Abstract
As obligate intracellular parasites, virus reproduction requires host cell functions. Despite variations in genome size and configuration, nucleic acid composition, and their repertoire of encoded functions, all viruses remain unconditionally dependent on the protein synthesis machinery resident within their cellular hosts to translate viral messenger RNAs (mRNAs). A complex signaling network responsive to physiological stress, including infection, regulates host translation factors and ribosome availability. Furthermore, access to the translation apparatus is patrolled by powerful host immune defenses programmed to restrict viral invaders. Here, we review the tactics and mechanisms used by viruses to appropriate control over host ribosomes, subvert host defenses, and dominate the infected cell translational landscape. These not only define aspects of infection biology paramount for virus reproduction, but continue to drive fundamental discoveries into how cellular protein synthesis is controlled in health and disease.
A defining attribute of virus infection biology is its absolute reliance on the host translation machinery to produce the polypeptides needed for virus reproduction. This feature cannot be overstated, as virus replication and spread depend on conscripting host ribosomes to translate viral messenger RNAs (mRNAs). Failure to engage ribosomes would have dire consequences for virus reproduction and evolution. To ensure translation of their mRNAs, virus-encoded functions dominate cell signaling pathways that control the host protein synthesis machinery. Commandeering these regulatory circuits preserves the functionality of components that recruit ribosomes to viral mRNAs. Although many host cell intrinsic immune defenses target translation factors to incapacitate the protein synthesis apparatus of the infected cell, viral factors have evolved to limit host antiviral responses. Exploiting virus model systems continues to reveal fundamental parameters governing how mRNA translation is controlled in infected and uninfected cells. Here, we review molecular interactions between select plant and animal viruses and their hosts that regulate protein synthesis in infected cells, and recent developments in the field are highlighted.
A PRIMER ON VIROLOGY AND VIRUS REPRODUCTION STRATEGIES
Viruses are vastly diverse, and although a comprehensive discussion of virus biology is not our purpose, an overview of select principles is warranted. Viruses are obligate intracellular parasites with varied lifestyles. Whereas acute infections are often cytolytic and destroy the cell, persistent infections can be tolerated. Some viruses establish permanent infections, in which viruses continually replicate or, alternatively, remain latent within cells and reproduce only periodically. These distinct reproduction strategies differentially impact the host translation machinery and translated mRNA landscape. Viruses replicate within and may remodel the cytoplasm or nucleus. Not all infections cause clinical disease, and infection outcome varies among organisms or cell types and is dependent on immune status. Finally, viruses have diverse genome structures comprised of single- or double-stranded (ds)DNA or RNA, and RNA virus genomes can be composed of single or multiple nucleic acid segments. Viral genome size varies from less than 10 Kb for small RNA viruses to greater than 200 Kb for large human DNA viruses and megabase genomes of DNA viruses that infect Acanthamoeba (Schultz et al. 2017). Notwithstanding genome size, viruses are under selective pressure to optimize coding capacity, and tactics like polyprotein processing, where proteases generate multiple proteins from one open reading frame (ORF; e.g., in picornaviruses and flaviviruses) and recoding (e.g., frameshifting as in retrovirus Gag-Pol) are frequently observed (Jan et al. 2016; Atkins et al. 2016).
Genome structure largely informs mRNA biogenesis and mRNA features impact translation. Single-strand RNA virus genomes having an identical polarity to mRNA, termed plus (+)-strand RNA viruses, function as mRNA and are directly translated on infection. RNA virus genomes of opposite polarity, designated minus (−)-strand RNA viruses, and dsRNA virus genomes require an RNA-dependent RNA polymerase to produce mRNA. Discrete structural elements within 5′ and 3′ untranslated regions (UTRs) support RNA virus genome replication and mRNA translation. Viral mRNA 5′ ends may be uncapped, protein-linked, modified by viral capping enzymes to contain a 5′ methyl-7-GTP (m7GTP) cap, or derived from host mRNA 5′ terminal fragments that are naturally capped (Decroly et al. 2012). Uncapped viral mRNAs deploy specialized genome elements to recruit 40S ribosomal subunits that support translation even when canonical cap-dependent translation is impaired (Kwan and Thompson 2018). Polyadenylated 3′ ends are generally template-encoded for RNA viruses or replaced by an element that recruits a viral 3′ terminal binding protein (Poon et al. 1999; Deo et al. 2002; Kempf and Barton 2015). After converting (+)-strand RNA genomes into dsDNA using reverse transcriptase, retroviruses produce mRNA using the host RNA polymerase II transcriptional machinery. Whereas most DNA viruses replicate within the nucleus and use host enzymes for mRNA biogenesis, others, including poxviruses and asfarviruses, replicate in the cytoplasm, encoding their own transcription, capping, and 3′ end processing machinery (Van Etten et al. 2010).
STRESS RESPONSES AND INFECTION BIOLOGY
Controlling gene expression by mRNA translation enables swift responses to environmental and physiological insults, including infection. Indeed, many host proteins and mechanisms regulating protein synthesis in response to stress in uninfected cells (reviewed by Wek 2018) play significant roles during infection. Likewise, features of translational control in virus-infected cells resemble and perhaps were co-opted from host cell stress responses.
Host Defenses and Antiviral Immunity
While hijacking ribosomes enables virus protein production, it also is a vulnerability exploited by the host. Significantly, many innate host defenses limit mRNA access to ribosomes by selectively or globally impairing the translation machinery. The struggle between viruses and their hosts to control ribosomes occupies the front lines in the innate immune response to virus infection.
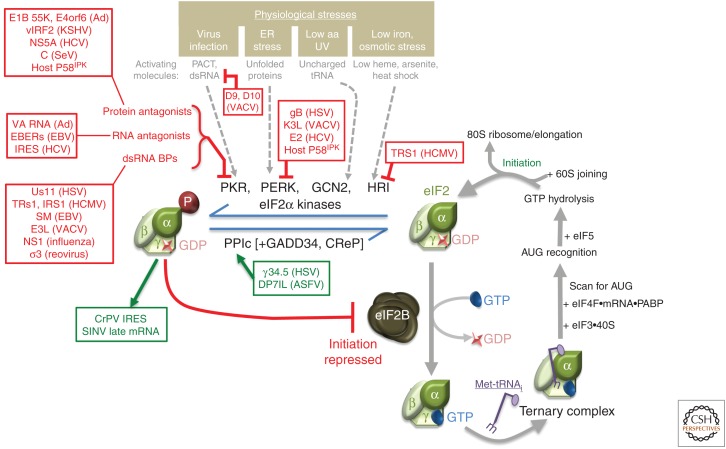
By detecting conserved viral nucleic acid features, including uncapped single-stranded RNA, dsRNA, or cytoplasmic DNA, host sentinel pattern recognition receptors trigger type I interferon (IFN) production (Chiang and Gack 2017). The resulting IFN-induced host proteins establish an “antiviral state” that is not permissive for virus reproduction. Many IFN-induced proteins impact translation and ribosome recruitment to mRNA (Diamond and Farzan 2013; Schoggins 2014). Central among these are PKR, a eukaryotic initiation factor (eIF)2α kinase, and oligoadenylate synthetase (OAS), which are both activated by dsRNA. Importantly, dsRNA accumulates in cells infected with RNA or DNA viruses. Although some viruses have dsRNA genomes, dsRNA is an obligatory intermediate in RNA virus genome replication and can be present as secondary structures in single-strand RNA. dsRNA also accumulates in cells infected with DNA viruses, which produce mRNAs from opposing strands of the viral genome. Upon activation by dsRNA, PKR phosphorylates the α subunit of eIF2 (Fig. 1). This inactivates eIF2, a GTP-regulated, three-subunit complex that loads 40S ribosomal subunits with initiator methionyl-transfer RNA (Met-tRNAiMet) (Merrick and Pavitt 2018; Wek 2018). Normally, GTP hydrolysis is stimulated by eIF5 following start codon recognition enabling 60S subunit joining. Subsequent eIF2•GDP recycling to the GTP-bound form requires guanine nucleoside exchange factor (GEF) eIF2B (Merrick and Pavitt 2018; Wek 2018). However, phosphorylated eIF2 binds tightly to eIF2B, inhibiting its GEF activity (Fig. 1). Because eIF2B is limiting, small changes in phosphorylated eIF2α have a large impact on protein synthesis and globally inhibit initiation.
Figure 1.
Targeting stress-responsive host defenses by viral functions controls eukaryotic initiation factor (eIF)2α phosphorylation and translation initiation. Composed of three subunits (α,β,γ), eIF2 is a GTP-binding translation initiation factor that loads eIF3-bound 40S ribosomal subunits with methionyl-transfer RNA (Met-tRNAi) (right panel). Subsequent mRNA recruitment, AUG recognition, and eIF5-stimulated GTP hydrolysis is followed by 60S subunit joining and translation initiation by the 80S ribosome. Recycling inactive eIF2•GDP to the active GTP-bound form requires the GEF eIF2B. Phosphorylation of eIF2α on S51 blocks initiation by binding to and inhibiting eIF2B, preventing GDP-GTP exchange. Four eIF2 kinases, each of which is activated by specific molecules that accumulate in response to a discrete physiological stress, and the protein phosphatase 1 catalytic subunit (PP1c), partnered with an inducible (GADD34) or constitutively active (CreP) regulatory subunit, control eIF2α phosphorylation. Viral functions that activate (green), respond to (green), or repress (red) the indicated host effectors are shown. aa, amino acid.
Whereas OAS is similarly activated by dsRNA, it interferes with protein synthesis via a different mechanism. OAS synthesizes short oligoadenylate (OA) polymers with a 2′–5′ linkage that stimulate RNase L, an endogenous ribonuclease whose activation is dependent on OA (Sadler and Williams 2008; Han et al. 2014). Active RNase L degrades ribosomal RNA (rRNA) and mRNA, inactivating both the translation machinery and its templates. RNase L cleavage products, which contain 2′–3′ cyclic phosphorylated termini, also stimulate inflammasome activation (Chakrabarti et al. 2015).
Viral Tactics to Counter Host Defenses
Viruses have acquired countermeasures that neutralize host dsRNA-activated defenses or allow their replication despite them. Coronavirus and rotavirus-encoded phosphodiesterases attack OA, limiting its accumulation and RNase L activation (Zhang et al. 2013). In addition, a picornavirus-encoded RNA and protein inhibit RNase L (Townsend et al. 2008; Sorgeloos et al. 2013), and Rift Valley fever virus (RVFV) triggers PKR degradation (Mudhasani et al. 2016). dsRNA-binding proteins encoded by a variety of viruses shield dsRNA from host sensors, foiling PKR and OAS activation (Fig. 1), and some physically interact with PKR to prevent its activation (Jan et al. 2016). Surprisingly, dsRNA accumulation and dsRNA-responsive immune effectors are regulated by the host Xrn1 mRNA exoribonuclease (Burgess and Mohr 2015). By accelerating RNA decay, viral enzymes like poxvirus decapping enzymes or coronavirus nsp15 endonuclease, which presumably produce Xrn1 substrates, limit dsRNA accumulation (Burgess and Mohr 2015; Liu et al. 2015; Deng et al. 2017; Kindler et al. 2017). Alternatively, an adenovirus (Ad) small RNA, VA RNAI, binds PKR but is not large enough to support PKR dimerization, which is needed for activation (Mathews and Shenk 1991; Launer-Felty et al. 2015).
Broader strategies protect eIF2α from phosphorylation not only by PKR, but by the other stress-activated eIF2α kinases PERK, HRI, and GCN2 (Fig. 1) (Pavio et al. 2003; Berlanga et al. 2006; Won et al. 2012; Vincent et al. 2017). Examples include the poxvirus-encoded eIF2α pseudo-substrate K3L (Sood et al. 2000; Seo et al. 2008), the HCMV TRS1 protein that binds and inhibits PKR and HRI (Hakki et al. 2006; Vincent et al. 2017), and a phosphatase regulatory subunit encoded by herpes simplex virus (HSV)-1 (γ34.5) and African swine fever virus (ASFV) DP71L, which recruits the host protein phosphatase 1α catalytic subunit to eIF2α where it counteracts the activity of eIF2α kinases (Rojas et al. 2015; Barber et al. 2017). Induction of host p58IPK by infection can also limit PKR, PERK, and GCN2 activation (Goodman et al. 2011; Roobol et al. 2015). Multiple functions that act synergistically to restrict eIF2α phosphorylation are encoded by HSV-1 and poxviruses, illustrating the importance of preserving functional eIF2 (Mulvey et al. 2003, 2007; Seo et al. 2008; Rice et al. 2011; Sciortino et al. 2013; Burgess and Mohr 2015; Liu et al. 2015). Mutant viruses deficient in functions that subvert host dsRNA-activated defenses often have an altered host range (Haller et al. 2014; Carpentier et al. 2016; Peng et al. 2016), are hypersensitive to IFNs and in some cases are profoundly attenuated (Mulvey et al. 2004; White and Jacobs 2012; Liu et al. 2015). Activation of the unfolded protein response and PERK by hepatitis C virus (HCV) and vesicular stomatitis virus (VSV), however, may also favor virus replication by accelerating type I IFN receptor degradation to attenuate IFN responses (Liu et al. 2009). Finally, several RNA viruses including cricket paralysis virus (CrPV) and Sindbis virus (SINV) contain cis-elements that obviate the requirement for eIF2 altogether (Wilson et al. 2000; Spahn et al. 2004; Kerr et al. 2016; Sanz et al. 2017). Although in vitro studies suggested a mechanism in which eIF2A or eIF2D supply Met-tRNAiMet, recent work using knockout cell lines shows that these factors are dispensable (Sanz et al. 2017; Gonzales-Almela et al. 2018). The role of RNA structures and contribution of other proteins to eIF2-independent initiation on CrPV and SINV remain to be established.
Stress responses, including those resulting from infection, impact mRNA and translation factor subcellular distribution. Stalled initiation complexes containing messenger ribonucleoproteins (mRNPs), mRNA, 40S subunits, initiation factors, and RNA-binding proteins (RBPs) accumulate in phase-dense, non-membrane-bound aggregates called stress granules (SGs) (Protter and Parker 2016; Ivanov et al. 2018). Although eIF2α phosphorylation often accompanies SG formation, implying a reversible way of regulating initiation, SG accumulation can be inhibited or viral mRNAs released from SGs regardless of eIF2α phosphorylation (Montero et al. 2008; Qin et al. 2011). SG core components can directly interface with viral RNA genomes or mRNAs, repressing their translation (Albornoz et al. 2014). Host cytoplasmic RNA sensors including MDA-5, RIG-I, and PKR can be recruited to SGs in response to eIF2α phosphorylation, suggesting SGs are platforms for viral RNA detection (Reineke and Lloyd 2015; Oh et al. 2016; Sánchez-Aparicio et al. 2017). Indeed, many viruses interfere with SG formation (Emara and Brinton 2007; White et al. 2007; Finnen et al. 2014; Khaperskyy et al. 2014; Dauber et al. 2016; Humoud et al. 2016; Nelson et al. 2016; Rabouw et al. 2016; Amorim et al. 2017; Basu et al. 2017; Choudhury et al. 2017; Khong et al. 2017; Le Sage et al. 2017). In cells infected with Middle East respiratory syndrome (MERS) coronavirus, Kaposi sarcoma–associated herpesvirus (KSHV), or HSV-1, this is achieved in part by preventing PKR activation (Rabouw et al. 2016; Sharma et al. 2017; Burgess and Mohr 2018). Even though SG formation is inhibited by dengue virus (DENV) and Zika virus, host protein synthesis remains suppressed, uncoupling these processes (Roth et al. 2017). Depleting key SG components like G3BP reduces Zika virus replication, however, indicating that host SG components may be repurposed to benefit virus reproduction without SG formation (Hou et al. 2017). Whereas SGs can be dynamic platforms for staging host antiviral responses (Tsai and Lloyd 2014; McCormick and Khaperskyy 2017), SG formation is stimulated by rabies virus (Nikolic et al. 2016) and enhances respiratory syncytial virus replication (Lindquist et al. 2010). The rationale underlying this seemingly opposite strategy is unknown.
Remodeling Host mRNA Translation in Infected Cells
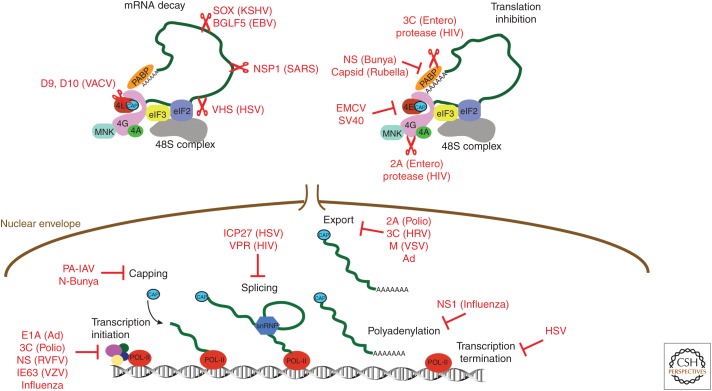
By interfering with host translation, which overwhelmingly is cap-dependent, viruses antagonize host defenses. In its extreme form, termed “host shut-off,” virus infection impairs ongoing host protein synthesis, limiting production of host defense molecules and allowing viral mRNAs to better compete for limiting factors (Mohr and Sonenberg 2012; Mohr 2016). Different mechanisms account for host shut-off (Fig. 2). Picornaviruses suppress host protein synthesis by inhibiting cap-dependent translation and often target eIF4F, a multisubunit initiation factor comprised of the cap-binding protein eIF4E, the eIF4G scaffold, and the DEAD-box-containing RNA helicase eIF4A, needed to load 40S subunits onto m7GTP-capped mRNAs. Poliovirus 2A proteinase cleaves eIF4G, separating the eIF4E-binding domain from the eIF3-binding region (Gradi et al. 1998), whereas 3C proteinases cleave the poly(A)-binding protein (PABP) (Rivera and Lloyd 2008; Kobayashi et al. 2012a). Cleavage of eIF4G by poliovirus or group A rhinovirus 2A protease is stimulated by eIF4E (Aumayr et al. 2017; Avanzino et al. 2017). During encephalomyocarditis virus (EMCV) infection, hypophosphorylated 4E-BP1 repressor accumulates, inhibiting eIF4E binding to eIF4G and limiting eIF4F assembly (Gingras et al. 1996). Enterovirus 71 (EV 71) instead induces host microRNA miR-141 expression, which reduces eIF4E abundance to suppress host protein synthesis (Ho et al. 2010).
Figure 2.
Viruses use multiple mechanisms to block host messenger RNA (mRNA) translation. Cellular mRNA biogenesis starts in the nucleus where RNA polymerase II generates primary transcripts that undergo several processing steps including 5′ capping, splicing, and polyadenylation before functional mRNAs are produced. Mature mRNAs are exported to the cytoplasm where they are translated. Initiation of cap-dependent mRNA translation typically requires recruitment of the multisubunit eukaryotic translation initiation factor (eIF)4F, which consists of the cap-binding protein eIF4E, the RNA helicase eIF4A, and the adaptor protein eIF4G. Binding of eIF4G to poly(A)-binding protein (PABP) mediates communication between 5′ and 3′ ends. Virus-encoded factors that degrade (scissors) cellular proteins and mRNAs or repress cellular functions are shown.
Remodeling the host mRNA pool available for translation by cytoplasmic ribosomes is another shut-off mechanism. By encoding virus factors that stimulate mRNA turnover, viral mRNAs, which are actively transcribed in acutely infected cells, dominate the mRNA pool. Accelerating mRNA decay also sculpts viral mRNA populations, facilitating temporal viral gene expression transitions (Jan et al. 2016). Viruses that produce mRNAs with m7GTP-capped 5′ termini, like herpesviruses and poxviruses, rely on mRNA decay to liberate host ribosomes without impairing cap-dependent translation. Viral functions subsequently can stimulate cap-dependent translation by subverting cell signaling (Walsh et al. 2005, 2008). To generate substrates for mRNA decay, some viruses encode endoribonucleases (influenza, HSV-1, coronavirus, KSHV) (Kamitani et al. 2009; Read 2013; Abernathy and Glaunsinger 2015; Khaperskyy et al. 2016), whereas others like poxviruses encode decapping enzymes (Parrish and Moss 2007). The HSV-1 endoribonuclease vhs interacts with eIF4B and eIF4H to target mRNA cleavage (Read 2013), the KSHV SOX protein directs host RNA decay components, including the exoribonuclease XRN1, to translating ribosomes (Abernathy and Glaunsinger 2015), and the coronavirus nuclease nsp1 associates with 40S ribosomal subunits (Kamitani et al. 2009). Viruses can also disrupt host mRNA biogenesis and nuclear export to remodel the host mRNA pool (Faria et al. 2005; Rutkowski et al. 2015; Gong et al. 2016).
Not all viruses impair ongoing host translation during their acute reproductive cycle, however. Notably, host protein synthesis proceeds in human cytomegalovirus (HCMV)-infected cells. Activation of the mechanistic target of rapamycin complex 1 (mTORC1) (see Proud 2018) by the viral UL38 protein promotes cap-dependent translation and stimulates synthesis of factors encoded by mRNAs containing a 5′ terminal oligopyrimidine (5′-TOP; see Proud 2018) sequence element including PABP (McKinney et al. 2012, 2014). Higher PABP levels allow HCMV to overcome a host response that increases PABP-interacting protein 2 (Paip2) abundance, which inhibits PABP binding to eIF4G and poly(A) RNA (Fig. 3). Interfering with the HCMV-induced PABP increase results in restriction of virus replication by Paip2, highlighting an unexpected antiviral role for Paip2 in host-cell-intrinsic defenses (McKinney et al. 2013). Avian herpesvirus microRNAs similarly repress Paip2 to stimulate translation of a specific viral mRNA (Tahiri-Alaoui et al. 2014). Additional host genes, like the IFN-induced C19orf66 (RyDEN) may also target PABP to restrict both DNA and RNA viruses, including DENV (Suzuki et al. 2016).
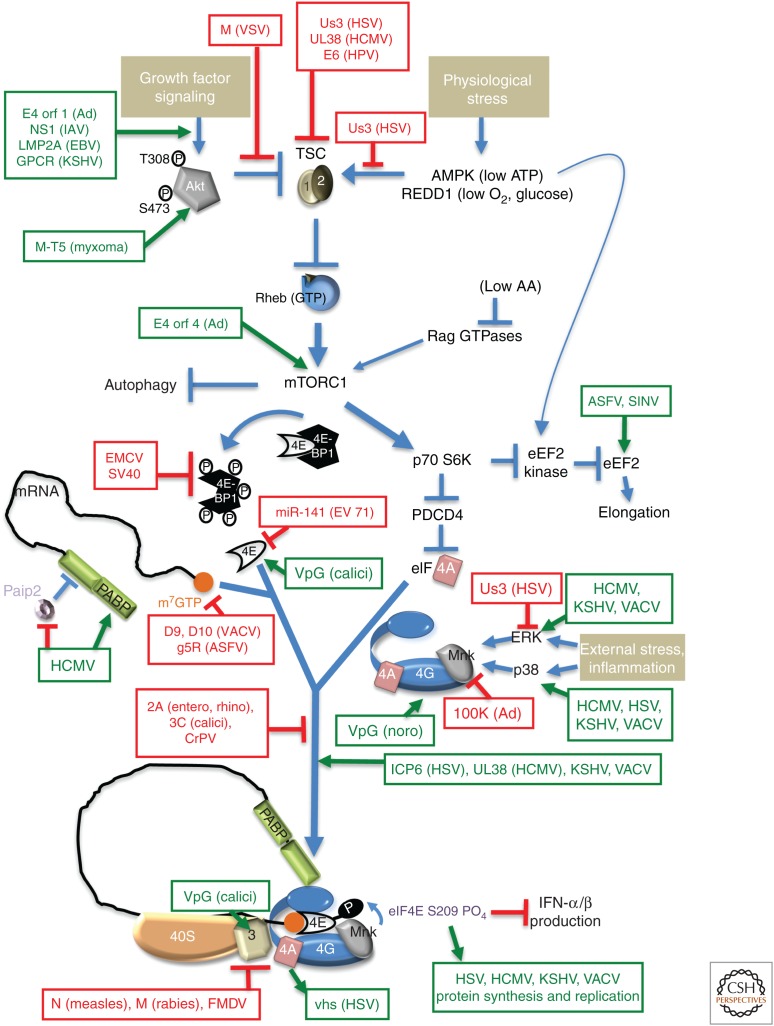
Figure 3.
Subverting mechanistic target of rapamycin complex 1 (mTORC1) signaling to control translation in virus-infected cells. Occupying a nexus of intersecting signaling networks, the cellular ser/thr kinase mTORC1 plays a critical role stimulating anabolic programs, like protein synthesis, and repressing catabolic outcomes like autophagy. Briefly, growth factor–induced Akt phosphorylation (T308, S473) and activation represses TSC1/2, which, in turn, allows Rheb-GTP to activate mTORC1. Nutrient, energy, amino acid (aa), or oxygen insufficiency (physiological stress) all repress mTORC1 through discrete effectors. Signaling through mTORC1 allows swift changes in translational output in response to differing environmental and physiological inputs by controlling initiation and elongation. Initiation is stimulated by phosphorylating and inactivating 4E-BP translational repressor family members (e.g., 4E-BP1), which bind the cap-binding protein eIF4E to prevent its interaction with eIF4G. Through its substrate p70S6K, mTORC1 controls the DEAD-box-containing RNA helicase eIF4A, which together with eukaryotic initiation factor (eIF)4E and eIF4G comprises the multisubunit initiation factor eIF4F. Regulated eIF4F assembly controls cap-dependent mRNA translation as eIF4F recruits 40S subunits to the mRNA capped 5′ end. By repressing eEF2 kinase, mTORC1 stimulates eEF2 and elongation. The impact of mTORC1 activation on translation initiation and elongation is shown, as are viral factors that stimulate (green) and repress (red) the indicated cellular effectors. SV40, simian virus 40; calici, calicivirus; noro, norovirus; entero, enterovirus; rhino, rhinovirus.
Regulating eIF4E phosphorylation impacts selective host mRNA translation and modulates IFN production, influencing host defenses. Coronaviruses, flaviviruses, noroviruses, and many large DNA viruses promote eIF4F assembly, positioning the eIF4G-associated kinase Mnk1 proximal to its substrate eIF4E (Fig. 3) (Mizutani et al. 2004; Walsh and Mohr 2004, 2006; Walsh et al. 2005, 2008; Royall et al. 2015; Roth et al. 2017; Proud 2018). This stimulates eIF4E S209 phosphorylation and translation of IκB mRNA, which encodes an NF-κB inhibitor (Herdy et al. 2012). Besides counteracting NF-κB-dependent IFN production, eIF4E phosphorylation stimulates viral mRNA translation (Walsh and Mohr 2004; Walsh et al. 2008). Significantly, eIF4E phosphorylation is dependent on eIF3 subunit e, showing how loading eIF3-bound 40S marks cap-bound eIF4F by phosphorylation to regulate selective mRNA translation (Walsh and Mohr 2014). Unphosphorylated eIF4E accumulates in cells infected with adenovirus and many RNA viruses (Jan et al. 2016), resulting in reduced translation of IκB-encoding mRNA, NF-κB activation, and IFN production.
Harnessing Stress Responses to Control Viral Persistence
Unlike acute infections, different viral lifestyles necessitate specialized interactions with host cells. Some, like herpesviruses, establish life-long latency where virus reproduction is suppressed in specific cell types. Periodically, latent infections reactivate and reenter the productive growth cycle, which allows virus reproduction and spread to new hosts. Reactivation is influenced by host defenses and physiological stress responses induced by disrupting homeostasis. KSHV stimulates eIF4F assembly on inducible reactivation, and eIF4E phosphorylation is needed for optimal production of the lytic activator RTA (Arias et al. 2009). In contrast, HSV-1 establishes latency in neurons, where inactivation of the translation repressor 4E-BP1 by persistent mTORC1-dependent phosphorylation in response to neurotrophic factor sufficiency promotes latency (Camarena et al. 2010). Conversely, preventing 4E-BP1 phosphorylation stimulates reactivation presumably by restricting translation of eIF4E-responsive mRNAs that are repressed by 4E-BP1 (Kobayashi et al. 2012b). Whereas the mRNA targets remain unknown, monitoring host mRNA translation can gauge homeostasis in long-term, latent infections to support latency or trigger virus reproduction.
VIRAL STRATEGIES TO CAPTURE RIBOSOMES
The absolute dependence of viruses on host ribosomes for protein production demands tactics to ensure their recruitment to viral mRNAs. Shut-off mechanisms suppress host cap-dependent translation and effectively remodel the translation-ready mRNA pool. Ribosome recruitment, however, cannot be left to chance given its vital role in virus reproduction.
Mechanisms of Cap-Independent Translation Used by Viruses
When eIF4F becomes limiting, cap-dependent translation is restricted. Cleavage of eIF4G by poliovirus 2A proteinase shows a severe example of inactivating eIF4F. Uncapped poliovirus mRNA contains a specialized, cis-acting RNA internal ribosome entry site (IRES) in the 5′UTR. In contrast to canonical cap-dependent initiation where 40S ribosomes load onto the mRNA 5′ end, highly structured IRES elements enable ribosome recruitment to specific internal sites within viral RNA (Kwan and Thompson 2018). IRESs were first discovered in poliovirus and EMCV where they allow translation initiation of uncapped, single-stranded (+)-sense RNA viral genomes immediately upon release into the cytoplasm (Jang et al. 1988; Pelletier and Sonenberg 1988). Subsequently, IRESs have been identified in many virus genomes including retroviruses like the human immunodeficiency virus (HIV)-1, which uses an IRES to express the Gag protein late in infection when cap-dependent initiation is impaired (Brasey et al. 2003; Amorim et al. 2014; Carvajal et al. 2016). Some DNA virus genomes like KSHV even contain IRES elements to facilitate translation of polycistronic mRNA (Othman et al. 2014).
Although they share the general capacity to direct initiation when canonical cap-dependent translation mechanisms are suppressed, different IRESs are structurally distinct and show varied functional requirements for eukaryotic initiation factors to load 40S subunits (Table 1) (Jan et al. 2016; Kwan and Thompson 2018). For example, the poliovirus IRES requires the eIF4G carboxy-terminal fragment, and selectively recruits 40S subunits to viral mRNA in a bona fide cap-independent manner (Sweeney et al. 2014). Even though the hepatitis A virus (HAV) RNA genome lacks an m7GTP cap, it contains the only IRES found to require the cap-binding protein eIF4E (Table 1). Stimulation of the HAV IRES by eIF4E is independent of m7GTP cap binding. Instead, eIF4E increases the affinity of eIF4G for several picornavirus IRESs, including HAV, and promotes IRES restructuring by the eIF4A helicase (Avanzino et al. 2017). In contrast, some IRESs, such as those in the CrPV intergenic region (IGR) and HCV, recruit 40S subunits without eIFs (Jan and Sarnow 2002; Pestova and Hellen 2003). Whereas the HCV IRES requires eIF3 and eIF2 to initiate translation (Fraser and Doudna 2007), the CrPV IGR can directly assemble 80S ribosomes and initiate translation in the absence of canonical initiation factors and even the initiator tRNA (Thompson et al. 2001; Jan et al. 2003; Pestova and Hellen 2003; Deniz et al. 2009).
Table 1.
Major types of IRESs
| IRES | Examples | Required initiation factorsa | References |
|---|---|---|---|
| Picornaviruses | |||
| I. Enterovirus | Poliovirus, EV71, bovine enterovirus | eIF1A, eIF2, eIF3, eIF4A, eIF4B, central domain eIF4G, ITAFs (PCBP1/2, PTB) | Martinez-Salas et al. 2015 |
| II. Cardio-/apthovirus | EMCV, FMDV | eIF2, eIF3, eIF4A, eIF4B, central domain eIF4G, ITAF PTB) | Martinez-Salas et al. 2015 |
| III. HAV-like | Hepatitis A virus | eIF2, eIF3, eIF4A, eIF4B, eIF4E, eIF4G, ITAF (PTB, PCBP2) | Martinez-Salas et al. 2015 |
| IV. HCV-like picornaviruses | Simian picornavirus type 9 (SPV9), porcine teschovirus I (PTV-1), Avian encephalomyelitis virus | eIF2, eIF3 | Pisarev et al. 2004; Chard et al. 2006; Martinez-Salas et al. 2015 |
| V. Aichivirus | Aichivirus | eIF2, eIF3, eIF4A, central domain eIF4G, ITAF (DHX29, PTB) | Yu et al. 2011 |
| HCV-like | HCV, CSFV | eIF2, eIF3, binds to 40S directly | Fraser and Doudna 2007 |
| Dicistrovirus intragenic (IGR) | CrPV, DCV, IAPV, PSIV | No initiation factors, binds 40S and assembles 80s directly | Hertz and Thompson 2011b |
| Retrovirus | HIV-1, HTLV-1, SIV, FIV | Central domain eIF4G, hypusine-eIF5A, eIF4A, likely other eIFs, ITAF (hnRNPA1, hRIP, DDX3), viral proteins (Rev) | Ohlmann et al. 2014; Plank et al. 2014; Caceres et al. 2016 |
| KSHV vFLIP | KSHV vFLIP | eIF4A, eIF4G, eIF4E, eIF3, eIF2 | Othman et al. 2014 |
aITAFs: IRES transacting factors.
In an alternative strategy, some RNA viruses like norovirus and calicivirus rely on a 5′ terminal protein covalently linked to the 5′ end of the genome to recruit eIF4E, eIF4G, and/or eIF3 (Daughenbaugh et al. 2003; Leen et al. 2016). Different cis elements mediating long-range RNA interactions have been identified in plant (+)-strand RNA viruses (barley yellow dwarf virus, maize necrotic streak virus, carnation Italian ringspot virus, tomato bushy stunt virus, saguaro cactus virus). These cap-independent translation enhancers (CITEs) reside within 3′UTRs where they recruit 40S ribosomes directly or via eIF4F subunits eIF4E or eIF4G (Simon and Miller 2013). Functional base-pairing subsequently repositions 3′-CITE-loaded ribosomes proximal to 5′UTR structural elements (Nicholson and White 2014). Multiple 3′-CITEs in pea enation mosaic virus recruit both 40S and 60S subunits (Gao et al. 2014, 2017). By adopting alternate conformations induced by viral RNA polymerase binding, CITEs also repress translation to facilitate RNA replication (Le et al. 2017).
Strategies and Mechanisms Used by Viruses that Produce m7GTP-Capped mRNAs
Viral mRNAs with a 5′-m7GTP-cap and 3′-poly(A) tail are indistinguishable from host mRNAs and are typically translated by cap-dependent processes. A number of different strategies that are used to commandeer control over the translational machinery by dominating host cell signaling pathways (reviewed by Proud 2018) have been defined (Fig. 3). Herpesviruses and poxviruses constitutively stimulate mTORC1 to ensure that the 4E-BP1 translational repressor is inactivated (Moorman et al. 2008; Walsh et al. 2008; Arias et al. 2009; Chuluunbaatar et al. 2010). By targeting the host tuberous sclerosis complex (TSC), the HSV1 Us3 ser/thr kinase not only enforces mTORC1 activation by mimicking the cellular kinase Akt (Fig. 3), but also subverts host AMPK-dependent responses to energy insufficiency and supports virus replication during stress (Vink et al. 2017). In addition to inactivating 4E-BP1, these viruses rely on diverse mechanisms to promote assembly of eIF4F, the multisubunit initiation factor that recruits 40S ribosomes to m7GTP-capped mRNA. These include an eIF4G-binding protein encoded by HSV-1 (ICP6) that stimulates eIF4E binding to eIF4G, promotes eIF4E phosphorylation, and stimulates viral mRNA translation (Walsh and Mohr 2006). To capitalize on limiting cytoplasmic PABP availability in HSV-1-infected cells, the viral ICP27 RNA-binding protein may stimulate initiation downstream of cap binding by harnessing PABP and eIF4G in a mechanism similar to that used by the cellular regulator Dazl (Deleted in azoospermia-like) (Smith et al. 2017). HCMV increases the overall abundance of eIFs and PABP. Moreover, preventing the virus-induced rise in PABP levels reduces eIF4F complex assembly and virus growth (McKinney et al. 2012). Instead of manipulating translation factor abundance, vaccinia virus (VACV) and ASFV, which replicate in the cytoplasm, increase the effective local concentration of eIFs by sequestering them within discrete replication compartments (Katsafanas and Moss 2007; Walsh et al. 2008; Castelló et al. 2009; Zaborowska et al. 2012; Desmet et al. 2014).
Following ribosome loading on capped viral mRNAs, AUG start codons are usually identified by 5′UTR scanning (Merrick and Pavitt 2018). However, some viruses use cis elements that allow ribosomes to bypass 5′UTR segments and resume AUG scanning further downstream. This nonlinear ribosome translocation, called ribosome shunting, is used to produce heat shock proteins in uninfected cells subjected to stress and involves base-pairing with 18S rRNA and mRNA cis elements (Yueh and Schneider 2000). In adenovirus-infected cells, ribosome shunting requires a cis-acting tripartite leader RNA sequence in late mRNAs, the virus-encoded 100K protein and eIF4G (Xi et al. 2004). Shunting also occurs in mRNAs encoded by cauliflower mosaic virus, human papillomavirus (HPV), DNA pararetrovirus, and a picorna-like virus (Remm et al. 1999; Pooggin et al. 2012).
Controlling Elongation
In comparison with initiation, our understanding of whether viruses control translation elongation is much less developed. Elongation factors are repurposed to function in RNA replication by many bacterial and plant viruses (Takeshita and Tomita 2012; Li et al. 2013). ASFV and SINV concentrate eEF2 within virus replication compartments (Castelló et al. 2009; Sanz et al. 2009). Whereas mTORC1 activation by many viruses likely inhibits eEF2K (Fig. 3), stimulating elongation, the impact on infection biology remains largely unknown. Finally, expression of prokaryote-like elongation factors encoded by the giant mimivirus changes in response to nutrient availability (Silva et al. 2015), hinting that viral translation factors may supplant or supplement those resident in the host.
TACKLING THE RIBOSOME DIRECTLY
Viruses depend on host ribosomes regardless of whether they use cap-dependent or noncanonical translation initiation mechanisms. Whereas ribosome loading onto mRNAs in eukaryotes is typically reliant on initiation factors, a more direct role for the ribosome itself in regulating translation is emerging.
Dicistrovirus IRESs
Dicistroviruses are (+)-sense, single-stranded RNA viruses with two ORFs that are translated by two different IRESs. The 5′-IRES drives expression of the nonstructural proteins at a low level early in infection, whereas the IGR IRES drives robust structural protein expression late in infection (Garrey et al. 2010). IGR IRESs assemble 80S ribosomes without assistance from eIFs and initiate from the ribosomal A site at a non-AUG codon by priming the ribosome for elongation (Sasaki and Nakashima 2000; Wilson et al. 2000; Jan and Sarnow 2002; Jan et al. 2003; Pestova et al. 2004; Cevallos and Sarnow 2005; Yamamoto et al. 2007; Deniz et al. 2009; Zhang et al. 2016). By dissociating eIF4G from eIF4E to impair host protein synthesis, dicistroviruses subvert host ribosomes to translate viral IRES-containing RNAs (Garrey et al. 2010).
Ribosomal Proteins Control Translation in Virus-Infected Cells
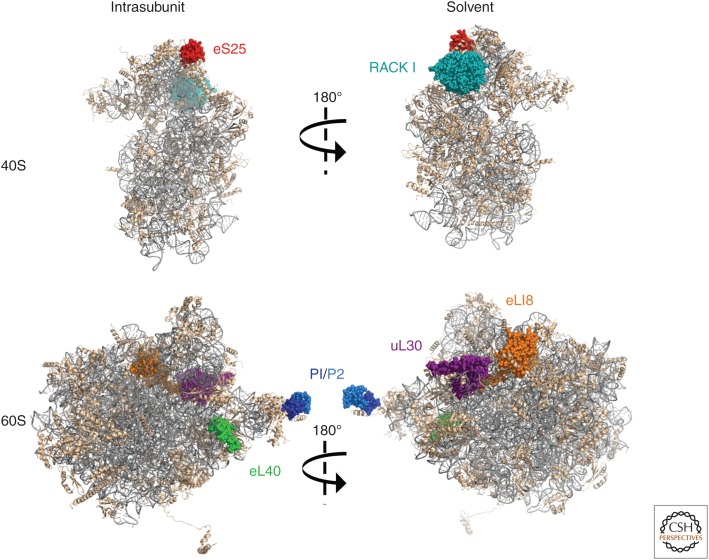
Through discrete functions and modifications of specific protein subunits, ribosomes themselves selectively control viral and host mRNA translation (Fig. 4). Indeed, significant insight into how ribosome composition impacts translation has been gleaned from viral models. Two 40S subunit-associated proteins (eS25, RACK1) dispensable for cap-dependent initiation, but required for IRES activity, have been identified (Landry et al. 2009; Hertz and Thompson 2011a; Majzoub et al. 2014; Olivares et al. 2014; Carvajal et al. 2016). Whereas RACK1 is required by specific IRESs (dicistroviral 5′-IRES, HCV), it is not compulsory for the IGR IRES (Majzoub et al. 2014). In contrast, eS25 is essential for 40S subunit recruitment by numerous IRESs, including the dicistrovirus IGR and picornavirus, flavivirus, retrovirus, and cellular IRESs (Landry et al. 2009; Hertz et al. 2013; Olivares et al. 2014; Carvajal et al. 2016). Unexpectedly, eS25 is also important for ribosomal shunting, suggesting a potential shared mechanism between these two noncanonical initiation strategies (Hertz et al. 2013). Although RACK1 is not essential for cap-dependent translation of cellular mRNAs, poxvirus late mRNAs, which have 5′UTR adenosine tracts, require phosphorylated RACK1 for translation. By expressing a viral kinase that phosphorylates a loop region on RACK1, poxviruses remodel host ribosomes to confer a translational advantage on viral mRNAs that contain adenosine repeats in their 5′UTR (Jha et al. 2017). Consistent with this, adenosine-rich leaders enhance translation in plants where the RACK1 loop region is naturally negatively charged, suggesting that poxviruses have exploited a conserved mechanism to preferentially translate certain capped mRNAs (Jha et al. 2017).
Figure 4.
Ribosomal proteins are required for translation of viral RNAs. Locations of ribosomal proteins on 40S and 60S subunits that are important for viral messenger RNA (mRNA) translation are shown on the ribosome structure derived from PDB entry 4v88. (Figure based on data in Ben-Shem et al. 2011.) Both intrasubunit (left) and solvent surfaces (right) are shown. Ribsomal RNA (rRNA) (gray) and ribosomal proteins (tan) are depicted with the proteins implicated in viral mRNA translation in color: eS25 (red), RACK1 (turquoise), eL40 (green), uL30 (purple), eL18 (orange), and P1/P2 (dark blue/light blue).
A 60S subunit protein (eL40/RPL40) is required for VSV, rabies, and measles virus mRNA translation (Lee et al. 2013). These viruses produce capped, polyadenylated transcripts like cellular mRNAs, yet viral mRNAs are efficiently translated by an unknown mechanism even though host protein synthesis is suppressed. A screen for eL40-dependent mRNAs in yeast revealed that 7% of the yeast mRNAs required eL40, suggesting that VSV has usurped a highly conserved mechanism for translating a subset of cap-dependent mRNAs (Lee et al. 2013).
Dengue virus NS1 protein associates with ribosomal proteins from both the small and large subunits (Cervantes-Salazar et al. 2015). Whereas the DENV RNA genome has a capped 5′ end, it lacks a poly(A) tail (Edgil et al. 2006). Two 60S ribosomal subunit proteins, eL18 and uL30 (previously called RPL18 and RPL7; Ban et al. 2014), were shown to relocalize with NS1 following DENV infection. Whether they play a direct role in translation is not known; however, knockdown of eL18 reduced viral titers by one log (Cervantes-Salazar et al. 2015). P1/P2 (RPLP1/2) are 60S subunit proteins that are part of the ribosome L1 stalk. They are also required for DENV protein expression early in infection (before replication) (Campos et al. 2017). Because the P1/P2 complex only associates with 80S ribosomes (not free subunits) and is important for eEF2 recruitment and GTPase activity, whether it effects elongation rather than initiation needs to be evaluated (Uchiumi et al. 2002; Bautista-Santos and Zinker 2014).
NEW INSIGHTS FROM GENOME-WIDE STRATEGIES
Recent progress using genome-wide, high-throughput technologies has reshaped our understanding of translational control. Besides uncovering new principles and mechanisms, these methodologies provide a global, unbiased view of the infected cell translational landscape.
Genome Annotation
Virus genomes contain densely packed coding information that is often accessed via specialized translation strategies. Ribosome profiling (RP) (sequencing ribosome-protected fragments) allows global translation analysis and direct, experimental annotation of translation events (Ingolia et al. 2009, 2018). It couples classic nuclease footprinting with deep sequencing and indicates ribosome position with single-nucleotide resolution. Besides identifying precise boundaries of translated regions, the three-nucleotide footprint periodicity (reflecting translocation steps) indicates which reading frame is being decoded. Translational start sites can be mapped using conditions that preferentially capture initiating ribosomes (Ingolia et al. 2011; Lee et al. 2012; Stern-Ginossar et al. 2012). These approaches allow transcriptome annotation in areas translated in two overlapping reading frames, a frequent occurrence in virus genomes. Two herpesvirus genomes (HCMV, KSHV) (Stern-Ginossar et al. 2012; Arias et al. 2014), VACV (Yang et al. 2015), and a murine coronavirus (Irigoyen et al. 2016) have been annotated using RP, identifying numerous novel, mostly short ORFs translated upstream of, or within, known virus coding regions. Ribosome pause sites were identified in coronavirus A59, but their significance remains unknown (Irigoyen et al. 2016). RP also accurately measures virus gene expression kinetics throughout infection. During EMCV infection, RP exposed a temporally regulated frameshifting event. Although negligible early in infection, frameshifting efficiency increased to 70% at late time points, suggesting a new mechanism to modulate relative levels of EMCV structural and nonstructural proteins (Napthine et al. 2017).
Host Shut-Off
Application of genome-wide technologies to investigate host shut-off has clarified our understanding of underlying mechanisms and their relative contribution to impairing host translation. Concurrent measurement of translation and mRNA levels throughout influenza A virus (IAV), VACV, and coronavirus infections revealed that genome-wide changes to the host translation landscape are primarily driven by remodeling the mRNA pool (Bercovich-Kinori et al. 2016; Irigoyen et al. 2016; Dai et al. 2017). Although many viruses encode endoribonucleases, their specificity varies. In α (HSV-1) and γ (Epstein–Barr virus [EBV], KSHV) herpesvirus subfamily members, viral and cellular mRNAs are degraded, whereas viral mRNAs are spared by coronavirus and IAV (Rivas et al. 2016). A degenerate sequence motif was identified that enables cleavage of numerous RNA targets by a γ-herpesvirus endoribonuclease (Gaglia et al. 2015). Finally, transcription termination of cellular, but not viral, genes was unexpectedly disrupted by HSV-1, broadening our perception of the range of processes that potentially impact host shut-off (Rutkowski et al. 2015). Furthermore, γ-herpesvirus-induced mRNA degradation reduced host but not viral mRNA transcription, suggesting this cellular feedback mechanism empowers viral gene expression (Abernathy et al. 2015).
Infected Cell Translational Landscape
The notion of host shut-off as a blunt, indiscriminate instrument to halt host gene expression has been revised by genome-wide studies. While host shut-off curtails host antiviral responses, impairing overall cellular protein production could adversely impact virus reproduction. Precisely how the infected cell translational landscape impacts viral propagation is just beginning to be shown. Significantly, cellular mRNAs encoding critical maintenance functions like oxidative phosphorylation are spared from host shut-off and translated in IAV- and VACV-infected cells (Bercovich-Kinori et al. 2016; Dai et al. 2017). Although ongoing production of these proteins requires stable mRNA expression in IAV-infected cells, translation of mRNAs encoding oxidative phosphorylation effector proteins is stimulated by VACV. Continuous oxidative phosphorylation is important for viral propagation in both cases. Discrimination among targets during host shut-off is likely greater than previously anticipated. Similarly, numerous host mRNAs escape KSHV-mediated shut-off and affect viral pathogenesis (Glaunsinger and Ganem 2004). A different tactic was observed in HCMV-infected cells, where host protein synthesis is not globally suppressed, so it was assumed that host protein synthesis proceeded uninterrupted. Using polysome profiling (McKinney et al. 2014) and RP (Tirosh et al. 2015), HCMV was shown to dramatically reshape the infected cell translation landscape. This translational reprogramming is dependent on mTOR activation, and expression of the virus-encoded UL38 mTORC1 activator in uninfected cells in part recapitulates these translational alterations. Importantly, interfering with the virus-induced activation of cellular mRNA translation can limit or enhance HCMV growth (McKinney et al. 2014). These examples show how genome-wide methodologies have revised our understanding of how viruses facilitate selective translation of host mRNAs needed for infection.
Synthetic Genome Recoding
Coding genes are biased in the relative frequencies of codons specifying the same amino acid. In some organisms, codon bias reflects optimization for specific tRNAs and elongation rates (Gingold and Pilpel 2011). Some synonymous codon pairs are used more or less frequently than expected, a phenomenon termed codon pair bias (Yarus and Folley 1985) that may influence translation efficiency (Tats et al. 2008). Indeed, altering codon pair frequencies reduced virus replication without affecting protein sequence (Coleman et al. 2008; Mueller et al. 2010; Martrus et al. 2013; Yang et al. 2013; Le Nouen et al. 2014). As the resulting virus attenuation depends on numerous mutations, reversion is markedly reduced, providing an exciting opportunity for live-attenuated vaccine development (Wimmer et al. 2009). However, in addition to codon pair bias and translation efficiency, other viral genome features, such as suppression of CpG and UpA dinucleotide frequencies, and variations in the propensity to mutate, which generate differential access to protein sequence space can contribute to the reduced replication phenotypes (Lauring et al. 2013; Tulloch et al. 2014; Kunec and Osterrieder 2016). Therefore, precisely how codon usage impacts virus attenuation and the relative contribution of translation repression requires further investigation.
RNA MODIFICATION AND INFECTION BIOLOGY
Although noncoding RNAs contain numerous modifications, only a few have been found in mRNAs. By far the most prevalent internal modified base on mRNAs is a methyl group on the N6 position of adenosine (m6A) (Yue et al. 2015; Peer et al. 2018).
Modulating Viral mRNA Metabolism by m6A
The m6A modification occurs predominantly in the nucleus and is mediated by the enzyme methyltransferase-like 3 (METTL3) together with METTL14, WTAP, KIAA1429, and RBM15/RBM15B. Following nuclear export, m6A is recognized by cytoplasmic “reader” proteins, YTHDF1, YTHDF2, and YTHDF3 (Yue et al. 2015; Peer et al. 2018). m6A modification reportedly influences mRNA splicing, export, translation and stability, all of which may impact virus biology.
Advances in genome-wide m6A mapping on mRNAs and identification of the m6A machinery fueled investigations into how this modification influences virus gene expression. m6A modification was reported to enhance HIV-1 replication by regulating viral mRNA nuclear export or by stimulating viral gene expression (Kennedy et al. 2016; Lichinchi et al. 2016a; Tirumuru et al. 2016). YTHDF m6A-reader proteins bind HIV-1 RNA at m6A sites but their suggested function varies from promoting viral transcript abundance and translation to suppressing reverse transcription. Several m6A-modified regions were identified in flaviviruses, such as HCV, Zika virus, DENV, yellow fever virus, and West Nile virus (Gokhale et al. 2016; Lichinchi et al. 2016b). Because these viruses replicate exclusively within the cytoplasm, the m6A methyltransferase machinery is apparently active in this compartment. Knockdown of the host m6A machinery enhances HCV and Zika virus production. HCV mRNA translation and replication are unaffected, but m6A inhibits HCV RNA packaging into viral particles (Gokhale et al. 2016). Although the mechanism(s) through which m6A act remain unclear, virus model systems could help clarify how m6A controls gene expression.
Discriminating Self versus Nonself mRNAs
Most eukaryotic mRNAs contain a Cap-0 (m7GpppN) structure with a methyl group at the guanine N-7 position. In higher eukaryotes, the mRNA cap is further modified by ribose 2′-O-methylation on the first and sometimes second cap-proximal nucleotides, resulting in Cap-1 (m7GpppNmN) or Cap-2 (m7GpppNmNm). Although 2′-O-methylation does not affect translation directly, it provides a molecular signature of “self.” Hence mRNAs that lack 2′-O-methylation are marked as “nonself,” triggering type I IFN production (Zust et al. 2011), which induces transcription of IFN-stimulated genes (ISGs). Among the most highly upregulated ISGs are those encoding IFIT (IFN-induced proteins with tetratricopeptide repeats) proteins (Fensterl and Sen 2015). Viral 2′-O-methyltransferases encoded by coronavirus, flaviviruses, and VACV prevent recognition by IFIT1 (Daffis et al. 2010; Szretter et al. 2012; Menachery et al. 2014) and cap-proximal structural elements in α virus Cap-0 mRNA restrict IFIT1 action (Hyde et al. 2014). Mechanistically, IFIT1 can sequester eIF4F, inhibiting viral Cap-0 mRNA translation (Habjan et al. 2013). However, N1 methylation (Cap-1) may be insufficient to protect all mRNAs from IFIT1, as the second N2 methylation (Cap-2) and secondary structure may also impact IFIT1 activity (Daugherty et al. 2016; Young et al. 2016; Abbas et al. 2017). Most mammals encode several IFIT proteins with varying affinities for distinct RNA ligands (Hyde and Diamond 2015), suggesting that additional RNA determinants are recognized. Besides competing with eIF4E for cap binding, IFIT1 binds eIF3 subunits, preventing ribosome recruitment, and may inhibit 48S complex formation (Hyde and Diamond 2015). Because IFITs alter translation in uninfected cells (Guo et al. 2000) and increase following infection, IFITs might remodel the infected cell-translation landscape.
CODING CAPACITY
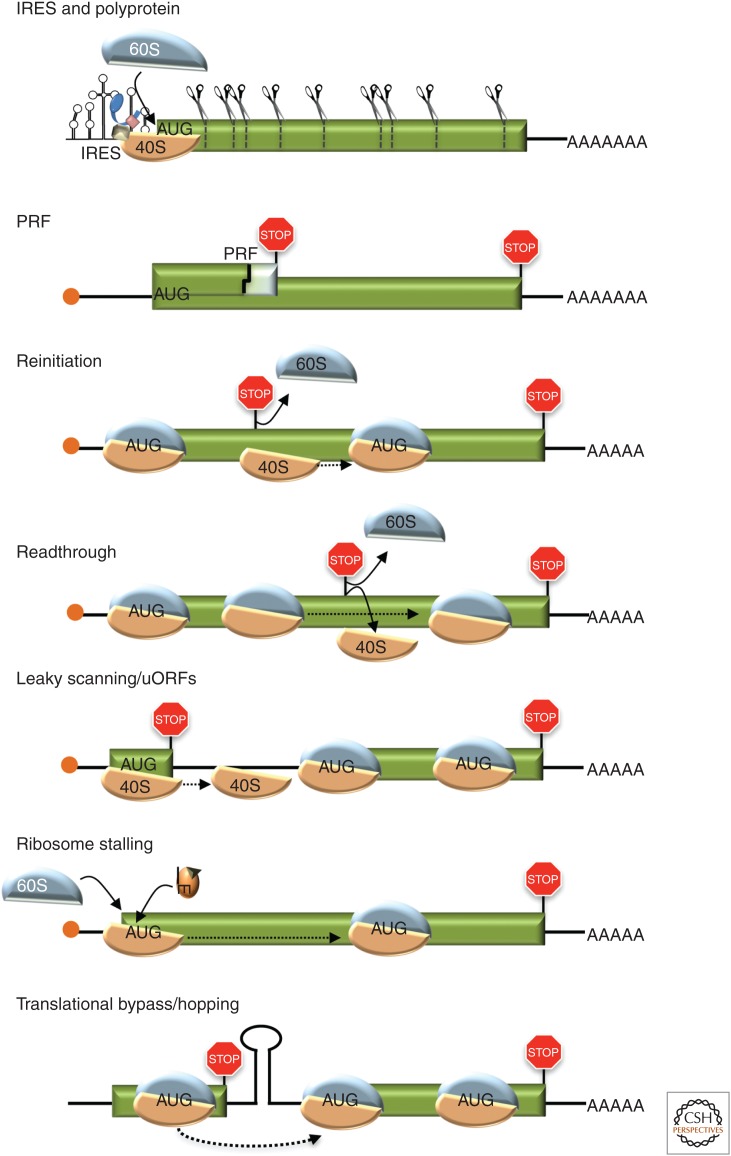
To maximize their genome coding capacity, viruses use multiple strategies, including leaky scanning, polyproteins, reinitiation, translational bypass (hopping), readthrough of stop codons, and programmed ribosomal frameshifting (PRF) (Fig. 5).
Figure 5.
Strategies to maximize viral genome coding capacity. Internal ribosome entry site (IRES)-driven polyprotein production, programmed ribosome frameshifting (PRF), reinitiation, stop-codon readthrough, leaky scanning, ribosome stalling, and translational bypass/ribosome hopping are shown in the cartoon. (See the text for a detailed description.) Stop codons (stop signs), proteolytic cleavage sites (scissors), 5′-cap structure (orange circle), open reading frames (green), ribosome subunits (brown and blue), and initiation factor loaded with transfer RNA (tRNA) are shown.
Coding and Recoding Strategies
PRF is a recoding event that shifts the reading frame of a translating ribosome one or two nucleotides in the + or − direction (Caliskan et al. 2015; Dever et al. 2018). PRF frequencies range from 1% to 80%, but are typically low. A 7-nucleotide slippery site sequence, the spacer region, and structural barriers that stall the ribosome, impact PRF efficiency. Retroviruses generate high levels of Gag and low levels of a Gag-Pol fusion from the same transcript via PRF or by readthrough of a stop codon (Hung et al. 1998; Csibra et al. 2014). Perturbations in this ratio affect viral assembly, RNA packaging and maturation. Mimiviruses use both PRF and readthrough to encode a polypeptide chain release factor homolog (Jeudy et al. 2012). West Nile virus, which encodes a single polyprotein, uses an efficient -1 PRF (∼30% to 70%) to generate additional structural proteins, which increases virus levels in birds and mosquitoes for efficient transmission (Melian et al. 2010, 2014). Dicistroviridae IGR IRESs induce a +1 frameshift during initiation by extending pseudoknot I by one base pair to translate a short protein, ORFx (Ren et al. 2012). In addition, +1 frameshifting in IAV gene segment 3, which encodes the viral RNA polymerase subunit PA, produces the PA-X endonuclease that coordinates host shut-off (Jagger et al. 2012). Rarer -2 PRF events, first reported in porcine reproductive and respiratory syndrome virus (PRRSV, an RNA virus), also occur (Fang et al. 2012). PRRSV -2 frameshifting is responsive to a viral protein transactivator that binds to a C-rich region downstream of the PRF (Li et al. 2014). Efficient +1/−2 PRF within repetitive sequence elements encoded by the DNA viruses EBV and KSHV also generates alternative reading frame versions of their respective latency proteins EBNA1 and LANA1, although their functions remain unknown (Kwun et al. 2014).
Polyproteins, which are proteolytically processed by viral and cellular proteases, are used by many (+)-stranded RNA viruses and allow for a single viral RNA to encode several different proteins. Alternatively, some DNA and RNA viruses express multiple proteins from a polycistronic transcript using a reinitiation mechanism (ribosomal termination-reinitiation or stop–start) (Wise et al. 2011; Kronstad et al. 2013, 2014; Royall and Locker 2016). For example, caliciviruses recruit ribosomes to the viral RNA 5′ end to translate an upstream ORF (uORF). Calicivirus RNAs contain a termination upstream ribosomal binding site (TURBS) that retains the 40S subunit after termination. By base pairing with 18S rRNA, TURBS facilitates efficient translation reinitiation at a nearby AUG or non-AUG codon (Luttermann and Meyers 2014; Royall and Locker 2016). In vitro, reinitiation by a terminated 40S ribosomal subunit requires eIF2, 1, and 1A but not eIF3 (Zinoviev et al. 2015).
To produce a single polypeptide from two discrete ORFs, bacteriophage T4 relies on a translational bypass or ribosome hopping. After translating a glycine codon that precedes a stop codon, the 80S ribosome “hops” over a stretch of sequences and resumes translation at a downstream glycine codon. This hopping mechanism likely requires a compact structure in the gap region to bring together the two glycine codons in the translating ribosome and to prevent the release factor from entering the A site (Todd and Walter 2013).
uORFs
Some DNA and RNA viral polycistronic transcripts contain upstream start codons that can translate short uORFs. The uORFs are a barrier to efficient initiation at a downstream ORF. Viruses modulate downstream ORF expression by leaky scanning, which transpires when the 40S subunit scans past an upstream AUG in a weak or moderate Kozak consensus sequence context before initiating at a downstream AUG (Wise et al. 2011; Kronstad et al. 2013, 2014). For example, members of the triple gene block (TGB) superfamily of movement proteins encoded by many plant viruses, which allow cell-to-cell movement and vascular spread needed to cause disease, are often produced by leaky scanning along a single mRNA transcript that contains overlapping ORFs (Lezzhov et al. 2015; Miras et al. 2017).
Ribosome Stalling
During cap-dependent initiation, the start codon is recognized when the Met-tRNAiMet base-pairs with the start codon. When eIF2 is phosphorylated, levels of ternary complex (eIF2•GTP•Met-tRNAiMet) are reduced, which shuts down protein synthesis globally. Alphaviruses use a stable RNA stem-loop structure located within their coding sequence to stall the ribosome on the initiation codon of the 26S mRNA when ternary complex levels are low and rely on an as-yet-unknown initiation mechanism to avoid a major antiviral defense (Toribio et al. 2016, 2018). Once the 60S subunit joins the stalled ribosome, it is released to translate the viral RNA.
CONCLUDING REMARKS
The categorical requirement for host ribosomes to translate viral mRNAs continues to provide powerful opportunities to investigate how protein synthesis is regulated. Indeed, exploiting virus model systems has defined fundamental features of the cellular protein synthesis machinery, how it is regulated in uninfected cells, and how it responds to physiological stress. Insights gleaned from investigating viral mechanisms have implications for understanding how different forms of acute and chronic stress impact translational control of gene expression in health and disease. As virus reproduction is dependent on protein synthesis, the identity and roles of leading molecular actors during infection have been revealed. RNA structural elements and modifications detected by host sentinel molecules coordinate powerful antiviral responses intended to restrict virus access to the translational apparatus. Further insights can be expected from functional analysis of the genes encoding certain translation system components in giant Acanthamoeba viruses (Abrahão et al. 2017; Schultz et al. 2017).
Virus–host interactions that control protein synthesis could potentially be valuable targets for new antiviral therapies and/or lead to new vaccines and biological treatments. Included among these are virus-encoded functions that counter host dsRNA-dependent, antiviral responses. By capitalizing on this biology, a replicating HSV-1 missing the ICP34.5 eIF2α phosphatase subunit, but producing a viral dsRNA-binding protein that inhibits PKR and OAS, was developed into the first oncolytic virus immunotherapy approved by U.S. and European regulatory agencies (Taneja et al. 2001; U.S. Food and Drug Administration 2015; European Medicines Agency 2016; Ribas et al. 2017). Potent innate defenses in normal cells, including IFN-induced functions like PKR and OAS, effectively limit replication of the oncolytic HSV-1 lacking the virus-encoded phosphatase subunit. However, impaired cell-intrinsic immune responses in cancer cells support preferential virus reproduction and spread through tumor tissue. This new class of biological therapeutics for cancer was an unexpected outgrowth of understanding and manipulating a translational control mechanism in virus-infected cells. New fundamental mechanisms of initiation and regulation have been revealed. In particular, roles for RNA modifications and the ribosome itself in regulating translation have emerged with discrete ribosomal proteins selectively controlling translation. Furthermore, application of genome-wide methodologies has revealed surprising insights into how host shut-off is achieved and how viruses that do not impair host protein synthesis globally impact the host translational landscape. While the underlying mechanisms remain unknown, viral systems continue to be instrumental in delineating how ribosomes differentially recruit or exclude messages and how discrete modifications of ribosomes and mRNAs comprehensively shape translation in response to physiological and environmental stress.
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abbas YM, Laudenbach BT, Martinez-Montero S, Cencic R, Habjan M, Pichlmair A, Damha MJ, Pelletier J, Nagar B. 2017. Structure of human IFIT1 with capped RNA reveals adaptable mRNA binding and mechanisms for sensing N1 and N2 ribose 2′-O methylations. Proc Natl Acad Sci 114: E2106–E2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abernathy E, Glaunsinger B. 2015. Emerging roles for RNA degradation in viral replication and antiviral defense. Virology 479–480: 600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abernathy E, Gilbertson S, Alla R, Glaunsinger B. 2015. Viral nucleases induce an mRNA degradation-transcription feedback loop in mammalian cells. Cell Host Microbe 18: 243–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahão JS, Araújo R, Colson P, La Scola B. 2017. The analysis of translation-related gene set boosts debates around origin and evolution of mimiviruses. PLoS Genet 13: e1006532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albornoz A, Carletti T, Corazza G, Marcello A. 2014. The stress granule component TIA-1 binds tick-borne encephalitis virus RNA and is recruited to peri-nuclear sites of viral replication to inhibit viral translation. J Virol 88: 6611–6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim R, Costa SM, Cavaleiro NP, da Silva EE, da Costa LJ. 2014. HIV-1 transcripts use IRES-initiation under conditions where cap-dependent translation is restricted by poliovirus 2A protease. PLoS ONE 9: e88619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim R, Temzi A, Griffin BD, Mouland AJ. 2017. Zika virus inhibits eIF2α-dependent stress granule assembly. PLoS Negl Trop Dis 11: e0005775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias C, Walsh D, Harbell J, Wilson AC, Mohr I. 2009. Activation of host translational control pathways by a viral developmental switch. PLOS Pathog 5: e1000334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias C, Weisburd B, Stern-Ginossar N, Mercier A, Madrid AS, Bellare P, Holdorf M, Weissman JS, Ganem D. 2014. KSHV 2.0: A comprehensive annotation of the Kaposi’s sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog 10: e1003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins JF, Loughran G, Bhatt PR, Firth AE, Baranov PV. 2016. Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res 44: 7007–7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aumayr M, Schrempf A, Uzulmez O, Olek KM, Skern T. 2017. Interaction of 2A protease of human rhinovirus genetic group A with eIF4E is required for eIF4G cleavage during infection. Virology 511: 123–134. [DOI] [PubMed] [Google Scholar]
- Avanzino BC, Fuchs G, Fraser CS. 2017. Cellular cap-binding protein, eIF4E, promotes picornavirus genome restructuring and translation. Proc Natl Acad Sci 114: 9611–9616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, et al. 2014. A new system for naming ribosomal proteins. Curr Opin Struct Biol 24: 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber C, Netherton C, Goatley L, Moon A, Goodbourn S, Dixon L. 2017. Identification of residues within the African swine fever virus DP71L protein required for dephosphorylation of translation initiation factor eIF2α and inhibiting activation of pro-apoptotic CHOP. Virology 504: 107–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu M, Courtney SC, Brinton MA. 2017. Arsenite-induced stress granule formation is inhibited by elevated levels of reduced glutathione in West Nile virus-infected cells. PLoS Pathog 13: e1006240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista-Santos A, Zinker S. 2014. The P1/P2 protein heterodimers assemble to the ribosomal stalk at the moment when the ribosome is committed to translation but not to the native 60S ribosomal subunit in Saccharomyces cerevisiae. Biochemistry 53: 4105–4112. [DOI] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. 2011. The structure of the eukaryotic ribosome at 3.0 Å resolution. Science 334: 1524–1529. [DOI] [PubMed] [Google Scholar]
- Bercovich-Kinori A, Tai J, Gelbart IA, Shitrit A, Ben-Moshe S, Drori Y, Itzkovitz S, Mandelboim M, Stern-Ginossar N. 2016. A systematic view on influenza induced host shutoff. eLife 5: e18311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanga JJ, Ventoso I, Harding HP, Deng J, Ron D, Sonenberg N, Carrasco L, de Haro C. 2006. Antiviral effect of the mammalian translation initiation factor 2α kinase GCN2 against RNA viruses. EMBO J 25: 1730–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasey A, Lopez-Lastra M, Ohlmann T, Beerens N, Berkhout B, Darlix JL, Sonenberg N. 2003. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J Virol 77: 3939–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess HM, Mohr I. 2015. Cellular 5′-3′ mRNA exonuclease Xrn1 controls double-stranded RNA accumulation and anti-viral responses. Cell Host Microbe 17: 332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess HM, Mohr I. 2018. Defining the role of stress granules in innate immune suppression by the HSV-1 endoribonuclease VHS. J Virol doi:10.1128/JVI.00829-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres CJ, Angulo J, Contreras N, Pino K, Vera-Otarola J, Lopez-Lastra M. 2016. Targeting deoxyhypusine hydroxylase activity impairs cap-independent translation initiation driven by the 5′untranslated region of the HIV-1, HTLV-1, and MMTV mRNAs. Antiviral Res 134: 192–206. [DOI] [PubMed] [Google Scholar]
- Caliskan N, Peske F, Rodnina MV. 2015. Changed in translation: mRNA recoding by -1 programmed ribosomal frameshifting. Trends Biochem Sci 40: 265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camarena V, Kobayashi M, Kim JY, Roehm P, Perez R, Gardiner J, Wilson AC, Mohr I, Chao MV. 2010. Nature and duration of growth factor signaling through receptor tyrosine kinases regulates HSV-1 latency in neurons. Cell Host Microbe 8: 320–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos RK, Wong B, Xie X, Lu YF, Shi PY, Pompon J, Garcia-Blanco MA, Bradrick SS. 2017. RPLP1 and RPLP2 are essential flavivirus host factors that promote early viral protein accumulation. J Virol 91: e01706-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpentier KS, Esparo NM, Child SJ, Geballe AP. 2016. A single amino acid dictates protein kinase R susceptibility to unrelated viral antagonists. PLoS Pathog 12: e1005966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvajal F, Vallejos M, Walters B, Contreras N, Hertz MI, Olivares E, Caceres CJ, Pino K, Letelier A, Thompson SR, et al. 2016. Structural domains within the HIV-1 mRNA and the ribosomal protein S25 influence cap-independent translation initiation. FEBS J 283: 2508–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelló A, Quintas A, Sánchez EG, Sabina P, Nogal M, Carrasco L, Revilla Y. 2009. Regulation of host translational machinery by African swine fever virus. PLOS Pathog 5: e1000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Salazar M, Angel-Ambrocio AH, Soto-Acosta R, Bautista-Carbajal P, Hurtado-Monzon AM, Alcaraz-Estrada SL, Ludert JE, Del Angel RM. 2015. Dengue virus NS1 protein interacts with the ribosomal protein RPL18: This interaction is required for viral translation and replication in Huh-7 cells. Virology 484: 113–126. [DOI] [PubMed] [Google Scholar]
- Cevallos RC, Sarnow P. 2005. Factor-independent assembly of elongation-competent ribosomes by an internal ribosome entry site located in an RNA virus that infects penaeid shrimp. J Virol 79: 677–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti A, Banerjee S, Franchi L, Loo YM, Gale M Jr, Nunez G, Silverman RH. 2015. RNase L activates the NLRP3 inflammasome during viral infections. Cell Host Microbe 17: 466–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chard LS, Bordeleau ME, Pelletier J, Tanaka J, Belsham GJ. 2006. Hepatitis C virus-related internal ribosome entry sites are found in multiple genera of the family Picornaviridae. J Gen Virol 87: 927–936. [DOI] [PubMed] [Google Scholar]
- Chiang C, Gack MU. 2017. Post-translational control of intracellular pathogen sensing pathways. Trends Immunol 38: 39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury P, Bussiere L, Miller CL. 2017. Mammalian orthoreovirus factories modulate stress granule protein localization by interaction with G3BP1. J Virol 10.1128/JVI.01298-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuluunbaatar U, Roller R, Feldman ME, Brown S, Shokat KM, Mohr I. 2010. Constitutive mTORC1 activation by a herpesvirus Akt surrogate stimulates mRNA translation and viral replication. Genes Dev 24: 2627–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman JR, Papamichail D, Skiena S, Futcher B, Wimmer E, Mueller S. 2008. Virus attenuation by genome-scale changes in codon pair bias. Science 320: 1784–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csibra E, Brierley I, Irigoyen N. 2014. Modulation of stop codon read-through efficiency and its effect on the replication of murine leukemia virus. J Virol 88: 10364–10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, et al. 2010. 2′-O-methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468: 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai A, Cao S, Dhungel P, Luan Y, Liu Y, Xie Z, Yang Z. 2017. Ribosome profiling reveals translational upregulation of cellular oxidative phosphorylation mRNAs during vaccinia virus-induced host shutoff. J Virol 91: e01858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauber B, Poon D, Dos Santos T, Duguay BA, Mehta N, Saffran HA, Smiley JR. 2016. The herpes simplex virus virion host shutoff protein enhances translation of viral true late mRNAs independently of suppressing protein kinase R and stress granule formation. J Virol 90: 6049–6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughenbaugh KF, Fraser CS, Hershey JW, Hardy ME. 2003. The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. EMBO J 22: 2852–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty MD, Schaller AM, Geballe AP, Malik HS. 2016. Evolution-guided functional analyses reveal diverse antiviral specificities encoded by IFIT1 genes in mammals. eLife 5: e14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decroly E, Ferron F, Lescar J, Canard B. 2012. Conventional and unconventional mechanisms for capping viral mRNA. Nat Rev Microbiol 10: 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Hackbart M, Mettelman RC, O'Brien A, Mielech AM, Yi G, Kao CC, Baker SC. 2017. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci 114: E4251–E4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deniz N, Lenarcic EM, Landry DM, Thompson SR. 2009. Translation initiation factors are not required for Dicistroviridae IRES function in vivo. RNA 15: 932–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deo RC, Groft CM, Rajashankar KR, Burley SK. 2002. Recognition of the rotavirus mRNA 3′ consensus by an asymmetric NSP3 homodimer. Cell 108: 71–81. [DOI] [PubMed] [Google Scholar]
- Desmet EA, Anguish LJ, Parker JS. 2014. Virus-mediated compartmentalization of the host translational machinery. MBio 5: e01463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Dever TE, Dinman JD, Green R. 2018. Translation elongation and recoding in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, Farzan M. 2013. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol 13: 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgil D, Polacek C, Harris E. 2006. Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited. J Virol 80: 2976–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara MM, Brinton MA. 2007. Interaction of TIA-1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc Natl Acad Sci 104: 9041–9046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Medicines Agency. 2016. Imlygic, www.ema.europa.eu. [Google Scholar]
- Fang Y, Treffers EE, Li Y, Tas A, Sun Z, van der Meer Y, de Ru AH, van Veelen PA, Atkins JF, Snijder EJ, et al. 2012. Efficient -2 frameshifting by mammalian ribosomes to synthesize an additional arterivirus protein. Proc Natl Acad Sci 109: E2920–E2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria PA, Chakraborty P, Levay A, Barber GN, Ezelle HJ, Enninga J, Arana C, van Deursen J, Fontoura BM. 2005. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell 17: 93–102. [DOI] [PubMed] [Google Scholar]
- Fensterl V, Sen GC. 2015. Interferon-induced Ifit proteins: Their role in viral pathogenesis. J Virol 89: 2462–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnen RL, Hay TJ, Dauber B, Smiley JR, Banfield BW. 2014. The herpes simplex virus 2 virion-associated ribonuclease vhs interferes with stress granule formation. J Virol 88: 12727–12739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CS, Doudna JA. 2007. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat Rev Microbiol 5: 29–38. [DOI] [PubMed] [Google Scholar]
- Gaglia MM, Rycroft CH, Glaunsinger BA. 2015. Transcriptome-wide cleavage site mapping on cellular mRNAs reveals features underlying sequence-specific cleavage by the viral ribonuclease SOX. PLoS Pathog 11: e1005305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Simon AE. 2017. Differential use of 3′CITEs by the subgenomic RNA of Pea enation mosaic virus 2. Virology 510: 194–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Kasprzak WK, Szarko C, Shapiro BA, Simon AE. 2014. The 3′ untranslated region of pea enationmosaic virus contains two T-shaped, ribosome-binding, cap-independent translation enhancers. J Virol 88: 11696–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrey JL, Lee YY, Au HH, Bushell M, Jan E. 2010. Host and viral translational mechanisms during cricket paralysis virus infection. J Virol 84: 1124–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingold H, Pilpel Y. 2011. Determinants of translation efficiency and accuracy. Mol Syst Biol 7: 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Svitkin Y, Belsham GJ, Pause A, Sonenberg N. 1996. Activation of the translational suppressor 4E-BP1 following infection with encephalomyocarditis virus and poliovirus. Proc Natl Acad Sci 93: 5578–5583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaunsinger B, Ganem D. 2004. Highly selective escape from KSHV-mediated host mRNA shutoff and its implications for viral pathogenesis. J Exp Med 200: 391–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale NS, McIntyre AB, McFadden MJ, Roder AE, Kennedy EM, Gandara JA, Hopcraft SE, Quicke KM, Vazquez C, Willer J, et al. 2016. N6-methyladenosine in flaviviridae viral RNA genomes regulates infection. Cell Host Microbe 20: 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong D, Kim YH, Xiao Y, Du Y, Xie Y, Lee KK, Feng J, Farhat N, Zhao D, Shu S, et al. 2016. A herpesvirus protein selectively inhibits cellular mRNA nuclear export. Cell Host Microbe 20: 642–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales-Almela E, Williams H, Sanz MA, Carrasco L. 2018. The initiation factors eIF2, eIF2A, eIF2D, eIF4A and eIF4G are not involved in translation driven by hepatitis C virus IRES in human cells. Front Microbiol 9: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman AG, Tanner BC, Chang ST, Esteban M, Katze MG. 2011. Virus infection rapidly activates the P58(IPK) pathway, delaying peak kinase activation to enhance viral replication. Virology 417: 27–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Hui DJ, Merrick WC, Sen GC. 2000. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J 19: 6891–6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradi A, Svitkin YV, Imataka H, Sonenberg N. 1998. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc Natl Acad Sci 95: 11089–11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habjan M, Hubel P, Lacerda L, Benda C, Holze C, Eberl CH, Mann A, Kindler E, Gil-Cruz C, Ziebuhr J, et al. 2013. Sequestration by IFIT1 impairs translation of 2′O-unmethylated capped RNA. PLoS Pathog 9: e1003663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakki M, Marshall EE, De Niro KL, Geballe AP. 2006. Binding and nuclear relocalization of protein kinase R by human cytomegalovirus TRS1. J Virol 80: 11817–11826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller SL, Peng C, McFadden G, Rothenburg S. 2014. Poxviruses and the evolution of host range and virulence. Infect Genet Evol 21: 15–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Donovan J, Rath S, Whitney G, Chitrakar A, Korennykh A. 2014. Structure of human RNase L reveals the basis for regulated RNA decay in the IFN response. Science 343: 1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdy B, Jaramillo M, Svitkin YV, Rosenfeld AB, Kobayashi M, Walsh D, Alain T, Sean P, Robichaud N, Topisirovic I, et al. 2012. Translational control of the activation of transcription factor NF-κB and production of type I interferon by phosphorylation of the translation factor eIF4E. Nat Immunol 13: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz MI, Thompson SR. 2011a. In vivo functional analysis of the Dicistroviridae intergenic region internal ribosome entry sites. Nucleic Acids Res 39: 7276–7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz MI, Thompson SR. 2011b. Mechanism of translation initiation by Dicistroviridae IGR IRESs. Virology 411: 355–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertz MI, Landry DM, Willis AE, Luo G, Thompson SR. 2013. Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol Cell Biol 33: 1016–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho BC, Yu SL, Chen JJ, Chang SY, Yan BS, Hong QS, Singh S, Kao CL, Chen HY, Su KY, et al. 2010. Enterovirus-induced miR-141 contributes to shutoff of host protein translation by targeting the translation initiation factor eIF4E. Cell Host Microbe 9: 58–69. [DOI] [PubMed] [Google Scholar]
- Hou S, Kumar A, Xu Z, Airo AM, Stryapunina I, Wong CP, Branton W, Tchesnokov E, Götte M, Power C, et al. 2017. Zika virus hijacks stress granule proteins and modulates the host stress response. J Virol 10.1128/JVI.00474-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humoud MN, Doyle N, Royall E, Willcocks MM, Sorgeloos F, van Kuppeveld F, Roberts LO, Goodfellow IG, Langereis MA, Locker N. 2016. Feline calicivirus infection disrupts assembly of cytoplasmic stress granules and induces g3bp1 cleavage. J Virol 90: 6489–6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung M, Patel P, Davis S, Green SR. 1998. Importance of ribosomal frameshifting for human immunodeficiency virus type 1 particle assembly and replication. J Virol 72: 4819–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JL, Diamond MS. 2015. Innate immune restriction and antagonism of viral RNA lacking 2-O methylation. Virology 479–480: 66–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde JL, Gardner CL, Kimura T, White JP, Liu G, Trobaugh DW, Huang C, Tonelli M, Paessler S, Takeda K, et al. 2014. A viral RNA structural element alters host recognition of nonself RNA. Science 343: 783–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. 2009. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324: 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Lareau LF, Weissman JS. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147: 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Ingolia NT, Hussmann JA, Weissman JS. 2018. Ribosome profiling: Global views of translation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irigoyen N, Firth AE, Jones JD, Chung BY, Siddell SG, Brierley I. 2016. High- resolution analysis of coronavirus gene expression by RNA sequencing and ribosome profiling. PLoS Pathog 12: e1005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Ivanov P, Kedersha N, Anderson P. 2018. Stress granules and processing bodies in translational control. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, et al. 2012. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science 337: 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E, Sarnow P. 2002. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol 324: 889–902. [DOI] [PubMed] [Google Scholar]
- Jan E, Kinzy TG, Sarnow P. 2003. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc Natl Acad Sci 100: 15410–15415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E, Mohr I, Walsh D. 2016. A cap-to-tail guide to mRNA translation strategies in virus-infected cells. Ann Rev Virol 3: 283–307. [DOI] [PubMed] [Google Scholar]
- Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 62: 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeudy S, Abergel C, Claverie JM, Legendre M. 2012. Translation in giant viruses: A unique mixture of bacterial and eukaryotic termination schemes. PLoS Genet 8: e1003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha S, Rollins MG, Fuchs G, Procter DJ, Hall EA, Cozzolino K, Sarnow P, Savas JN, Walsh D. 2017. Trans-kingdom mimicry underlies ribosome customization by a poxvirus kinase. Nature 546: 651–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani W, Huang C, Narayanan K, Lokugamage KG, Makino S. 2009. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat Struct Mol Biol 16: 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsafanas GC, Moss B. 2007. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2: 221–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf BJ, Barton DJ. 2015. Picornavirus RNA polyadenylation by 3D(pol), the viral RNA-dependent RNA polymerase. Virus Res 206: 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy EM, Bogerd HP, Kornepati AV, Kang D, Ghoshal D, Marshall JB, Poling BC, Tsai K, Gokhale NS, Horner SM, et al. 2016. Posttranscriptional m6A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe 19: 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr CH, Ma ZW, Jang CJ, Thompson SR, Jan E. 2016. Molecular analysis of the factorless internal ribosome entry site in Cricket Paralysis virus infection. J Sci Rep 6: 37319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaperskyy DA, Emara MM, Johnston BP, Anderson P, Hatchette TF, McCormick C. 2014. Influenza a virus host shutoff disables antiviral stress-induced translation arrest. PLoS Pathog 10: e1004217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaperskyy DA, Schmaling S, Larkins-Ford J, McCormick C, Gaglia MM. 2016. Selective degradation of host RNA polymerase II transcripts by influenza A virus PA-X host shutoff protein. PLoS Pathog 12: e1005427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Kerr CH, Yeung CH, Keatings K, Nayak A, Allan DW, Jan E. 2017. Disruption of stress granule formation by the multifunctional cricket paralysis virus 1a protein. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E, Gil-Cruz C, Spanier J, Li Y, Wilhelm J, Rabouw HH, Züst R, Hwang M, V’kovski P, Stalder H, et al. 2017. Early endonuclease-mediated evasion of RNA sensing ensures efficient coronavirus replication. PLoS Pathog 13: e1006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Arias C, Garabedian A, Palmenberg AC, Mohr I. 2012a. Site-specific cleavage of the host poly(A) binding protein by the encephalomyocarditis virus 3C proteinase stimulates viral replication. J Virol 86: 10686–10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M, Wilson AC, Chao MV, Mohr I. 2012b. Control of viral latency in neurons by axonal mTOR signaling and the 4E-BP translation repressor. Genes Dev 26: 1527–1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad LM, Brulois KF, Jung JU, Glaunsinger BA. 2013. Dual short upstream open reading frames control translation of a herpesviral polycistronic mRNA. PLoS Pathog 9: e1003156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad LM, Brulois KF, Jung JU, Glaunsinger BA. 2014. Reinitiation after translation of two upstream open reading frames (ORF) governs expression of the ORF35-37 Kaposi’s sarcoma-associated herpesvirus polycistronic mRNA. J Virol 88: 6512–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunec D, Osterrieder N. 2016. Codon pair bias is a direct consequence of dinucleotide bias. Cell Rep 14: 55–67. [DOI] [PubMed] [Google Scholar]
- *.Kwan T, Thompson SR. 2018. Noncanonical translation initiation in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwun HJ, Toptan T, Ramos da Silva S, Atkins JF, Moore PS, Chang Y. 2014. Human DNA tumor viruses generate alternative reading frame proteins through repeat sequence recoding. Proc Natl Acad Sci 111: E4342–E4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry DM, Hertz MI, Thompson SR. 2009. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev 23: 2753–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Launer-Felty K, Wong CJ, Cole JL. 2015. Structural analysis of adenovirus VAI RNA defines the mechanism of inhibition of PKR. Biophys J 108: 748–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring AS, Frydman J, Andino R. 2013. The role of mutational robustness in RNA virus evolution. Nat Rev Microbiol 11: 327–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le MT, Kasprzak WK, Kim T, Gao F, Young MY, Yuan X, Shapiro BA, Seog J, Simon AE. 2017. Folding behavior of a T-shaped, ribosome-binding translation enhancer implicated in a wide-spread conformational switch. eLife 13: e22883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Liu B, Lee S, Huang SX, Shen B, Qian SB. 2012. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci 109: E2424–E2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AS, Burdeinick-Kerr R, Whelan SP. 2013. A ribosome-specialized translation initiation pathway is required for cap-dependent translation of vesicular stomatitis virus mRNAs. Proc Natl Acad Sci 110: 324–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leen EN, Sorgeloos F, Correia S, Chaudhry Y, Cannac F, et al. 2016. A conserved interaction between a C-terminal motif in norovirus VPg and the HEAT-1 domain of eIF4G is essential for translation initiation. PLOS Pathog 12: e1005379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Nouen C, Brock LG, Luongo C, McCarty T, Yang L, Mehedi M, Wimmer E, Mueller S, Collins PL, Buchholz UJ, et al. 2014. Attenuation of human respiratory syncytial virus by genome-scale codon-pair deoptimization. Proc Natl Acad Sci 111: 13169–13174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Sage V, Cinti A, McCarthy S, Amorim R, Rao S, Daino GL, Tramontano E, Branch DR, Mouland AJ. 2017. Ebola virus VP35 blocks stress granule assembly. Virology 502: 73–83. [DOI] [PubMed] [Google Scholar]
- Lezzhov AA, Gushchin VA, Lazareva EA, Vishnichenko VK, Morozov SY, Solovyev AG. 2015. Translation of the shallot virus X TGB3 gene depends on non-AUG initiation and leaky scanning. J Gen Virol 96: 3159–3164. [DOI] [PubMed] [Google Scholar]
- Li D, Wei T, Abbott CM, Harrich D. 2013. The unexpected roles of eukaryotic translation elongation factors in RNA virus replication and pathogenesis. Microbiol Mol Biol Rev 77: 253–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Treffers EE, Napthine S, Tas A, Zhu L, Sun Z, Bell S, Mark BL, van Veelen PA, van Hemert MJ, et al. 2014. Transactivation of programmed ribosomal frameshifting by a viral protein. Proc Natl Acad Sci 111: E2172–E2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist ME, Lifland AW, Utley TJ, Santangelo PJ, Crowe JE Jr. 2010. Respiratory syncytial virus induces host RNA stress granules to facilitate viral replication. J Virol 84: 12274–12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G, Gao S, Saletore Y, Gonzalez GM, Bansal V, Wang Y, Mason CE, Rana TM. 2016a. Dynamics of the human and viral m6A RNA methylomes during HIV-1 infection of T cells. Nat Microbiol 1: 16011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G, Zhao BS, Wu Y, Lu Z, Qin Y, He C, Rana TM. 2016b. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe 20: 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, HuangFu WC, Kumar KG, Qian J, Casey JP, Hamanaka RB, Grigoriadou C, Aldabe R, Diehl JA, Fuchs SY. 2009. Virus-induced unfolded protein response attenuates antiviral defenses via phosphorylation-dependent degradation of the type I interferon receptor. Cell Host Microbe 5: 72–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SW, Katsafanas GC, Liu R, Wyatt LS, Moss B. 2015. Poxvirus decapping enzymes enhance virulence by preventing the accumulation of dsRNA and the induction of innate antiviral responses. Cell Host Microbe 17: 320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttermann C, Meyers G. 2014. Two alternative ways of start site selection in human norovirus reinitiation of translation. J Biol Chem 289: 11739–11754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majzoub K, Hafirassou ML, Meignin C, Goto A, Marzi S, Fedorova A, Verdier Y, Vinh J, Hoffmann JA, Martin F, et al. 2014. RACK1 controls IRES-mediated translation of viruses. Cell 159: 1086–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Salas E, Francisco-Velilla R, Fernandez-Chamorro J, Lozano G, Diaz-Toledano R. 2015. Picornavirus IRES elements: RNA structure and host protein interactions. Virus Res 206: 62–73. [DOI] [PubMed] [Google Scholar]
- Martrus G, Nevot M, Andres C, Clotet B, Martinez MA. 2013. Changes in codon-pair bias of human immunodeficiency virus type 1 have profound effects on virus replication in cell culture. Retrovirology 10: 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews MB, Shenk T. 1991. Adenovirus virus-associated RNA and translational control. J Virol 65: 5657–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick C, Khaperskyy DA. 2017. Translation inhibition and stress granules in the antiviral immune response. Nat Rev Immunol 10.1038/nri.2017.63. [DOI] [PubMed] [Google Scholar]
- McKinney C, Perez C, Mohr I. 2012. Poly(A) binding protein abundance regulates eukaryotic translation initiation factor 4F assembly in human cytomegalovirus-infected cells. Proc Natl Acad Sci 109: 5627–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C, Yu D, Mohr I. 2013. A new role for the cellular PABP repressor Paip2 as an innate restriction factor capable of limiting productive cytomegalovirus replication. Genes Dev 27: 1809–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney C, Zavadil J, Bianco C, Shiflett L, Brown S, Mohr I. 2014. Global reprogramming of the cellular translational landscape facilitates cytomegalovirus replication. Cell Rep 6: 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melian EB, Hinzman E, Nagasaki T, Firth AE, Wills NM, Nouwens AS, Blitvich BJ, Leung J, Funk A, Atkins JF, et al. 2010. NS1′ of flaviviruses in the Japanese encephalitis virus serogroup is a product of ribosomal frameshifting and plays a role in viral neuroinvasiveness. J Virol 84: 1641–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melian EB, Hall-Mendelin S, Du F, Owens N, Bosco-Lauth AM, Nagasaki T, Rudd S, Brault AC, Bowen RA, Hall RA, et al. 2014. Programmed ribosomal frameshift alters expression of West Nile virus genes and facilitates virus replication in birds and mosquitoes. PLoS Pathog 10: e1004447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery VD, Yount BL Jr, Josset L, Gralinski LE, Scobey T, Agnihothram S, Katze MG, Baric RS. 2014. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-O-methyltransferase activity. J Virol 88: 4251–4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Merrick WC, Pavitt GD. 2018. Protein synthesis initiation in eukaryotic cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miras M, Miller WA, Truniger V, Aranda MA. 2017. Non-canonical translation in plant RNA viruses. Front Plant Sci 8: 494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani T, Fukushi S, Saijo M, Kurane I, Morikawa S. 2004. Phosphorylation of p38 MAPK and its downstream targets in SARS coronavirus-infected cells. Biochem Biophys Res Commun 319: 1228–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr I. 2016. Closing in on the causes of host shutoff. eLife 5: e20755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr I, Sonenberg N. 2012. Host translation at the nexus of infection and immunity. Cell Host Microbe 12: 470–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero H, Rojas M, Arias CF, Lopez S. 2008. Rotavirus infection induces the phosphorylation of eIF2α but prevents the formation of stress granules. J Virol 82: 1496–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorman NJ, Cristea IM, Terhune SS, Rout MP, Chait BT, Shenk T. 2008. Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3: 253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudhasani R, Tran JP, Retterer C, Kota KP, Whitehouse CA, Bavari S. 2016. Protein kinase R degradation is essential for Rift Valley fever virus infection and is regulated by SKP1-CUL1-F-box(SCF)FBXW11-NSs E3 ligase. PLOS Pathog 12: e1005437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S, Coleman JR, Papamichail D, Ward CB, Nimnual A, Futcher B, Skiena S, Wimmer E. 2010. Live attenuated influenza virus vaccines by computer-aided rational design. Nat Biotechnol 28: 723–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M, Poppers J, Sternberg D, Mohr I. 2003. Regulation of eIF2α phosphorylation by different functions that act during discrete phases in the herpes simplex virus type 1 life cycle. J Virol 77: 10917–10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M, Camarena V, Mohr I. 2004. Full resistance of HSV-1 infected primary human cells to interferon α require both the Us11 and γ134.5 gene products. J Virol 78: 10193–10196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M, Arias C, Mohr I. 2007. Maintenance of endoplasmic reticulum (ER) homeostasis in herpes simplex virus type 1-infected cells through the association of a viral glycoprotein with PERK, a cellular ER stress sensor. J Virol 81: 3377–3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napthine S, Ling R, Finch LK, Jones JD, Bell S, Brierley I, Firth AE. 2017. Protein-directed ribosomal frameshifting temporally regulates gene expression. Nat Commun 8: 15582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EV, Schmidt KM, Deflubé LR, Doğanay S, Banadyga L, Olejnik J, Hume AJ, Ryabchikova E, Ebihara H, Kedersha N, et al. 2016. Ebola virus does not induce stress granule formation during infection and sequesters stress granule proteins within viral inclusions. J Virol 90: 7268–7268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson BL, White KA. 2014. Functional long-range RNA–RNA interactions in positive-strand RNA viruses. Nat Rev Microbiol 12: 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolic J, Civas A, Lama Z, Lagaudrière-Gesbert C, Blondel D. 2016. Rabies virus infection induces the formation of stress granules closely connected to the viral factories. PLoS Pathog 12: e1005942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Onomoto K, Wakimoto M, Onoguchi K, Ishidate F, Fujiwara T, Yoneyama M, Kato H, Fujita T. 2016. Leader-containing uncapped viral transcript activates RIG-I in antiviral stress granules. PLoS Pathog 12: e1005444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlmann T, Mengardi C, Lopez-Lastra M. 2014. Translation initiation of the HIV-1 mRNA. Translation (Austin) 2: e960242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares E, Landry DM, Caceres CJ, Pino K, Rossi F, Navarrete C, Huidobro-Toro JP, Thompson SR, Lopez-Lastra M. 2014. The 5′ untranslated region of the human T-cell lymphotropic virus type 1 mRNA enables cap-independent translation initiation. J Virol 88: 5936–5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Othman Z, Sulaiman MK, Willcocks MM, Ulryck N, Blackbourn DJ, Sargueil B, Roberts LO, Locker N. 2014. Functional analysis of Kaposi’s sarcoma-associated herpesvirus vFLIP expression reveals a new mode of IRES-mediated translation. RNA 20: 1803–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish S, Moss B. 2007. Characterization of a second vaccinia virus mRNA-decapping enzyme conserved in poxviruses. J Virol 81: 12973–12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavio N, Romano PR, Graczyk TM, Feinstone SM, Taylor DR. 2003. Protein synthesis and endoplasmic reticulum stress can be modulated by the hepatitis C virus envelope protein E2 through the eukaryotic initiation factor 2α kinase PERK. J Virol 77: 3578–3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Peer E, Moshitch-Moshkovitz S, Rechavi G, Dominissini D. 2018. The epitranscriptome in translation regulation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334: 320–325. [DOI] [PubMed] [Google Scholar]
- Peng C, Haller SL, Rahman MM, McFadden G, Rothenburg S. 2016. Myxoma virus M156 is a specific inhibitor of rabbit PKR but contains a loss-of-function mutation in Australian virus isolates. Proc Natl Acad Sci 113: 3855–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU. 2003. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev 17: 181–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Hellen CU. 2004. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep 5: 906–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Chard LS, Kaku Y, Johns HL, Shatsky IN, Belsham GJ. 2004. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J Virol 78: 4487–4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank TD, Whitehurst JT, Cencic R, Pelletier J, Kieft JS. 2014. Internal translation initiation from HIV-1 transcripts is conferred by a common RNA structure. Translation (Austin) 2: e27694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooggin MM, Rajeswaran R, Schepetilnikov MV, Ryabova LA. 2012. Short ORF-dependent ribosome shunting operates in an RNA picorna-like virus and a DNA pararetrovirus that cause rice tungro disease. PLOS Pathog 8: e1002568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon LL, Pritlove DC, Fodor E, Brownlee GG. 1999. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J Virol 73: 3473–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DS, Parker R. 2016. Principles and properties of stress granules. Trends Cell Biol 26: 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Proud CG. 2018. Phosphorylation and signal transduction pathways in translational control. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q, Carroll K, Hastings C, Miller CL. 2011. Mammalian orthoreovirus escape from host translational shutoff correlates with stress granule disruption and is independent of eIF2α phosphorylation and PKR. J Virol 85: 8798–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouw HH, Langereis MA, Knaap RC, Dalebout TJ, Canton J, Sola I, Enjuanes L, Bredenbeek PJ, Kikkert M, de Groot RJ, et al. 2016. Middle East respiratory coronavirus accessory protein 4a inhibits PKR-mediated antiviral stress responses. PLoS Pathog 12: e1005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Read GS. 2013. Virus-encoded endonucleases: Expected and novel functions. RNA 4: 693–708. [DOI] [PubMed] [Google Scholar]
- Reineke LC, Lloyd RE. 2015. The stress granule protein G3BP1 recruits protein kinase R to promote multiple innate immune antiviral responses. J Virol 89: 2575–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M, Remm A, Ustav M. 1999. Human papillomavirus type 18 E1 protein is translated from polycistronic mRNA by a discontinuous scanning mechanism. J Virol 73: 3062–3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Q, Wang QS, Firth AE, Chan MM, Gouw JW, Guarna MM, Foster LJ, Atkins JF, Jan E. 2012. Alternative reading frame selection mediated by a tRNA-like domain of an internal ribosome entry site. Proc Natl Acad Sci 109: E630–E639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribas A, Dummer R, Puzanov I, VanderWalde A, Andtbacka RHI, Michielin O, Olszanski AJ, Malvehy J, Cebon J, Fernandez E, et al. 2017. Oncolytic virotherapy promotes intratumoral T cell infiltration and improves anti-PD-1 immunotherapy. Cell 170: 1109–1119.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice AD, Turner PC, Embury JE, Moldawer LL, Baker HV, Moyer RW. 2011. Roles of vaccinia virus genes E3L and K3L and host genes PKR and RNase L during intratracheal infection of C57BL/6 mice. J Virol 85: 550–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas HG, Schmaling SK, Gaglia MM. 2016. Shutoff of host gene expression in influenza A virus and herpesviruses: Similar mechanisms and common themes. Viruses 8: 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera CI, Lloyd RE. 2008. Modulation of enteroviral proteinase cleavage of poly(A)-binding protein (PABP) by conformation and PABP-associated factors. Virology 375: 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas M, Vasconcelos G, Dever TE. 2015. An eIF2α-binding motif in protein phosphatase 1 subunitGADD34 and its viral orthologs is required to promote dephosphorylation of eIF2α. Proc Natl Acad Sci 112: E3466–E3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roobol A, Roobol J, Bastide A, Knight JR, Willis AE, Smales CM. 2015. p58IPK is an inhibitor of the eIF2α kinase GCN2 and its localization and expression underpin protein synthesis and ER processing capacity. Biochem J 465: 213–225. [DOI] [PubMed] [Google Scholar]
- Roth H, Magg V, Uch F, Mutz P, Klein P, Haneke K, Lohmann V, Bartenschlager R, Fackler OT, Locker N, et al. 2017. Flavivirus infection uncouples translation suppression from cellular stress responses. MBio 8: e00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royall E, Locker N. 2016. Translational control during calicivirus infection. Viruses 8: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royall E, Doyle N, Abdul-Wahab A, Emmott E, Morley SJ, Goodfellow I, Roberts LO, Locker N. 2015. Murine norovirus 1 (MNV1) replication induces translational control of the host by regulating eIF4E activity during infection. J Biol Chem 290: 4748–4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutkowski AJ, Erhard F, L'Hernault A, Bonfert T, Schilhabel M, Crump C, Rosenstiel P, Efstathiou S, Zimmer R, Friedel CC, et al. 2015. Widespread disruption of host transcription termination in HSV-1 infection. Nat Commun 6: 7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol 8: 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Aparicio MT, Ayllón J, Leo-Macias A, Wolff T, García-Sastre A. 2017. Subcellular localizations of RIG-I, TRIM25, and MAVS complexes. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz MA, Castelló A, Ventoso I, Berlanga JJ, Carrasco L. 2009. Dual mechanism for the translation of subgenomic mRNA from Sindbis virus in infected and uninfected cells. PLoS ONE 4: e4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz MA, Almela EG, Carrasco L. 2017. Translation of sindbis subgenomic mRNA is independent of eIF2, eIF2A and eIF2D. Sci Rep 7: 43876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki J, Nakashima N. 2000. Methionine-independent initiation of translation in the capsid protein of an insect RNA virus. Proc Natl Acad Sci 97: 1512–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW. 2014. Interferon-stimulated genes: Roles in viral pathogenesis. Curr Opin Virol 6: 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz F, Yutin N, Ivanova NN, Ortega DR, Lee TK, Vierheilig J, Daims H, Horn M, Wagner M, Jensen GJ, et al. 2017. Giant viruses with an expanded complement of translation system components. Science 356: 82–85. [DOI] [PubMed] [Google Scholar]
- Sciortino MT, Parisi T, Siracusano G, Mastino A, Taddeo B, Roizman B. 2013. The virion host shutoff RNase plays a key role in blocking the activation of protein kinase R in cells infected with herpes simplex virus 1. J Virol 87: 3271–3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo EJ, Liu F, Kawagishi-Kobayashi M, Ung TL, Cao C, Dar AC, Sicheri F, Dever TE. 2008. Protein kinase PKR mutants resistant to the poxvirus pseudosubstrate K3L protein. Proc Natl Acad Sci 105: 16894–16899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma NR, Majerciak V, Kruhlak MJ, Zheng ZM. 2017. KSHV inhibits stress granule formation by viral ORF57 blocking PKR activation. PLoS Pathog 13: e1006677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva LC, Almeida GM, Assis FL, Albarnaz JD, Boratto PV, Dornas FP, Andrade KR, La Scola B, Kroon EG, da Fonseca FG, et al. 2015. Modulation of the expression of mimivirus-encoded translation-related genes in response to nutrient availability during Acanthamoeba castellanii infection. Front Microbiol 6: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon AE, Miller WA. 2013. 3′-cap–independent translation enhancers of plant viruses. Annu Rev Microbiol 67: 21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RWP, Anderson RC, Larralde O, Smith JWS, Gorgoni B, Richardson WA, Malik P, Graham SV, Gray NK. 2017. Viral and cellular mRNA-specific activators harness PABP and eIF4G to promote translation initiation downstream of cap binding. Proc Natl Acad Sci 114: 6310–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood R, Porter AC, Ma K, Quilliam LA, Wek RC. 2000. Pancreatic eukaryotic initiation factor-2α kinase (PEK) homologues in humans, Drosophila melanogaster and Caenorhabditis elegans that mediate translational control in response to endoplasmic reticulum stress. Biochem J 346: 281–293. [PMC free article] [PubMed] [Google Scholar]
- Sorgeloos F, Jha BK, Silverman RH, Michiels T. 2013. Evasion of antiviral innate immunity by Theiler’s virus L* protein through direct inhibition of RNase L. PLOS Pathog 9: e1003474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. 2004. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: The IRES functions as an RNA-based translation factor. Cell 118: 465–475. [DOI] [PubMed] [Google Scholar]
- Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, et al. 2012. Decoding human cytomegalovirus. Science 338: 1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Chin WX, Han Q, Ichiyama K, Lee CH, Eyo ZW, Takahashi H, Takahashi C, Tan BH, Hishiki T, et al. 2016. Characterization of RyDEN (C19orf66) as an interferon stimulated cellular inhibitor against dengue virus replication. PLoS Pathog 12: e1005357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney TR, Abaeva IS, Pestova TV, Hellen CU. 2014. The mechanism of translation initiation on Type 1 picornavirus IRESs. EMBO J 33: 76–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szretter KJ, Daniels BP, Cho H, Gainey MD, Yokoyama WM, Gale M Jr, Virgin HW, Klein RS, Sen GC, Diamond MS. 2012. 2′-O methylation of the viral mRNA cap by West Nile virus evades ifit1-dependent and -independent mechanisms of host restriction in vivo. PLoS Pathog 8: e1002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiri-Alaoui A, Zhao Y, Sadigh Y, Popplestone J, Kgosana L, Smith LP, Nair V. 2014. Poly(A) binding protein 1 enhances cap-dependent translation initiation of neurovirulence factor from avian herpesvirus. PLoS ONE 9: e114466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita D, Tomita K. 2012. Molecular basis for RNA polymerization by Qβ replicase. Nat Struct Mol Biol 9: 229–237. [DOI] [PubMed] [Google Scholar]
- Taneja S, MacGregor J, Markus S, Ha S, Mohr I. 2001. Enhanced antitumor efficacy of a herpes simplex virus mutant isolated by genetic selection in cancer cells. Proc Natl Acad Sci 98: 8804–8808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tats A, Tenson T, Remm M. 2008. Preferred and avoided codon pairs in three domains of life. BMC Genomics 9: 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SR, Gulyas KD, Sarnow P. 2001. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc Natl Acad Sci 98: 12972–12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh O, Cohen Y, Shitrit A, Shani O, Le-Trilling VT, Trilling M, Friedlander G, Tanenbaum M, Stern-Ginossar N. 2015. The transcription and translation landscapes during human cytomegalovirus infection reveal novel host-pathogen interactions. PLoS Pathog 11: e1005288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirumuru N, Zhao BS, Lu W, Lu Z, He C, Wu L. 2016. N6-methyladenosine of HIV-1 RNA regulates viral infection and HIV-1 Gag protein expression. eLife 5: e15528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd GC, Walter NG. 2013. Secondary structure of bacteriophage T4 gene 60 mRNA: Implications for translational bypassing. RNA 19: 685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toribio R, Diaz-Lopez I, Boskovic J, Ventoso I. 2016. An RNA trapping mechanism in Alphavirus mRNA promotes ribosome stalling and translation initiation. Nucleic Acids Res 44: 4368–4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toribio R, Diaz-Lopez I, Boskovic J, Ventoso I. 2018. Translation initiation of αvirus mRNA reveals new insights into the topology of the 48S initiation complex. Nucleic Acids Res 10.1093/nar/gky071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend HL, Jha BK, Han JQ, Maluf NK, Silverman RH, Barton DJ. 2008. A viral RNA competitively inhibits the antiviral endoribonuclease domain of RNase L. RNA 14: 1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WC, Lloyd RE. 2014. Cytoplasmic RNA granules and viral infection. Ann Rev Virol 1: 147–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulloch F, Atkinson NJ, Evans DJ, Ryan MD, Simmonds P. 2014. RNA virus attenuation by codon pair deoptimisation is an artefact of increases in CpG/UpA dinucleotide frequencies. eLife 3: e04531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiumi T, Honma S, Nomura T, Dabbs ER, Hachimori A. 2002. Translation elongation by a hybrid ribosome in which proteins at the GTPase center of the Escherichia coli ribosome are replaced with rat counterparts. J Biol Chem 277: 3857–3862. [DOI] [PubMed] [Google Scholar]
- U.S. Food and Drug Administration. 2015. Imlygic, www.fda.gov. [Google Scholar]
- Van Etten JL, Lane LC, Dunigan DD. 2010. DNA viruses: The really big ones (giruses). Annu Rev Microbiol 64: 83–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent HA, Ziehr B, Moorman NJ. 2017. Mechanism of protein kinase R inhibition by human cytomegalovirus pTRS1. J Virol 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink EI, Smiley JR, Mohr I. 2017. Subversion of host responses to energy insufficiency by Us3 supports herpes simplex virus replication during stress. J Virol 91: e00295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mohr I. 2004. Phosphorylation of eIF4E by Mnk-1 enhances HSV-1 translation and replication in quiescent cells. Genes Dev 18: 660–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mohr I. 2006. Assembly of an active translation initiation factor complex by a viral protein. Genes Dev 20: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Mohr I. 2014. Coupling 40S ribosome recruitment to modification of a cap-binding initiation factor by eIF3 subunit e. Genes Dev 28: 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Perez C, Notary J, Mohr I. 2005. Regulation of the translation initiation factor eIF4F by multiple mechanisms in human cytomegalovirus-infected cells. J Virol 79: 8057–8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh D, Arias C, Perez C, Halladin D, Escandon M, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Mohr I. 2008. EukaryoticEukk translation initiation factor 4F architectural alterations accompany translation initiation factor redistributions in poxvirus-infected cells. Mol Cell Biol 28: 2648–2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wek RC. 2018. Role of eIF2α kinases and translational control and adaptation to cellular stresses. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SD, Jacobs BL. 2012. The amino terminus of the vaccinia virus E3 protein is necessary to inhibit the interferon response. J Virol 86: 5895–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JP, Cardenas AM, Marissen WE, Lloyd RE. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe 2: 295–305. [DOI] [PubMed] [Google Scholar]
- Wilson JE, Pestova TV, Hellen CU, Sarnow P. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102: 511–520. [DOI] [PubMed] [Google Scholar]
- Wimmer E, Mueller S, Tumpey TM, Taubenberger JK. 2009. Synthetic viruses: A new opportunity to understand and prevent viral disease. Nat Biotechnol 27: 1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise HM, Barbezange C, Jagger BW, Dalton RM, Gog JR, Curran MD, Taubenberger JK, Anderson EC, Digard P. 2011. Overlapping signals for translational regulation and packaging of influenza A virus segment 2. Nucleic Acids Res 39: 7775–7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won S, Eidenschenk C, Arnold CN, Siggs OM, Sun L, Brandl K, Mullen TM, Nemerow GR, Moresco EM, Beutler B. 2012. Increased susceptibility to DNA virus infection in mice with a GCN2 mutation. J Virol 86: 1802–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Q, Cuesta R, Schneider RJ. 2004. Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes Dev 18: 1997–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Nakashima N, Ikeda Y, Uchiumi T. 2007. Binding mode of the first aminoacyl-tRNA in translation initiation mediated by Plautia stali intestine virus internal ribosome entry site. J Biol Chem 282: 7770–7776. [DOI] [PubMed] [Google Scholar]
- Yang C, Skiena S, Futcher B, Mueller S, Wimmer E. 2013. Deliberate reduction of hemagglutinin and neuraminidase expression of influenza virus leads to an ultraprotective live vaccine in mice. Proc Natl Acad Sci 110: 9481–9486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Cao S, Martens CA, Porcella SF, Xie Z, Ma M, Shen B, Moss B. 2015. Deciphering poxvirus gene expression by RNA sequencing and ribosome profiling. J Virol 89: 6874–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus M, Folley LS. 1985. Sense codons are found in specific contexts. J Mol Biol 182: 529–540. [DOI] [PubMed] [Google Scholar]
- Young DF, Andrejeva J, Li X, Inesta-Vaquera F, Dong C, Cowling VH, Goodbourn S, Randall RE. 2016. Human IFIT1 inhibits mRNA translation of rubulaviruses but not other members of the paramyxoviridae family. J Virol 90: 9446–9456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Sweeney TR, Kafasla P, Jackson RJ, Pestova TV, Hellen CU. 2011. The mechanism of translation initiation on Aichivirus RNA mediated by a novel type of picornavirus IRES. EMBO J 30: 4423–4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Liu J, He C. 2015. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev 29: 1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yueh A, Schneider RJ. 2000. Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev 14: 414–421. [PMC free article] [PubMed] [Google Scholar]
- Zaborowska I, Kellner K, Henry M, Meleady P, Walsh D. 2012. Recruitment of host translation initiation factor eIF4G by the vaccinia virus ssDNA binding protein I3. Virology 425: 11–22. [DOI] [PubMed] [Google Scholar]
- Zhang R, Jha BK, Ogden KM, Dong B, Zhao L, Elliott R, Patton JT, Silverman RH, Weiss SR. 2013. Homologous 2′,5′-phosphodiesterases from disparate RNA viruses antagonize antiviral innate immunity. Proc Natl Acad Sci 110: 13114–13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Ng MY, Chen Y, Cooperman BS. 2016. Kinetics of initiating polypeptide elongation in an IRES-dependent system. eLife 5: e13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoviev A, Hellen CU, Pestova TV. 2015. Multiple mechanisms of reinitiation on bicistronic calicivirus mRNAs. Mol Cell 57: 1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust R, Cervantes-Barragan L, Habjan M, Maier R, Neuman BW, Ziebuhr J, Szretter KJ, Baker SC, Barchet W, Diamond MS, et al. 2011. Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat Immunol 12: 137–143. [DOI] [PMC free article] [PubMed] [Google Scholar]