Abstract
Ras-specific GTPase-activating proteins (RasGAPs) down-regulate the biological activity of Ras proteins by accelerating their intrinsic rate of GTP hydrolysis, basically by a transition state stabilizing mechanism. Oncogenic Ras is commonly not sensitive to RasGAPs caused by interference of mutants with the electronic or steric requirements of the transition state, resulting in up-regulation of activated Ras in respective cells. RasGAPs are modular proteins containing a helical catalytic RasGAP module surrounded by smaller domains that are frequently involved in the subcellular localization or contributing to regulatory features of their host proteins. In this review, we summarize current knowledge about RasGAP structure, mechanism, regulation, and dual-substrate specificity and discuss in some detail neurofibromin, one of the most important negative Ras regulators in cellular growth control and neuronal function.
GTPase-activating proteins (GAPs) are commonly thought to act as negative regulators of guanine nucleotide-binding proteins (GNBPs) (Bourne et al. 1990, 1991; Vetter and Wittinghofer 2001). They function by accelerating GNBP-mediated GTP hydrolysis thereby down-regulating the biological activity of their targets (Scheffzek and Ahmadian 2005; Mishra and Lambright 2016). Although they appear to be family specific in the sense that each GNBP family has its cognate GAP module, substrate promiscuous variants are known (Krapivinsky et al. 2004; Kupzig et al. 2006). Currently there are 14 Ras-specific GTPase-activating protein (RasGAP) homologous sequences known, the majority of which result in RasGAP active proteins (Bernards 2003). The discovery of p120GAP/RASA1 by Trahey and McCormick (1987; Trahey et al. 1988; Vogel et al. 1988) not only marked the first and Ras-directed GTPase-activating protein (RasGAP), but most importantly opened up a new concept of how G proteins are regulated in the cell (Boguski and McCormick 1993). Today we know that virtually every G protein has a cognate GAP as well as a cognate guanine nucleotide exchange factor (GEF) (Bos et al. 2007; Cherfils and Zeghouf 2013). In this review, we look back over 30 years of RasGAP research, here focusing on structural and biochemical aspects. For functional and physiological aspects, the reader is referred to respective excellent review articles published over the last decade (Donovan et al. 2002; Yarwood et al. 2006; Iwashita and Song 2008; Grewal et al. 2011; Ligeti et al. 2012; King et al. 2013; Maertens and Cichowski 2014).
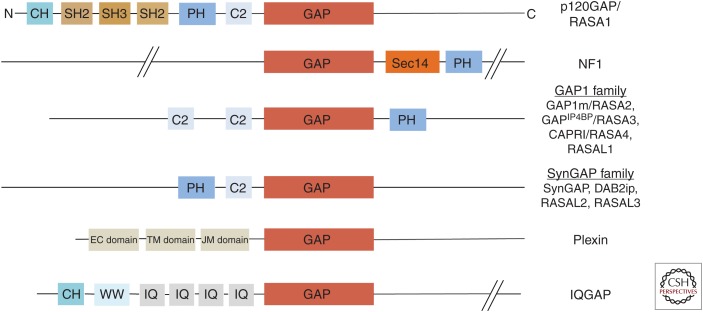
RasGAPs typically display a modular architecture of variable size, typically ranging from 100 to 300 kDa. They share a catalytic RasGAP module (35 kDa) (see Figs. 1 and 2) commonly flanked by C2 (Cho and Stahelin 2006), pleckstrin homology (PH) (Lemmon 2004; Scheffzek and Welti 2012), calponin homology (CH) (Banuelos et al. 1998), or Sec14-like domains (Bankaitis et al. 2010; Ghosh and Bankaitis 2011) with variable positioning in the coding sequence (Bernards 2003). Some canonical RasGAPs show at least dual-target specificity inactivating Ras and Rap family members (Krapivinsky et al. 2004; Kupzig et al. 2006). Members of the IQGAP group show no detectable RasGAP activity but bind to Rho family members (Kurella et al. 2009; LeCour et al. 2016). Generally, accessory domains have been implicated in protein localization as well as in regulatory features (Pamonsinlapatham et al. 2009).
Figure 1.
Sculpture of the canonical Ras-specific GTPase-activating protein (RasGAP) module showing the catalyitic portion central domain (GAPc) in red and the extra domain (GAPex) in green. (Artwork, by G. Haeusser, was created using foam rubber strips and acrylic on hard board [original size: 48″ × 35.4″].)
Figure 2.
Domain architecture of Ras-specific GTPase-activating proteins (RasGAPs). Representative RasGAPs are schematically shown with emphasis on their domain structure. Red boxes indicate GAP-(related) domains (GRDs). Sec14 represented in orange, src homology (SH, bronze), pleckstrin homology (PH, blue). Other domains like Ca2+-dependent phospholipid-binding/conserved region 2 (C2), isoleucine-glutamine repeat region (IQ), WW (conserved tryptophan), calponin homology (CH), juxtamembrane (JM), transmembrane (TM), and extracellular (EC) domains are also shown.
From a systematic viewpoint, six groups of mammalian RasGAP homologous sequences are commonly distinguished (see Fig. 2):
P120GAP/RASA1 (120 kDa) contains a carboxy-terminal RasGAP module (Marshall et al. 1989) preceded by a PH and C2 domain and an SH2/SH3/SH2 motif (Pamonsinlapatham et al. 2009). It is ubiquitously expressed and is essential for embryonic development (Henkemeyer et al. 1995). Germline mutations in the RASA1 gene are associated with the genetic disorder capillary vascular malformation-arteriovenous malformation (CVM-AM) with most of the lesions implying loss of function of the protein (Boon et al. 2005).
Neurofibromin (320 kDa), the protein product of the neurofibromatosis type 1 (NF1) tumor suppressor gene NF1, contains a central GAP-related domain (GRD) (Ballester et al. 1990; Martin et al. 1990; Xu et al. 1990b) followed by a bipartite glycerophospholipid-binding Sec14-PH-like module (D’Angelo et al. 2006; Welti et al. 2007). Neurofibromin is ubiquitously expressed with highest levels in neuronal cells in the adult. It is essential for embryonic development and mice lacking a functional NF1 gene die early from cardiovascular defects (Henkemeyer et al. 1995). NF1 patients have germline alterations in the NF1 gene causative for the disease (Riccardi 1992; Cichowski and Jacks 2001; Zhu and Parada 2001) contributing to a rather complex clinical picture that include growth regulatory as well as brain function defects. The NF1 gene is also genetically altered in several sporadic human malignancies, unrelated to NF1 (Philpott et al. 2017). See separate section below for a more detailed description of neurofibromin.
GAP1 subfamily members (ca. 95 kDa) contain a canonical RasGAP module preceded by two C2 domains followed by a PH domain. The group comprises GAP1m/RASA2, GAPIP4BP/RASA3, CAPRI/RASA4 and RASAL1. Dual-target specificity inactivating Ras and Rap family members has been shown for GAPIP4BP, CAPRI, and RASAL1 (Kupzig et al. 2006), Ca2+-dependent membrane translocation has been seen in CAPRI and RASAL representatives (Yarwood et al. 2006).
SynGAP family members (ca. 140 kDa) contain a canonical RasGAP module preceded by a C2 and PH domain, comprising the representatives SynGAP, DAB2ip, RASAL2, and RASAL3. SynGAP is present only in neuronal tissues (Chen et al. 1998; Kim et al. 1998; Jeyabalan and Clement 2016). Different from p120GAP/RASA1, the PH-C2-RasGAP triple modules are located at the amino-terminus of the protein. Germline mutations in SynGAP encoding genes have been associated with intellectual disabilities and autism (Jeyabalan and Clement 2016).
Plexins are transmembrane receptors (70 kDa) for semaphorins, small proteins triggering neuronal axon guidance and other processes, after receptor binding (Hota and Buck 2012). In these proteins, a transmembrane span separates an extracellular receptor from an intracellular RasGAP-like domain regulating the nucleotide-binding state of Rap proteins, mechanistically potentially similar to dual-target specificity RasGAPs (Wang et al. 2012; Pascoe et al. 2015).
IQGAPs (108–190 kDa) are cytoskeletal scaffolding proteins that share a carboxy-terminal structural RasGAP module accompanied by amino-terminal CH, a WW domain, and calmodulin-binding IQ motifs (Brown and Sacks 2006). Although IQGAPs are devoid of RasGAP activity (Kurella et al. 2009), they bind Rho family GNBPs (LeCour et al. 2016), presumably to coordinate processes involving cytoskeletal architecture and signaling (Hedman et al. 2015; Smith et al. 2015).
RasGAP STRUCTURE AND MECHANISM OF GTPase ACTIVATION
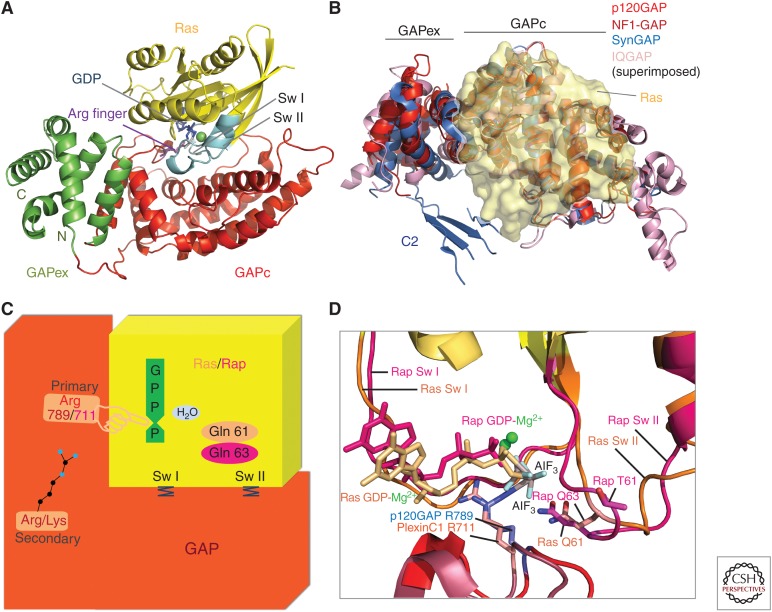
The structure of the canonical RasGAP module as originally identified in p120GAP (Marshall et al. 1989) shows a largely helical protein that is composed of two subdomains: a central domain (GAPc) that carries the catalytic amino acid equipment required for RasGAP function and an extra domain (GAPex) (see Fig. 3) (Scheffzek et al. 1996, 1998a; Pena et al. 2008) that is dispensable for GAP activity (Ahmadian et al. 1996). It is conserved in canonical RasGAP structures (Scheffzek et al. 1996, 1998a; Scheffzek and Ahmadian 2005; Pena et al. 2008) and also in the GAP inactive IQGAP (Kurella et al. 2009) but its function has been largely unclear (see below).
Figure 3.
Ras-specific GTPase-activating protein (RasGAP) structure and mechanism. (A) Ribbon representation of the Ras–RasGAP complex as derived from the structure of the catalytic domain of p120GAP/RASA1 (red and green) bound to Ras (yellow) and GDP-aluminum fluoride (AlF3) (blue) (Scheffzek et al. 1997a), switch regions are shown in cyan, and arginine finger motif in violet. (B) Ribbon representation of different RasGAPs superimposed and modeled onto Ras shown in transparent yellow. View is rotated approximately 90° about a horizontal axis as compared to RasGAP in panel A representing a top view on the modeled complex. C2 domain fragment of SynGAP (blue) appears in proximity of the Ras-binding region in SynGAP. (C) Cartoon representation of the Ras–RasGAP complex including dual-specificity functionalities. Ras/Rap is shown in yellow and GAP in red. Upon binding of Ras/Rap to GAP, the GTPase activity is strongly enhanced by the complementation of the active site, delivering the catalytic arginine, 789 (p120GAP), or 711(PlexinC1). This is further stabilized by interaction with a secondary Arg/Lys in the GAP domain. Glutamine 61 of Ras or Glutamine 63 of Rap contributes to the catalysis reaction by positioning phosphate accepting water molecule for nucleophilic attack. Additional residues and motifs in GAP stabilize the switch I and switch II regions of Ras/Rap, supporting a conformation that is favorable for efficient GTP hydrolysis. (D) Close-up view of the Ras/Rap active site bound to GAP showing conformational concordance or variations in the switch regions on Ras-p120GAP and Rap-PlexinC1GAP. Components of the nucleotide-binding site are shown for Ras (beige) and Rap (pink). The catalytic arginines in PlexinC1GAP and p120GAP are included. (Structural visualizations in panels A, B, and D were done with The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.)
A seminal contribution to the elucidation of the RasGAP mechanism was the discovery that GDP-bound Ras forms a stable complex with RasGAP in the presence of aluminum fluoride (Mittal et al. 1996), a compound that when bound to GDP can be viewed as a transition state mimic of the phosphoryl transfer reaction (Wittinghofer 1997). Its binding to GDP-bound Ras in the presence of RasGAP suggested a transition state-stabilizing mechanism of the GAP-catalyzed GTP hydrolysis reaction (Mittal et al. 1996). Such a complex was not formed with representative oncogenic Ras mutants in which GTPase activity is impaired or in the presence of Ras effectors (Mittal et al. 1996; Gremer et al. 2008). In the Ras–RasGAP complex derived from the catalytic domain of p120GAP/RASA1 (Scheffzek et al. 1996) bound to the putative transition state mimic GDP-aluminum fluoride (GDP-AlF3), Ras binds to a shallow grove in the GAPc module that is lined by conserved residues that interact with conserved regions in Ras, primarily the switch and P-loop regions (Scheffzek et al. 1997b). A major feature of the nucleotide-binding site situation is the presence of a conserved arginine (Arg789 in p120GAP/RASA1, Arg1276 in neurofibromin), termed arginine finger, contributed from RasGAP that contacts the β/γ phosphate region and is believed to stabilize the transition state of the phosphoryl transfer reaction (see Fig. 3) (Mittal et al. 1996; Ahmadian et al. 1997c; Sermon et al. 1998). The arginine-containing finger loop is stabilized by residues derived from the FLRV (Phe-Leu-Arg-Val) motif of RasGAPs (Hettich and Marshall 1994; Miao et al. 1996). Its site-directed mutagenesis confirms the impact of the catalytic arginine, as even conservative substitution to lysine reduces GAP activity by three orders of magnitude (Ahmadian et al. 1997c; Sermon et al. 1998). Additional features include the stabilization of the switch regions that are found frequently flexible in isolated Ras structures (Pai et al. 1989; Milburn et al. 1990; Wittinghofer and Vetter 2011). Along this line, particularly, the critical Gln61 is oriented to stabilize a water molecule for nucleophilic attack (see Fig. 3) (Scheffzek et al. 1997a). A so-called variable loop has been implicated in mediating substrate specificity (Scheffzek et al. 1997a; Ahmadian et al. 2003).
The structure of the Ras–RasGAP complex provided an explanation why transforming Gly12/Gln61-substitutions in Ras (Seeburg et al. 1984; Der et al. 1986) render Ras insensitive to RasGAPs; they would interfere with the geometric or electronic requirements of the transition state as represented by the complex bound to GDP-AlF3 (Scheffzek et al. 1997b). The Gly12Pro substitution is nontransforming, has a slightly increased rate of GTP hydrolysis with respect to cellular Ras, and is yet insensitive to RasGAP (Franken et al. 1993), suggesting that potentially a slight increase in the GTPase rate with respect to oncogenic Ras might be sufficient to overcome the Ras-activating effects believed to be prevalent in oncogenic Ras (Wittinghofer and Waldmann 2000).
The RasGAP mechanism has been investigated in detail using Fourier transform infrared (FTIR) spectroscopic methods and computer simulation approaches (Martin-Garcia et al. 2012; Gerwert et al. 2017). In these studies, the authors define three rate constants in the catalytic complex. In a first step, the protein adopts the GTP-bound ON-state; in the second step, the catalytic arginine side chain moves into the active site to complete the active site for GTP hydrolysis occurring simultaneously and forming a protein-bound Pi intermediate; the last step is characterized by release of Pi into the solvent, switch regions returning to the OFF-state conformation, and the catalytic arginine finger leaving the nucleotide-binding pocket (Kotting et al. 2008; Rudack et al. 2012; reviewed in Gerwert et al. 2017).
The GAP mechanism outlined above appears to be essentially valid also for Rho/RhoGAP systems (Rittinger et al. 1997; Nassar et al. 1998) that also share similar structural folds (Bax 1998; Gamblin and Smerdon 1998; Scheffzek et al. 1998b), although individual effects of mutations on the formation of AlF3/4 complexes and thus catalysis may be variable (Gremer et al. 2008). It is interesting to note that the arginine finger mechanism appears to be shared also by bacterial toxins that unfold their toxic activity by acting as Rho family–specific GAPs but seem to have a minimal and entirely different three-dimensional structure (Stebbins and Galan 2000; Wurtele et al. 2001; Evdokimov et al. 2002). Different from initial considerations, the arginine finger mechanism does not seem to be generally valid for GAPs targeting other small GNBPs (Scheffzek et al. 1998b; Scheffzek and Ahmadian 2005; Mishra and Lambright 2016), although the concept of transition state stabilization is supported in most of the structures.
The Ras-related Rap protein is generally regulated by its cognate Rap1GAP (Rubinfeld et al. 1991; Bos et al. 2007). Structural and biochemical investigations have revealed that these Rap-specific GAPs do not use a catalytic arginine “finger,” like in RasGAPs. Instead, they provide an asparagine “thumb” as catalytic residue, presumably to replace the GNBP conserved Gln 61 that structurally corresponds to a noncatalytic threonine (Thr 61) in Rap proteins (Daumke et al. 2004; Scrima et al. 2008). Dual Ras/Rap-specific RasGAPs (sharing the canonical RasGAP fold as shown in Figs. 1 and 3) seem to follow a mechanism employing a catalytic arginine (see Fig. 3) (Pena et al. 2008; Kupzig et al. 2009) associated with a glutamine residue (Gln 63) of the switch II region (Sot et al. 2010) (see below). A dual in trans mechanism employing catalytic arginine and glutamine fingers contributed by the GAP component seems to be implemented in the RabGAP Gyp1p and potentially in other RabGAPs (Pan et al. 2006; Gavriljuk et al. 2012).
MECHANISMS OF MEMBRANE TRANSLOCATION
The subcellular localization of RasGAPs is certainly influenced and modulated by their requirement to meet Ras at cellular membranes. Post-translational regulation of Ras signaling commonly requires translocation of respective protein components to membrane compartments where they can interact with membrane-associated Ras species to regulate their biological activity (i.e., the level of their output signals). Translocation to the membrane environment is thought to increase the local two-dimensional concentration to be equivalent to an increase in binding constants of five orders of magnitude compared with the cytosolic concentration (Simanshu et al. 2017). The modular architecture of RasGAPs containing lipid-binding domains suggests that such domains may be involved in localization and targeting of their respective protein hosts, as has been observed for the PH domains of GAP1 family members GAP1m and GAPIP4BP that mediate constitutive or stimulus-dependent membrane association (Cozier et al. 2000; Yarwood et al. 2006). Another mechanism of GAP localization may be via adaptor proteins such as Annexins (Gerke and Moss 2002). Annexin A6 has been shown to promote membrane binding of p120GAP/RASA1 in vitro and membrane localization in living cells (Grewal et al. 2005; Grewal and Enrich 2006). The C2 domains of CAPRI/RASA4 and RASAL1 mediate membrane association after increase of intracellular calcium concentration (Liu et al. 2005). In the case of CAPRI/RASA4 this association is stable. RASAL1 senses calcium oscillations shuttling between membrane and cytoplasm. In the case of CAPRI/RASA4 and RASAL1, detection of RasGAP activity requires calcium-dependent interaction of the C2 domain with membrane structures and glycerophospholipids (Sot et al. 2013). Translocation to the sites of GAP regulation may also be mediated by protein–protein interaction of GAP and receptor tyrosine kinase components (Kaplan et al. 1990; Fantl et al. 1992; Kashishian et al. 1992; Cooper and Kashishian 1993), and in the case of p120GAP/RASA1 may involve SH2/SH3 domains (Rojas et al. 2007; Pamonsinlapatham et al. 2009).
Interestingly, membrane translocation of neurofibromin is mediated by binding of the membrane targeting Sprouty-related protein (Bundschu et al. 2007) Spred1 (Stowe et al. 2012) to the GAP-related domain (Dunzendorfer-Matt et al. 2016; Hirata et al. 2016) (see separate section below). Sec14- and PH-like domains have been detected in a variety of signal regulatory proteins that harbor GEF or GAP domains (Lemmon 2004; Bankaitis et al. 2009; Ghosh and Bankaitis 2011; Scheffzek and Welti 2012). They have been found to control cellular localization of their host proteins, presumably by protein–protein interaction mechanisms (Fig 4) (Kostenko et al. 2004; Sirokmany et al. 2005). With neurofibromin, such localization has not been reported but cannot be excluded at the moment.
Figure 4.
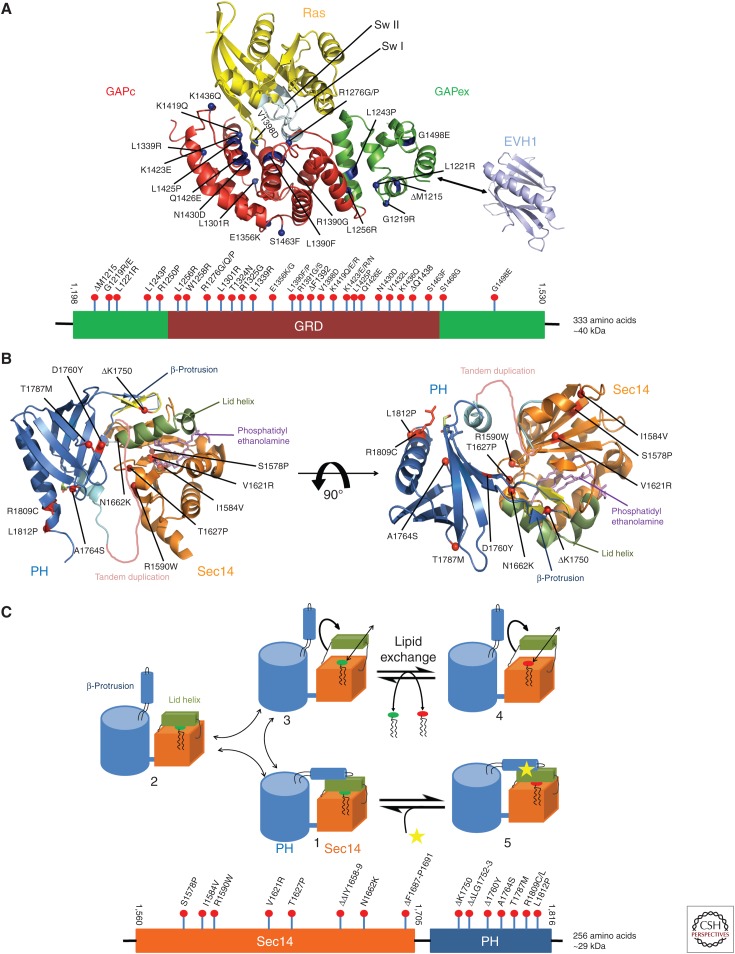
Structural components of neurofibromin. (A) Ribbon representation of a putative Ras–GRD complex, modeled on the basis of the GAP-related domain (GRD) structure (Scheffzek et al. 1998a) and the Ras-p120GAP complex (Scheffzek et al. 1997a). Blue dots indicate the location of missense mutations found in neurofibromatosis type 1 (NF1) patients. The EVH1 domain of Spred1 shown to interact with extra domain (GAPex) is depicted in cyan. Below is the domain scheme of GRD of neurofibromin with amino-terminal and carboxy-terminal extra domains with nontruncating patient-derived mutations included (see text). (B) Ribbon diagram of the Sec14-PH module bound to lipid with approximately 90° rotation about a horizontal axis. Orange ribbon represents Sec14-like domain bound to lipid (phosphatidyl ethanolamine) (in violet). Pleckstrin homology (PH)-like domain is shown in blue. Helix represented in green is a part of Sec14 that forms lid helix. Red dots indicate the location of missense mutations found in NF1 patients. A tandem duplication and ΔK1750 are also shown. Below is the domain scheme of Sec14-PH of neurofibromin with nontruncating mutations identified in NF1 patients (see text). (C) Cartoon of the Sec14-PH module, illustrating the proposed structural changes associated with the exchange of glycerophospholipid ligands. Starting from the observed structure (1), lid helix (in green), and β-protrusion helix can move either stepwise (1 > 2 > 3) or in a concerted fashion (1 > 3) into an open conformation, which allows the exchange of lipid molecules between Sec14-PH and membrane (4). Binding of modulator to the lid helix or the β-protrusion appears to prevent the lipid exchange reaction (5). (Structural visualizations in panels A and B were done with The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC.)
REGULATION OF GAP ACTIVITY
Regulatory mechanisms other than protein translocation have been investigated with GAPs in general, including protein–protein interaction, phosphorylation, proteasomal degradation, and regulation by small molecules such as lipids, as reviewed earlier (Bernards and Settleman 2004).
The fact that RasGAPs apparently have to interact with membrane-associated GNBP/Ras targets (Simanshu et al. 2017) raises the question whether membrane lipids may affect GAP activity via competitive or noncompetitive mechanisms. Early studies reported modulation of p120GAP/RASA1 or neurofibromin GAP activity by lipid compounds, including arachidonic acid, eicosanoids, fatty acids, and phospholipids with conflicting results (Tsai et al. 1989; Bollag and McCormick 1991; Han et al. 1991; Serth et al. 1991; Sermon et al. 1996) potentially reflecting challenges in defining the experimental conditions under which measurements were performed. The structural Ras–RasGAP interface does not offer a particularly hydrophobic amino acid distribution (Ahmadian et al. 2003) to suggest lipid binding in the Ras-binding region.
A GAP regulatory mechanism proposed for neurofibromin has been proteasomal degradation (followed by resynthesis) upon growth factor stimulation (Cichowski et al. 2003) mediated by the Cullin 3–associated complex (Hollstein and Cichowski 2013) or in a developmental context by the SAG/RBX2/ROC2 protein, an essential RING component of SCF E3 ubiquitin ligase (Dai et al. 2011). Little is known about the mechanistic details of the underlying protein interactions and in some cell types or conditions growth factor–dependent degradation (and resynthesis) may be inhibited (McClatchey and Cichowski 2012). Another study showed ETEA/UBXD8-mediated ubiquitination of the GRD of neurofibromin in vitro, correlating with increased levels of activated Ras after silencing of ETEA expression (Phan et al. 2010).
Protein phosphorylation is an important regulatory post-translational modification and has been reported to regulate GAP activity with some GNBP systems (Bernards and Settleman 2004). Protein kinase A (PKA) has been shown to phosphorylate the carboxy-terminal region of neurofibromin. Phosphorylation promoted its association with a 14-3-3 protein followed by down-regulation of neurofibromin-mediated GAP activity (Feng et al. 2004). Phosphorylation of SynGAP by Ca2+/calmodulin-dependent protein kinase II (CamKII) was reported to increase RasGAP activity by ∼90% potentially by an allosteric mechanism because phosphorylated residues are located outside the GAP domain (Oh et al. 2004). Another phosphorylation process involving cyclin-dependent kinase 5 (CDK5) and CamKII appears to control the ratio of Ras- and RapGAP activities of SynGAP (Walkup et al. 2015) (see below). Allosteric positive regulation of neurofibromin GAP activity after protein kinase C (PKC)-mediated phosphorylation of the cysteine serine-rich domain (CSRD) was proposed to act like a molecular switch in epidermal growth factor (EGF) receptor signaling (Mangoura et al. 2006).
Protein–protein interaction may represent another strategy/mechanism of RasGAP regulation (Bernards and Settleman 2004, 2005). Along this line, the isolated PH domain of p120GAP/RASA1 has been reported to interact specifically with its GAP domain and to inhibit interaction between activated Ras and p120GAP/RASA1 (Drugan et al. 2000). The physiological relevance of this finding remained unclear as such an interaction would have to be shown also in the full-length context of p120GAP/RASA1.
Neurofibromin has been shown to directly interact with Gβ/γ subunits linking opioid receptors to Ras signaling in the brain. Gβ/γ interacts with the Sec14-like domain and reduces GAP activity of neurofibromin in a construct composed of GRD and the Sec14-PH modules (Xie et al. 2016). To fully evaluate this observation the measurement would have to be performed also with full-length neurofibromin.
DUAL-TARGET SPECIFICITY AND MECHANISM VARIATION
The GAP mechanism implemented in the canonical RasGAP module is found modified in systems showing dual Ras/Rap specificity as in members of the GAP1 (GAPIP4BP, RASAL1, CAPRI) and SynGAP families. Although the isolated RasGAP module commonly mediates RasGAP activity, surrounding domains are required for RapGAP activity (Krapivinsky et al. 2004; Kupzig et al. 2006). The specificity switch may be achieved by different yet complementary mechanisms.
In the case of CAPRI/RASA4, Ca2+-dependent monomer/dimer formation has been shown to switch the protein between RasGAP and RapGAP activities (Dai et al. 2011), although a detailed mechanism or structures were not presented in the respective paper. SynGAP requires the RasGAP module preceding C2 domain to unfold RapGAP activity (Pena et al. 2008) and has otherwise very poor RasGAP activity (Krapivinsky et al. 2004; Pena et al. 2008). In the structure of a C2-GAP construct of SynGAP the C2 domain is partly disordered with only a few β-strands recognizable. Their positions are in the vicinity of the putative Ras/Rap-binding sites but did not allow mechanistic conclusions regarding RapGAP activity (see Fig. 3) (Pena et al. 2008). It has been reported that CamKII increases SynGAP activity (Oh et al. 2004) and along with CDK5-mediated phosphorylation of SynGAP modulates the ratio of its GAP activities toward Ras and Rap targets (see above) (Walkup et al. 2015).
The semaphorin receptor plexin transduces signals regulating axon guidance in neurons and other systems (Pascoe et al. 2015). It contains an extracellular receptor domain binding the protein semaphorin and an intracellular RasGAP-like module that acts on Rap proteins upon dimerization (Wang et al. 2012) has been reported to exhibit GAP activity toward R- and M-Ras in cellular systems (Oinuma et al. 2004; Saito et al. 2009), and in pure preparations only as a RapGAP (Wang et al. 2012). Upon semaphorin binding, the intracellular GAP module dimerizes thereby inducing a GAP active conformation that interacts with activated Rap proteins to accelerate GTP hydrolysis via an arginine finger-dependent mechanism in which the role of Gln61 of Ras is taken by Gln63 of Rap (see Fig. 3). A similar mechanism has been proposed by Sot et al. (2010) for GAPIP4BP using biophysical approaches that did not include a three-dimensional structure of a respective Rap-RasGAP complex. It appears that the RapGAP mechanisms implemented in canonical RasGAP modules require an arginine finger for catalysis (Pena et al. 2008; Kupzig et al. 2009). Interestingly, the Rab family–specific RabGAP Gyp1p employs a dual-finger mechanism comprising a catalytic arginine along with a catalytic glutamine to promote catalysis. Importantly, the RasGln61 analogous glutamine does not stabilize the transition state (Pan et al. 2006; Gavriljuk et al. 2012). Recently, the topology of RASAL1 has been presented using negative stain electron microscopy. The authors show the relative organization of the individual domains but higher resolution will be required to obtain detailed information about the mechanism of dual-target specificity (Cuellar et al. 2017).
RasGAPS IN HUMAN DISEASES
Given the importance of Ras signaling pathways in numerous cellular functions and their association with various diseases, most importantly with malignant cell transformation and cancer, it is not surprising that molecular damages do occur not only in Ras itself but also in Ras-regulating protein components. The most prominent example in this context is neurofibromin and will be described in more detail below. Its encoding gene is altered in the germline of NF1 patients (Upadhyaya and Cooper 2012) and also in a large number of sporadic malignancies unrelated to NF1 (Philpott et al. 2017). Up-regulation of Ras is believed to represent at least one mechanism of pathogenicity in respective cases. Genetic alterations in the p120GAP/RASA1 encoding gene have been reported to be responsible for capillary malformation-arteriovenous malformation (CVM-AM), a genetic disorder characterized by vascular development defects (Boon et al. 2005; Revencu et al. 2008, 2013). Most alterations would likely be incompatible with a folded protein suggesting loss of RasGAP activity as at least one mechanism of pathogenicity. Tumor-suppressive functions have been shown for DAB2ip (McLaughlin et al. 2013; Bellazzo et al. 2017) and for members of the GAP1 family, suggesting mechanisms potentially involving transcriptional deregulation (Jin et al. 2007) including DNA hypermethylation. Germline mutations in SynGAP encoding genes have been associated with intellectual disabilities and autism (Jeyabalan and Clement 2016). Semaphorin signaling via neuropilin and plexin receptors has been implicated in tumor progression, including metastasis and tumor angiogenesis (Neufeld and Kessler 2008).
NEUROFIBROMIN—A MAJOR RasGAP IN GROWTH CONTROL AND NEURONAL FUNCTION
NF1 (von Recklinghausen 1882; Riccardi 1992) was the first RASopathy identified (Rauen 2013; Ratner and Miller 2015). NF1 patients have an increased risk to develop the disease-typical neurofibroma, benign and malignant tumors of the nervous system. Additional features include pigment anomalies of the skin, bone deformations, and learning disabilities in more than 10% of adolescent patients. Further medical complications contribute to a clinical picture that is far from being well understood. Although the disease was described in 1882 (von Recklinghausen 1882; Riccardi 1992; Upadhyaya and Cooper 2012), the gene responsible was identified about 100 years later (Cawthon et al. 1990; Viskochil et al. 1990; Wallace et al. 1990). Alterations in the tumor suppressor gene NF1 are responsible for the pathology of NF1 (Riccardi 1992; Cichowski and Jacks 2001; Zhu and Parada 2001) and are found also in a large number of aggressive sporadic malignancies unrelated to NF1 (Cancer Genome Atlas Research Network 2008, 2014; Sangha et al. 2008; Patch et al. 2015; Philpott et al. 2017). The NF1 gene shows sequence similarity to the previously discovered p120GAP/RASA1 and the related IRA1/IRA2 proteins in yeast and encodes a large cytoplasmic protein, termed neurofibromin (320 kDa) (DeClue et al. 1991; Gutmann et al. 1991). Neurofibromin is highly conserved in vertebrates with sequence identity of >90% in the evolutionarily old Fugu and Zebrafish proteins (Kehrer-Sawatzki et al. 1998; Padmanabhan et al. 2009) and thus seems to be an essential vertebrate protein. Homologous sequences have been found in fly (The et al. 1997) and can be detected in honey bee (Honeybee Genome Sequencing 2006), hydra (Chapman et al. 2010), and slime mold (Eichinger et al. 2005). In yeast strains lacking IRA1/2 neurofibromin can complement a missing IRA function (Xu et al. 1990a).
A central region, GRD, shows sequence similarity to the carboxy-terminal catalytic domain of p120GAP/RASA1 (Marshall et al. 1989) and displayed RasGAP activity (Ballester et al. 1990; Martin et al. 1990; Xu et al. 1990b), defining neurofibromin as a RasGAP. The cellular impact of neurofibromin on Ras regulation is evident from studies in which loss of neurofibromin is associated with increased intracellular levels of activated Ras (Basu et al. 1992; DeClue et al. 1992; Bollag et al. 1996). Conversely, expression of the GRD in otherwise NF1−/− cells can restore the cellular phenotypes (Hiatt et al. 2001; Thomas et al. 2006). Neuronal studies suggest that neurofibromin contributes to more than 90% of Ras inactivation in dendritic spines (Oliveira and Yasuda 2014). Nontruncating gene alterations (currently >400 identified) cover the whole coding sequence suggesting important functions represented in regions outside the GRD (Kaufmann 2008). The GRD is preceded by a 90-residue peptide segment that has been reported to be important for the interaction with the cytoskeletal protein tubulin (Bollag et al. 1993). Mutations in the GRD region abolish tubulin binding (Xu and Gutmann 1997). At its carboxy-terminal end, it is followed by a bipartite glycerophospholipid-binding module that is composed of an amino-terminal Sec14- and a carboxy-terminal PH-like domain (see below) (D’Angelo et al. 2006; Welti et al. 2007). The so-called cysteine serine-rich domain (CSRD) (Izawa et al. 1996) appears to cluster a number of mutations found in NF1 patients (see Fig. 5) (Fahsold et al. 2000; Mattocks et al. 2004), but has not been structurally validated so far.
Figure 5.
Interaction partners of neurofibromin. Domain scheme of neurofibromin with reported interaction partners and structurally validated domains colored. Red dots at the bottom part of the scheme indicate nontruncating mutations found in patients and blue and yellow dots on the top part of the scheme represent potential phosphorylation sites for protein kinase A (PKA) and protein kinase C (PKC), respectively. Two ubiquitination sites are indicated as green boxes. Several potential caveolin-binding domains (CBDs) are also indicated. The GAP-related domain (GRD) part indicated in dark red is the minimal GAP domain required for the GAP activity and binds to Ras. Amino-terminal and carboxy-terminal extensions, indicated in light green are overlapping with the tubulin-binding region and the Sec14-PH domain, respectively, and interact with the Sprouty-related protein with an EVH1 domain 1 (Spred1). DDAH1, dimethylarginine dimethylaminohydrolase 1; LRPPRC, leucine-rich pentatricopeptide repeat-containing protein; APP, amyloid precursor protein; FAF2, Fas-associated factor 2; ETEA, expressed in T cells and eosinophils in atopic dermatitis; VCP, valosin-containing protein; LIMK2, LIM domain kinase 2; DPYSL2, dihydropyrimidinase-related protein 2; CRMP2/4, collapsin response mediator protein 2/4; SCF, Skp-Cullin-F box-containing complex; FAK, focal adhesion kinase; CASK, calcium/calmodulin-dependent serine protein kinase (see text).
Structure of Neurofibromin
Bioinformatics sequence analysis indicates a highly helical protein with few flexible segments in the carboxy-terminal regions. From a structural perspective, the domain composition/architecture is poorly defined and include only the GRD (Scheffzek et al. 1998a) and the neighboring Sec14-PH module (D’Angelo et al. 2006; Welti et al. 2007). These account for approximately 25% of the total molecular weight. In the following sections, we describe the structures of the respective modules and try to address potential functional implications also considering mutational studies.
The GAP-Related Domain (GRD)
The classic GRD corresponding to the catalytic domain of p120GAP comprises a segment ranging from residues 1198 to 1530, which for crystallographic structure determination has been proteolytically treated to facilitate crystallization. The structure of the GRD module (see Fig. 4A) (Scheffzek et al. 1998a) confirms the canonical helical RasGAP fold with few differences to the respective module of p120GAP/RASA1 (Scheffzek et al. 1996). They can be located essentially in loop regions connecting helices.
The differentially spliced and GAP activity modulating exon 23a (Andersen et al. 1993) would insert a 21-residue peptide segment in a helix that is in a structural neighborhood near the active site region (Scheffzek et al. 1998a). A structure derived from this type II transcript has not been reported so far. GAP-accelerated GTP hydrolysis has been characterized in detail using kinetic approaches (Ahmadian et al. 1997a,b, 2003; Sermon et al. 1998). Nontruncating mutations found in NF1 patients have different effects on the catalytic properties of the GRD (see Fig. 4). A most striking alteration has been found earlier in a patient with severe tumor burden where Arg1276 (the arginine finger of neurofibromin) was replaced by a proline residue. Structural modeling of this mutation indicates loss of a catalytic residue for which there is no functionally alternative residue in the vicinity of the site of GTP hydrolysis, confirmed by biochemical investigation of the associated enzyme kinetics yielding an 8000-fold decrease in catalytic efficiency for the mutated protein (Klose et al. 1998). Numerous mutations have been identified in the GRD. Several are indicated in Figure 4A.
Those and other mutations include ΔM1215, G1219R/E, L1221R, L1243P, R1250P, L1256R, W1258R, R1276G/Q/P, L1301R, T1324N, R1325G, L1339R, E1356K/G, L1390F/P, R1391G/S, ΔF1392, V1398D, K1419Q/E/R, K1423E/R/N, L1425P, Q1426E, N1430D, V1432L, K1436Q, ΔQ1438, S1463F, S1468G, and G1498E (see Fig. 4A) (Li et al. 1992; Upadhyaya et al. 1997b; Fahsold et al. 2000; Han et al. 2001; Ars et al. 2003; De Luca et al. 2003; Mattocks et al. 2004; Lee et al. 2006; Kaufmann 2008; Thomas et al. 2012; Bianchessi et al. 2015). The HGMD (Institute of Medical Genetics, Cardiff, Wales, UK, www.hgmd.cf.ac.uk/ac/index.php) and LOVD (Leiden Open Variation Database, www.LOVD.nl/NF1) (Fokkema et al. 2011; van Minkelen et al. 2014) databases were interrogated to summarize patient-derived mutations.
The Sec14-PH Module
Using structural biology approaches we have discovered earlier a bipartite structural module composed of an amino-terminal Sec14- and a carboxy-terminal PH-like domain (D’Angelo et al. 2006), the former binding glycerophospholipids (Welti et al. 2007). The structure of the Sec14-PH module in neurofibromin allows definition of glycerophospholipid binding in a hydrophobic cage of the Sec14-like portion that appears to be modulated/regulated by a hairpin-like protrusion derived from the PH-like portion (see Fig. 4B). This protrusion appears to keep the lid structure closed in the respective crystal structure thereby preventing access of lipid ligands to the interior of the Sec14 portion. In a hypothetical mechanistic model, two ligands would be involved in Sec14-PH function/regulation. Ligand 1 would induce conformational changes in the hairpin region thereby releasing structural constraints of the lid helix to allow binding of ligand 2 (a glycerophospholipid) to the lipid-binding cage (see Fig. 4B and C). Given the abundance of phosphatidyl ethanolamine (PE) in eukaryotic cells, it appears likely that PE represents at least one physiological ligand of the Sec14-like portion in neurofibromin (Welti et al. 2007). Binding of immobilized PIPs in overlay assays has been shown for the Sec14-PH module and is modulated by mutations of conserved residues in the vicinity of the hairpin protrusion (D’Angelo et al. 2006; Welti et al. 2007). The isolated Sec14-PH module does not seem to localize to specific cellular structures/compartments (S Welti and K Scheffzek, unpubl.). Structural consequences of prominent alterations have been investigated, suggesting structural rearrangements in at least one of the three cases investigated (Welti et al. 2011). We speculate that they affect interactions between neurofibromin and/or small molecules or proteins.
A particularly interesting alteration has been identified at the carboxy-terminal end of the PH-like domain. Arg1809 was found substituted by cysteine in a set of six NF1 patients in a cohort of patients presenting with a mild NF1 phenotype but without neurofibroma (Ekvall et al. 2014; Rojnueangnit et al. 2015; Santoro et al. 2015). The genetic analysis of the mutation along with the common symptoms in patients gives one of the rare examples for a genotype–phenotype relationship of which only two have been reported previously (Cnossen et al. 1997; Upadhyaya et al. 2007). In the Sec14-PH structure representing the cellular protein sequence Arg1809 interacts with Ser1738 in the amino-terminal region of the PH β-sheet (D’Angelo et al. 2006). Mutation of Arg1809 to cysteine (or other amino acids observed) is likely to abolish the interaction between the two structural elements of the PH-like portion. The structural impact of this interaction is difficult to assess because in contrast to Arg1809, Ser1738 shows little conservation in NF1-related sequences (D’Angelo et al. 2006). To evaluate the cellular consequences of the observed alteration, its investigation in the full-length context of neurofibromin within relevant cell lines is required, potentially comparing protein mass spectrometry-guided interaction profiles with those of the wild-type protein. In any case, one would expect that alterations in the respective sequence position would converge on a common biochemical/cellular feature of neurofibromin function.
Other nontruncating mutations in the Sec14-PH module include S1578P, I1584V, R1590W, V1621R, T1627P, ΔΔIY1658-9, N1662K, ΔF1687-P1691, ΔK1750, ΔΔLG1752-3, Δ1760Y, A1764S, T1787M, R1809C/L, and L1812P (Tassabehji et al. 1993; Upadhyaya et al. 1997a; Wu et al. 1999; Fahsold et al. 2000; Jeong et al. 2006; Welti et al. 2011; Bianchessi et al. 2015). The HGMD (Institute of Medical Genetics, Cardiff, Wales, UK, www.hgmd.cf.ac.uk/ac/index.php) and LOVD (Leiden Open Variation Database, www.LOVD.nl/NF1) (Fokkema et al. 2011; van Minkelen et al. 2014) databases were interrogated to summarize patient mutations.
What is the function of the Sec14-PH module? The module has been discovered in search for novel soluble domains of neurofibromin (D’Angelo et al. 2006; Bonneau et al. 2009). While the Sec14-like portion has been anticipated based on bioinformatics analyses (Aravind et al. 1999), the PH-like portion was entirely unexpected in the structure of neurofibromin as was the seemingly mechanistic interaction between the two domains (D’Angelo et al. 2006). PH-like domains commonly share a sandwich-like fold with halves formed by two β-sheets closed by a carboxy-terminal α-helix (Scheffzek and Welti 2012). Depending on the detailed amino acid equipment, they may bind various ligand including phospholipids, phosphotyrosine peptides, or proteins including Rho family GNBPs (Lemmon 2004; Scheffzek and Welti 2012). Sec14- and PH-like domains occur as stand-alone modules or as parts of signal regulatory proteins, such as RhoGEFs, RhoGAPs, or phosphotyrosine phosphatases (PTPs) (Saito et al. 2007; Bankaitis et al. 2010). Their function in these proteins is not entirely understood. Some studies nevertheless suggest targeting functions in p50RhoGAP and the RhoGEF Dbs (Kostenko et al. 2004; Sirokmany et al. 2005). It is so far unclear whether this function is associated with lipid-binding properties of the proteins.
Full-Length Neurofibromin
The structure of the entire neurofibromin is not known so far. The classic method for structure determination of biomolecular species is X-ray crystallography. However, this method requires the availability of high-quality protein crystals to yield detailed information of the crystalline protein. To obtain such crystals the protein has to be available in pure and homogeneous quality (Shi 2014). Obtaining high-quality neurofibromin for structural investigation has long been a rate-limiting step in neurofibromin structural biology. With the routine availability of gene synthesis, including sequence optimization for protein expression (Bonneau et al. 2009), neurofibromin overexpression in insect cells became a powerful method to obtain pure and at least partially homogeneous neurofibromin from such preparations (Dunzendorfer-Matt et al. 2016). Recombinant neurofibromin has been used in protein transduction experiments demonstrating down-regulation of the Ras/MAPK pathway in NF1-depleted cells, as shown by monitoring phospho-ERK decrease (Mellert et al. 2018). To our knowledge, crystalline neurofibromin has so far not been obtained. An alternative method with increasingly high potential is cryoelectron microscopy (cryoEM) (Carroni and Saibil 2016). With this method, the molecular species is preserved in a native-like vitrified state that does not require crystallinity for structural analysis (Earl et al. 2017). Many large macromolecular and signaling species have been analyzed by cryoEM over the past years (Merk et al. 2016). Initial and preliminary analyses of such specimens show neurofibromin at low resolution (>20 Å), forming compact dimeric particles of unclear molecular oligomerization state (J Briggs, H Stark, and K Scheffzek, unpubl.). Further efforts will be necessary to improve sample and data quality of cryoEM approaches to obtain a high-resolution view of full-length neurofibromin.
Nontruncating mutations can be found along the entire coding sequence of neurofibromin (Figs. 4 and 5), suggesting the whole sequence to be essential for protein function. Their assessment in terms of impact on protein function may be difficult depending on structural and biochemical details. From a structural perspective, nonconservative substitutions are likely to have a more severe effect on the structure or function of the protein than conservative exchanges. Such substitutions are likely to change the local environment in a manner that may not be compatible with the native neurofibromin structure. In general, the availability of 3D structural information contributes significantly to the assessment of nontruncating mutations found in patients.
Interaction Partners of Neurofibromin
Meanwhile, a large number of interaction partners beyond activated Ras have been reported (see Fig. 5) (Ratner and Miller 2015). They include kinesin-1 (Hakimi et al. 2002), DDAH (Tokuo et al. 2001), PKA (Feng et al. 2004), tubulin (Bollag et al. 1993), LRPPRC (Arun et al. 2013), amyloid precursor protein (APP) (De Schepper et al. 2006), Spred1 (Stowe et al. 2012; Dunzendorfer-Matt et al. 2016; Hirata et al. 2016), syndecans (Hsueh et al. 2001), phospholipids (D’Angelo et al. 2006; Welti et al. 2007), G proteins or components thereof (Tong et al. 2002; Xie et al. 2016), p97/vcp (Wang et al. 2011), LIMK2 (Vallee et al. 2012), serotonin receptor (Deraredj Nadim et al. 2016), caveolin-1 (Boyanapalli et al. 2006), CRMP2/4 (Lin and Hsueh 2008; Patrakitkomjorn et al. 2008), 14-3-3 (Feng et al. 2004), focal adhesion kinase (FAK) (Kweh et al. 2009), SCF (Tan et al. 2011), Cullin-3 (Hollstein and Cichowski 2013), and CASK (Volta et al. 2010). Technically, they were identified in a variety of approaches including yeast two-hybrid screening, coimmunoprecipitation or pull-down-like experiments. The biological significance of the underlying protein–protein interactions has not been clearly established in most of the reported interactions. We describe here in more detail selected examples where direct interaction and/or a functional link appear explored or established.
Spred1
A most insightful observation came from experiments uncovering the interaction between neurofibromin and Sprouty-related proteins with an EVH1 domain 1 (Spred1) (Stowe et al. 2012).
SPRED domain proteins (Bundschu et al. 2007) have been reported to act as negative regulators of the Ras/MAPK pathway. Although interaction with Ras has been reported, the mechanism of Ras inhibition remains unclear (Wakioka et al. 2001). Genetic alterations in the Spred1 gene are associated with Legius syndrome (LS), a rare genetic disorder that shares mild symptoms with NF1 (Brems et al. 2007) in the absence of severe clinical phenotypes. Stowe et al. (2012) have found that Spred1 down-regulates the Ras/MAPK pathway by interaction with neurofibromin. In a respective model, Spred1 uses its amino-terminal EVH1 domain to bind neurofibromin and is recruited to the cellular membrane by the carboxy-terminal SPY-domain of Spred1. Membrane recruitment enables neurofibromin to interact with Ras, and by its RasGAP activity, down-regulate the biological activity of Ras at the cellular membrane. These observations provide a molecular link between NF1 and LS both being associated with deregulation of the Ras/MAPK signaling pathway (McClatchey and Cichowski 2012; Stowe et al. 2012). In a collaborative follow-up study with the laboratory of F. McCormick, we were able to map the EVH1-binding site to the GRD of neurofibromin (Dunzendorfer-Matt et al. 2016). EVH1 binding to the GRD was compatible with binding of activated Ras and did not influence RasGAP activity. Refined mapping experiments finally allowed us to identify the extra domain of the GRD (GAPex) as the EVH1-binding site on the GRD (Dunzendorfer-Matt et al. 2016). Importantly, EVH1 binding to the GRD did not interfere with binding of activated Ras or with GAP activity or with the formation of a transition state–like complex (Dunzendorfer-Matt et al. 2016). Pathogenic mutations found in LS patients impaired the EVH1–GRD binding as has also been found by the group of A. Yoshimura who mapped the EVH1-binding site based on yeast two-hybrid experiments (Hirata et al. 2016). In addition, a pathogenic single residue deletion of the GAPex retained RasGAP activity but failed to translocate to the membrane for interaction with Ras (Dunzendorfer-Matt et al. 2016). Although it is tempting to speculate that GAPex provides a general docking platform for proteins targeting RasGAPs to membrane compartments, it has to be noted that at least p120GAP/RASA1 is not membrane targeted by Spred proteins (Stowe et al. 2012).
P97/VCP
P97/VCP is a cytoplasmic abundant AAA (pronounced “triple A”) ATPase that is associated with various cellular activities including proteasome-mediated protein degradation. Based on immunoprecipitation studies, Wang et al. (2011) identified the protein as a neurofibromin-binding protein and mapped the binding region to a portion comprising Sec14-PH and amino- and carboxy-terminal extensions thereof. Employing pull-down experiments from mouse brain lysates using glutathione-S-transferase (GST)-fused Sec14-PH as bait we identified p97/VCP as an interaction partner of the protein (M Schwarz, F Bonneau, and K Scheffzek, unpubl.) indicating that the Sec14-PH module is sufficient to mediate the underlying protein–protein interaction. The physiological relevance of the p97/VCP neurofibromin interaction was proposed and experimentally supported to negatively regulate dendritic spine density. Mutations found in NF1 patients indeed interfered with spine density (Wang et al. 2011). Further studies are necessary to investigate the mechanistic aspects of this protein–protein interaction.
LIM Kinase2
Using yeast two-hybrid approaches to screen for interaction partners of the Sec14-PH module, Vallee et al. (2012) identified LIMK2 to interact with Sec14-PH. Sec14-PH binds to LIMK2 but not to the closely related LIMK1 and inhibits its kinase activity with respect to cofilin phosphorylation by preventing ROCK activation. Little information has been presented about the specific mechanism that causes ROCK activation to be inhibited (Vallee et al. 2012).
Cross Talk with GPCR Signaling
Potential cross talk with G protein-coupled receptor signaling (GPCR) has been identified with neurofibromin (Tong et al. 2002). The serotonin 5-hydroxytryptamin 6 (5-HT6) receptor constitutively activates the Gs/adenylyl cyclase pathway in several cell types including neurons. Silencing the expression of neurofibromin strongly reduces the constitutive activity of the receptor (Deraredj Nadim et al. 2016). This process apparently requires the interaction with the PH-like domain of neurofibromin. Interestingly, disease-associated mutations found in NF1 patients blocked the interaction with the receptor.
Another GPCR cross talk has been described recently when Xie et al. (2016) described a mechanism linking opioid receptors to Ras signaling in the brain. Most importantly, Gβ/γ subunits of the µ-opioid receptor interact directly with neurofibromin. The authors mapped a Gβ/γ-binding site to the Sec14-PH portion of neurofibromin and using a GST-fused GRD-Sec14-PH construct showed that Gβ/γ inhibits neurofibromin-mediated GAP activity (Xie et al. 2016). It will be important to perform GAP assays in the context of full-length neurofibromin.
CONCLUDING REMARKS
Thirty years after the discovery of the first RasGAP, the concept of G protein regulation by GEFs, and GTPase-activating proteins is well established and a plethora of research papers investigating the details of RasGAP-mediated signal regulation has accumulated in the scientific literature and beyond. Given that Ras and its major regulator neurofibromin play major roles in cellular growth control and cancer development, understanding the GAP mechanism and the roles of individual RasGAPs including neurofibromin in more detail, and also for each functionally relevant mutant, will be among the important challenges in the future.
ACKNOWLEDGMENTS
We thank our colleagues for sharing research enthusiasm, scientific results, and stimulating discussions over decades of RasGAP investigation. We are grateful to Gabi Haeusser (SAP-SE, Walldorf, Germany) for the artwork in Figure 1.
Footnotes
Editors: Linda Van Aelst, Julian Downward, and Frank McCormick
Additional Perspectives on Ras and Cancer in the 21st Century available at www.perspectivesinmedicine.org
REFERENCES
- Ahmadian MR, Wiesmuller L, Lautwein A, Bischoff FR, Wittinghofer A. 1996. Structural differences in the minimal catalytic domains of the GTPase-activating proteins p120GAP and neurofibromin. J Biol Chem 271: 16409–16415. [DOI] [PubMed] [Google Scholar]
- Ahmadian MR, Hoffmann U, Goody RS, Wittinghofer A. 1997a. Individual rate constants for the interaction of Ras proteins with GTPase-activating proteins determined by fluorescence spectroscopy. Biochemistry 36: 4535–4541. [DOI] [PubMed] [Google Scholar]
- Ahmadian MR, Mittal R, Hall A, Wittinghofer A. 1997b. Aluminum fluoride associates with the small guanine nucleotide binding proteins. FEBS Lett 408: 315–318. [DOI] [PubMed] [Google Scholar]
- Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A. 1997c. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol 4: 686–689. [DOI] [PubMed] [Google Scholar]
- Ahmadian MR, Kiel C, Stege P, Scheffzek K. 2003. Structural fingerprints of the Ras-GTPase activating proteins neurofibromin and p120GAP. J Mol Biol 329: 699–710. [DOI] [PubMed] [Google Scholar]
- Andersen LB, Ballester R, Marchuk DA, Chang E, Gutmann DH, Saulino AM, Camonis J, Wigler M, Collins FS. 1993. A conserved alternative splice in the von Recklinghausen neurofibromatosis (NF1) gene produces two neurofibromin isoforms, both of which have GTPase-activating protein activity. Mol Cell Biol 13: 487–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Neuwald AF, Ponting CP. 1999. Sec14p-like domains in NF1 and Dbl-like proteins indicate lipid regulation of Ras and Rho signaling. Curr Biol 9: R195–R197. [DOI] [PubMed] [Google Scholar]
- Ars E, Kruyer H, Morell M, Pros E, Serra E, Ravella A, Estivill X, Lazaro C. 2003. Recurrent mutations in the NF1 gene are common among neurofibromatosis type 1 patients. J Med Genet 40: e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun V, Wiley JC, Kaur H, Kaplan DR, Guha A. 2013. A novel neurofibromin (NF1) interaction with the leucine-rich pentatricopeptide repeat motif-containing protein links neurofibromatosis type 1 and the French Canadian variant of Leigh’s syndrome in a common molecular complex. J Neurosci Res 91: 494–505. [DOI] [PubMed] [Google Scholar]
- Ballester R, Marchuk D, Boguski M, Saulino A, Letcher R, Wigler M, Collins F. 1990. The NF1 locus encodes a protein functionally related to mammalian GAP and yeast IRA proteins. Cell 63: 851–859. [DOI] [PubMed] [Google Scholar]
- Bankaitis VA, Mousley CJ, Schaaf G. 2009. The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem Sci 35: 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankaitis VA, Mousley CJ, Schaaf G. 2010. The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem Sci 35: 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuelos S, Saraste M, Djinovic Carugo K. 1998. Structural comparisons of calponin homology domains: Implications for actin binding. Structure 6: 1419–1431. [DOI] [PubMed] [Google Scholar]
- Basu TN, Gutmann DH, Fletcher JA, Glover TW, Collins FS, Downward J. 1992. Aberrant regulation of ras proteins in malignant tumour cells from type 1 neurofibromatosis patients. Nature 356: 713–715. [DOI] [PubMed] [Google Scholar]
- Bax B. 1998. Domains of rasGAP and rhoGAP are related. Nature 392: 447–448. [DOI] [PubMed] [Google Scholar]
- Bellazzo A, Di Minin G, Collavin L. 2017. Block one, unleash a hundred. Mechanisms of DAB2IP inactivation in cancer. Cell Death Differ 24: 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards A. 2003. GAPs galore! A survey of putative Ras superfamily GTPase activating proteins in man and Drosophila. Biochim Biophys Acta 1603: 47–82. [DOI] [PubMed] [Google Scholar]
- Bernards A, Settleman J. 2004. GAP control: Regulating the regulators of small GTPases. Trends Cell Biol 14: 377–385. [DOI] [PubMed] [Google Scholar]
- Bernards A, Settleman J. 2005. GAPs in growth factor signalling. Growth Factors 23: 143–149. [DOI] [PubMed] [Google Scholar]
- Bianchessi D, Morosini S, Saletti V, Ibba MC, Natacci F, Esposito S, Cesaretti C, Riva D, Finocchiaro G, Eoli M. 2015. 126 novel mutations in Italian patients with neurofibromatosis type 1. Mol Genet Genomic Med 3: 513–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski MS, McCormick F. 1993. Proteins regulating Ras and its relatives. Nature 366: 643–654. [DOI] [PubMed] [Google Scholar]
- Bollag G, McCormick F. 1991. Differential regulation of rasGAP and neurofibromatosis gene product activities. Nature 351: 576–579. [DOI] [PubMed] [Google Scholar]
- Bollag G, McCormick F, Clark R. 1993. Characterization of full-length neurofibromin: Tubulin inhibits Ras GAP activity. EMBO J 12: 1923–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollag G, Adler F, elMasry N, McCabe PC, Conner E Jr, Thompson P, McCormick F, Shannon K. 1996. Biochemical characterization of a novel KRAS insertion mutation from a human leukemia. J Biol Chem 271: 32491–32494. [DOI] [PubMed] [Google Scholar]
- Bonneau F, Lenherr ED, Pena V, Hart DJ, Scheffzek K. 2009. Solubility survey of fragments of the neurofibromatosis type 1 protein neurofibromin. Protein Expr Purif 65: 30–37. [DOI] [PubMed] [Google Scholar]
- Boon LM, Mulliken JB, Vikkula M. 2005. RASA1: Variable phenotype with capillary and arteriovenous malformations. Curr Opin Genet Dev 15: 265–269. [DOI] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. 2007. GEFs and GAPs: Critical elements in the control of small G proteins. Cell 129: 865–877. [DOI] [PubMed] [Google Scholar]
- Bourne HR, Sanders DA, McCormick F. 1990. The GTPase superfamily: A conserved switch for diverse cell functions. Nature 348: 125–132. [DOI] [PubMed] [Google Scholar]
- Bourne HR, Sander DA, McCormick F. 1991. The GTPase superfamily: Conserved structure and molecular mechanism. Nature 349: 117–127. [DOI] [PubMed] [Google Scholar]
- Boyanapalli M, Lahoud OB, Messiaen L, Kim B, Anderle de Sylor MS, Duckett SJ, Somara S, Mikol DD. 2006. Neurofibromin binds to caveolin-1 and regulates ras, FAK, and Akt. Biochem Biophys Res Commun 340: 1200–1208. [DOI] [PubMed] [Google Scholar]
- Brems H, Chmara M, Sahbatou M, Denayer E, Taniguchi K, Kato R, Somers R, Messiaen L, De Schepper S, Fryns JP, et al. 2007. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1-like phenotype. Nat Genet 39: 1120–1126. [DOI] [PubMed] [Google Scholar]
- Brown MD, Sacks DB. 2006. IQGAP1 in cellular signaling: Bridging the GAP. Trends Cell Biol 16: 242–249. [DOI] [PubMed] [Google Scholar]
- Bundschu K, Walter U, Schuh K. 2007. Getting a first clue about SPRED functions. Bioessays 29: 897–907. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2008. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455: 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. 2014. Comprehensive molecular profiling of lung adenocarcinoma. Nature 511: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroni M, Saibil HR. 2016. Cryo electron microscopy to determine the structure of macromolecular complexes. Methods 95: 78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawthon RM, O’Connell P, Buchberg AM, Viskochil D, Weiss RB, Culver M, Stevens J, Jenkins NA, Copeland NG, White R. 1990. Identification and characterization of transcripts from the neurofibromatosis 1 region: The sequence and genomic structure of EVI2 and mapping of other transcripts. Genomics 7: 555–565. [DOI] [PubMed] [Google Scholar]
- Chapman JA, Kirkness EF, Simakov O, Hampson SE, Mitros T, Weinmaier T, Rattei T, Balasubramanian PG, Borman J, Busam D, et al. 2010. The dynamic genome of Hydra. Nature 464: 592–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HJ, Rojas-Soto M, Oguni A, Kennedy MB. 1998. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron 20: 895–904. [DOI] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M. 2013. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 93: 269–309. [DOI] [PubMed] [Google Scholar]
- Cho W, Stahelin RV. 2006. Membrane binding and subcellular targeting of C2 domains. Biochim Biophys Acta 1761: 838–849. [DOI] [PubMed] [Google Scholar]
- Cichowski K, Jacks T. 2001. NF1 tumor suppressor gene function: Narrowing the GAP. Cell 104: 593–604. [DOI] [PubMed] [Google Scholar]
- Cichowski K, Santiago S, Jardim M, Johnson BW, Jacks T. 2003. Dynamic regulation of the Ras pathway via proteolysis of the NF1 tumor suppressor. Genes Dev 17: 449–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cnossen MH, van der Est MN, Breuning MH, van Asperen CJ, Breslau-Siderius EJ, van der Ploeg AT, de Goede-Bolder A, van den Ouweland AM, Halley DJ, Niermeijer MF. 1997. Deletions spanning the neurofibromatosis type 1 gene: Implications for genotype-phenotype correlations in neurofibromatosis type 1? Hum Mutat 9: 458–464. [DOI] [PubMed] [Google Scholar]
- Cooper JA, Kashishian A. 1993. In vivo binding properties of SH2 domains from GTPase-activating protein and phosphatidylinositol 3-kinase. Mol Cell Biol 13: 1737–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozier GE, Lockyer PJ, Reynolds JS, Kupzig S, Bottomley JR, Millard TH, Banting G, Cullen PJ. 2000. GAP1IP4BP contains a novel group I pleckstrin homology domain that directs constitutive plasma membrane association. J Biol Chem 275: 28261–28268. [DOI] [PubMed] [Google Scholar]
- Cuellar J, Valpuesta JM, Wittinghofer A, Sot B. 2017. Domain topology of human Rasal. Biol Chem 399: 63–72. [DOI] [PubMed] [Google Scholar]
- Dai Y, Walker SA, de Vet E, Cook S, Welch HC, Lockyer PJ. 2011. Ca2+-dependent monomer and dimer formation switches CAPRI Protein between Ras GTPase-activating protein (GAP) and RapGAP activities. J Biol Chem 286: 19905–19916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo I, Welti S, Bonneau F, Scheffzek K. 2006. A novel bipartite phospholipid-binding module in the neurofibromatosis type 1 protein. EMBO Rep 7: 174–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daumke O, Weyand M, Chakrabarti PP, Vetter IR, Wittinghofer A. 2004. The GTPase-activating protein Rap1GAP uses a catalytic asparagine. Nature 429: 197–201. [DOI] [PubMed] [Google Scholar]
- DeClue JE, Cohen BD, Lowy DR. 1991. Identification and characterization of the neurofibromatosis type 1 protein product. Proc Natl Acad Sci 88: 9914–9918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeClue JE, Papageorge AG, Fletcher JA, Diehl SR, Ratner N, Vass WC, Lowy DR. 1992. Abnormal regulation of mammalian p21ras contributes to malignant tumor growth in von Recklinghausen (type 1) neurofibromatosis. Cell 69: 265–273. [DOI] [PubMed] [Google Scholar]
- De Luca A, Buccino A, Gianni D, Mangino M, Giustini S, Richetta A, Divona L, Calvieri S, Mingarelli R, Dallapiccola B. 2003. NF1 gene analysis based on DHPLC. Hum Mutat 21: 171–172. [DOI] [PubMed] [Google Scholar]
- Der CJ, Finkel T, Cooper GM. 1986. Biological and biochemical properties of human rasH genes mutated at codon 61. Cell 44: 167–176. [DOI] [PubMed] [Google Scholar]
- Deraredj Nadim W, Chaumont-Dubel S, Madouri F, Cobret L, De Tauzia ML, Zajdel P, Benedetti H, Marin P, Morisset-Lopez S. 2016. Physical interaction between neurofibromin and serotonin 5-HT6 receptor promotes receptor constitutive activity. Proc Natl Acad Sci 113: 12310–12315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schepper S, Boucneau JM, Westbroek W, Mommaas M, Onderwater J, Messiaen L, Naeyaert JM, Lambert JL. 2006. Neurofibromatosis type 1 protein and amyloid precursor protein interact in normal human melanocytes and colocalize with melanosomes. J Invest Dermatol 126: 653–659. [DOI] [PubMed] [Google Scholar]
- Donovan S, Shannon KM, Bollag G. 2002. GTPase activating proteins: Critical regulators of intracellular signaling. Biochim Biophys Acta 1602: 23–45. [DOI] [PubMed] [Google Scholar]
- Drugan JK, Rogers-Graham K, Gilmer T, Campbell S, Clark GJ. 2000. The Ras/p120 GTPase-activating protein (GAP) interaction is regulated by the p120 GAP pleckstrin homology domain. J Biol Chem 275: 35021–35027. [DOI] [PubMed] [Google Scholar]
- Dunzendorfer-Matt T, Mercado EL, Maly K, McCormick F, Scheffzek K. 2016. The neurofibromin recruitment factor Spred1 binds to the GAP related domain without affecting Ras inactivation. Proc Natl Acad Sci 113: 7497–7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl LA, Falconieri V, Milne JL, Subramaniam S. 2017. Cryo-EM: Beyond the microscope. Curr Opin Struct Biol 46: 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435: 43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekvall S, Sjors K, Jonzon A, Vihinen M, Anneren G, Bondeson ML. 2014. Novel association of neurofibromatosis type 1-causing mutations in families with neurofibromatosis-Noonan syndrome. Am J Med Genet A 164A: 579–587. [DOI] [PubMed] [Google Scholar]
- Evdokimov AG, Tropea JE, Routzahn KM, Waugh DS. 2002. Crystal structure of the Yersinia pestis GTPase activator YopE. Protein Sci 11: 401–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahsold R, Hoffmeyer S, Mischung C, Gille C, Ehlers C, Kucukceylan N, Abdel-Nour M, Gewies A, Peters H, Kaufmann D, et al. 2000. Minor lesion mutational spectrum of the entire NF1 gene does not explain its high mutability but points to a functional domain upstream of the GAP-related domain. Am J Hum Genet 66: 790–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fantl WJ, Escobedo JA, Martin GA, Turck CW, del Rosario M, McCormick F, Williams LT. 1992. Distinct phosphotyrosines on a growth factor receptor bind to specific molecules that mediate different signaling pathways. Cell 69: 413–423. [DOI] [PubMed] [Google Scholar]
- Feng L, Yunoue S, Tokuo H, Ozawa T, Zhang D, Patrakitkomjorn S, Ichimura T, Saya H, Araki N. 2004. PKA phosphorylation and 14-3-3 interaction regulate the function of neurofibromatosis type I tumor suppressor, neurofibromin. FEBS Lett 557: 275–282. [DOI] [PubMed] [Google Scholar]
- Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. 2011. LOVD v.2.0: The next generation in gene variant databases. Hum Mutat 32: 557–563. [DOI] [PubMed] [Google Scholar]
- Franken SM, Scheidig AJ, Krengel U, Rensland H, Lautwein A, Geyer M, Scheffzek K, Goody RS, Kalbitzer HR, Pai EF, et al. 1993. Three-dimensional structures and properties of a transforming and a nontransforming glycine-12 mutant of p21H–ras. Biochemistry 32: 8411–8420. [DOI] [PubMed] [Google Scholar]
- Gamblin SJ, Smerdon SJ. 1998. GTPase-activating proteins and their complexes. Curr Opin Struct Biol 8: 195–201. [DOI] [PubMed] [Google Scholar]
- Gavriljuk K, Gazdag EM, Itzen A, Kotting C, Goody RS, Gerwert K. 2012. Catalytic mechanism of a mammalian Rab·RabGAP complex in atomic detail. Proc Natl Acad Sci 109: 21348–21353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke V, Moss SE. 2002. Annexins: From structure to function. Physiol Rev 82: 331–371. [DOI] [PubMed] [Google Scholar]
- Gerwert K, Mann D, Kotting C. 2017. Common mechanisms of catalysis in small and heterotrimeric GTPases and their respective GAPs. Biol Chem 398: 523–533. [DOI] [PubMed] [Google Scholar]
- Ghosh R, Bankaitis VA. 2011. Phosphatidylinositol transfer proteins: Negotiating the regulatory interface between lipid metabolism and lipid signaling in diverse cellular processes. Biofactors 37: 290–308. [DOI] [PubMed] [Google Scholar]
- Gremer L, Gilsbach B, Ahmadian MR, Wittinghofer A. 2008. Fluoride complexes of oncogenic Ras mutants to study the Ras–RasGap interaction. Biol Chem 389: 1163–1171. [DOI] [PubMed] [Google Scholar]
- Grewal T, Enrich C. 2006. Molecular mechanisms involved in Ras inactivation: The annexin A6-p120GAP complex. Bioessays 28: 1211–1220. [DOI] [PubMed] [Google Scholar]
- Grewal T, Evans R, Rentero C, Tebar F, Cubells L, de Diego I, Kirchhoff MF, Hughes WE, Heeren J, Rye KA, et al. 2005. Annexin A6 stimulates the membrane recruitment of p120GAP to modulate Ras and Raf-1 activity. Oncogene 24: 5809–5820. [DOI] [PubMed] [Google Scholar]
- Grewal T, Koese M, Tebar F, Enrich C. 2011. Differential regulation of RasGAPs in cancer. Genes Cancer 2: 288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann DH, Wood DL, Collins FS. 1991. Identification of the neurofibromatosis type 1 gene product. Proc Natl Acad Sci 88: 9658–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakimi MA, Speicher DW, Shiekhattar R. 2002. The motor protein kinesin-1 links neurofibromin and merlin in a common cellular pathway of neurofibromatosis. J Biol Chem 277: 36909–36912. [DOI] [PubMed] [Google Scholar]
- Han JW, McCormick F, Macara IG. 1991. Regulation of Ras-GAP and the neurofibromatosis-1 gene product by eicosanoids. Science 252: 576–579. [DOI] [PubMed] [Google Scholar]
- Han SS, Cooper DN, Upadhyaya MN. 2001. Evaluation of denaturing high performance liquid chromatography (DHPLC) for the mutational analysis of the neurofibromatosis type 1 (NF1) gene. Hum Genet 109: 487–497. [DOI] [PubMed] [Google Scholar]
- Hedman AC, Smith JM, Sacks DB. 2015. The biology of IQGAP proteins: Beyond the cytoskeleton. EMBO Rep 16: 427–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henkemeyer M, Rossi DJ, Holmyard DP, Puri MC, Mbamalu G, Harpal K, Shih TS, Jacks T, Pawson T. 1995. Vascular system defects and neuronal apoptosis in mice lacking ras GTPase-activating protein. Nature 377: 695–701. [DOI] [PubMed] [Google Scholar]
- Hettich L, Marshall M. 1994. Structural analysis of the Ras GTPase activating protein catalytic domain by semirandom mutagenesis: Implications for a mechanism of interaction with Ras-GTP. Cancer Res 54: 5438–5444. [PubMed] [Google Scholar]
- Hiatt KK, Ingram DA, Zhang Y, Bollag G, Clapp DW. 2001. Neurofibromin GTPase-activating protein-related domains restore normal growth in Nf1−/− cells. J Biol Chem 276: 7240–7245. [DOI] [PubMed] [Google Scholar]
- Hirata Y, Brems H, Suzuki M, Kanamori M, Okada M, Morita R, Llano-Rivas I, Ose T, Messiaen L, Legius E, et al. 2016. Interaction between a domain of the negative regulator of the Ras-ERK pathway, SPRED1 protein, and the GTPase-activating protein-related domain of neurofibromin is implicated in Legius syndrome and neurofibromatosis type 1. J Biol Chem 291: 3124–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollstein PE, Cichowski K. 2013. Identifying the ubiquitin ligase complex that regulates the NF1 tumor suppressor and Ras. Cancer Discov 3: 880–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeybee Genome Sequencing C. 2006. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443: 931–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hota PK, Buck M. 2012. Plexin structures are coming: Opportunities for multilevel investigations of semaphorin guidance receptors, their cell signaling mechanisms, and functions. Cell Mol Life Sci 69: 3765–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsueh YP, Roberts AM, Volta M, Sheng M, Roberts RG. 2001. Bipartite interaction between neurofibromatosis type I protein (neurofibromin) and syndecan transmembrane heparan sulfate proteoglycans. J Neurosci 21: 3764–3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwashita S, Song SY. 2008. RasGAPs: A crucial regulator of extracellular stimuli for homeostasis of cellular functions. Mol Biosyst 4: 213–222. [DOI] [PubMed] [Google Scholar]
- Izawa I, Tamaki N, Saya H. 1996. Phosphorylation of neurofibromatosis type 1 gene product (neurofibromin) by cAMP-dependent protein kinase. FEBS Lett 382: 53–59. [DOI] [PubMed] [Google Scholar]
- Jeong SY, Park SJ, Kim HJ. 2006. The spectrum of NF1 mutations in Korean patients with neurofibromatosis type 1. J Korean Med Sci 21: 107–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyabalan N, Clement JP. 2016. SYNGAP1: Mind the gap. Front Cell Neurosci 10: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Wang X, Ying J, Wong AH, Cui Y, Srivastava G, Shen ZY, Li EM, Zhang Q, Jin J, et al. 2007. Epigenetic silencing of a Ca2+-regulated Ras GTPase-activating protein RASAL defines a new mechanism of Ras activation in human cancers. Proc Natl Acad Sci 104: 12353–12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DR, Morrison DK, Wong G, McCormick F, Williams LT. 1990. PDGF β-receptor stimulates tyrosine phosphorylation of GAP and association of GAP with a signaling complex. Cell 61: 125–133. [DOI] [PubMed] [Google Scholar]
- Kashishian A, Kazlauskas A, Cooper JA. 1992. Phosphorylation sites in the PDGF receptor with different specificities for binding GAP and PI3 kinase in vivo. EMBO J 11: 1373–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann D. (ed.). 2008. Neurofibromatoses. Monographs in human genetics, Vol. 16 S. Karger, Basel, Switzerland. [Google Scholar]
- Kehrer-Sawatzki H, Maier C, Moschgath E, Elgar G, Krone W. 1998. Genomic characterization of the neurofibromatosis type 1 gene of Fugu rubripes. Gene 222: 145–153. [DOI] [PubMed] [Google Scholar]
- Kim JH, Liao D, Lau LF, Huganir RL. 1998. SynGAP: A synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron 20: 683–691. [DOI] [PubMed] [Google Scholar]
- King PD, Lubeck BA, Lapinski PE. 2013. Nonredundant functions for Ras GTPase-activating proteins in tissue homeostasis. Sci Signal 6: re1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose A, Ahmadian MR, Schuelke M, Scheffzek K, Hoffmeyer S, Gewies A, Schmitz F, Kaufmann D, Peters H, Wittinghofer A, et al. 1998. Selective disactivation of neurofibromin GAP activity in neurofibromatosis type 1. Hum Mol Genet 7: 1261–1268. [DOI] [PubMed] [Google Scholar]
- Kostenko EV, Mahon GM, Cheng L, Whitehead IP. 2004. The Sec14 homology domain regulates the cellular distribution and transforming activity of the Rho-specific guanine nucleotide exchange factor, Dbs. J Biol Chem 280: 2807–2817. [DOI] [PubMed] [Google Scholar]
- Kotting C, Kallenbach A, Suveyzdis Y, Wittinghofer A, Gerwert K. 2008. The GAP arginine finger movement into the catalytic site of Ras increases the activation entropy. Proc Natl Acad Sci 105: 6260–6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapivinsky G, Medina I, Krapivinsky L, Gapon S, Clapham DE. 2004. SynGAP-MUPP1-CaMKII synaptic complexes regulate P38 MAP kinase activity and NMDA receptor-dependent synaptic AMPA receptor potentiation. Neuron 43: 563–574. [DOI] [PubMed] [Google Scholar]
- Kupzig S, Deaconescu D, Bouyoucef D, Walker SA, Liu Q, Polte CL, Daumke O, Ishizaki T, Lockyer PJ, Wittinghofer A, et al. 2006. GAP1 family members constitute bifunctional Ras and Rap GTPase-activating proteins. J Biol Chem 281: 9891–9900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupzig S, Bouyoucef-Cherchalli D, Yarwood S, Sessions R, Cullen PJ. 2009. The ability of GAP1IP4BP to function as a Rap1 GTPase-activating protein (GAP) requires its Ras GAP-related domain and an arginine finger rather than an asparagine thumb. Mol Cell Biol 29: 3929–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurella VB, Richard JM, Parke CL, Lecour LF Jr, Bellamy HD, Worthylake DK. 2009. Crystal structure of the GTPase-activating protein-related domain from IQGAP1. J Biol Chem 284: 14857–14865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweh F, Zheng M, Kurenova E, Wallace M, Golubovskaya V, Cance WG. 2009. Neurofibromin physically interacts with the N-terminal domain of focal adhesion kinase. Mol Carcinog 48: 1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCour L Jr, Boyapati VK, Liu J, Li Z, Sacks DB, Worthylake DK. 2016. The structural basis for Cdc42-induced dimerization of IQGAPs. Structure 24: 1499–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Su YN, You HL, Chiou SC, Lin LC, Yang CC, Lee WC, Hwu WL, Hsieh FJ, Stephenson DA, et al. 2006. Identification of forty-five novel and twenty-three known NF1 mutations in Chinese patients with neurofibromatosis type 1. Hum Mutat 27: 832. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. 2004. Pleckstrin homology domains: Not just for phosphoinositides. Biochem Soc Trans 32: 707–711. [DOI] [PubMed] [Google Scholar]
- Li Y, Bollag G, Clark R, Stevens J, Conroy L, Fults D, Ward K, Friedman E, Samowitz W, Robertson M, et al. 1992. Somatic mutations in the neurofibromatosis 1 gene in human tumors. Cell 69: 275–281. [DOI] [PubMed] [Google Scholar]
- Ligeti E, Welti S, Scheffzek K. 2012. Inhibition and termination of physiological responses by GTPase activating proteins. Physiol Rev 92: 237–272. [DOI] [PubMed] [Google Scholar]
- Lin YL, Hsueh YP. 2008. Neurofibromin interacts with CRMP-2 and CRMP-4 in rat brain. Biochem Biophys Res Commun 369: 747–752. [DOI] [PubMed] [Google Scholar]
- Liu Q, Walker SA, Gao D, Taylor JA, Dai YF, Arkell RS, Bootman MD, Roderick HL, Cullen PJ, Lockyer PJ. 2005. CAPRI and RASAL impose different modes of information processing on Ras due to contrasting temporal filtering of Ca2+. J Cell Biol 170: 183–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens O, Cichowski K. 2014. An expanding role for RAS GTPase activating proteins (RAS GAPs) in cancer. Adv Biol Regul 55: 1–14. [DOI] [PubMed] [Google Scholar]
- Mangoura D, Sun Y, Li C, Singh D, Gutmann DH, Flores A, Ahmed M, Vallianatos G. 2006. Phosphorylation of neurofibromin by PKC is a possible molecular switch in EGF receptor signaling in neural cells. Oncogene 25: 735–745. [DOI] [PubMed] [Google Scholar]
- Marshall MS, Hill WS, Ng AS, Vogel US, Schaber MD, Scolnick EM, Dixon RA, Sigal IS, Gibbs JB. 1989. A C-terminal domain of GAP is sufficient to stimulate ras p21 GTPase activity. EMBO J 8: 1105–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GA, Viskochil D, Bollag G, McCabe PC, Crosier WJ, Haubruck H, Conroy L, Clark R, O’Connell P, Cawthon RM, et al. 1990. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell 63: 843–849. [DOI] [PubMed] [Google Scholar]
- Martin-Garcia F, Mendieta-Moreno JI, Lopez-Vinas E, Gomez-Puertas P, Mendieta J. 2012. The role of Gln61 in HRas GTP hydrolysis: A quantum mechanics/molecular mechanics study. Biophys J 102: 152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattocks C, Baralle D, Tarpey P, ffrench-Constant C, Bobrow M, Whittaker J. 2004. Automated comparative sequence analysis identifies mutations in 89% of NF1 patients and confirms a mutation cluster in exons 11-17 distinct from the GAP related domain. J Med Genet 41: e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClatchey AI, Cichowski K. 2012. SPRED proteins provide a NF-ty link to Ras suppression. Genes Dev 26: 1515–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin SK, Olsen SN, Dake B, De Raedt T, Lim E, Bronson RT, Beroukhim R, Polyak K, Brown M, Kuperwasser C, et al. 2013. The RasGAP gene, RASAL2, is a tumor and metastasis suppressor. Cancer Cell 24: 365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellert K, Lechner S, Ludeke M, Lamla M, Moller P, Kemkemer R, Scheffzek K, Kaufmann D. 2018. Restoring functional neurofibromin by protein transduction. Sci Rep 8: 6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merk A, Bartesaghi A, Banerjee S, Falconieri V, Rao P, Davis MI, Pragani R, Boxer MB, Earl LA, Milne JLS, et al. 2016. Breaking cryo-EM resolution barriers to facilitate drug discovery. Cell 165: 1698–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao W, Eichelberger L, Baker L, Marshall MS. 1996. p120 Ras GTPase-activating protein interacts with Ras-GTP through specific conserved residues. J Biol Chem 271: 15322–15329. [DOI] [PubMed] [Google Scholar]
- Milburn MV, Tong L, deVos AM, Brunger A, Yamaizumi Z, Nishimura S, Kim SH. 1990. Molecular switch for signal transduction: Structural differences between active and inactive forms of protooncogenic ras proteins. Science 247: 939–945. [DOI] [PubMed] [Google Scholar]
- Mishra AK, Lambright DG. 2016. Invited review: Small GTPases and their GAPs. Biopolymers 105: 431–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal R, Ahmadian MR, Goody RS, Wittinghofer A. 1996. Formation of a transition-state analog of the Ras GTPase reaction by Ras-GDP, tetrafluoroaluminate, and GTPase-activating proteins. Science 273: 115–117. [DOI] [PubMed] [Google Scholar]
- Nassar N, Hoffman GR, Manor D, Clardy JC, Cerione RA. 1998. Structures of Cdc42 bound to the active and catalytically compromised forms of Cdc42GAP. Nat Struct Biol 5: 1047–1052. [DOI] [PubMed] [Google Scholar]
- Neufeld G, Kessler O. 2008. The semaphorins: Versatile regulators of tumour progression and tumour angiogenesis. Nat Rev Cancer 8: 632–645. [DOI] [PubMed] [Google Scholar]
- Oh JS, Manzerra P, Kennedy MB. 2004. Regulation of the neuron-specific Ras GTPase-activating protein, synGAP, by Ca2+/calmodulin-dependent protein kinase II. J Biol Chem 279: 17980–17988. [DOI] [PubMed] [Google Scholar]
- Oinuma I, Ishikawa Y, Katoh H, Negishi M. 2004. The Semaphorin 4D receptor Plexin-B1 is a GTPase activating protein for R-Ras. Science 305: 862–865. [DOI] [PubMed] [Google Scholar]
- Oliveira AF, Yasuda R. 2014. Neurofibromin is the major Ras inactivator in dendritic spines. J Neurosci 34: 776–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan A, Lee JS, Ismat FA, Lu MM, Lawson ND, Kanki JP, Look AT, Epstein JA. 2009. Cardiac and vascular functions of the zebrafish orthologues of the type I neurofibromatosis gene NFI. Proc Natl Acad Sci 106: 22305–22310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai EF, Kabsch W, Krengel U, Holmes KC, John J, Wittinghofer A. 1989. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature 341: 209–214. [DOI] [PubMed] [Google Scholar]
- Pamonsinlapatham P, Hadj-Slimane R, Lepelletier Y, Allain B, Toccafondi M, Garbay C, Raynaud F. 2009. p120–Ras GTPase activating protein (RasGAP): A multi-interacting protein in downstream signaling. Biochimie 91: 320–328. [DOI] [PubMed] [Google Scholar]
- Pan X, Eathiraj S, Munson M, Lambright DG. 2006. TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature 442: 303–306. [DOI] [PubMed] [Google Scholar]
- Pascoe HG, Wang Y, Zhang X. 2015. Structural mechanisms of plexin signaling. Prog Biophys Mol Biol 118: 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patch AM, Christie EL, Etemadmoghadam D, Garsed DW, George J, Fereday S, Nones K, Cowin P, Alsop K, Bailey PJ, et al. 2015. Whole-genome characterization of chemoresistant ovarian cancer. Nature 521: 489–494. [DOI] [PubMed] [Google Scholar]
- Patrakitkomjorn S, Kobayashi D, Morikawa T, Wilson MM, Tsubota N, Irie A, Ozawa T, Aoki M, Arimura N, Kaibuchi K, et al. 2008. Neurofibromatosis type 1 (NF1) tumor suppressor, neurofibromin, regulates the neuronal differentiation of PC12 cells via its associating protein, CRMP-2. J Biol Chem 283: 9399–9413. [DOI] [PubMed] [Google Scholar]
- Pena V, Hothorn M, Eberth A, Kaschau N, Parret A, Gremer L, Bonneau F, Ahmadian MR, Scheffzek K. 2008. The C2 domain of SynGAP is essential for stimulation of the Rap GTPase reaction. EMBO Rep 9: 350–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan VT, Ding VW, Li F, Chalkley RJ, Burlingame A, McCormick F. 2010. The RasGAP proteins Ira2 and neurofibromin are negatively regulated by Gpb1 in yeast and ETEA in humans. Mol Cell Biol 30: 2264–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott C, Tovell H, Frayling IM, Cooper DN, Upadhyaya M. 2017. The NF1 somatic mutational landscape in sporadic human cancers. Hum Genomics 11: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner N, Miller SJ. 2015. A RASopathy gene commonly mutated in cancer: The neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer 15: 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauen KA. 2013. The RASopathies. Annu Rev Genomics Hum Genet 14: 355–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revencu N, Boon LM, Mulliken JB, Enjolras O, Cordisco MR, Burrows PE, Clapuyt P, Hammer F, Dubois J, Baselga E, et al. 2008. Parkes Weber syndrome, vein of Galen aneurysmal malformation, and other fast-flow vascular anomalies are caused by RASA1 mutations. Hum Mutat 29: 959–965. [DOI] [PubMed] [Google Scholar]
- Revencu N, Boon LM, Mendola A, Cordisco MR, Dubois J, Clapuyt P, Hammer F, Amor DJ, Irvine AD, Baselga E, et al. 2013. RASA1 mutations and associated phenotypes in 68 families with capillary malformation-arteriovenous malformation. Hum Mutat 34: 1632–1641. [DOI] [PubMed] [Google Scholar]
- Riccardi VM. 1992. Neurofibromatosis: Phenotype natural history, and pathogenesis, 2nd ed. The Johns Hopkins University Press, Baltimore, MD. [Google Scholar]
- Rittinger K, Walker PA, Eccleston JF, Nurmahomed K, Owen D, Laue E, Gamblin SJ, Smerdon SJ. 1997. Crystal structure of a small G protein in complex with the GTPase-activating protein ρGAP. Nature 388: 693–697. [DOI] [PubMed] [Google Scholar]
- Rojas RJ, Yohe ME, Gershburg S, Kawano T, Kozasa T, Sondek J. 2007. Galphaq directly activates p63ρGEF and Trio via a conserved extension of the Dbl homology-associated pleckstrin homology domain. J Biol Chem 282: 29201–29210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojnueangnit K, Xie J, Gomes A, Sharp A, Callens T, Chen Y, Liu Y, Cochran M, Abbott MA, Atkin J, et al. 2015. High incidence of Noonan syndrome features including short stature and pulmonic stenosis in patients carrying NF1 missense mutations affecting p.Arg1809: Genotype-phenotype correlation. Hum Mutat 36: 1052–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinfeld B, Munemitsu S, Clark R, Conroy L, Watt K, Crosier WJ, McCormick F, Polakis P. 1991. Molecular cloning of a GTPase activating protein specific for the Krev-1 protein p21rap1. Cell 65: 1033–1042. [DOI] [PubMed] [Google Scholar]
- Rudack T, Xia F, Schlitter J, Kotting C, Gerwert K. 2012. Ras and GTPase-activating protein (GAP) drive GTP into a precatalytic state as revealed by combining FTIR and biomolecular simulations. Proc Natl Acad Sci 109: 15295–15300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Tautz L, Mustelin T. 2007. The lipid-binding SEC14 domain. Biochim Biophys Acta 1771: 719–726. [DOI] [PubMed] [Google Scholar]
- Saito Y, Oinuma I, Fujimoto S, Negishi M. 2009. Plexin-B1 is a GTPase activating protein for M-Ras, remodelling dendrite morphology. EMBO Rep 10: 614–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangha N, Wu R, Kuick R, Powers S, Mu D, Fiander D, Yuen K, Katabuchi H, Tashiro H, Fearon ER, et al. 2008. Neurofibromin 1 (NF1) defects are common in human ovarian serous carcinomas and co-occur with TP53 mutations. Neoplasia 10: 1362–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro C, Maietta A, Giugliano T, Melis D, Perrotta S, Nigro V, Piluso G. 2015. Arg(1809) substitution in neurofibromin: Further evidence of a genotype-phenotype correlation in neurofibromatosis type 1. Eur J Hum Genet 23: 1460–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR. 2005. GTPase activating proteins: Structural and functional insights 18 years after discovery. Cell Mol Life Sci 62: 3014–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Welti S. 2012. Pleckstrin homology (PH) like domains—Versatile modules in protein–protein interaction platforms. FEBS Lett 586: 2662–2673. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Lautwein A, Kabsch W, Ahmadian MR, Wittinghofer A. 1996. Crystal structure of the GTPase-activating domain of human p120GAP and implications for the interaction with Ras. Nature 384: 591–596. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Kabsch W, Wiesmuller L, Lautwein A, Schmitz F, Wittinghofer A. 1997a. The Ras-RasGAP complex: Structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277: 333–338. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Lautwein A, Scherer A, Franken S, Wittinghofer A. 1997b. Crystallization and preliminary X-ray crystallographic study of the Ras- GTPase-activating domain of human p120GAP. Proteins 27: 315–318. [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Wiesmuller L, Kabsch W, Stege P, Schmitz F, Wittinghofer A. 1998a. Structural analysis of the GAP-related domain from neurofibromin and its implications. EMBO J 17: 4313–4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Wittinghofer A. 1998b. GTPase-activating proteins: Helping hands to complement an active site. Trends Biochem Sci 23: 257–262. [DOI] [PubMed] [Google Scholar]
- Scrima A, Thomas C, Deaconescu D, Wittinghofer A. 2008. The Rap–RapGAP complex: GTP hydrolysis without catalytic glutamine and arginine residues. EMBO J 27: 1145–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeburg PH, Colby WW, Capon DJ, Goeddel DV, Levinson AD. 1984. Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature 312: 71–75. [DOI] [PubMed] [Google Scholar]
- Sermon BA, Eccleston JF, Skinner RH, Lowe PN. 1996. Mechanism of inhibition by arachidonic acid of the catalytic activity of Ras GTPase-activating proteins. J Biol Chem 271: 1566–1572. [DOI] [PubMed] [Google Scholar]
- Sermon BA, Lowe PN, Strom M, Eccleston JF. 1998. The importance of two conserved arginine residues for catalysis by the Ras GTPase-activating protein, neurofibromin. J Biol Chem 273: 9480–9485. [DOI] [PubMed] [Google Scholar]
- Serth J, Lautwein A, Frech M, Wittinghofer A, Pingoud A. 1991. The inhibition of the GTPase activating protein–Ha-ras interaction by acidic lipids is due to physical association of the C-terminal domain of the GTPase activating protein with micellar structures. EMBO J 10: 1325–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. 2014. A glimpse of structural biology through X-ray crystallography. Cell 159: 995–1014. [DOI] [PubMed] [Google Scholar]
- Simanshu DK, Nissley DV, McCormick F. 2017. RAS proteins and their regulators in human Disease. Cell 170: 17–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirokmany G, Szidonya L, Kaldi K, Gaborik Z, Ligeti E, Geiszt M. 2005. Sec14 homology domain targets p50RhoGAP to endosomes and provides a link between Rab- and Rho GTPases. J Biol Chem 281: 6096–6105. [DOI] [PubMed] [Google Scholar]
- Smith JM, Hedman AC, Sacks DB. 2015. IQGAPs choreograph cellular signaling from the membrane to the nucleus. Trends Cell Biol 25: 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sot B, Kotting C, Deaconescu D, Suveyzdis Y, Gerwert K, Wittinghofer A. 2010. Unravelling the mechanism of dual-specificity GAPs. EMBO J 29: 1205–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sot B, Behrmann E, Raunser S, Wittinghofer A. 2013. Ras GTPase activating (RasGAP) activity of the dual specificity GAP protein Rasal requires colocalization and C2 domain binding to lipid membranes. Proc Natl Acad Sci 110: 111–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins CE, Galan JE. 2000. Modulation of host signaling by a bacterial mimic: Structure of the Salmonella effector SptP bound to Rac1. Mol Cell 6: 1449–1460. [DOI] [PubMed] [Google Scholar]
- Stowe IB, Mercado EL, Stowe TR, Bell EL, Oses-Prieto JA, Hernandez H, Burlingame AL, McCormick F. 2012. A shared molecular mechanism underlies the human rasopathies Legius syndrome and neurofibromatosis-1. Genes Dev 26: 1421–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Zhao Y, Kim SJ, Liu M, Jia L, Saunders TL, Zhu Y, Sun Y. 2011. SAG/RBX2/ROC2 E3 ubiquitin ligase is essential for vascular and neural development by targeting NF1 for degradation. Dev Cell 21: 1062–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassabehji M, Strachan T, Sharland M, Colley A, Donnai D, Harris R, Thakker N. 1993. Tandem duplication within a neurofibromatosis type 1 (NF1) gene exon in a family with features of Watson syndrome and Noonan syndrome. Am J Hum Genet 53: 90–95. [PMC free article] [PubMed] [Google Scholar]
- The I, Hannigan GE, Cowley GS, Reginald S, Zhong Y, Gusella JF, Hariharan IK, Bernards A. 1997. Rescue of a Drosophila NF1 mutant phenotype by protein kinase A. Science 276: 791–794. [DOI] [PubMed] [Google Scholar]
- Thomas SL, Deadwyler GD, Tang J, Stubbs EB Jr, Muir D, Hiatt KK, Clapp DW, De Vries GH. 2006. Reconstitution of the NF1 GAP-related domain in NF1-deficient human Schwann cells. Biochem Biophys Res Commun 348: 971–980. [DOI] [PubMed] [Google Scholar]
- Thomas L, Richards M, Mort M, Dunlop E, Cooper DN, Upadhyaya M. 2012. Assessment of the potential pathogenicity of missense mutations identified in the GTPase-activating protein (GAP)-related domain of the neurofibromatosis type-1 (NF1) gene. Hum Mutat 33: 1687–1696. [DOI] [PubMed] [Google Scholar]
- Tokuo H, Yunoue S, Feng L, Kimoto M, Tsuji H, Ono T, Saya H, Araki N. 2001. Phosphorylation of neurofibromin by cAMP-dependent protein kinase is regulated via a cellular association of NG,NG-dimethylarginine dimethylaminohydrolase. FEBS Lett 494: 48–53. [DOI] [PubMed] [Google Scholar]
- Tong J, Hannan F, Zhu Y, Bernards A, Zhong Y. 2002. Neurofibromin regulates G protein-stimulated adenylyl cyclase activity. Nat Neurosci 5: 95–96. [DOI] [PubMed] [Google Scholar]
- Trahey M, McCormick F. 1987. A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science 238: 542–545. [DOI] [PubMed] [Google Scholar]
- Trahey M, Wong G, Halenbeck R, Rubinfeld B, Martin GA, Ladner M, Long CM, Crosier WJ, Watt K, Koths K, et al. 1988. Molecular cloning of two types of GAP complementary DNA from human placenta. Science 242: 1697–1700. [DOI] [PubMed] [Google Scholar]
- Tsai MH, Yu CL, Wei FS, Stacey DW. 1989. The effect of GTPase activating protein upon ras is inhibited by mitogenically responsive lipids. Science 243: 522–526. [DOI] [PubMed] [Google Scholar]
- Upadhyaya M, Cooper DN., ed. 2012. Neurofibromatosis type 1—Molecular and cellular biology. Springer, New York. [Google Scholar]
- Upadhyaya M, Maynard J, Osborn M, Harper PS. 1997a. Six novel mutations in the neurofibromatosis type 1 (NF1) gene. Hum Mutat 10: 248–250. [DOI] [PubMed] [Google Scholar]
- Upadhyaya M, Osborn MJ, Maynard J, Kim MR, Tamanoi F, Cooper DN. 1997b. Mutational and functional analysis of the neurofibromatosis type 1 (NF1) gene. Hum Genet 99: 88–92. [DOI] [PubMed] [Google Scholar]
- Upadhyaya M, Huson SM, Davies M, Thomas N, Chuzhanova N, Giovannini S, Evans DG, Howard E, Kerr B, Griffiths S, et al. 2007. An absence of cutaneous neurofibromas associated with a 3-bp inframe deletion in exon 17 of the NF1 gene (c.2970-2972 delAAT): Evidence of a clinically significant NF1 genotype-phenotype correlation. Am J Hum Genet 80: 140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallee B, Doudeau M, Godin F, Gombault A, Tchalikian A, de Tauzia ML, Benedetti H. 2012. Nf1 RasGAP inhibition of LIMK2 mediates a new cross-talk between Ras and ρ pathways. PLoS ONE 7: e47283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Minkelen R, van Bever Y, Kromosoeto JN, Withagen-Hermans CJ, Nieuwlaat A, Halley DJ, van den Ouweland AM. 2014. A clinical and genetic overview of 18 years neurofibromatosis type 1 molecular diagnostics in the Netherlands. Clin Genet 85: 318–327. [DOI] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A. 2001. The guanine nucleotide-binding switch in three dimensions. Science 294: 1299–1304. [DOI] [PubMed] [Google Scholar]
- Viskochil D, Buchberg AM, Xu G, Cawthon RM, Stevens J, Wolff RK, Culver M, Carey JC, Copeland NG, Jenkins NA, et al. 1990. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell 62: 187–192. [DOI] [PubMed] [Google Scholar]
- Vogel US, Dixon RA, Schaber MD, Diehl RE, Marshall MS, Scolnick EM, Sigal IS, Gibbs JB. 1988. Cloning of bovine GAP and its interaction with oncogenic ras p21. Nature 335: 90–93. [DOI] [PubMed] [Google Scholar]
- Volta M, Calza S, Roberts AM, Roberts RG. 2010. Characterisation of the interaction between syndecan-2, neurofibromin and CASK: Dependence of interaction on syndecan dimerization. Biochem Biophys Res Commun 391: 1216–1221. [DOI] [PubMed] [Google Scholar]
- von Recklinghausen FD. 1882. Über die multiplen Fibrome der Haut und ihre Beziehung zu den multiplen Neuromen. Hirschwald, Berlin. [PubMed] [Google Scholar]
- Wakioka T, Sasaki A, Kato R, Shouda T, Matsumoto A, Miyoshi K, Tsuneoka M, Komiya S, Baron R, Yoshimura A. 2001. Spred is a Sprouty-related suppressor of Ras signalling. Nature 412: 647–651. [DOI] [PubMed] [Google Scholar]
- Walkup WGt, Washburn L, Sweredoski MJ, Carlisle HJ, Graham RL, Hess S, Kennedy MB. 2015. Phosphorylation of synaptic GTPase-activating protein (synGAP) by Ca2+/calmodulin-dependent protein kinase II (CaMKII) and cyclin-dependent kinase 5 (CDK5) alters the ratio of its GAP activity toward Ras and Rap GTPases. J Biol Chem 290: 4908–4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace MR, Marchuk DA, Andersen LB, Letcher R, Odeh HM, Saulino AM, Fountain JW, Brereton A, Nicholson J, Mitchell AL, et al. 1990. Type 1 neurofibromatosis gene: Identification of a large transcript disrupted in three NF1 patients. Science 249: 181–186. [DOI] [PubMed] [Google Scholar]
- Wang HF, Shih YT, Chen CY, Chao HW, Lee MJ, Hsueh YP. 2011. Valosin-containing protein and neurofibromin interact to regulate dendritic spine density. J Clin Invest 121: 4820–4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, He H, Srivastava N, Vikarunnessa S, Chen YB, Jiang J, Cowan CW, Zhang X. 2012. Plexins are GTPase-activating proteins for Rap and are activated by induced dimerization. Sci Signal 5: ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welti S, Fraterman S, D’Angelo I, Wilm M, Scheffzek K. 2007. The sec14 homology module of neurofibromin binds cellular glycerophospholipids: Mass spectrometry and structure of a lipid complex. J Mol Biol 366: 551–562. [DOI] [PubMed] [Google Scholar]
- Welti S, Kuhn S, D’Angelo I, Brugger B, Kaufmann D, Scheffzek K. 2011. Structural and biochemical consequences of NF1 associated nontruncating mutations in the Sec14-PH module of neurofibromin. Hum Mutat 32: 191–197. [DOI] [PubMed] [Google Scholar]
- Wittinghofer A. 1997. Signaling mechanistics: Aluminum fluoride for molecule of the year. Curr Biol 7: R682–R685. [DOI] [PubMed] [Google Scholar]
- Wittinghofer A, Vetter IR. 2011. Structure–function relationships of the G domain, a canonical switch motif. Annu Rev Biochem 80: 943–971. [DOI] [PubMed] [Google Scholar]
- Wittinghofer A, Waldmann H. 2000. Ras—A molecular switch involved in tumor formation. Angewandte Chemie 39: 4173–4390. [DOI] [PubMed] [Google Scholar]
- Wu R, Lopez-Correa C, Rutkowski JL, Baumbach LL, Glover TW, Legius E. 1999. Germline mutations in NF1 patients with malignancies. Genes Chromosomes Cancer 26: 376–380. [PubMed] [Google Scholar]
- Wurtele M, Renault L, Barbieri JT, Wittinghofer A, Wolf E. 2001. Structure of the ExoS GTPase activating domain. FEBS Lett 491: 26–29. [DOI] [PubMed] [Google Scholar]
- Xie K, Colgan LA, Dao MT, Muntean BS, Sutton LP, Orlandi C, Boye SL, Boye SE, Shih CC, Li Y, et al. 2016. nf1 is a direct g protein effector essential for opioid signaling to Ras in the striatum. Curr Biol 26: 2992–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Gutmann DH. 1997. Mutations in the GAP-related domain impair the ability of neurofibromin to associate with microtubules. Brain Res 759: 149–152. [DOI] [PubMed] [Google Scholar]
- Xu GF, Lin B, Tanaka K, Dunn D, Wood D, Gesteland R, White R, Weiss R, Tamanoi F. 1990a. The catalytic domain of the neurofibromatosis type 1 gene product stimulates ras GTPase and complements ira mutants of S. cerevisiae. Cell 63: 835–841. [DOI] [PubMed] [Google Scholar]
- Xu GF, O’Connell P, Viskochil D, Cawthon R, Robertson M, Culver M, Dunn D, Stevens J, Gesteland R, White R, et al. 1990b. The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell 62: 599–608. [DOI] [PubMed] [Google Scholar]
- Yarwood S, Bouyoucef-Cherchalli D, Cullen PJ, Kupzig S. 2006. The GAP1 family of GTPase-activating proteins: Spatial and temporal regulators of small GTPase signalling. Biochem Soc Trans 34: 846–850. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Parada LF. 2001. Neurofibromin, a tumor suppressor in the nervous system. Exp Cell Res 264: 19–28. [DOI] [PubMed] [Google Scholar]