buffer-exchange into a total volume of 80 μL of 50 mM Tris–HCl buffer containing 0.1% SDS, using a 10 K-cutoff Amicon Ultra-0.5 Centrifugal filter device. GalNAz-labeling of prior DTT/IAM-treated cell lysate was carried out using a Click-iT™ O-GlcNAc Enzymatic Labeling Kit (Invitrogen) as described previously. Gal-NAz-labeled or unlabeled C. elegans lysate (or HeLa cell lysate) prior treated with DTT/IAM was then reacted with various azide-reactive reagents as described above. Reaction results were determined by gel electrophoresis and Western blot analysis as described before.
3.3.10. Evaluation of adduct formation between Biotin–DIFO and sulfhydryl-containing small molecules
To 298 μL of PBS, were added 1 μL of 10 mM Biotin–DIFO (2) and 1 μL of 10 mM thiol-containing small molecule, namely DTT, l-cysteine, β-mercaptoethanol, or glutathione. Reaction mixture was stirred at room temperature for 1 h and analyzed by ESI Mass spectrometric analysis.
Acknowledgements
This work was supported by the NIDDK intramural funds (NIH) and the National Research Foundation of Korea (2011–0027257).
References
- 1.Mahal LK; Yarema KJ; Bertozzi CR Science 1997, 276, 1125–1128. [DOI] [PubMed] [Google Scholar]
- 2.Hang BC; Bertozzi CR J. Am. Chem. Soc 2001, 123, 1242–1243. [DOI] [PubMed] [Google Scholar]
- 3.Saxon E; Bertozzi CR Science 2000, 287, 2007–2010. [DOI] [PubMed] [Google Scholar]
- 4.Vocadlo DJ; Hang HC; Kim EJ; Hanover JA; Bertozzi CR Proc. Natl. Acad. Sci. U.S.A 2003, 100, 9116–9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rabuka D; Hubbard SC; Laughlin ST; Argade SP; Bertozzi CR J. Am. Chem. Soc 2006, 128, 12078–12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sampathkumar SG; Jones MB; Yarema KJ Nat. Protoc 2006, 1, 1840–1851. [DOI] [PubMed] [Google Scholar]
- 7.Wang Z; Udeshi ND; O’Malley M; Shabanowitz J; Hunt DF; Hart GW Mol. Cell. Proteomics 2010, 9, 153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Laughlin ST; Agard NJ; Baskin JM; Carrico IS; Chang PV; Ganguli AS; Hangauer MJ; Lo A; Prescher JA; Bertozzi CR Methods Enzymol 2006, 415, 230–250. [DOI] [PubMed] [Google Scholar]
- 9.Blackman ML; Royzen M; Fox JM J. Am. Chem. Soc 2008, 2, 2930–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prescher JA; Dube DH; Bertozzi CR Nature 2004, 130, 13518–13519. [DOI] [PubMed] [Google Scholar]
- 11.Weisbrod SH; Baccaro A; Marx A Methods Mol. Biol 2011, 751, 195–207. [DOI] [PubMed] [Google Scholar]
- 12.Tornøe CW; Christensen C; Meldal MJ Org. Chem 2002, 67, 3057–3064. [DOI] [PubMed] [Google Scholar]
- 13.Rostovtsev VV; Green LG; Fokin VV; Sharpless KB Angew. Chem., Int. Ed 2002, 41, 2596–2599. [DOI] [PubMed] [Google Scholar]
- 14.Kolb HC; Finn MG; Sharpless KB Angew. Chem., Int. Ed 2001, 40, 2004–2021. [DOI] [PubMed] [Google Scholar]
- 15.Wang Q; Chan TR; Hilgraf R; Fokin VV; Sharpless KB; Finn MG J. Am. Chem. Soc 2003, 125, 3192–3193. [DOI] [PubMed] [Google Scholar]
- 16.Link AJ; Tirrell DA J. Am. Chem. Soc 2003, 125, 11164–11165. [DOI] [PubMed] [Google Scholar]
- 17.Baskin JM; Bertozzi CR QSAR Comb. Sci 2007, 26, 1211–1219. [Google Scholar]
- 18.Best MD Biochemistry 2009, 48, 6571–6584. [DOI] [PubMed] [Google Scholar]
- 19.Requena JR; Chao C; Levine RL; Stadtman ER Proc. Natl. Acad. Sci. U.S.A 2001, 98, 69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burrows CJ; Muller JG Chem. Rev 1998, 98, 1109–1152. [DOI] [PubMed] [Google Scholar]
- 21.Sigman DS; Mazumder A; Perrin DM Chem. Rev 1993, 93, 2295–2316. [Google Scholar]
- 22.Agard NJ; Prescher JA; Bertozzi CR J. Am. Chem. Soc 2004, 126, 15046–15047. [DOI] [PubMed] [Google Scholar]
- 23.Poloukhtine AA; Mbua NE; Wolfert MA; Boons G-J; Popik VV J. Am. Chem. Soc 2009, 131, 15769–15776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Becer CR; Hoogenboom R; Schubert US Angew. Chem., Int. Ed 2009, 48, 4900–4908. [DOI] [PubMed] [Google Scholar]
- 25.Agard NJ; Baskin JM; Prescher JA; Bertozzi CR ACS Chem. Biol 2006, 1, 644–648. [DOI] [PubMed] [Google Scholar]
- 26.Baskin JM; Prescher JA; Laughlin ST; Agard NJ; Chang PV; Miller IA; Codelli JA; Bertozzi CR Proc. Nat. Acad. Sci. U.S.A 2007, 104, 16793–16799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Codelli JA; Baskin JM; Agard NJ; Bertozzi CR J. Am. Chem. Soc 2008, 130, 11486–11493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jewett JC; Sletten EM; Bertozzi CR J. Am. Chem. Soc 2010, 132, 3688–3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beatty KE; Fisk JD; Smart BP; Lu YY; Szychowski J; Hangauer MJ; Baskin JM; Bertozzi CR; Tirrell DA ChemBioChem 2010, 11, 2092–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laughlin ST; Baskin JM; Amacher SL; Bertozzi CR Science 2008, 320, 664–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dieterich DC; Hodas JJ; Gouzer G; Shadrin IY; Ngo JT; Triller A; Tirrell DA; Schuman EM Nat. Neurosci 2010, 13, 897–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bernardin A; Cazet AI; Guyon L; Delannoy P; Vinet F; Bonnaffe D; Texier I Bioconjugate Chem 2010, 21, 583–588. [DOI] [PubMed] [Google Scholar]
- 33.Ess DH; Jones GO; Houk KN Org. Lett 2008, 10, 1633–1636. [DOI] [PubMed] [Google Scholar]
- 34.van Geel R; Pruijn GJM; van Delft FL; Boelens WC Bioconjugate Chem 2012, 23, 392–398. [DOI] [PubMed] [Google Scholar]
- 35.Finzi C Gazz. Chim. Ital 1930, 60, 798–811. [Google Scholar]
- 36.Fairbanks BD; Scott TF; Kloxin CJ; Anseth KS; Bowman CN Macromolecules 2009, 42, 211–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fairbanks BD; Sims EA; Anseth KS; Bowman CN Macromolecules 2010, 43, 4113–4119. [Google Scholar]
- 38.Minozzi M; Monesi A; Nanni D; Spagnolo P; Marchetti N; Massi AJ Org. Chem 2011, 76, 450–459. [DOI] [PubMed] [Google Scholar]
- 39.Conte ML; Staderini S; Marra A; Sanchez-Navarro M; Davis BG; Dondoni A Chem. Commun 2011, 11086–11088. [DOI] [PubMed] [Google Scholar]