Abstract
Recent genome-wide analysis of neuroblastoma (NBL) revealed amplification and heterozygous mutation of anaplastic lymphoma kinase (ALK) are responsible for oncogenicity, frequently observed during relapses. A 3-year-old girl with relapsed high-risk NBL had a heterozygous ALK F1245L mutation at diagnosis, which became homozygous due to uniparental disomy (UPD) of the entire chromosome 2, confirmed by single nucleotide polymorphism array and variant allele frequency of this mutation. The ALK inhibitor, crizotinib, failed to control the tumor and the patient died of the disease. Further genomic analysis using targeted capture sequencing for 381 genes related to pediatric cancers identified more alterations acquired at relapse, such as TSC complex subunit 2 and protein tyrosine phosphatase receptor type D. In addition to these several acquired mutations, this extremely rare duplication of ALK mutation might explain the aggressive clinical course after relapse, because acquired UPD, resulting in the duplication of an oncogenic mutation, has been reported for various neoplasms. Although a clinical benefit of ALK inhibitors in patients with NBL has not been confirmed yet, a treatment based on the ALK mutation status will be promising in future using more potent next-generation ALK inhibitors.
Keywords: neuroblastoma, ALK, crizotinib, uniparental disomy, pediatric cancer, relapse
Introduction
Neuroblastoma (NBL) is a common solid tumor in children that originates from primordial neural crest cells (1). Despite multidisciplinary therapies, patients with high-risk NBL have a dismal prognosis. Recent genome-wide analyses have revealed anaplastic lymphoma kinase (ALK) gene alterations, amplification, and activating heterozygous mutations in the tyrosine kinase domain (TKD) in approximately 8% of primary NBLs (2–5). ALK mutations, often detected in heterozygous, are involved in cell proliferation and invasion and more frequently occur in high-risk and relapsed cases (1,6), while homozygous ALK mutations are quite rare (4,7). The discovery of ALK alterations provides the first tractable oncogenic target in NBL and facilitates the clinical application of ALK inhibitors, such as crizotinib (8). Here we present the case of a female patient with relapsed high-risk NBL harboring a heterozygous ALK F1245L mutation at the time of the initial diagnosis. This mutation became homozygous at relapse due to uniparental disomy (UPD), which was associated with aggressive tumor behavior after relapse. We further analyzed genetic alterations that may explain the molecular mechanism for NBL relapse.
Materials and methods
Samples
Biopsy samples at primary and relapse were subjected to this analysis after receiving written, informed consent according to protocols approved by the Human Genome, Gene Analysis Research Ethics Committee of the University of Tokyo and St. Luke's International Hospital. No matched normal sample was available. Genomic DNA of each sample was isolated by NucleoSpin DNA RapidLyse kit (Macherey-Nagel Gmbh & Co., Düren, Germany) according to manufacturer's protocol.
Direct sequencing
Direct sequencing of ALK was performed according to manufacturer's protocol with the use of an ABI 3500 Genetic Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The following set of primers were used: ALK-24_F: GCTGCCCATGTTTACAGAATGC; ALK-24_R: TCCATCGAGGAACTTGCTAC; ALK-25_F: CTGGTGTAGCTGCATGTTC; ALK-25_R: CATCCTACATCCAAATGGCTC; ALK-26_F: AATCCTAGTGATGGCCGTTG; ALK-26_R: GGAGATGATGTAAGGGACAAGC.
Targeted capture sequencing
Targeted capture was performed using a SureSelect custom kit (Agilent Technologies, Inc., Santa Clara, CA, USA) according to the manufacturer's protocol (9–11). The custom bait library was designed to include: i) all coding exons of 367 genes; ii) untranslated regions and introns of 16 genes (CD274, CTNNB1, ERG, ETV1, ETV4, EWSR1, FEV, FLI1, FOXO1, FUS, INO80D, NCOA1, NCOA2, NOTCH1, PAX3, and PAX7) for detecting breakpoints of structural variations; iii) 110,000 bases surrounding TERT for detecting TERT rearrangement; iv) promoter and enhancer regions of FGFR3, MYC and TERT; v) 11 microRNA genes (MIR100, MIRLET7A1, MIRLET7A2, MIRLET7A3, MIRLET7B, MIRLET7C, MIRLET7D, MIRLET7E, MIRLET7F1, MIRLET7F2, MIRLET7G), and vi) 3,527 SNP positions for copy number analysis. The total of 381 targeted genes and regions besides the SNP positions were selected to include: i) genes adopted in more than one of the following existing gene panels: MSK-IMPACT (12) CMS400 (Thermo Fisher Scientific, Inc.), FoundationOne (Foundation Medicine, Cambridge, MA, USA), Human Comprehensive Cancer Panel (Qiagen GmbH, Hilden, Germany); ii) the most frequently mutated 20 genes in each type of malignancy according to Catalogue of Somatic Mutations in Cancer (COSMIC) v78, and iii) genes that were recurrently affected in pediatric malignancies including neuroblastoma (6,13,14), hepatoblastoma (15,16), pleuropulmonary blastoma (17,18), rhabdomyosarcoma (19,20), Ewing's sarcoma (21,22), and germ cell tumor (23).
Captured targets were subjected to sequencing using a HiSeq 2000 (Illumina, Inc., San Diego, CA, USA) with a standard 125-bp pairedend read protocol according to the manufacturer's instructions (24). With mean depths of 383 and 381 in paired primary-relapse specimens, respectively, sequence alignment and mutation calling was performed using our in-house pipeline ‘Genomon v.2.5.0’, as previously described (25) in which reads that had either a mapping quality score of <25, a base quality score of <30, or five or more mismatched bases were excluded from the analysis. Candidate mutations with i) a variant allele frequency in tumor samples ≥0.1; and ii) a EBcall (26) (Empirical Bayesian mutation calling) P ≤1 ×10−10 was adopted and filtered by excluding i) synonymous mutations and variants without complete ORF information; ii) known variants listed in the 1000 Genomes Project (May 2011 release); NCBI SNP database (dbSNP) build 131; National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP) 5400; the Human Genome Variation Database (HGVD; October 2013 release); or our in-house SNP database; iii) variants present only in unidirectional reads; iv) variants occurring in repetitive genomic regions; v) variants with <5 supporting reads in tumor samples; and vi) all variants found in nonpaired normal samples (n=30) showing an allele frequency of >0.0025. Putative structural variations were manually curated and further filtered by removing those with: i) <20 supporting reads in tumor samples; ii) a VAF of <0.10 in tumor samples; and iii) all variants found in normal samples. Finally, mapping errors were removed by visual inspection on the Integrative Genomics Viewer (IGV) browser (27).
SNP Array Analysis
SNP array analysis was performed using GeneChip Human Mapping Cytoscan HD (Affymetrix; Thermo Fisher Scientific, Inc.). Genome-wide detection of copy number abnormalities or allelic imbalances was processed using Copy Number Analysis for Affymetrix GeneChip (CNAG)/Allele-Specific Copy Number using Anonymous References (AsCNAR) v3.5.1 software (28).
Results
Aggressive clinical course in a relapsed NBL patient
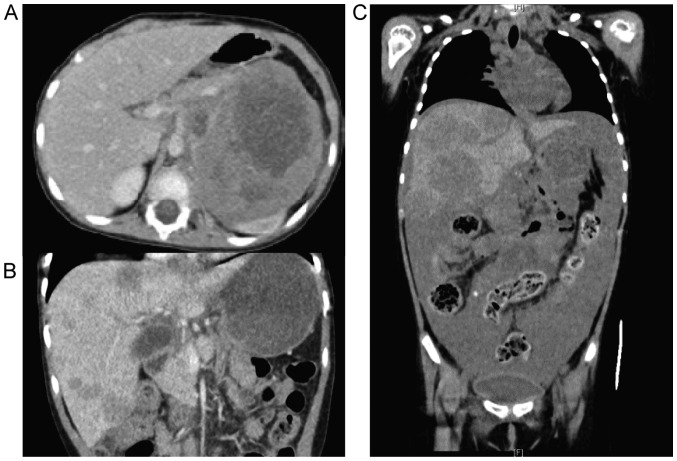
A 3-year-old girl was diagnosed with high-risk NBL of the left adrenal gland with bone and marrow metastases. Neuron specific enolase (NSE) and urine vanillylmandelic acid (VMA) levels were elevated (215 µg/l and 8.4 mg/l, respectively), and MYCN amplification was present. Immunohistochemical analyses revealed tumor cells positive for ALK. The patient received five courses of chemotherapy followed by high dose chemotherapy (busulfan and melphalan) with autologous hematopoietic stem cell transplantation, surgical gross total resection of the primary tumor, and radiation therapy to the tumor bed (19.8 Gy). The combination therapy successfully induced complete remission. However, 12 months after the diagnosis, she suffered from local tumor recurrence at the primary site. Two cycles of topotecan and cyclophosphamide (TC) and three of ifosfamide, cisplatin, and etoposide (ICE) gradually reduced the tumor size. After a fourth course of ICE, the tumor rapidly grew and urgent radiotherapy, with a combination of vincristine and irinotecan, was required. After obtaining approval from the institutional review board, crizotinib was administered for 28 days. Unfortunately, the tumor progressed and she succumbed to the disease (Fig. 1).
Figure 1.
Computed tomography imaging during the clinical course of the present case, (A) at diagnosis of relapsed neuroblastoma, (B) before administration of crizotinib, and (C) 28 d after the initiation of crizotinib.
Molecular analysis of paired diagnosis-relapse samples
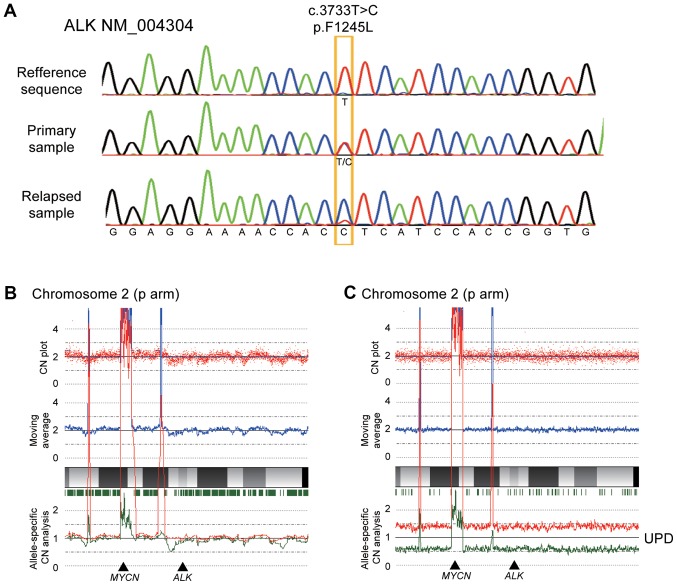
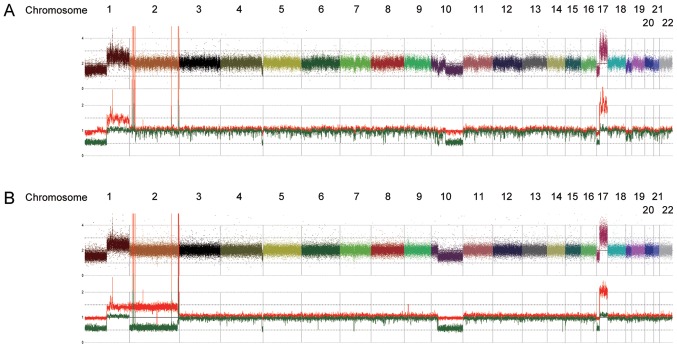
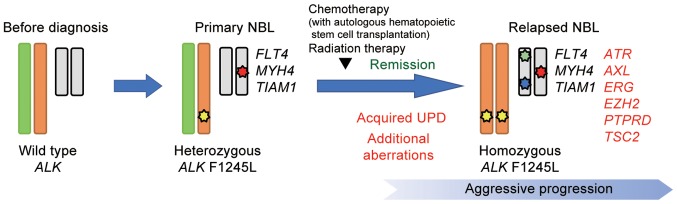
After obtaining informed consent from her guardians, DNA was extracted from biopsy samples of the adrenal tumor both at diagnosis and after relapse. Direct sequencing was performed to detect ALK mutations, and a F1245L mutation (c.T3733C; TTC>TCC) was identified in both the sample at diagnosis and relapse, which is a common activating mutation in NBL. Intriguingly, the relapse sample exclusively contained the mutated allele, whereas the diagnosis sample clearly showed a heterozygous change (Fig. 2A). In accordance with this, SNP arrays analysis in the relapse sample revealed acquired UPD of chromosome 2, including the ALK locus, in which the wild-type allele was likely to be lost by chromosomal mis-segregation with duplication of the mutated allele (Fig. 2B and C). We also detected several shared copy number alterations in both the diagnosis and relapse samples, including a focal amplification of 2p24.2–24.3 comprising the MYCN locus (Fig. 3). We further analyzed other genetic alterations, which might have been associated with disease recurrence and aggressiveness, using targeted capture sequencing (TCS) for 381 genes related to pediatric cancers (U-Tokyo Onco-panel v1). Because only tumor samples were analyzed, candidate mutations were filtered referring to our previous study (25), where we have confirmed the somatic mutations detected in TCS by using matched normal samples. We identified four and ten variants in the diagnosis and relapse samples, respectively (Tables I and II). All mutations detected at diagnosis were also detected in relapse sample. The ALK F1245L mutation was detected in both the diagnosis (variant allele frequency (VAF)=0.47) and relapse samples (VAF=0.83), which is consistent with other results showing duplication of ALK at relapse. Furthermore, alterations in TSC2 (p.P1092fs), PTPRD (translocation to chromosome 6), ATR (p.T1310S), AXL (p.A549S), ERG (p.S315X), and EZH2 (p.E204X) were acquired at relapse (Fig. 4).
Figure 2.
(A) ALK (NM_004304) F1245L mutation status by direct sequencing at diagnosis (heterozygous) and after relapse (homozygous). Copy number analysis using a SNP array for chromosome 2 including the (B) ALK gene region in diagnosis and (C) relapse samples. Red dots at the top represent the signal from each probe (raw data), and blue line indicates the moving average of red dots. Red and green lines at the bottom represent allele-specific copy number. UPD was observed in the relapse sample. UPD, niparental disomy.
Figure 3.
Results of SNP array analysis in (A) primary sample and (B) relapsed sample. Colorful dots at the top represent signal of each probe (raw data). Red and green lines at the bottom show allele specific copy number.
Table I.
Mutation calls of targeted capture sequencing.
| Sample | Chr | Start | End | Ref | Alt | Gene | Mutation type | Amino acid change | VAF |
|---|---|---|---|---|---|---|---|---|---|
| Primary | 2 | 29436860 | 29436860 | A | G | ALK | SNV | NM_004304:exon24:c.T3733C:p.F1245L | 0.469 |
| Relapse | 2 | 29436860 | 29436860 | A | G | ALK | SNV | NM_004304:exon24:c.T3733C:p.F1245L | 0.825 |
| Relapse | 3 | 142253938 | 142253938 | G | C | ATR | SNV | NM_001184:exon21:c.C3929G:p.T1310S | 0.409 |
| Primary | 5 | 180043943 | 180043943 | C | G | FLT4 | SNV | NM_182925:exon22:c.G3053C:p.S1018T | 0.425 |
| Relapse | 5 | 180043943 | 180043943 | C | G | FLT4 | SNV | NM_182925:exon22:c.G3053C:p.S1018T | 0.496 |
| Relapse | 7 | 148525847 | 148525847 | C | A | EZH2 | Stopgain | NM_001203247:exon6:c.G610T:p.E204X | 0.387 |
| Relapse | 16 | 2129420 | 2129420 | − | G | TSC2 | Frameshift | NM_000548:exon28:c.3276dupG:p. P1092fs | 0.245 |
| Primary | 17 | 10361019 | 10361019 | C | A | MYH4 | Stopgain | NM_017533:exon16:c.G1615T:p.E539X | 1 |
| Relapse | 17 | 10361019 | 10361019 | C | A | MYH4 | Stopgain | NM_017533:exon16:c.G1615T:p.E539X | 0.7 |
| Relapse | 19 | 41754659 | 41754660 | CG | TC | AXL | SNV | NM_021913:exon14:c.CG1644_1645TC: p.A549S | 0.429 |
| Primary | 21 | 32493125 | 32493125 | C | G | TIAM1 | SNV | NM_003253:exon29:c.G4337C:p.R1446T | 0.506 |
| Relapse | 21 | 39772318 | 39772318 | G | T | ERG | Stopgain | NM_001291391:exon8:c.C944A:p.S315X | 0.43 |
| Relapse | 21 | 32493125 | 32493125 | C | G | TIAM1 | SNV | NM_003253:exon29:c.G4337C:p.R1446T | 0.552 |
Chr, chromosome; SNV, single nucleotide variant; VAF, variant allele frequency
Table II.
Structural variant call of targeted capture sequencing.
| Sample | Chr Gene 1 | Position Gene 1 | Direction Gene 1 | Chr Gene 2 | Position Gene 2 | Direction Gene 2 | Variant type | Gene 1 | Gene 2 | VAF |
|---|---|---|---|---|---|---|---|---|---|---|
| Relapse | 6 | 138269463 | − | 9 | 8338848 | − | Translocation | − | PTPRD | 0.5745 |
Chr, chromosome; VAF, variant allele frequency
Figure 4.
Results of the genomic analysis and hypothetical model of aggressive progression in this case. A heterozygous ALK F1245 mutation was detected at diagnosis. This mutation became homozygous at relapse due to acquired UPD at chromosome 2, leading to both the duplication of the mutant allele and loss of the wild type allele. The duplication might be due to clonal evolution, acquired later in the process of relapse. UPD, uniparental disomy.
Discussion
The ALK mutation hotspots are located within the intracellular TKD at codons F1174, R1275, and F1245, accounting for 85% of the ALK mutations found in NBL (2–5). Germline ALK mutations, such as R1275, have also been identified as the major cause of hereditary NBL; however, their penetrance has been considered relatively low (29–31). In contrast, other hotspot mutations (F1174 and F1245) are rarely observed in familial cases. These F1174 and F1245 germline mutations have been linked to severe neurocognitive defects and abnormalities (30), which is consistent with reports that mutations in F1174 and F1245 have strong effects on the nonphosphorylated ALK TKD activity (5). Because both F1174 and F1245 are located in the region inhibiting ligand-independent activation, they impair self-inhibition and enhance the tyrosine kinase activity (32).
We detected a heterozygous ALK F1245 mutation at diagnosis that became homozygous at relapse in the present case. Homozygous ALK mutations have been rarely reported as far (4,7). This duplication at relapse may lead to growth advantage of tumor cells and be associated with disease progression. In fact, acquired UPD, resulting in the duplication of an oncogenic mutation, has been reported for various neoplasms. We have previously shown that the aggressive transformation of juvenile myelomonocytic leukemia is associated with the duplication of the oncogenic KRAS due to acquired UPD (33). Further, a previous report has revealed that the homozygous ALK F1174S mutation due to acquired UPD during the disease course may be responsible for the very rapid progress of NBL and resistance to chemotherapy (7). An enhanced oncogenic potential has also been reported in homozygous EGFR mutations on TKD in patients with non-small cell lung cancer, particularly in metastatic cases (34). Thus, homozygous TKD mutations are associated with an aggressive tumor progression in patients. In addition to the strong effects of the F1245 mutation on the ALK TKD activity, the bi-allelic presence of the ALK oncogenic mutation due to acquired UPD may have conferred further aggressiveness to the tumor during the relapse in this case.
In our comprehensive analysis, in addition to the ALK F1245 mutation and MYCN amplification, we also detected somatic oncogenic alterations comprising PTPRD (chromosomal rearrangement) and TSC2 (p.P1092fs) at relapse. PTPRD is a member of a large family of protein tyrosine phosphatases and functions as a tumor suppressor by destabilizing aurora kinase A (35). The low expression of PTPRD is associated with high-risk NBL with MYCN amplification (36). TSC2 is a tumor suppressor gene that is responsible for tuberous sclerosis, which is an autosomal dominant disorder characterized by the formation of hamartomas in various organs. The inactivation of TSC2 results in TSC1-TSC2 dissociation and markedly impairs the ability of TSC2 to inhibit mTOR signals, leading to cell proliferation and disease progression (37). A TSC2 mutation has not been reported in patients with NBL. However, because alterations of PTPRD and TSC2 occurred in a heterozygous setting, the functional relevance of these genes in the tumor progression of our case remains unclear. We also identified a truncated mutation of EZH2 (p.E204X), which is an essential component of the polycomb repressive complex 2. The pathogenic role of the truncated mutation of EZH2 is unclear because most EZH2 mutations reported in NBL cases were gain of function mutations mediating the epigenetic silencing of a tumor suppressor (38).
In the present case, all detected single nucleotide variants were observed in the major clone (VAF >0.35). None of acquired alterations at relapse were detected in primary sample even as the minor clone (VAF <0.35), although there was a technical limitation on our TCS, which cannot detect mutations with low allele frequency (VAF <0.05). Because no minor variants were detected, intratumor heterogeneity was not defined in the present case. Intratumor heterogeneity leads to selective outgrowth of clones that have a phenotypic advantage (39). The occurrence or presence of bi-allelic change of the ALK mutation due to acquired UPD and other acquired mutations such as TSC2 and PTPRD might be involved in tumor relapse and chemo-resistance in the present cases.
We report a patient with relapsed NBL harboring ALK F1245 mutation, which was heterozygous at diagnosis and became homozygous at relapse with resistance to crizotinib. This duplication of ALK mutation was extremely rare, and may be associated with aggressive behavior after relapse in this case. As previously reported, the ALK F1245 mutations would be suggested to be relatively crizotinib-resistant (5). Although a clinical benefit of ALK inhibitors in patients with NBL has not been confirmed, more potent next-generation ALK inhibitors have been developed and clinical trials to estimate their efficacy are currently underway (NCT03107988) (40,41). In future, a treatment based on the ALK mutation status will be promising.
Acknowledgements
The authors would like to thank Ms. M Matsumura, Ms. K Yin, and Ms. F Saito (The University of Tokyo) for their technical assistance; Dr T Okunushi and Dr M Hino (Chiba University) for collecting samples; and Dr K Yoshida, Dr Y Fujii, Dr K Kataoka (Kyoto University), and Dr Y Shiraishi (The University of Tokyo) for help with SNP array and next-generation sequencing.
Funding
The present study was supported by KAKENHI (grant no. 17H04224; received by JT) from the Japan Society of Promotion of Science and Project for Cancer Research and Therapeutic Evolution (P-CREATE) (grant no. 16cm0106509h001; received by JT) by the Japan Agency for Medical Research and Development (AMED).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
SK, DH, MSeki, MSekig, JT and AM wrote the manuscript; SK, DH, MSeki, YY, AD, SH, YH, and AM collected and analyzed data; SK, MSekig, and SM performed experiments; MK, SO, JT and AM conceived the study; JT designed the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the University of Tokyo and St. Luke's International Hospital.
Patient consent for publication
Prior written informed consent was obtained from the patient's guardians.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Cheung NK, Dyer MA. Neuroblastoma: Developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer. 2013;13:397–411. doi: 10.1038/nrc3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.George RE, Sanda T, Hanna M, Fröhling S, Luther W, II, Zhang J, Ahn Y, Zhou W, London WB, McGrady P, et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–978. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janoueix-Lerosey I, Lequin D, Brugières L, Ribeiro A, de Pontual L, Combaret V, Raynal V, Puisieux A, Schleiermacher G, Pierron G, et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature. 2008;455:967–970. doi: 10.1038/nature07398. [DOI] [PubMed] [Google Scholar]
- 4.Chen Y, Takita J, Choi YL, Kato M, Ohira M, Sanada M, Wang L, Soda M, Kikuchi A, Igarashi T, et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature. 2008;455:971–974. doi: 10.1038/nature07399. [DOI] [PubMed] [Google Scholar]
- 5.Bresler SC, Weiser DA, Huwe PJ, Park JH, Krytska K, Ryles H, Laudenslager M, Rappaport EF, Wood AC, McGrady PW, et al. ALK mutations confer differential oncogenic activation and sensitivity to ALK inhibition therapy in neuroblastoma. Cancer Cell. 2014;26:682–694. doi: 10.1016/j.ccell.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pugh TJ, Morozova O, Attiyeh EF, Asgharzadeh S, Wei JS, Auclair D, Carter SL, Cibulskis K, Hanna M, Kiezun A, et al. The genetic landscape of high-risk neuroblastoma. Nat Genet. 2013;45:279–284. doi: 10.1038/ng.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martinsson T, Eriksson T, Abrahamsson J, Caren H, Hansson M, Kogner P, Kamaraj S, Schönherr C, Weinmar J, Ruuth K, et al. Appearance of the novel activating F1174S ALK mutation in neuroblastoma correlates with aggressive tumor progression and unresponsiveness to therapy. Cancer Res. 2011;71:98–105. doi: 10.1158/0008-5472.CAN-10-2366. [DOI] [PubMed] [Google Scholar]
- 8.Mossé YP, Lim MS, Voss SD, Wilner K, Ruffner K, Laliberte J, Rolland D, Balis FM, Maris JM, Weigel BJ, et al. Safety and activity of crizotinib for paediatric patients with refractory solid tumours or anaplastic large-cell lymphoma: A children's oncology group phase 1 consortium study. Lancet Oncol. 2013;14:472–480. doi: 10.1016/S1470-2045(13)70095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kataoka K, Nagata Y, Kitanaka A, Shiraishi Y, Shimamura T, Yasunaga J, Totoki Y, Chiba K, Sato-Otsubo A, Nagae G, et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat Genet. 2015;47:1304–1315. doi: 10.1038/ng.3415. [DOI] [PubMed] [Google Scholar]
- 10.Haferlach T, Nagata Y, Grossmann V, Okuno Y, Bacher U, Nagae G, Schnittger S, Sanada M, Kon A, Alpermann T, et al. Landscape of genetic lesions in 944 patients with myelodysplastic syndromes. Leukemia. 2014;28:241–247. doi: 10.1038/leu.2013.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y, Shimamura T, Niida A, Motomura K, Ohka F, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47:458–468. doi: 10.1038/ng.3273. [DOI] [PubMed] [Google Scholar]
- 12.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, et al. Memorial sloan kettering-integrated mutation profiling of actionable cancer targets (MSK-IMPACT): A hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- 14.Valentijn LJ, Koster J, Zwijnenburg DA, Hasselt NE, van Sluis P, Volckmann R, van Noesel MM, George RE, Tytgat GA, Molenaar JJ, Versteeg R. TERT rearrangements are frequent in neuroblastoma and identify aggressive tumors. Nat Genet. 2015;47:1411–1414. doi: 10.1038/ng.3438. [DOI] [PubMed] [Google Scholar]
- 15.Eichenmüller M, Trippel F, Kreuder M, Beck A, Schwarzmayr T, Häberle B, Cairo S, Leuschner I, von Schweinitz D, Strom TM, Kappler R. The genomic landscape of hepatoblastoma and their progenies with HCC-like features. J Hepatol. 2014;61:1312–1320. doi: 10.1016/j.jhep.2014.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Sumazin P, Chen Y, Treviño LR, Sarabia SF, Hampton OA, Patel K, Mistretta TA, Zorman B, Thompson P, Heczey A, et al. Genomic analysis of hepatoblastoma identifies distinct molecular and prognostic subgroups. Hepatology. 2017;65:104–121. doi: 10.1002/hep.28888. [DOI] [PubMed] [Google Scholar]
- 17.Seki M, Yoshida K, Shiraishi Y, Shimamura T, Sato Y, Nishimura R, Okuno Y, Chiba K, Tanaka H, Kato K, et al. Biallelic DICER1 mutations in sporadic pleuropulmonary blastoma. Cancer Res. 2014;74:2742–2749. doi: 10.1158/0008-5472.CAN-13-2470. [DOI] [PubMed] [Google Scholar]
- 18.Pugh TJ, Yu W, Yang J, Field AL, Ambrogio L, Carter SL, Cibulskis K, Giannikopoulos P, Kiezun A, Kim J, et al. Exome sequencing of pleuropulmonary blastoma reveals frequent biallelic loss of TP53 and two hits in DICER1 resulting in retention of 5p-derived miRNA hairpin loop sequences. Oncogene. 2014;33:5295–5302. doi: 10.1038/onc.2014.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seki M, Nishimura R, Yoshida K, Shimamura T, Shiraishi Y, Sato Y, Kato M, Chiba K, Tanaka H, Hoshino N, et al. Integrated genetic and epigenetic analysis defines novel molecular subgroups in rhabdomyosarcoma. Nat Commun. 2015;6:7557. doi: 10.1038/ncomms8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shern JF, Chen L, Chmielecki J, Wei JS, Patidar R, Rosenberg M, Ambrogio L, Auclair D, Wang J, Song YK, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov. 2014;4:216–231. doi: 10.1158/2159-8290.CD-13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crompton BD, Stewart C, Taylor-Weiner A, Alexe G, Kurek KC, Calicchio ML, Kiezun A, Carter SL, Shukla SA, Mehta SS, et al. The genomic landscape of pediatric Ewing sarcoma. Cancer Discov. 2014;4:1326–1341. doi: 10.1158/2159-8290.CD-13-1037. [DOI] [PubMed] [Google Scholar]
- 22.Tirode F, Surdez D, Ma X, Parker M, Le Deley MC, Bahrami A, Zhang Z, Lapouble E, Grossetête-Lalami S, Rusch M, et al. Genomic landscape of Ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014;4:1342–1353. doi: 10.1158/2159-8290.CD-14-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor-Weiner A, Zack T, O'Donnell E, Guerriero JL, Bernard B, Reddy A, Han GC, Al Dubayan S, Amin-Mansour A, Schumacher SE, et al. Genomic evolution and chemoresistance in germ-cell tumours. Nature. 2016;540:114–118. doi: 10.1038/nature20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki H, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–867. doi: 10.1038/ng.2699. [DOI] [PubMed] [Google Scholar]
- 25.Seki M, Kimura S, Isobe T, Yoshida K, Ueno H, Nakajima-Takagi Y, Wang C, Lin L, Kon A, Suzuki H, et al. Recurrent SPI1 (PU.1) fusions in high-risk pediatric T cell acute lymphoblastic leukemia. Nat Genet. 2017;49:1274–1281. doi: 10.1038/ng.3900. [DOI] [PubMed] [Google Scholar]
- 26.Shiraishi Y, Sato Y, Chiba K, Okuno Y, Nagata Y, Yoshida K, Shiba N, Hayashi Y, Kume H, Homma Y, et al. An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res. 2013;41:e89. doi: 10.1093/nar/gkt126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto G, Nannya Y, Kato M, Sanada M, Levine RL, Kawamata N, Hangaishi A, Kurokawa M, Chiba S, Gilliland DG, et al. Highly sensitive method for genomewide detection of allelic composition in nonpaired, primary tumor specimens by use of affymetrix single-nucleotide-polymorphism genotyping microarrays. Am J Hum Genet. 2007;81:114–126. doi: 10.1086/518809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mossé YP, Laudenslager M, Longo L, Cole KA, Wood A, Attiyeh EF, Laquaglia MJ, Sennett R, Lynch JE, Perri P, et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature. 2008;455:930–935. doi: 10.1038/nature07261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Pontual L, Kettaneh D, Gordon CT, Oufadem M, Boddaert N, Lees M, Balu L, Lachassinne E, Petros A, Mollet J, et al. Germline gain-of-function mutations of ALK disrupt central nervous system development. Hum Mutat. 2011;32:272–276. doi: 10.1002/humu.21442. [DOI] [PubMed] [Google Scholar]
- 31.Bourdeaut F, Ferrand S, Brugières L, Hilbert M, Ribeiro A, Lacroix L, Bénard J, Combaret V, Michon J, Valteau-Couanet D, et al. ALK germline mutations in patients with neuroblastoma: A rare and weakly penetrant syndrome. Eur J Hum Genet. 2011;20:291–297. doi: 10.1038/ejhg.2011.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee CC, Jia Y, Li N, Sun X, Ng K, Ambing E, Gao MY, Hua S, Chen C, Kim S, et al. Crystal structure of the ALK (anaplastic lymphoma kinase) catalytic domain. Biochem J. 2010;430:425–437. doi: 10.1042/BJ20100609. [DOI] [PubMed] [Google Scholar]
- 33.Kato M, Yasui N, Seki M, Kishimoto H, Sato-Otsubo A, Hasegawa D, Kiyokawa N, Hanada R, Ogawa S, Manabe A, et al. Aggressive transformation of juvenile myelomonocytic leukemia associated with duplication of oncogenic KRAS due to acquired uniparental disomy. J Pediatr. 2013;162:1285–1288. doi: 10.1016/j.jpeds.2013.01.003. 1288.e1. [DOI] [PubMed] [Google Scholar]
- 34.Ma ES, Wong CL, Siu D, Chan WK. Amplification, mutation and loss of heterozygosity of the EGFR gene in metastatic lung cancer. Int J Cancer. 2007;120:1828–1831. doi: 10.1002/ijc.22506. [DOI] [PubMed] [Google Scholar]
- 35.Meehan M, Parthasarathi L, Moran N, Jefferies CA, Foley N, Lazzari E, Murphy D, Ryan J, Ortiz B, Fabius AW, et al. Protein tyrosine phosphatase receptor delta acts as a neuroblastoma tumor suppressor by destabilizing the aurora kinase a oncogene. Mol Cancer. 2012;11:6. doi: 10.1186/1476-4598-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nair P, De Preter K, Vandesompele J, Speleman F, Stallings RL. Aberrant splicing of the PTPRD gene mimics microdeletions identified at this locus in neuroblastomas. Genes Chromosomes Cancer. 2008;47:197–202. doi: 10.1002/gcc.20521. [DOI] [PubMed] [Google Scholar]
- 37.Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179–193. doi: 10.1016/j.cell.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 38.Wang C, Liu Z, Woo CW, Li Z, Wang L, Wei JS, Marquez VE, Bates SE, Jin Q, Khan J, et al. EZH2 mediates epigenetic silencing of neuroblastoma suppressor genes CASZ1, CLU, RUNX3, and NGFR. Cancer Res. 2012;72:315–324. doi: 10.1158/0008-5472.CAN-11-0961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- 40.Sakamoto H, Tsukaguchi T, Hiroshima S, Kodama T, Kobayashi T, Fukami TA, Oikawa N, Tsukuda T, Ishii N, Aoki Y. CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell. 2011;19:679–690. doi: 10.1016/j.ccr.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Infarinato NR, Park JH, Krytska K, Ryles HT, Sano R, Szigety KM, Li Y, Zou HY, Lee NV, Smeal T, et al. The ALK/ROS1 inhibitor PF-06463922 overcomes primary resistance to crizotinib in alk-driven neuroblastoma. Cancer Discov. 2016;6:96–107. doi: 10.1158/2159-8290.CD-15-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.