Abstract
Aneuploidy that arises during meiosis and/or mitosis is a major contributor to early embryo loss. We previously showed that human preimplantation embryos encapsulate missegregated chromosomes into micronuclei while undergoing cellular fragmentation and that fragments can contain chromosomal material, but the source of this DNA was unknown. Here, we leveraged the use of a nonhuman primate model and single-cell DNA-sequencing (scDNA-seq) to examine the chromosomal content of 471 individual samples comprising 254 blastomeres, 42 polar bodies, and 175 cellular fragments from a large number (N = 50) of disassembled rhesus cleavage-stage embryos. Our analysis revealed that the aneuploidy and micronucleation frequency is conserved between humans and macaques, and that fragments encapsulate whole and/or partial chromosomes lost from blastomeres. Single-cell/fragment genotyping showed that these chromosome-containing cellular fragments (CCFs) can be maternally or paternally derived and display double-stranded DNA breaks. DNA breakage was further indicated by reciprocal subchromosomal losses/gains between blastomeres and large segmental errors primarily detected at the terminal ends of chromosomes. By combining time-lapse imaging with scDNA-seq, we determined that multipolar divisions at the zygote or two-cell stage were associated with CCFs and generated a random mixture of chromosomally normal and abnormal blastomeres with uniparental or biparental origins. Despite frequent chromosome missegregation at the cleavage-stage, we show that CCFs and nondividing aneuploid blastomeres showing extensive DNA damage are prevented from incorporation into blastocysts. These findings suggest that embryos respond to chromosomal errors by encapsulation into micronuclei, elimination via cellular fragmentation, and selection against highly aneuploid blastomeres to overcome chromosome instability during preimplantation development.
The demand for human in vitro fertilization (IVF) increases each year, but success rates as measured by live birth(s) have remained only ∼30%–35% for decades (cdc.gov/art). One of the leading causes of IVF failure and embryo loss is the presence of unbalanced whole chromosome(s), or aneuploidy. Estimates of aneuploidy in IVF embryos via high-resolution techniques are 50%–80%, including those from young, fertile couples, regardless of embryonic stage (Vanneste et al. 2009a; Johnson et al. 2010; Chavez et al. 2012; Chow et al. 2014; Huang et al. 2014; Minasi et al. 2016). A similar efficiency (∼30%–35%) is thought to arise from natural human pregnancies, with up to 70% of spontaneous miscarriages diagnosed as aneuploid (Miller et al. 1980; Wilcox et al. 1995; Zinaman et al. 1996; Ogasawara et al. 2000). Chromosomal missegregation in oocytes during meiosis has long been considered the primary reason for aneuploidy, especially in cases of advanced maternal age (Nagaoka et al. 2012). However, recent studies using comprehensive chromosome screening of all blastomeres in cleavage-stage embryos established that mitotic errors occur at an equal or greater frequency and irrespective of maternal age (Vanneste et al. 2009a,b; Johnson et al. 2010; Chavez et al. 2012; Chow et al. 2014; McCoy et al. 2015). Mitotic chromosome missegregation not only leads to aneuploidy but also gives rise to a mosaic embryo with different chromosomal copy number among cells. Euploid–aneuploid mosaicism can still result in the birth of healthy offspring upon embryo transfer (Greco et al. 2015; Bolton et al. 2016; Fragouli et al. 2017), which suggests that corrective mechanisms exist to overcome chromosomal instability (CIN).
Another factor in the capacity of an IVF embryo to successfully implant is the timing and degree of cellular fragmentation, whereby cytoplasmic bodies pinch off of blastomeres during cytokinesis (Alikani et al. 1999; Antczak and Van Blerkom 1999). Distinct from cell death–induced DNA fragmentation (Hardy et al. 2001; Xu et al. 2001), cellular fragmentation also occurs naturally following in vivo human conceptions (Pereda and Croxatto 1978; Buster et al. 1985) and is not associated with maternal age (Wu et al. 2011). We previously showed that cellular fragments can contain chromosomal material and that missegregated chromosomes are encapsulated into micronuclei during mitotic divisions (Chavez et al. 2012), but whether it originated from blastomeres and what the parental source of this DNA is were unknown. Chromosomes within somatic cell micronuclei display an increased propensity to undergo double-stranded breaks and structural rearrangements, which may be the result of asynchrony in DNA replication timing between micronuclei and the primary nucleus (Crasta et al. 2012; Liu et al. 2018). A similar phenomenon has been proposed to occur in micronuclei of human embryos (Pellestor 2014; Pellestor et al. 2014), but a recent report suggests that mouse embryonic micronuclei do not rejoin the primary nucleus and instead undergo perpetual unilateral inheritance (Vázquez-Diez et al. 2016). Unlike humans, early cleavage-stage mouse embryos rarely show aneuploidy, micronuclei, and cellular fragmentation even in suboptimal culture conditions (Winston and Johnson 1992; Dozortsev et al. 1998; Lightfoot et al. 2006; Chavez et al. 2012; Macaulay et al. 2015; Bolton et al. 2016; Treff et al. 2016; Vázquez-Diez et al. 2016), and when micronuclei are induced experimentally, mouse embryos undergo cell lysis rather than fragmentation (Chavez et al. 2014). At the late cleavage (morula) stage, however, ∼10% of mouse embryos have been shown to contain micronuclei, and a similar number appeared between in vivo and IVF-derived embryos (Vázquez-Diez et al. 2016) to suggest that micronuclei formation is not a consequence of in vitro culture.
Previous studies with rhesus macaque embryos using DNA–fluorescent in situ hybridization (DNA-FISH) probes to human Chromosomes (Chr) 13, 16, 18, X, and Y indicated that the incidence of aneuploidy in rhesus embryos is more comparable to human than mouse (Dupont et al. 2009a,b, 2010). Given that only a few chromosomes were analyzed by low-resolution techniques, however, the actual percentage of rhesus embryos carrying chromosomal aberrations was unknown. Here, we used single-cell DNA-sequencing (scDNA-seq) to establish the frequency of whole and segmental chromosomal errors in 50 rhesus cleavage-stage embryos from the two-cell to 14-cell stage. By reconstructing the chromosomal content of each cell and fragment, we investigated whether whole or partial chromosomes lost from blastomeres are sequestered into cellular fragments. We also examined the fate of cellular fragments beyond the cleavage stage as well as embryo imaging parameters or morphological features that might lead to their formation via time-lapse monitoring (TLM) of preimplantation development.
Results
Incidence of micronucleation and cellular fragmentation is conserved between primates
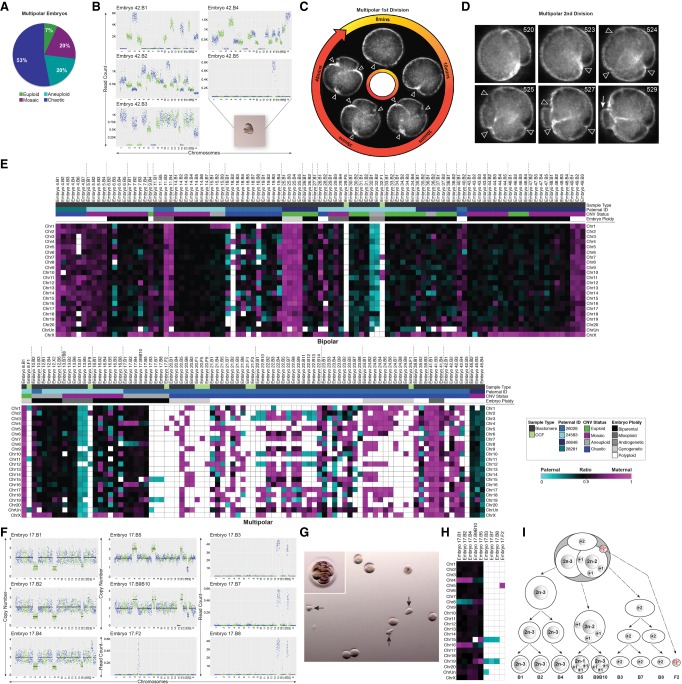
To determine the aneuploidy and micronucleation frequency in rhesus cleavage-stage embryos, we developed an experimental approach using scDNA-seq and TLM to noninvasively assess preimplantation development (Fig. 1A). Mature metaphase II (MII) oocytes underwent conventional IVF, and presumed zygotes with two polar bodies and/or pronuclei were analyzed by TLM to evaluate mitotic divisions, the absence or presence of cellular fragmentation (Fig. 1B), and other imaging parameters and/or morphological features indicative of embryo chromosomal status. After ∼24–96 h, cleavage-stage embryos (N = 50) were disassembled into individual blastomeres, cellular fragments, and polar bodies if still present (Fig. 1C) for chromosomal copy number variation (CNV) analysis and single-nucleotide polymorphism (SNP) genotyping. Another subset of intact embryos (N = 25) between the zygote and blastocyst stage was fixed and subjected to multi-color confocal imaging to assess micronuclei formation and DNA sequestration by cellular fragments. Lastly, an additional 92 rhesus embryos were allowed to proceed in development to evaluate the impact of micronuclei, fragmentation, and aneuploidy on embryonic arrest versus successful progression to the blastocyst stage (Fig. 1D).
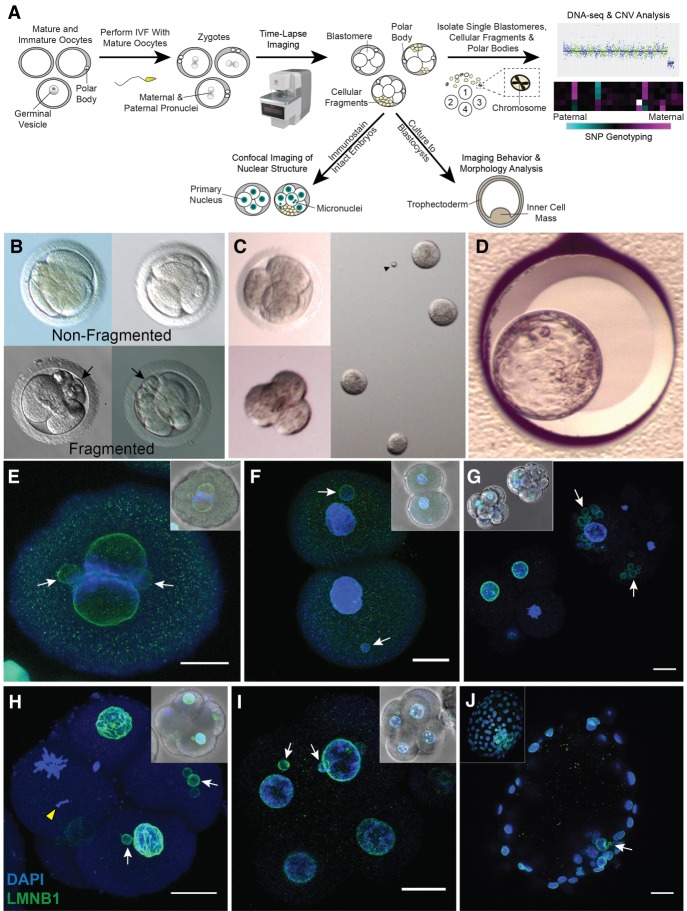
Figure 1.
Approach for assessing micronuclei, fragmentation, and CNV in rhesus embryos. (A) Mature and immature oocytes were obtained from female rhesus macaques undergoing controlled ovarian stimulations. MII oocytes displaying one polar body were fertilized by conventional IVF with sperm from rhesus males. (B) Early mitotic divisions and the incidence of cellular fragmentation in presumptive zygotes (identified by two pronuclei and/or polar bodies) were analyzed by time-lapse imaging. (C) Cleavage-stage embryos were disassembled into individual blastomeres, cellular fragments, and polar bodies for CNV and SNP analysis by scDNA-seq (N = 50). A subset of intact cleavage-stage embryos was fixed and immunostained for confocal imaging (N = 25). (D) Another group of embryos were cultured up to the blastocyst stage (N = 92). (E) Immunostaining of a zygote undergoing syngamy with two micronuclei (white arrows) shown by the nuclear envelope marker, LMNB1 (green); DAPI (blue) = DNA. (F) Two-cell embryo with one micronucleus in each blastomere. (G) Comparison of a fragmented (white arrowheads) cleavage-stage embryo with multiple micronuclei (right) and a nonfragmented seven-cell embryo (left). (H) Single imaging plane of a Z-stacked five-cell embryo with a missegregated chromosome (yellow arrowhead) and micronuclei, as well as a (I) nine-cell embryo showing micronuclei in two blastomeres, but no visible cellular fragmentation. Insets show a brightfield image for reference. Scale bars, 25 µm. (J) Blastocyst with two micronuclei in the ICM; the inset shows the maximum intensity projection of the embryo.
Upon fixation and immunolabeling with the nuclear envelope marker, LMNB1, we show that rhesus embryos contain micronuclei as early as the zygote (Fig. 1E) or two-cell stage (Fig. 1F) and that the emergence of micronuclei was often concomitant with cellular fragmentation by the four-cell stage (Fig. 1G). Some of these fragments encapsulate nuclear DNA positive for DAPI staining that is inconsistent with polar bodies, which contain a membrane-bound nucleus and/or condensed chromosomes that are thought to degenerate within 24 h of formation (Zamboni et al. 1966; Wang et al. 2014). Chromosomal material in cellular fragments was never detected in the absence of micronuclei, suggesting that micronuclei formation precedes and seems to be necessary for chromosome sequestration (Fig. 1E–G). Although micronuclei were not apparent until the five- to nine-cell stage or later in only a few embryos (Fig. 1H,I), we determined that the inner cell mass (ICM) of blastocysts might retain micronuclei (Fig. 1J). We also observed one embryo with a condensed chromosome separated from the mitotic spindle that lacked nuclear envelope (Fig. 1H), suggesting that missegregated chromosomes in embryonic micronuclei undergo condensation with nuclear envelope breakdown similar to chromosomes in primary nuclei. When all rhesus embryos were evaluated at high magnification before fixation or disassembly, we determined that >65% (N = 129/196) of cleavage-stage embryos show some degree of cellular fragmentation (Supplemental Movie S1).
Rhesus cleavage-stage embryos are often aneuploid or mosaic caused by mitotic errors
Embryos were disassembled into single cells and cellular fragments and the DNA in each sample was amplified, labeled with custom barcodes, PCR-validated using adapter sequences, and pooled for multiplex scDNA-seq (Supplemental Table S1). To detect CNV, we developed a bioinformatics pipeline that compares read counts in contiguous windows across the genome between embryonic samples and rhesus female euploid (42,XX) fibroblasts using a combination of variable nonoverlapping windows and circular binary segmentation (CBS) called VNOWC (Supplemental Fig. S1). We then used a second custom bioinformatics pipeline that incorporated the hidden Markov model (HMM) (Knouse et al. 2016) called CBS/HMM intersect (CHI) to further validate CNV calls. By using the dual-pipeline bioinformatics strategy, we sequenced 471 individual samples from 50 whole rhesus cleavage-stage embryos up to the 14-cell stage (Supplemental Table S2), 49 of which contained DNA that successfully amplified (Fig. 2A). This included 254 blastomeres and 175 cellular fragments as well as a large proportion of polar bodies (N = 42/471) confirmed by SNP analysis as described below. Each blastomere or polar body was classified as euploid or aneuploid and the type of chromosomal error determined by the following criteria: (1) Meiotic errors were identified by an aneuploid polar body, and in the absence of polar bodies or the presence of only one euploid polar body, it was considered meiotic if the same chromosome was affected in all sister blastomeres; (2) mitotic errors were defined as different and/or reciprocal chromosome losses and gains between blastomeres with euploid polar bodies; and (3) chaotic aneuploidy was characterized by multiple (five or more) random chromosome losses and gains in one or more blastomeres (Delhanty et al. 1997). Based on the above criteria, 26.5% (N = 13/49) of the embryos were composed of only euploid blastomeres with no segmental errors, whereas 73.5% (N = 36/49) contained at least one blastomere with whole and/or partial chromosome losses and gains (Fig. 2A; Supplemental Table S2). Further analysis revealed that 40.8% (N = 20/49) of the embryos consisted of blastomeres that were all affected, whereas 20.4% (N = 13/49) showed euploid–aneuploid mosaicism and 12.2% (N = 3/49) were mosaic with segmental errors only. Both polar bodies were obtained from ∼20% (N = 10/49) of the embryos and primarily at the early cleavage stages, but ∼74% (N = 31/42) of those isolated were euploid. Thus, we were able to confidently call the inheritance of meiotic errors in 25% (N = 9/36) and the occurrence of solely mitotic errors in 41.7% (N = 15/36) of embryos, with the remaining 33.3% (N = 12/36) either incurring both types of errors or being unknown because of the complexity of chromosomal mosaicism (Supplemental Table S3). The incidence of chaotic aneuploidy was 28.6% (N = 14/49), and this appeared to be mostly confined to embryos fertilized by a particular sperm donor (N = 11/14). Representative examples of genome-wide chromosomal CNV plots from embryos with euploid, mosaic, aneuploid, and/or chaotic aneuploid blastomeres are shown in Supplemental Figure S2.
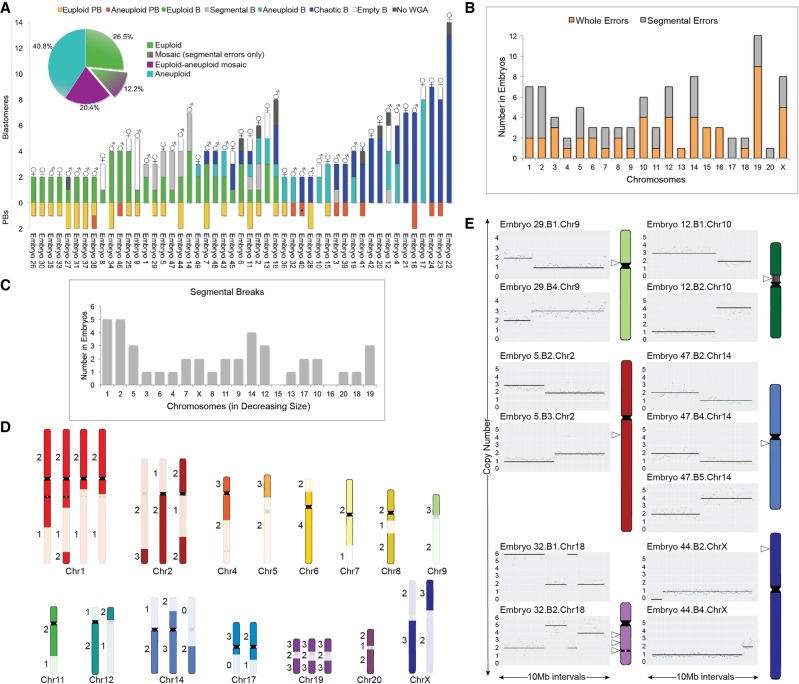
Figure 2.
Assessment of whole and subchromosomal instability in rhesus embryos. (A) CNV summary of rhesus embryos (N = 49) from the two- to 14-cell stage analyzed by scDNA-seq. Stacked bars represent euploid (yellow) and aneuploid (orange) polar bodies (PB); euploid (green), aneuploid (light blue), segmental aneuploid-only (light gray), and chaotic aneuploid (dark blue) blastomeres (B); no WGA (dark gray); and empty blastomeres (white) detectable by high mitochondrial (mtDNA) but no genomic DNA reads (N = 296 samples). Aneuploid PB containing only segmental errors labeled with asterisk. (♂) Chr Y present; (♀) two Chr X present. Percentage of euploid, aneuploid, or mosaic embryos with or without solely segmental errors is shown in the pie chart (upper left). (B) Number of times chromosomes were affected by whole (orange) or segmental (gray) losses or gains. (C) Graph showing that there was no significant association (P-value = 0.1475) between the number of segmental breaks and chromosome size (Spearman's correlation = 0.3273). (D) Location of chromosomal breaks in embryos with segmental aneuploidy. Numbers to the left represent the copy number state of blastomeres. (E) CNV plots of six embryos, in which chromosomal breakage resulted in a reciprocal loss and gain of chromosome segments between blastomeres (left). Chromosome ideograms showing the approximate breakpoint locations (right; white arrowheads) of each embryo with reciprocal breaks. Vertical lines in Chr 10 delineate the nucleolus organizer region adjacent to the centromere (black), and the gray circle in Chr 18 designates the ancestral inactivated centromere.
Reciprocal subchromosomal deletions and duplications indicate chromosome breakage
Excluding chaotic samples, we then assessed the frequency of whole and segmental errors by chromosome in the embryos (Fig. 2B). Chr 1 and Chr 2 were the most highly susceptible to aneuploidy because of DNA breakage, which was not the result of chromosome size (Fig. 2C), whereas Chr 19 experienced the greatest incidence of whole CIN. However, this chromosome is GC rich, and when combined with scDNA-seq as previously shown for human Chr 19 (Knouse et al. 2016), whole chromosomal losses and gains are difficult to distinguish from large segmental CNVs. Chr 20 (corresponding to human Chr 16) was the least frequently affected by aneuploidy, and large segmental deletions, duplications, and amplifications were predominantly located at the terminal ends of chromosome arms (N = 33/37) (Fig. 2D). In a small proportion of embryos (16.7%; N = 6/36), chromosomes underwent unbalanced rearrangements, in which the reciprocal chromosome segments were found in a sister blastomere (Fig. 2E). We determined that several of these breakpoints localized near existing centromeres or, in the case of Chr 18, an inactivated ancient centromere (Ventura et al. 2007). The approximate location of breaks in Chr 10 and Chr 14 also aligned with corresponding fission or inversion evolutionary breakpoints, respectively, in the common primate ancestor.
Cellular fragments may contain whole or partial chromosomes lost from blastomeres
Although reciprocal exchange of chromosomes between blastomeres was observed in some embryos, chromosome(s) were entirely lost in the majority of cases (83.3%; N = 30/36), and 86.7% (N = 26/30) of these embryos showed cellular fragmentation. Based on our previous findings of chromosomal material in cellular fragments from human embryos (Chavez et al. 2012), we hypothesized that the missing chromosome(s) had been sequestered during fragmentation. To test this, we sequenced 175 single fragments obtained from each fragmented rhesus embryo and found an instance in which both copies of Chr 9 and Chr 12 lost from blastomeres were located in one of the cellular fragments (Fig. 3A). By similarly reconstructing the chromosomal content of each embryo via single-cell/-fragment DNA-seq, we observed additional examples of individual, multiple, and/or partial chromosomes in the fragments of other embryos (Fig. 3B). Maternal versus paternal SNP allele genotyping analysis, which is described in more detail below, revealed that chromosome-containing cellular fragments (CCFs) can originate from either the mother or father (Fig. 3C). There did not appear to be preferential sequestering of particular chromosomes, as both small and large chromosomes were affected and the partial chromosomes identified in fragments ranged in size from 6 to 85 Mb (Fig. 3D). Overall, we confirmed the presence of entire or portions of chromosomes in one or more CCFs in ∼18% (N = 8/45) of fragmented embryos, and ∼88% (N = 7/8) of these embryos were aneuploid to varying degrees. However, only ∼6.3% (N = 11/175) of the cellular fragments examined contained chromosomal material (Fig. 3E). Thus, missegregated chromosomes encapsulated within embryonic micronuclei not only persist or rejoin the primary nucleus but may also be eliminated from the embryo upon cytoplasmic pinching of cellular fragments from blastomeres.
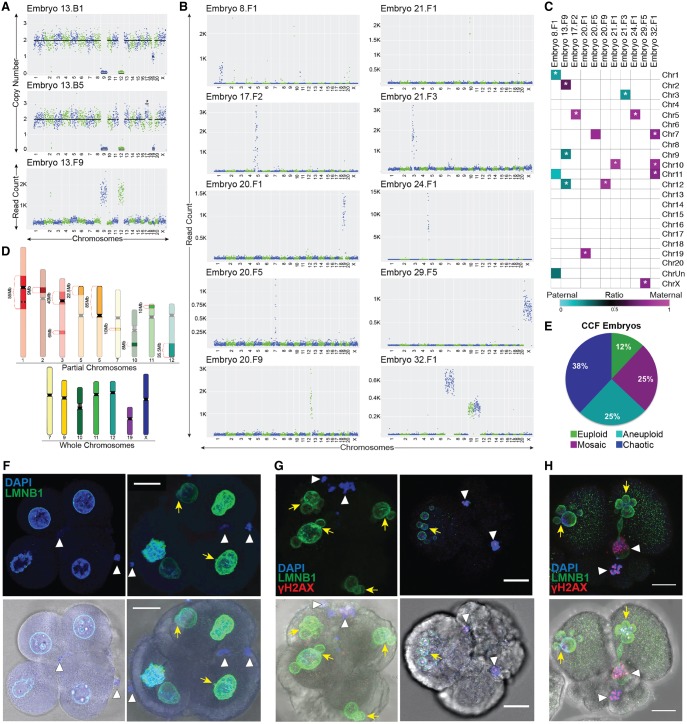
Figure 3.
Chromosomes are eliminated via cellular fragmentation and are susceptible to DNA damage. (A) CNV or read count plots demonstrating that Chr 9 and Chr 12 lost from two blastomeres (top and middle; also missing one to two copies of Chr 19) were detected in a cellular fragment (bottom) from the same embryo. (B) Additional examples of individual, multiple, and/or partial chromosomes in fragments of rhesus embryos. (C) Heat map of maternal versus paternal SNP genotyping ratios showing that CCFs can originate from the mother or father. White asterisk demarcates significant P-values (P < 9.1 × 10−6) for cumulative binomial test with Bonferroni correction. (D) Rhesus ideograms representing whole (bottom) and partial (top) chromosomes with approximate sizes highlighted that were detected in fragments. (E) Percentage of embryos with CCFs (N = 8 embryos) that were chaotic (blue), aneuploid (turquoise), mosaic (magenta), and euploid (green). (F) Five-cell embryos with normal-appearing blastomeres containing micronuclei (yellow arrows) and CCFs (white arrowheads) identified by DAPI (blue) and LMNB1 (green). Brightfield images (bottom) provided for reference. (G) Other cleavage-stage embryos with multiple micronuclei and CCFs also immunostained for the double-stranded DNA break marker, γH2A.X (red), showed that the chromosomes within fragments are unstable and damaged. (H) One embryo also contained a micronucleus that appeared to be in the process of CCF sequestration. Scale bar, 25 µm.
Chromosomes within cellular fragments are susceptible to DNA breaks and damage
Based on findings of whole chromosomes and/or chromosomal segments in cellular fragments from a minority of embryos and several reports of DNA fragility within the micronuclei of somatic cells (Crasta et al. 2012; Hatch et al. 2013; Liu et al. 2018), we next sought to determine whether the CCFs were susceptible to DNA damage and rapid degradation once separated from the primary nucleus. To accomplish this, we immunostained fragmented cleavage-stage embryos with LMNB1 and γ-H2A.X, the serine phosphorylated form of H2AFX and a marker of DNA damage and double-stranded breaks (Rogakou et al. 1998). Positive DAPI staining confirmed the presence of DNA in cellular fragments from rhesus embryos (Fig. 3F), but these CCFs appeared to lack a nuclear envelope or to have a defective nuclear envelope as described for somatic cell micronuclei (Crasta et al. 2012; Hatch et al. 2013; Liu et al. 2018) that was also inconsistent with polar body identity. Multiple γ-H2A.X foci were detected in the micronuclei of blastomeres as well as in the DNA of cellular fragments, and DNA damage appeared markedly increased in CCFs and micronuclei compared with primary nuclei (Fig. 3G–H). One of these embryos also contained a micronucleus that seemed to be in the process of CCF sequestration from a blastomere (Fig. 3H). Because DNA degradation and double-stranded DNA breaks are not distinguishable by γ-H2A.X immunosignals, however, it is also possible that subchromosomal segments were initially sequestered into cellular fragments rather than the product of DNA degradation.
Parental contribution to aneuploidy is revealed by SNP allele genotyping analysis
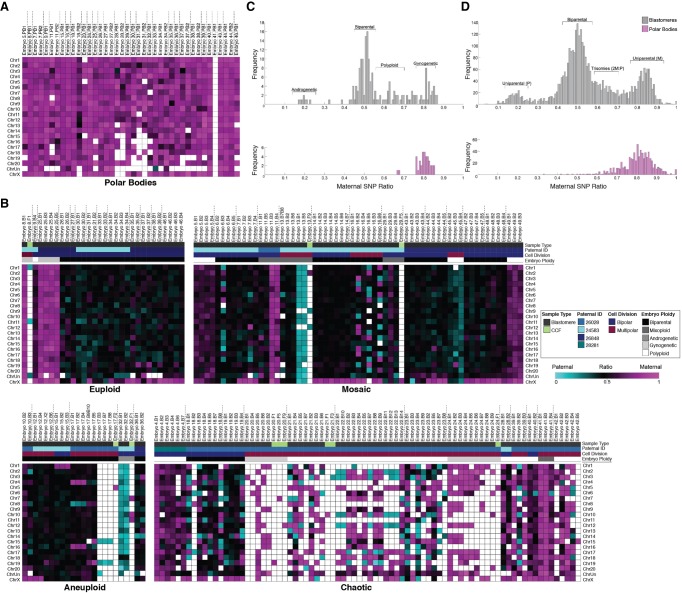
It is generally accepted that polar bodies degenerate within 24 h of extrusion from the oocyte or zygote (Schmerler and Wessel 2011), but we unexpectedly identified polar bodies in several embryos beyond the two- to four-cell stage by scDNA-seq (Fig. 2A). To validate their identity and further distinguish them from cellular fragments, we isolated DNA from each of the parents whose gametes were used for IVF and performed whole-genome DNA-seq (Supplemental Table S4) for comparison of maternal versus paternal SNP alleles in all embryonic samples (Supplemental Table S5). The proportion of maternal alleles was significantly different (P < 1.88 × 10−4, binomial test) from the expected 50%, with an average of 80% of alleles identified as maternal in origin, confirming polar body identity (Fig. 4A). SNP genotyping was also used to assess the parental origins of all chromosomes in each blastomere by simultaneously analyzing multiple SNPs with only opposite homozygous genotypes in the parents across each chromosome. Although the majority of embryos initially classified as euploid were biparental (76.9%; N = 10/13), three of these embryos contained blastomeres with chromosomes that were entirely from the mother (Fig. 4B) and either gynogenetic (embryos 8 and 25) or digynic triploid (embryo 9). In contrast, only ∼45% of the aneuploid embryos were biparental in origin (N = 15/33), and the remaining embryos were gynogenetic (N = 2/33), androgenetic (N = 1/33), polyploid (N = 11/33) or contained a mixture of uniparental, biparental, and triploid cells termed mixoploid (N = 4/33; Fig. 4C; Supplemental Fig S3; Supplemental Table S5). Further analysis revealed at least one case of a paternally contributed meiotic error (embryo 15; Chr 1 monosomy) and that the triploid embryos were composed of two copies of maternal chromosomes and one copy of each paternal chromosome (Fig. 4D). When we compared the CCFs observed in Figure 3E to the embryos from which they arose via SNP genotyping, we determined that the blastomeres were biparental (N = 1/8), gynogenetic (N = 3/8), androgenetic (N = 1/8), triploid (N = 1/8), or mixoploid (N = 2/8), suggesting that the production of CCFs is not associated with a certain type of chromosomal abnormality.
Figure 4.
SNP profiling confirms polar bodies and reveals complexity in parental contribution to aneuploidy. (A) Heat map of SNP allele parentage ratios in polar bodies confirming their maternal origins and (B) heat maps of euploid, aneuploid, mosaic, or chaotic aneuploid embryos. Each embryo is separated by vertical dotted lines. Samples were further sorted based on the paternal donor, cell type, mitotic divisions, and overall embryo ploidy. Pink, blue, and black boxes indicate maternal, paternal, and biparental inheritance, respectively. White boxes show that either the chromosome was not detected or it could not be called with high confidence. (C) Histograms showing the distribution of SNP allele ratios across blastomeres (dark gray) and polar bodies (pink) revealed that the majority of rhesus embryos were biparental, but a small proportion were androgenetic, polyploid, or gynogenetic. (D) Frequency of SNP allele ratios in histograms stratified at an individual chromosome level.
Multipolar divisions often lead to chromosome loss and chaotic aneuploidy
By combining scDNA-seq with TLM of embryos, we next sought to determine whether there were imaging parameters indicative of chromosome loss from blastomeres and sequestration by cellular fragments. Indeed, the majority (N = 6/8) of embryos with CCFs showed multipolar divisions at the one- or two-cell stage followed by cellular fragmentation (Supplemental Movie S2). When we evaluated higher-order mitotic divisions in all embryos, we determined that although only one of the 15 embryos with multipolar divisions was euploid, the remaining embryos were chromosomally abnormal and mainly chaotic aneuploid (N = 8/15) (Fig. 5A), with almost every blastomere affected (Fig. 5B). The multipolar division most often occurred at the zygote stage (11/15) (Fig. 5C), but there were some embryos that showed a multipolar division at the two-cell stage (N = 4/15) (Fig. 5D). SNP analysis of parental ratios showed that all of the multipolar embryos with chaotic aneuploidy originated from the same sperm donor (Supplemental Fig. S4) and that multipolar divisions often resulted in a random mixture of chromosomally normal and abnormal blastomeres with biparental or uniparental origins regardless of which male was used (Fig. 5E). In one of the multipolar zygotes, we identified a loss of Chr 4, Chr 8, and Chr 16 in three blastomeres and the reciprocal copies in two other blastomeres from the same embryo (Fig. 5F). These two blastomeres also contained only a single copy of Chr 19 and/or a complete loss of Chr 15, which were detected in additional cells that appeared unusual in shape and size upon disassembly (Fig. 5G). We determined that this chromosomal complexity was because of a nondisjunction event that likely occurred during the multipolar division at the zygote stage based on the distribution of paternal-only, maternal-only, or biparental contribution of certain chromosomes after one or more normal subsequent cell divisions (Fig. 5H), all of which is depicted in Figure 5I.
Figure 5.
Multipolar divisions in embryos often result in chromosome loss and chaotic aneuploidy. (A) Ploidy status of rhesus embryos (N = 15) with multipolar divisions at the one- or two-cell stage. (B) CNV plots of blastomeres from an embryo, which underwent a multipolar first division, showing chaotic aneuploidy in almost every cell. Inset is a stereomicroscope image of blastomere 5 containing only Chr 15 with a cellular fragment-like protrusion. Darkfield time-lapse images of a zygote (C) and two-cell embryo undergoing a tripolar division (D). Arrowheads point to three simultaneous cleavage furrows. An arrow designates a small blastomere/large fragment produced from the multipolar second division, and the number refers to the frame. (E) Heat map of SNP allele parentage ratios in embryos that underwent bipolar (top) or multipolar (bottom) cleavage during the first three cell divisions. (F) The CNV plots of blastomeres from the tripolar zygote showing multiple reciprocal chromosome losses and gains. Note that blastomeres 3, 7, and 8 each contained only two chromosomes. (G) Stereomicroscope image of this embryo still intact (inset) and then disassembled. Arrows indicate the irregularly shaped blastomeres that had only two chromosomes. Blastomere 6 lysed and is demarcated with an asterisk. (H) Heat map of maternal versus paternal SNP allele ratios for the tripolar zygote delineates parental inheritance. (I) Schematic of the chromosome copy number state of this embryo based on the imaging and CNV analysis.
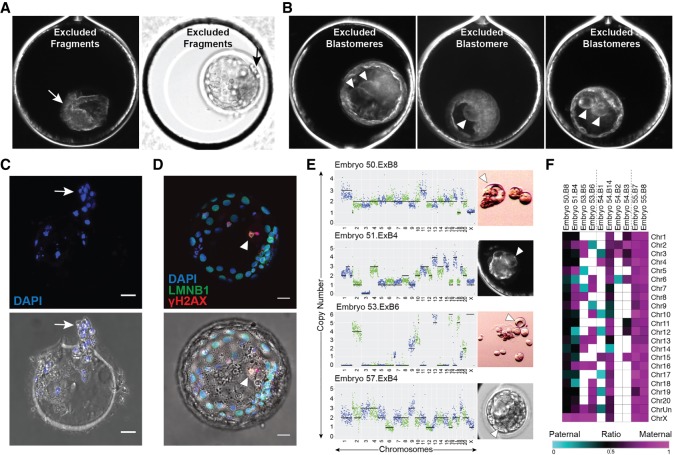
Cellular fragments and nondividing aneuploid blastomeres are excluded during blastocyst formation
To determine the impact of multipolar divisions and/or cellular fragmentation on subsequent preimplantation development, we monitored an additional 92 rhesus zygotes by TLM up to the blastocyst stage. Although 42 of these embryos arrested before day 7 (Supplemental Movie S3, right), the remaining embryos formed blastocysts (Supplemental Movie S3, left), resulting in a blastocyst formation rate of ∼54% (N = 50/92). Moreover, ∼18.5% (17/92) of the embryos underwent a multipolar division, and ∼88% (N = 15/17) of those arrested directly following the abnormal cytokinesis. The two multipolar embryos that still formed blastocysts showed a unique one- to four-cell symmetrical multipolar division without cellular fragmentation at the one- or two-cell stage. Of the blastomeres produced from these tetrapolar divisions, at least one large cell ceased dividing as shown by time-lapse imaging and was confined to the blastocoel cavity upon blastocyst formation (Supplemental Movie S4, right). We also observed the confinement of cellular fragments produced during the early cleavage stages to the perivitelline space of some blastocysts (Supplemental Movie S4, left). Overall, we documented 10 out of the 50 embryos with excluded cellular fragments (Fig. 6A) and/or blastomeres (Fig. 6B) that appeared at the two- to eight-cell stage and persisted during the morula-to-blastocyst transition. Numerous DAPI-positive nuclei were detected in the zona pellucida (ZP) of blastocysts that showed exclusion of cellular fragments after hatching (Fig. 6C). A large binucleated cell with extensive DNA damage was also detected in one the blastocysts with excluded blastomere(s) via LMNB1 and γH2A.X immunostaining (Fig. 6D). Because it was difficult to separate the excluded blastomeres from blastocysts once the blastocoel cavity formed, we disassembled another four embryos with large nondividing cells before or during morula compaction for scDNA-seq analysis. We determined that these excluded blastomeres were highly chaotic with multiple chromosomal losses and gains (Fig. 6E). SNP genotyping also showed that the chromosomes in excluded blastomeres were both maternal and paternal in origin (Fig. 6F). Because these blastomeres remained similar in size from their first appearance at two- to eight-cells up to the blastocyst stage and never divided again, this suggests that blastomere exclusion represents one mechanism by which an embryo can select against aneuploid cells during preimplantation development.
Figure 6.
Cellular fragments and aneuploid blastomeres are excluded upon blastocyst formation. Time-lapse image frames from two rhesus blastocysts showing exclusion of several cellular fragments to the perivitelline space of the embryo (A; arrow) or of three rhesus blastocysts with one to two nondividing excluded blastomeres in the blastocoel cavity (B; arrowheads). (C) The zona pellucida of the blastocyst that showed cellular fragment exclusion with remaining DNA positive for DAPI (blue) staining following hatching (white arrow). (D) A blastocyst with blastomere exclusion immunostained for LMNB1 (green) and γH2A.X (red) using DAPI as a marker for DNA. The large excluded blastomere appeared binucleated with strong γH2A.X signals (white arrowhead), indicating that double-stranded DNA breaks had occurred. Brightfield image with immunofluorescence overlay provided below. (E) Additional examples of large excluded blastomeres (right; white arrowheads) collected during the morula-to-blastocyst transition for sequencing. CNV analysis (left) determined that each excluded blastomere had chaotic aneuploidy. Scale bars, 25 µm. (F) Heat map of maternal versus paternal SNP allele ratios in excluded blastomeres shows parental origins.
Discussion
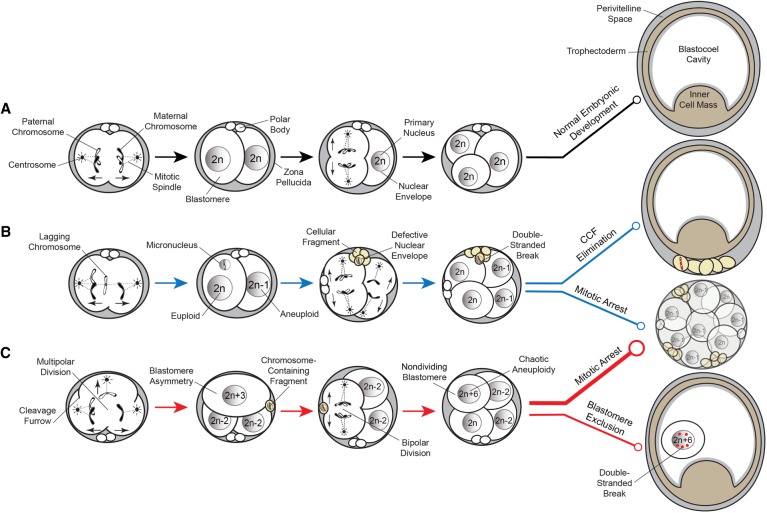
Established estimates of aneuploidy in human IVF embryos via whole-genome methods are 50%–80% regardless of maternal age, fertility status, or embryonic stage and largely contribute to embryo arrest before the blastocyst stage (Vanneste et al. 2009a,b; Johnson et al. 2010; Chavez et al. 2012; Chow et al. 2014; McCoy et al. 2015; Minasi et al. 2016). Here, we show that rhesus preimplantation embryos also have a high incidence of aneuploidy and chromosomal mosaicism because of meiotic and/or mitotic errors. Besides aneuploidy, we show that rhesus cleavage-stage embryos exhibit micronuclei formation, cellular fragmentation, and multipolar divisions at an equivalent frequency to human embryos (Alikani et al. 1999; Antczak and Van Blerkom 1999; Chavez et al. 2012; Hlinka et al. 2012; Chamayou et al. 2013; Ottolini et al. 2017). An examination of the rhesus time-mated breeding colony at our center during the same timeframe as this study (November–May, 2013–2017) revealed that of the confirmed ovulation and mating cases, 73.5% (N = 200/272) did not result in a live birth. Given the equal percentage (73.5%) of aneuploidy observed in rhesus IVF embryos here, we propose that CIN likely contributes to such low success rates following natural conceptions. Based on all of the above, we also argue that the rhesus monkey represents an ideal surrogate for studying the effects of human embryonic aneuploidy on normal preimplantation development (Fig. 7A).
Figure 7.
Proposed model of aneuploidy generation and potential resolution in embryos. (A) Simplified model of normal embryo development, whereby a euploid zygote undergoes proper chromosome segregation with bipolar cell divisions devoid of cellular fragmentation and blastomere exclusion (black lines). (B) A euploid zygote that contains a lagging chromosome from merotelic attachments during the first mitotic division becomes encapsulated in a micronucleus. Through the process of cellular fragmentation, the chromosome is eliminated from the blastomere, where it undergoes DNA damage in the form of double-stranded breaks because of a defective nuclear envelope. This mosaic embryo is more likely to undergo mitotic arrest, but if it is able to reach the blastocyst stage, the CCF may be sequestered to the perivitelline space (blue lines). (C) Multipolar cell divisions, including a tripolar cleavage occurring at the zygote stage, may also generate CCFs as well as blastomere asymmetry and a mosaic embryo with chaotic aneuploidy. The vast majority of these embryos will eventually arrest upon embryonic genome activation when a critical number of euploid blastomeres is not achieved (thick red line). Alternatively, mosaic embryos may progress beyond the approximately eight-cell stage and aneuploid blastomeres that fail to divide during this time will sustain DNA damage and become excluded to the blastocoel cavity upon blastocyst formation (thin red line).
To our knowledge, this is the first study to show that whole and/or partial chromosomes lost from blastomeres are encapsulated within cellular fragments and are highly unstable. Once separated from the primary nucleus, chromosomes within somatic cell micronuclei undergo DNA damage and double-stranded breaks because of defective nuclear envelope assembly (Crasta et al. 2012; Hatch et al. 2013; Liu et al. 2018). Because CCFs lacked nuclear envelope and we observed the greatest DNA damage in the sequestered chromosomes, this may explain why only a small number of cellular fragments contained intact DNA detectable by scDNA-seq. Chromothripsis, whereby chromosomes are “shattered” and rearranged in a single catastrophic event, also arises in somatic cell micronuclei following DNA damage (Zhang et al. 2015). The occurrence of chromothripsis in embryos has been suggested (Pellestor 2014; Pellestor et al. 2014), but not yet confirmed, because of the depth of genome coverage and large amplicon size required to accurately call structural variants by scDNA-seq (de Bourcy et al. 2014). We were limited by the same factors in this study but did identify large segmental losses, duplications, and amplifications at the terminal ends of chromosome arms in rhesus blastomeres analogous to observations of terminal chromosome imbalances and rearrangements in human embryos (Vanneste et al. 2009a). Additional sequencing and bioinformatics approaches are required to delineate if there are structural differences in the chromosomes from embryonic micronuclei, cellular fragments, and excluded blastomeres, the latter of which may be more susceptible to chromothripsis. This assumption arises from the apparent requirement that the damaged chromosome(s) within somatic cell micronuclei be exposed to nucleoplasm of the primary nucleus before undergoing DNA repair and rearrangement (Zhang et al. 2015). Even if embryos do not undergo chromothripsis, we speculate that severely damaged DNA indicated by the extensive γH2A.X signals in CCFs and excluded blastomeres is selectively eliminated from the embryo to prevent further propagation of highly unstable chromosomes (Fig. 7B). Given the parallels between embryonic and somatic cell micronuclei as well as recent evidence that polyploid giant cancer cells may represent the somatic equivalent of blastomeres (Niu et al. 2017), additional scDNA-seq embryo studies may also inform the cancer field.
When we evaluated whether there were imaging parameters indicative of chromosome sequestration by cellular fragments or blastomeres in embryos, there was a clear association between these two events and multipolar divisions. In certain cases, the multipolar division resulted in the production of small blastomeres or large fragments that could only be distinguished by examining DNA content and determining whether a full or partial genome was contained within. These observations are consistent with other cell models, whereby the formation of several cytokinetic furrows during a multipolar mitosis leads to the formation of microcells containing small amounts of chromatin (Sherwood et al. 1994; Gisselsson et al. 2008). TLM has shown that ∼12% of human zygotes undergo multipolar divisions (Chamayou et al. 2013) and are less likely to form blastocysts and implant (Hlinka et al. 2012). Multipolar divisions occurring later in preimplantation development are also highly correlated with human embryo arrest (Ottolini et al. 2017). Almost all of the rhesus embryos with higher-order divisions here arrested before forming blastocysts, and the two embryos that did progress underwent a one- to four-cell symmetrical cell division without fragmentation. The same two embryos also showed blastomere exclusion during the morula-to-blastocyst transition to suggest that multipolar divisions might provide a mechanism to overcome aneuploidy under certain circumstances (Fig. 7C). This is supported by findings that some of the embryos with higher-order divisions were either euploid with adjacent empty blastomeres or chromosomally mosaic. We determined that the most prevalent type of chromosomal abnormality observed in multipolar embryos was chaotic aneuploidy, and all of these embryos shared a common sperm donor. Because the centrosome for the first mitotic division(s) is paternally inherited in most mammalian species except rodents (Sathananthan et al. 1991; Schatten et al. 1991), defective or supernumerary centrosomes from the sperm likely contributed to the higher-order divisions. Moreover, sperm are also responsible for the activation of oocytes during fertilization (Whitaker 2006; Yoon et al. 2008), suggesting that premature oocyte activation might have also been a factor. Regardless of the underlying mechanism(s) and which male was used, multipolar divisions often generated a random mixture of euploid, aneuploid, and chaotic blastomeres with biparental or uniparental origins. Further investigation is required to determine if the abnormal cytokinesis resulting in the appearance of CCFs or excluded blastomeres exacerbates CIN or whether such events are a deliberate attempt to eliminate aberrant chromosomes and blastomeres from the embryo.
One of the most intriguing findings from the SNP analysis was the identification of a few euploid and a relatively large proportion of aneuploid embryos with cells that were derived from only one parent. This phenomenon, called uniparental genome segregation, has been described in bovine embryos at the zygote stage and was thought to be a consequence of in vitro oocyte maturation for fertilization (Destouni et al. 2016; Tšuiko et al. 2017). Our study is the first to show that uniparental genome segregation as well as mixoploidy also occurs in embryos following the maturation of oocytes in vivo, which is used in >98% of human IVF cycles (https://www.cdc.gov/art) and beyond the zygote stage. Although it is fairly well established that gynogenetic and androgenetic embryos can result from IVF, we speculate that a similar percentage of human embryos with uniparental origins has not yet been reported given that current preimplantation genetic screening methods do not examine parental origins of aneuploidy unless SNP arrays are used. However, SNP arrays are rarely used by clinics for CNV analysis because of high allele drop-out rates and the need to include parental DNA to interpret SNPs (McCoy et al. 2015, 2018). To determine parentage, we used next-generation sequencing (NGS) and a filtering strategy to combine information from several highly informative SNPs across large chromosomal segments, which permitted us to confidently determine parentage despite the low confidence of individual SNP calls from scDNA-seq coverage. We note that some polyploid embryos were initially classified as euploid by CNV analysis, and because both array-based and NGS approaches can only identify polyploidy by the ratio of sex chromosomes, this might help explain why seemingly euploid embryos fail to implant (Maxwell et al. 2016). In summary, we show that chromosomal loss from primate preimplantation embryos is because of sequestration by cellular fragments and/or nondividing blastomeres, which may denote mechanisms to surpass aneuploidy as embryos undergo implantation and continue in development. Additional work is necessary to capture the formation and fate of micronuclei, CCFs, and excluded blastomeres in real-time and at the single chromosome level to determine the molecular mechanisms underlying their production and resolution.
Methods
Rhesus oocytes and sperm
Oocytes were collected from cycling adult female macaques of average maternal age (9.2 ± 2.3 yr old) undergoing controlled ovarian stimulations (COSs) according to the protocol from a previous study (Stouffer and Zelinski-Wooten 2004). Briefly, multiple ovarian follicles were induced to simultaneously develop by injecting exogenous hormones as described in more detail in the Extended Methods of the Supplemental Material. Follicular aspirations were laparoscopically performed on anesthetized animals to obtain cumulus–oocyte complexes (COCs). Each oocyte was denuded of cumulus cells and only mature MI and MII oocytes collected. Fresh semen was obtained from one of four adult male rhesus monkeys of average paternal age (9.4 ± 1.5 yr old) to minimize variability between sperm donors the same day as oocyte retrieval. Mature MII oocytes underwent conventional IVF at 37°C with 5% CO2 for 14–16 h with sperm diluted as previously described (Lanzendorf et al. 1990). Excess sperm was removed and fertilized oocytes visually assessed for two pronuclei and/or two polar bodies. The collection and preparation of oocytes and sperm were performed according to the approved Institutional Animal Care and Use Committee (IACUC) Assisted Reproductive Technologies (ART) Support Core protocol 0095 entitled, “Assisted Reproduction in Macaques.”
Time-lapse imaging
Confirmed zygotes were transferred to Eeva 12-well polystyrene petri dishes (Progyny, formerly Auxogyn) and cultured in 100 µL of commercial media supplemented with 10% serum protein (LifeGlobal) under mineral oil (CopperSurgical) at 37°C with 6% CO2, 5% O2, and 89% N2. Embryos were monitored with an Eeva darkfield 2.2.1 or bimodal (darkfield-brightfield) 2.3.5 time-lapse microscope system (Progyny) in a trigas incubator (Panasonic Healthcare) as previously described (Vera-Rodriguez et al. 2015). Images were taken every 5 min with a 0.6 sec (sec.) exposure time, and each image was time stamped with a frame number. All images were compiled into an AVI movie using Fiji software version 2.0.0 (Schindelin et al. 2012).
Embryo disassembly
The ZP was removed from each embryo by exposure to acidified Tyrode's solution (EMD Millipore) and washed with Ca2+- and Mg2+-free phosphate buffered saline (PBS). Cleavage-stage embryos were disaggregated into single cells, polar bodies, and cellular fragments if present with Quinn's advantage Ca2+- and Mg2+-free medium with HEPES plus 10% human albumin (CooperSurgical) and 0.05% trypsin-EDTA (Thermo Fisher Scientific) as necessary. Each blastomere, polar body, and cellular fragment was washed with Ca2+- and Mg2+-free PBS and collected individually for transfer to a sterile UltraFlux PCR tube (VWR). All of the above was performed under a stereomicroscope equipped with a digital camera (Leica Microsystems), which has movie-making capabilities, to document the collection of every sample. Samples were put into tubes, flash frozen on dry ice, and stored at −80°C. Only embryos for which the disassembly process occurred effectively with no apparent loss of material were carried forward for library preparation and sequencing.
Somatic cells
Human B lymphocytes (GM12878, Coriell Institute) were used for CNV analysis as previously described (Vitak et al. 2017). Female human skin fibroblasts from patients with monosomy X or trisomy 21 (GM10179 and AG05024, respectively; Coriell Institute) as well as karyotypically normal female and male rhesus skin fibroblasts (AG08312 and AG08305, respectively; Coriell Institute) were obtained and grown in DMEM F12 medium (GIBCO) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich). Cells were trypsinized and the cell suspension serially diluted in Ca2+/Mg2+-free PBS until single cells were detected in drops for freezing in the low-retention PCR tubes. Karyotyping of the human and rhesus primary fibroblasts (N = 50 metaphase spreads per cell line) was performed by the OHSU Research Cytogenetics Laboratory. All cell lines showed low levels of karyotypic heterogeneity (Supplemental Fig. S1A–D), and a Chr 19 pericentric inversion was detected in the rhesus male fibroblasts (Supplemental Fig. S1E).
DNA library preparation
Individual samples underwent DNA extraction and whole-genome amplification (WGA) using the PicoPLEX single-cell WGA kit (Rubicon Genomics) according to the manufacturer's instructions with slight modifications described in the Extended Methods of the Supplemental Material. Libraries were quantified by Qubit high sensitivity (HS) DNA assay (Life Technologies) and validated for sequencing by PCR amplification of the adapter sequence. Only libraries with DNA quantities greater than the no-template controls were included in sequencing. Fifty nanograms of DNA was prepared from each blastomere or fibroblast and 25 ng from both polar bodies and CCFs. Pooled libraries were purified with AMPure XP beads (Beckman Coulter) and requantified by the Qubit HS DNA kit. Quality was assessed with a 2200 TapeStation and/or a 2100 Bioanalyzer (Agilent).
Multiplex DNA sequencing
Pooled libraries were sequenced on Illumina platforms using primarily a 75-cycle kit with a modified single-end workflow that incorporated 14 dark cycles at the start of the first read before the imaged cycles. This step excluded the quasi-random priming sequences that are G-rich and lack a fluorophore for the two-color chemistry used by Illumina during cluster assignment as well as recently published custom indices for multiplexing (Vitak et al. 2017). All raw sample reads were demultiplexed and sequencing quality assessed with FastQC as previously described (Krueger et al. 2011). Illumina adapters were removed from raw reads with the sequence grooming tool, Cutadapt (Chen et al. 2014), resulting in reads of 120 bp on average. Trimmed reads were aligned to the most recent rhesus genome reference, rheMac8 (Zimin et al. 2014), using the BWA-MEM option of the Burrows–Wheeler alignment tool (Salavert Torres et al. 2012). To avoid read pile-ups owing to common repeats, all repeat sequences were “masked” (converted to an “N”) using RepeatMasker (Tarailo-Graovac and Chen 2009). Resulting BAM files were filtered to remove alignments with quality scores below 30 (Q < 30) as well as alignment duplicates with the SAMtools suite (Ramirez-Gonzalez et al. 2012). The average number of raw reads (2.7 × 106), trimmed and deduplicated reads (2.0 × 106), and uniquely aligned reads that passed Q > 30 (1.5 × 106) obtained from each blastomere are shown in Supplemental Table S1. Because CCFs contained only a partial genome and polar bodies may be either degraded or haploid (Schmerler and Wessel 2011), these samples were not included in the calculations.
Parental DNA library construction and sequencing
Whole blood was obtained from the male and female rhesus macaque parents in K2EDTA vacutainer collection tubes (BD Diagnostics) by the Colony Genetics Resource Core within the Primate Genetics Program at Oregon National Primate Research Center (ONPRC). Parental DNA was extracted using the Gentra Puregene blood kit (Qiagen) according to the manufacturer's protocol and stored at −80°C. Samples (1 µg) were fragmented using the Diagenode Bioruptor pico for a 300- to 400-bp size selection. The NEBNext DNA library prep master mix set and multiplex oligos (NEB) were then used to generate Illumina whole-genome sequencing libraries. Libraries were quantified with the Qubit HS DNA kit and size distribution assessed with the 2100 Bioanalyzer. Multiplexed libraries were sequenced at the Oregon State University Center for Genomic Research and Biocomputing on the HiSeq 3000 platform using the 150 paired-end protocol for a total of 2.84 × 109 reads (1.56 × 108 reads/sample). One parental sample (ID: 26129) was sequenced on the Illumina NextSeq using our custom 75-bp paired-end protocol for a total of approximately 3.50 × 108 reads.
Chromosome copy number calling
CNV was determined in each embryonic sample by integrating a newly developed bioinformatics pipeline called VNOWC and the previously published pipeline, CHI (Vitak et al. 2017). The VNOWC pipeline generates variable-sized windows with a constant number of expected reads per window and uses CBS to identify putative copy number changes between windows across each chromosome (Olshen et al. 2004). Before applying this approach to embryos, the pipeline was trained and tested on rhesus male euploid (42,XY) cells, as well as human fibroblasts carrying known aneuploidies (trisomy 21 or monosomy X). As shown in Supplemental Figure S1F, our bioinformatics pipeline was able to successfully detect chromosome losses and gains in all rhesus and human fibroblast samples, including single cells. Each call was confirmed with the use of Ginkgo (http://qb.cshl.edu/ginkgo), an open-source web tool for evaluating CNV in single cells (Garvin et al. 2015). To correct for GC bias across the genome, we also implemented the CHI pipeline, which uses the HMM (Ha et al. 2012) based on parameters determined previously (Knouse et al. 2016). Because other studies have shown that CNV can be accurately assessed with a genome coverage of 0.5–1× at a 15-Mb resolution (Lee et al. 2013; Zhou et al. 2018), we used this cut-off to reliably call subchromosomal losses and gains. We estimated false-positive calls following integration of the two pipelines and determined that the average counts from unexpected whole and segmental chromosome CNV calls depended on the number of mapped reads (Supplemental Fig. S1G,H). The percentage of samples with expected whole-chromosome CNV calls was also calculated and window sizes containing 4000 reads produced highly accurate CNV calling (Supplemental Fig. S1I). Thus, this window size was applied to the rhesus embryo samples, and all calls from the VNOWC and CHI methods generated variable sized windows that were manually intersected on a window-by-window basis as described in more detail in the Extended Methods of the Supplemental Material. Although the majority of CNV calls were shared between pipelines (N = 150/177; 84.7%), there were discordant calls detected between the VNOWC (N = 18/177; 10.2%) and CHI (N = 9/177; 5.1%) pipelines, but they were primarily subchromosomal differences (Supplemental Fig. S1J). Approximate DNA breakpoint locations were identified in rhesus chromosome ideograms adapted from Ventura et al. (2007), http://www.biologia.uniba.it/macaque/, and http://www.biologia.uniba.it/primates/2-OWM/MMU/MMU.html, and by identifying the syntenic g-band interval in the UCSC human assembly, GRCh38.
SNP parentage analysis
Parental assignment by the summation of information from multiple SNPs across large chromosomal segments used SNP calls from a pipeline based on the Broad Institute's Genome Analysis Toolkit (GATK) (McKenna et al. 2010; Van der Auwera et al. 2013) but adapted for rhesus macaque. Briefly, reads were trimmed using Trimmomatic (Bolger et al. 2014) and aligned to the Mmul_8.0.1 reference genome using BWA-MEM (Li and Durbin 2010). BAM postprocessing included local realignment around indels and marking of duplicate reads using Picard tools (http://broadinstitute.github.io/picard). GATK's HaplotypeCaller was used to produce VCF files, and genotypes were called via GenotypeGVCFs. All data were processed using DISCVR-seq (https://github.com/bbimber/discvr-seq/wiki), a LabKey Server-based system (Nelson et al. 2011). SNPs identified in repetitive regions and those samples that had fewer than 10 SNPs per chromosome (with the exception of CCFs) were removed. To further restrict the set of SNPs to only those of higher confidence, we required that there be at least two reads for each SNP and only used SNPs with opposite homozygous genotypes in the parents. Combining information from several highly informative SNPs across large chromosomal segments permitted us to confidently determine parentage despite the low confidence of individual SNP calls owing to scDNA-seq coverage (∼0.05× on average). Unlike typical genotyping of individual SNPs by NGS, this multiple SNP approach does not require a high level of sequencing depth to assign parentage as recently shown in other contexts (Gorjanc et al. 2017; Whalen et al. 2018). The ratio of maternal to paternal alleles was used to assess parental inheritance and was visualized in heat maps by Morpheus (Broad Institute) and in histograms (MATLAB R2016a).
Immunofluorescence confocal imaging
ZP-free embryos were fixed with 4% paraformaldehyde in PBS (Alfa Aesar) for 20 min at room temperature (RT). Once fixed and washed in PBS with 0.1% BSA and 0.1% Tween-20 (PBS-T; Calbiochem), embryos were permeabilized in 1% Triton X-100 (Calbiochem) for 1 h at RT. To block nonspecific binding, embryos were transferred to a 7% donkey serum (Jackson ImmunoResearch Laboratories)/PBS-T solution overnight at 4°C. LMNB1 (rabbit monoclonal; Abcam ab16048) and γH2A.X (mouse monoclonal; EMD Millipore 05-636) antibodies were diluted 1:1000 and 1:100, respectively, in PBS-T with 1% donkey serum and embryos sequentially stained overnight at 4°C. Primary antibodies were detected using 488- or 647-conjugated donkey Alexa Fluor secondary antibodies (Thermo Fisher Scientific catalog A21206 and A31571) to the appropriate species at a 1:250 dilution with 1% donkey serum in PBS-T at RT for 1 h in the dark. The DNA was stained with 1 µg/mL DAPI for 15 min and immunofluorescence visualized using glass bottom Petri dishes (Mattek) and a Leica SP5 AOBS spectral confocal system. Z-stacks 1–5 µM apart were imaged one fluorophore at a time to avoid spectral overlap between channels. Stacked images and individual channels for each color were combined into composite images using Fiji (Schindelin et al. 2012).
Statistical methods
Significance of SNP allele parental ratios was examined using the cumulative binomial test with Bonferroni correction at P < 0.05. Such a significant agreement across multiple alleles is unlikely to arise by chance. Chromosome size versus number of segmental breaks was evaluated by Spearman's correlation.
Data access
All DNA-seq data from this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject) under accession number PRJNA415642. All scripts and pipelines generated in this study are available as Supplemental Code. The VNOWC pipeline was also uploaded on GitHub (https://github.com/nathanlazar/Oocyte_CN).
Supplementary Material
Acknowledgments
We thank the ONPRC ART Core for their assistance with oocyte and sperm collection, Colony Genetics Resource Core for the parental DNA, Molecular and Cellular Biology Core for the MiSeq runs, and Imaging and Morphology Core for confocal microscopy (supported by grant S10 RR024585), all under the auspices of the NIH/OD ONPRC core grant (P51 OD011092). The TLM part of the study would not be possible without the generosity and support from Auxogyn. We also thank the OHSU ExaCloud Cluster Computational Resource, which allowed us to perform the intensive large-scale data workflows. We give special thanks to Drs. C. Bishop, C. Hanna, and J. Hennebold, as well as C. Ramsey, for rhesus samples and/or embryology expertise; S. Vitak and A. Fields for sequencing support; and G. Schau for assistance in CNV pipeline development. We also thank members of the Chavez and Carbone laboratories for insightful discussions. B.L.D. was supported by the P.E.O. Scholar Award, N.L. Tartar Research Fellowship, and T32 Reproductive Biology NIH Training Grant (T32 HD007133). J.L.R. was supported by the Collins Medical Trust Foundation, Glenn/AFAR Scholarship for Research in the Biology of Aging, and the NIH/NICHD (F31HD094472). N.H.L. was supported by a fellowship from the National Library of Medicine Biomedical Informatics Training Grant (T15LM007088). This work was supported by the NIH/NICHD (R01HD086073-A1), National Centers for Translational Research in Reproduction and Infertility (NCTRI) pilot funds (NIH, Eunice Kennedy Shriver National Institute of Child Health and Human Development, P50 HD071836), Howard & Georgeanna Jones Foundation for Reproductive Medicine, Medical Research Foundation of Oregon, and Collins Medical Trust (to S.L.C.).
Author contributions: B.L.D. and S.L.C. designed the study, performed experiments, analyzed data, and wrote the manuscript. J.L.R. developed the single-cell library preparation and multiplex scDNA-seq approach. N.H.L. created and implemented the VNOWC CNV pipeline. S.S.F. aligned and genotyped the samples and generated SNP parentage summaries. N.R. provided support for the embryology and imaging experiments. K.A.T. developed and implemented the CHI CNV pipeline. A.A. provided scDNA-seq expertise, the NextSeq runs, and human B-lymphocyte data. M.Y. determined the expected/unexpected CNV calls and prepared the SRA submission. L.G. and B.P. provided biostatistical analysis for the SNP data and segmental breaks. K.A.N. prepared sequencing libraries from the parental DNA. L.C. assisted in the study design and interpretation of the scDNA-seq results. All authors were involved in editing the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.239830.118.
References
- Alikani M, Cohen J, Tomkin G, Garrisi GJ, Mack C, Scott RT. 1999. Human embryo fragmentation in vitro and its implications for pregnancy and implantation. Fertil Steril 71: 836–842. 10.1016/S0015-0282(99)00092-8 [DOI] [PubMed] [Google Scholar]
- Antczak M, Van Blerkom J. 1999. Temporal and spatial aspects of fragmentation in early human embryos: possible effects on developmental competence and association with the differential elimination of regulatory proteins from polarized domains. Hum Reprod 14: 429–447. 10.1093/humrep/14.2.429 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton H, Graham SJ, Van der Aa N, Kumar P, Theunis K, Fernandez Gallardo E, Voet T, Zernicka-Goetz M. 2016. Mouse model of chromosome mosaicism reveals lineage-specific depletion of aneuploid cells and normal developmental potential. Nat Commun 7: 11165 10.1038/ncomms11165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buster JE, Bustillo M, Rodi IA, Cohen SW, Hamilton M, Simon JA, Thorneycroft IH, Marshall JR. 1985. Biologic and morphologic development of donated human ova recovered by nonsurgical uterine lavage. Am J Obstet Gynecol 153: 211–217. 10.1016/0002-9378(85)90116-4 [DOI] [PubMed] [Google Scholar]
- Chamayou S, Patrizio P, Storaci G, Tomaselli V, Alecci C, Ragolia C, Crescenzo C, Guglielmino A. 2013. The use of morphokinetic parameters to select all embryos with full capacity to implant. J Assist Reprod Genet 30: 703–710. 10.1007/s10815-013-9992-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez SL, Loewke KE, Han J, Moussavi F, Colls P, Munne S, Behr B, Reijo Pera RA. 2012. Dynamic blastomere behaviour reflects human embryo ploidy by the four-cell stage. Nat Commun 3: 1251 10.1038/ncomms2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez SL, McElroy SL, Bossert NL, De Jonge CJ, Rodriguez MV, Leong DE, Behr B, Westphal LM, Reijo Pera RA. 2014. Comparison of epigenetic mediator expression and function in mouse and human embryonic blastomeres. Hum Mol Genet 23: 4970–4984. 10.1093/hmg/ddu212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Khaleel SS, Huang H, Wu CH. 2014. Software for pre-processing Illumina next-generation sequencing short read sequences. Source Code Biol Med 9: 8 10.1186/1751-0473-9-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow JF, Yeung WS, Lau EY, Lee VC, Ng EH, Ho PC. 2014. Array comparative genomic hybridization analyses of all blastomeres of a cohort of embryos from young IVF patients revealed significant contribution of mitotic errors to embryo mosaicism at the cleavage stage. Reprod Biol Endocrinol 12: 105 10.1186/1477-7827-12-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482: 53–58. 10.1038/nature10802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bourcy CF, De Vlaminck I, Kanbar JN, Wang J, Gawad C, Quake SR. 2014. A quantitative comparison of single-cell whole genome amplification methods. PLoS One 9: e105585 10.1371/journal.pone.0105585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhanty JD, Harper JC, Ao A, Handyside AH, Winston RM. 1997. Multicolour FISH detects frequent chromosomal mosaicism and chaotic division in normal preimplantation embryos from fertile patients. Hum Genet 99: 755–760. 10.1007/s004390050443 [DOI] [PubMed] [Google Scholar]
- Destouni A, Zamani Esteki M, Catteeuw M, Tšuiko O, Dimitriadou E, Smits K, Kurg A, Salumets A, Van Soom A, Voet T, et al. 2016. Zygotes segregate entire parental genomes in distinct blastomere lineages causing cleavage-stage chimerism and mixoploidy. Genome Res 26: 567–578. 10.1101/gr.200527.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dozortsev D, Ermilov A, El-Mowafi DM, Diamond M. 1998. The impact of cellular fragmentation induced experimentally at different stages of mouse preimplantation development. Hum Reprod 13: 1307–1311. 10.1093/humrep/13.5.1307 [DOI] [PubMed] [Google Scholar]
- Dupont C, Bavister BD, Armant DR, Brenner CA. 2009a. Rhesus macaque embryos derived from MI oocytes maturing after retrieval display high rates of chromosomal anomalies. Hum Reprod 24: 929–935. 10.1093/humrep/den429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont C, Froenicke L, Lyons LA, Bavister BD, Brenner CA. 2009b. Chromosomal instability in rhesus macaque preimplantation embryos. Fertil Steril 91: 1230–1237. 10.1016/j.fertnstert.2008.01.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont C, Segars J, DeCherney A, Bavister BD, Armant DR, Brenner CA. 2010. Incidence of chromosomal mosaicism in morphologically normal nonhuman primate preimplantation embryos. Fertil Steril 93: 2545–2550. 10.1016/j.fertnstert.2009.06.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragouli E, Alfarawati S, Spath K, Babariya D, Tarozzi N, Borini A, Wells D. 2017. Analysis of implantation and ongoing pregnancy rates following the transfer of mosaic diploid–aneuploid blastocysts. Hum Genet 136: 805–819. 10.1007/s00439-017-1797-4 [DOI] [PubMed] [Google Scholar]
- Garvin T, Aboukhalil R, Kendall J, Baslan T, Atwal GS, Hicks J, Wigler M, Schatz MC. 2015. Interactive analysis and assessment of single-cell copy-number variations. Nat Methods 12: 1058–1060. 10.1038/nmeth.3578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson D, Håkanson U, Stoller P, Marti D, Jin Y, Rosengren AH, Stewénius Y, Kahl F, Panagopoulos I. 2008. When the genome plays dice: circumvention of the spindle assembly checkpoint and near-random chromosome segregation in multipolar cancer cell mitoses. PLoS One 3: e1871 10.1371/journal.pone.0001871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorjanc G, Dumasy J-F, Gonen S, Gaynor RC, Antolin R, Hickey JM. 2017. Potential of low-coverage genotyping-by-sequencing and imputation for cost-effective genomic selection in biparental segregating populations. Crop Science 57: 1404–1420. 10.2135/cropsci2016.08.0675 [DOI] [Google Scholar]
- Greco E, Minasi MG, Fiorentino F. 2015. Healthy babies after intrauterine transfer of mosaic aneuploid blastocysts. N Engl J Med 373: 2089–2090. 10.1056/NEJMc1500421 [DOI] [PubMed] [Google Scholar]
- Ha G, Roth A, Lai D, Bashashati A, Ding J, Goya R, Giuliany R, Rosner J, Oloumi A, Shumansky K, et al. 2012. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res 22: 1995–2007. 10.1101/gr.137570.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy K, Spanos S, Becker D, Iannelli P, Winston RM, Stark J. 2001. From cell death to embryo arrest: mathematical models of human preimplantation embryo development. Proc Natl Acad Sci 98: 1655–1660. 10.1073/pnas.98.4.1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. 2013. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154: 47–60. 10.1016/j.cell.2013.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlinka D, Kalatova B, Uhrinova I, Dolinska S, Rutarova J, Rezacova J, Lazarovska S, Dudas M. 2012. Time-lapse cleavage rating predicts human embryo viability. Physiol Res 61: 513–525. [DOI] [PubMed] [Google Scholar]
- Huang J, Yan L, Fan W, Zhao N, Zhang Y, Tang F, Xie XS, Qiao J. 2014. Validation of multiple annealing and looping-based amplification cycle sequencing for 24-chromosome aneuploidy screening of cleavage-stage embryos. Fertil Steril 102: 1685–1691. 10.1016/j.fertnstert.2014.08.015 [DOI] [PubMed] [Google Scholar]
- Johnson DS, Gemelos G, Baner J, Ryan A, Cinnioglu C, Banjevic M, Ross R, Alper M, Barrett B, Frederick J, et al. 2010. Preclinical validation of a microarray method for full molecular karyotyping of blastomeres in a 24-h protocol. Hum Reprod 25: 1066–1075. 10.1093/humrep/dep452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouse KA, Wu J, Amon A. 2016. Assessment of megabase-scale somatic copy number variation using single-cell sequencing. Genome Res 26: 376–384. 10.1101/gr.198937.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR, Osborne CS. 2011. Large scale loss of data in low-diversity Illumina sequencing libraries can be recovered by deferred cluster calling. PLoS One 6: e16607 10.1371/journal.pone.0016607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzendorf SE, Gliessman PM, Archibong AE, Alexander M, Wolf DP. 1990. Collection and quality of rhesus monkey semen. Mol Reprod Dev 25: 61–66. 10.1002/mrd.1080250111 [DOI] [PubMed] [Google Scholar]
- Lee J, Lee U, Kim B, Yoon J. 2013. A computational method for detecting copy number variations using scale-space filtering. BMC Bioinformatics 14: 57 10.1186/1471-2105-14-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26: 589–595. 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lightfoot DA, Kouznetsova A, Mahdy E, Wilbertz J, Höög C. 2006. The fate of mosaic aneuploid embryos during mouse development. Dev Biol 289: 384–394. 10.1016/j.ydbio.2005.11.001 [DOI] [PubMed] [Google Scholar]
- Liu S, Kwon M, Mannino M, Yang N, Renda F, Khodjakov A, Pellman D. 2018. Nuclear envelope assembly defects link mitotic errors to chromothripsis. Nature 561: 551–555. 10.1038/s41586-018-0534-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay IC, Haerty W, Kumar P, Li YI, Hu TX, Teng MJ, Goolam M, Saurat N, Coupland P, Shirley LM, et al. 2015. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat Methods 12: 519–522. 10.1038/nmeth.3370 [DOI] [PubMed] [Google Scholar]
- Maxwell SM, Colls P, Hodes-Wertz B, McCulloh DH, McCaffrey C, Wells D, Munné S, Grifo JA. 2016. Why do euploid embryos miscarry? A case-control study comparing the rate of aneuploidy within presumed euploid embryos that resulted in miscarriage or live birth using next-generation sequencing. Fertil Steril 106: 1414–1419.e5. 10.1016/j.fertnstert.2016.08.017 [DOI] [PubMed] [Google Scholar]
- McCoy RC, Demko Z, Ryan A, Banjevic M, Hill M, Sigurjonsson S, Rabinowitz M, Fraser HB, Petrov DA. 2015. Common variants spanning PLK4 are associated with mitotic-origin aneuploidy in human embryos. Science 348: 235–238. 10.1126/science.aaa3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy RC, Newnham LJ, Ottolini CS, Hoffmann ER, Chatzimeletiou K, Cornejo OE, Zhan Q, Zaninovic N, Rosenwaks Z, Petrov DA, et al. 2018. Tripolar chromosome segregation drives the association between maternal genotype at variants spanning PLK4 and aneuploidy in human preimplantation embryos. Hum Mol Genet 27: 2573–2585. 10.1093/hmg/ddy147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JF, Williamson E, Glue J, Gordon YB, Grudzinskas JG, Sykes A. 1980. Fetal loss after implantation: a prospective study. Lancet 2: 554–556. 10.1016/S0140-6736(80)91991-1 [DOI] [PubMed] [Google Scholar]
- Minasi MG, Colasante A, Riccio T, Ruberti A, Casciani V, Scarselli F, Spinella F, Fiorentino F, Varricchio MT, Greco E. 2016. Correlation between aneuploidy, standard morphology evaluation and morphokinetic development in 1730 biopsied blastocysts: a consecutive case series study. Hum Reprod 31: 2245–2254. 10.1093/humrep/dew183 [DOI] [PubMed] [Google Scholar]
- Nagaoka SI, Hassold TJ, Hunt PA. 2012. Human aneuploidy: mechanisms and new insights into an age-old problem. Nat Rev Genet 13: 493–504. 10.1038/nrg3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EK, Piehler B, Eckels J, Rauch A, Bellew M, Hussey P, Ramsay S, Nathe C, Lum K, Krouse K, et al. 2011. LabKey Server: an open source platform for scientific data integration, analysis and collaboration. BMC Bioinformatics 12: 71 10.1186/1471-2105-12-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu N, Mercado-Uribe I, Liu J. 2017. Dedifferentiation into blastomere-like cancer stem cells via formation of polyploid giant cancer cells. Oncogene 36: 4887–4900. 10.1038/onc.2017.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara M, Aoki K, Okada S, Suzumori K. 2000. Embryonic karyotype of abortuses in relation to the number of previous miscarriages. Fertil Steril 73: 300–304. 10.1016/S0015-0282(99)00495-1 [DOI] [PubMed] [Google Scholar]
- Olshen AB, Venkatraman ES, Lucito R, Wigler M. 2004. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 5: 557–572. 10.1093/biostatistics/kxh008 [DOI] [PubMed] [Google Scholar]
- Ottolini CS, Kitchen J, Xanthopoulou L, Gordon T, Summers MC, Handyside AH. 2017. Tripolar mitosis and partitioning of the genome arrests human preimplantation development in vitro. Sci Rep 7: 9744 10.1038/s41598-017-09693-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellestor F. 2014. Chromothripsis: how does such a catastrophic event impact human reproduction? Hum Reprod 29: 388–393. 10.1093/humrep/deu003 [DOI] [PubMed] [Google Scholar]
- Pellestor F, Gatinois V, Puechberty J, Geneviève D, Lefort G. 2014. Chromothripsis: potential origin in gametogenesis and preimplantation cell divisions. A review. Fertil Steril 102: 1785–1796. 10.1016/j.fertnstert.2014.09.006 [DOI] [PubMed] [Google Scholar]
- Pereda J, Croxatto HB. 1978. Ultrastructure of a seven-cell human embryo. Biol Reprod 18: 481–489. 10.1095/biolreprod18.3.481 [DOI] [PubMed] [Google Scholar]
- Ramirez-Gonzalez RH, Bonnal R, Caccamo M, Maclean D. 2012. Bio-samtools: Ruby bindings for SAMtools, a library for accessing BAM files containing high-throughput sequence alignments. Source Code Biol Med 7: 6 10.1186/1751-0473-7-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273: 5858–5868. 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Salavert Torres J, Espert IB, Dominguez AT, Hernandez Garcia V, Medina Castello I, Tarraga Gimenez J, Dopazo Blazquez J. 2012. Using GPUs for the exact alignment of short-read genetic sequences by means of the Burrows-Wheeler transform. IEEE/ACM Trans Comput Biol Bioinform 9: 1245–1256. 10.1109/TCBB.2012.49 [DOI] [PubMed] [Google Scholar]
- Sathananthan AH, Kola I, Osborne J, Trounson A, Ng SC, Bongso A, Ratnam SS. 1991. Centrioles in the beginning of human development. Proc Natl Acad Sci 88: 4806–4810. 10.1073/pnas.88.11.4806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatten G, Simerly C, Schatten H. 1991. Maternal inheritance of centrosomes in mammals? Studies on parthenogenesis and polyspermy in mice. Proc Natl Acad Sci 88: 6785–6789. 10.1073/pnas.88.15.6785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmerler S, Wessel GM. 2011. Polar bodies: more a lack of understanding than a lack of respect. Mol Reprod Dev 78: 3–8. 10.1002/mrd.21266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood SW, Sheridan JP, Schimke RT. 1994. Induction of apoptosis by the anti-tubulin drug colcemid: relationship of mitotic checkpoint control to the induction of apoptosis in HeLa S3 cells. Exp Cell Res 215: 373–379. 10.1006/excr.1994.1354 [DOI] [PubMed] [Google Scholar]
- Stouffer RL, Zelinski-Wooten MB. 2004. Overriding follicle selection in controlled ovarian stimulation protocols: quality vs quantity. Reprod Biol Endocrinol 2: 32 10.1186/1477-7827-2-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarailo-Graovac M, Chen N. 2009. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics Chapter 4: Unit 4 10 10.1002/0471250953.bi0410s25 [DOI] [PubMed] [Google Scholar]
- Treff NR, Krisher RL, Tao X, Garnsey H, Bohrer C, Silva E, Landis J, Taylor D, Scott RT, Woodruff TK, et al. 2016. Next generation sequencing-based comprehensive chromosome screening in mouse polar bodies, oocytes, and embryos. Biol Reprod 94: 76 10.1095/biolreprod.115.135483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tšuiko O, Catteeuw M, Zamani Esteki M, Destouni A, Bogado Pascottini O, Besenfelder U, Havlicek V, Smits K, Kurg A, Salumets A, et al. 2017. Genome stability of bovine in vivo-conceived cleavage-stage embryos is higher compared to in vitro-produced embryos. Hum Reprod 32: 2348–2357. 10.1093/humrep/dex286 [DOI] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43: 11.10.11–11.10.33. 10.1002/0471250953.bi1110s43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste E, Voet T, Le Caignec C, Ampe M, Konings P, Melotte C, Debrock S, Amyere M, Vikkula M, Schuit F, et al. 2009a. Chromosome instability is common in human cleavage-stage embryos. Nat Med 15: 577–583. 10.1038/nm.1924 [DOI] [PubMed] [Google Scholar]
- Vanneste E, Voet T, Melotte C, Debrock S, Sermon K, Staessen C, Liebaers I, Fryns JP, D'Hooghe T, Vermeesch JR. 2009b. What next for preimplantation genetic screening? High mitotic chromosome instability rate provides the biological basis for the low success rate. Hum Reprod 24: 2679–2682. 10.1093/humrep/dep266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Diez C, Yamagata K, Trivedi S, Haverfield J, FitzHarris G. 2016. Micronucleus formation causes perpetual unilateral chromosome inheritance in mouse embryos. Proc Natl Acad Sci 113: 626–631. 10.1073/pnas.1517628112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Antonacci F, Cardone MF, Stanyon R, D'Addabbo P, Cellamare A, Sprague LJ, Eichler EE, Archidiacono N, Rocchi M. 2007. Evolutionary formation of new centromeres in macaque. Science 316: 243–246. 10.1126/science.1140615 [DOI] [PubMed] [Google Scholar]
- Vera-Rodriguez M, Chavez SL, Rubio C, Reijo Pera RA, Simon C. 2015. Prediction model for aneuploidy in early human embryo development revealed by single-cell analysis. Nat Commun 6: 7601 10.1038/ncomms8601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitak SA, Torkenczy KA, Rosenkrantz JL, Fields AJ, Christiansen L, Wong MH, Carbone L, Steemers FJ, Adey A. 2017. Sequencing thousands of single-cell genomes with combinatorial indexing. Nat Methods 14: 302–308. 10.1038/nmeth.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Sha H, Ji D, Zhang HL, Chen D, Cao Y, Zhu J. 2014. Polar body genome transfer for preventing the transmission of inherited mitochondrial diseases. Cell 157: 1591–1604. 10.1016/j.cell.2014.04.042 [DOI] [PubMed] [Google Scholar]
- Whalen A, Gorjanc G, Hickey JM. 2018. Parentage assignment with low density array data and low coverage sequence data. bioRxiv 10.1101/270561. [DOI]
- Whitaker M. 2006. Calcium at fertilization and in early development. Physiol Rev 86: 25–88. 10.1152/physrev.00023.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox AJ, Weinberg CR, Baird DD. 1995. Timing of sexual intercourse in relation to ovulation: effects on the probability of conception, survival of the pregnancy, and sex of the baby. N Engl J Med 333: 1517–1521. 10.1056/NEJM199512073332301 [DOI] [PubMed] [Google Scholar]
- Winston NJ, Johnson MH. 1992. Can the mouse embryo provide a good model for the study of abnormal cellular development seen in human embryos? Hum Reprod 7: 1291–1296. 10.1093/oxfordjournals.humrep.a137844 [DOI] [PubMed] [Google Scholar]
- Wu DH, Reynolds K, Maxwell R, Lindheim SR, Aubuchon M, Thomas MA. 2011. Age does not influence the effect of embryo fragmentation on successful blastocyst development. Fertil Steril 95: 2778–2780. 10.1016/j.fertnstert.2011.05.024 [DOI] [PubMed] [Google Scholar]
- Xu J, Cheung T, Chan ST, Ho P, Yeung WS. 2001. The incidence of cytoplasmic fragmentation in mouse embryos in vitro is not affected by inhibition of caspase activity. Fertil Steril 75: 986–991. 10.1016/S0015-0282(01)01687-9 [DOI] [PubMed] [Google Scholar]
- Yoon SY, Jellerette T, Salicioni AM, Lee HC, Yoo MS, Coward K, Parrington J, Grow D, Cibelli JB, Visconti PE, et al. 2008. Human sperm devoid of PLC, ζ1 fail to induce Ca2+ release and are unable to initiate the first step of embryo development. J Clin Invest 118: 3671–3681. 10.1172/JCI36942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni L, Mishell DR Jr, Bell JH, Baca M. 1966. Fine structure of the human ovum in the pronuclear stage. J Cell Biol 30: 579–600. 10.1083/jcb.30.3.579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang CZ, Spektor A, Cornils H, Francis JM, Jackson EK, Liu S, Meyerson M, Pellman D. 2015. Chromothripsis from DNA damage in micronuclei. Nature 522: 179–184. 10.1038/nature14493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Ho SS, Zhang X, Pattni R, Haraksingh RR, Urban AE. 2018. Whole-genome sequencing analysis of CNV using low-coverage and paired-end strategies is efficient and outperforms array-based CNV analysis. J Med Genet 55: 735–743. 10.1136/jmedgenet-2018-105272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, Cornish AS, Maudhoo MD, Gibbs RM, Zhang X, Pandey S, Meehan DT, Wipfler K, Bosinger SE, Johnson ZP, et al. 2014. A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biol Direct 9: 20 10.1186/1745-6150-9-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinaman MJ, Clegg ED, Brown CC, O'Connor J, Selevan SG. 1996. Estimates of human fertility and pregnancy loss. Fertil Steril 65: 503–509. 10.1016/S0015-0282(16)58144-8 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.