Abstract
Most yeast genes have a nucleosome-depleted region (NDR) at the promoter and an array of regularly spaced nucleosomes phased relative to the transcription start site. We have examined the interplay between RSC (a conserved essential SWI/SNF-type complex that determines NDR size) and the ISW1, CHD1, and ISW2 nucleosome spacing enzymes in chromatin organization and transcription, using isogenic strains lacking all combinations of these enzymes. The contributions of these remodelers to chromatin organization are largely combinatorial, distinct, and nonredundant, supporting a model in which the +1 nucleosome is positioned by RSC and then used as a reference nucleosome by the spacing enzymes. Defective chromatin organization correlates with altered RNA polymerase II (Pol II) distribution. RSC-depleted cells exhibit low levels of elongating Pol II and high levels of terminating Pol II, consistent with defects in both termination and initiation, suggesting that RSC facilitates both. Cells lacking both ISW1 and CHD1 show the opposite Pol II distribution, suggesting elongation and termination defects. These cells have extremely disrupted chromatin, with high levels of closely packed dinucleosomes involving the second (+2) nucleosome. We propose that ISW1 and CHD1 facilitate Pol II elongation by separating closely packed nucleosomes.
Eukaryotic DNA is packaged into the nucleus in the form of chromatin. The basic structural unit of chromatin is the nucleosome, which contains ∼147 bp of DNA wrapped almost twice around a central core histone octamer containing two molecules each of histones H2A, H2B, H3, and H4 (Luger et al. 1997). Nucleosomes are compact, stable structures that inhibit DNA-dependent processes such as transcription. Cells possess chromatin remodeling enzymes that manipulate the repressive properties of chromatin to regulate genes. Chromatin remodeling enzymes use the free energy obtained from ATP hydrolysis to assemble, evict, slide or restructure nucleosomes (Clapier and Cairns 2009; Narlikar et al. 2013; Bartholomew 2014; Swygert and Peterson 2014). Mutations in genes encoding subunits of human remodeling complexes are strongly associated with cancer and other diseases (Narlikar et al. 2013).
Genome-wide nucleosome maps for budding yeast reveal that most promoters are located in nucleosome-depleted regions (NDRs) and that nucleosomes are regularly spaced and phased relative to the transcription start site (TSS) (Lee et al. 2004; Yuan et al. 2005; Jiang and Pugh 2009). RSC is an essential SWI/SNF-type complex homologous to human PBAF (Clapier and Cairns 2009). In vitro, RSC can evict, mobilize, or remodel nucleosomes, and create promoter NDRs in reconstituted chromatin (Lorch et al. 2001; Saha et al. 2002; Shukla et al. 2010; Krietenstein et al. 2016). Depletion of an essential RSC subunit results in growth arrest (Treich and Carlson 1997), major changes in gene expression (Ng et al. 2002), and much reduced overall levels of transcription by all three RNA polymerases (Parnell et al. 2008). In RSC-depleted cells, nucleosomes shift into the promoter, resulting in narrowing and partial filling of the NDR (Hartley and Madhani 2009; Ganguli et al. 2014; Rawal et al. 2018a). RSC is preferentially associated with the promoter-flanking −1 and +1 nucleosomes and, to a lesser extent, with the +2 and +3 nucleosomes (Yen et al. 2012). RSC is modestly enriched on highly active genes and at tRNA genes (Ng et al. 2002; Floer et al. 2010; Ganguli et al. 2014; Spain et al. 2014; Rawal et al. 2018a).
The ISW1, CHD1, and ISW2 remodelers space nucleosomes in vitro, such that CHD1 spaces nucleosomes closer together than ISW1, whereas ISW2 spaces them farther apart (Ito et al. 1997; Tsukiyama et al. 1999; Lusser et al. 2005; Stockdale et al. 2006). In vivo, deletion of both ISW1 and CHD1 results in a major disruption of spacing and phasing (Gkikopoulos et al. 2011; Ocampo et al. 2016). We have proposed that the three spacing enzymes compete to set the spacing on individual genes, and the global average spacing reflects the outcome of this competition (Ocampo et al. 2016). In addition to playing a major role in global chromatin organization, ISW1 and CHD1 suppress histone exchange and cryptic initiation (Cheung et al. 2008; Quan and Hartzog 2010; Tirosh et al. 2010; Gkikopoulos et al. 2011; Pointner et al. 2012; Radman-Livaja et al. 2012; Smolle et al. 2012). Single-gene studies show that CHD1 and ISW1 are involved in transcription through chromatin. CHD1 associates with Pol II elongation factors (Simic et al. 2003) and acts as a termination factor, often in concert with ISW1 or ISW2 (Alén et al. 2002). Two different ISW1 complexes (ISW1a and ISW1b) (Vary et al. 2003) coordinate transcription of the MET16 gene, such that ISW1a positions nucleosomes at the promoter to regulate initiation, whereas ISW1b acts in the coding region to facilitate Pol II elongation and termination (Alén et al. 2002; Morillon et al. 2003). The third spacing enzyme, ISW2, also limits cryptic initiation, but its contribution to chromatin organization is restricted to inactive genes (Goldmark et al. 2000; Kent et al. 2001; Whitehouse et al. 2007; Ocampo et al. 2016).
Much less is known about the interplay between the SWI/SNF-type complexes and the nucleosome spacing enzymes. ISW1 and RSC cooperate to remodel the PHO5 promoter (Musladin et al. 2014), but they behave antagonistically at other promoters, where ISW1 partly reverses RSC-mediated widening of the NDR (Parnell et al. 2015). Here, we report a systematic study of the roles of RSC and the three nucleosome spacing enzymes in chromatin organization and transcription by analyzing all possible mutant combinations.
Results
Combinatorial contributions of the RSC, ISW1, CHD1, and ISW2 remodelers to chromatin organization
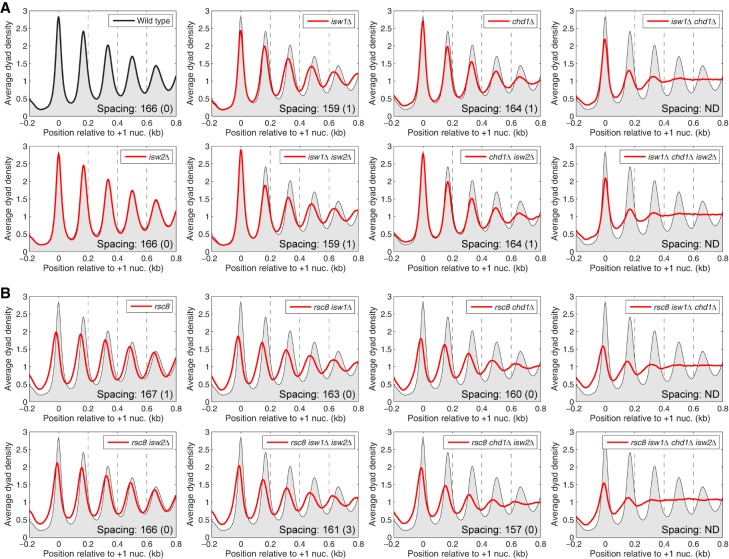
Previously, we reported the effects on chromatin organization of all combinations of null mutations in ISW1, CHD1, and ISW2, using high resolution nucleosome maps obtained by micrococcal nuclease (MNase) digestion and paired-end sequencing of nucleosomal DNA (MNase-seq; GEO accession number GSE69400) (Ocampo et al. 2016). To investigate the interplay between RSC and the spacing enzymes, we constructed a second set of eight strains to complete the set of 16 isogenic strains corresponding to all possible combinations of RSC depletion with null mutants in the three spacing enzymes. For simplicity, we will refer to Rsc8-depleted cells as rsc8 cells. Nucleosome phasing analysis relative to the +1 nucleosome on all ∼5000 yeast genes was performed on data from all 16 strains (Fig. 1). The use of the +1 nucleosome as a reference point should give more accurate measurements of nucleosome spacing (defined as the average distance between the centers of neighboring nucleosomes) than using the TSS, although the results are only slightly different (Supplemental Fig. S1).
Figure 1.
Combinatorial effects of RSC, ISW1, CHD1, and ISW2 remodelers on the chromatin organization of yeast genes. Average nucleosome phasing on all yeast genes relative to the dyad of the +1 nucleosome in wild-type cells. (A) Phasing in wild type, isw1Δ, chd1Δ, and isw2Δ mutants (from Ocampo et al. 2016). (B) Phasing in rsc8 mutants. For comparison, the phasing profile for wild-type cells is shown in all plots (gray shading). Inset values: average spacing determined by regression analysis of the first five peaks, with standard deviation (two biological replicate experiments); (ND) not determined because phasing is poor. (Note: Nucleosome spacing refers to the average distance between the centers of neighboring nucleosomes. An equivalent way of estimating the space between neighboring nucleosomes, or nucleosome repeat length, is by analyzing the periodicity of the bands observed after gel electrophoresis of MNase-digested chromatin.) The sequencing depths of all samples were normalized to one read per bp. See Supplemental Figure S1 for alignment on the TSS.
The data we reported previously (Ocampo et al. 2016) are summarized in Figure 1A. In wild-type cells, there is a deep NDR at the promoter with strong nucleosome phasing on the gene. Both chd1Δ and isw1Δ cells show broader peaks with reduced amplitudes on gene bodies, indicating weaker nucleosome phasing, whereas isw2Δ cells show no obvious changes in chromatin structure. Nucleosome spacing is reduced from 166 bp in wild-type cells to 159 bp in isw1Δ cells: The +1 nucleosome is in the same position as in wild-type cells, but the downstream nucleosomes are shifted toward the promoter. The isw1Δ isw2Δ and chd1Δ isw2Δ double mutants resemble the isw1Δ and chd1Δ single mutants, respectively, consistent with a minimal contribution from the isw2Δ mutation. The isw1Δ chd1Δ double mutant shows a major perturbation of nucleosome organization (Gkikopoulos et al. 2011). Both double mutants involving isw1Δ show the reduced nucleosome spacing observed in the single mutant, although the effect is not as clear in the isw1Δ chd1Δ mutant because the phasing is very poor. Finally, the isw1Δ chd1Δ isw2Δ triple mutant is essentially the same as the isw1Δ chd1Δ double mutant, again consistent with a minimal global contribution from ISW2. Thus, ISW1 and CHD1 are the major phasing and spacing enzymes. Because the spacing in isw1Δ cells is short, we inferred previously that CHD1 sets short spacing and that ISW1 makes it longer (Ocampo et al. 2016). We also proposed that ISW1 activity may be partially dependent on CHD1 activity, in order to explain the fact that spacing in chd1Δ cells is not longer than that of wild type, as predicted by our simple model.
The new data, for the rsc8 strains, are presented in Figure 1B. In rsc8 cells, as expected from previous studies, all of the nucleosomes in the upstream and downstream arrays shift toward the promoter with consequent narrowing and filling of the NDR, with no change in spacing (Hartley and Madhani 2009; Ganguli et al. 2014; Rawal et al. 2018a). All eight of the rsc8 strains show the same narrowing of the NDR and shift of the +1 nucleosome, indicating that RSC is required for setting the correct positions of the +1 and −1 nucleosomes. Furthermore, the nucleosomal array shifts toward the promoter in all of the rsc8 strains for which spacing can be measured (i.e., all strains except those with both isw1Δ and chd1Δ, in which phasing is heavily disrupted).
The contributions of the spacing enzymes to chromatin structure in rsc8 strains are also mostly as expected from the single mutants. The rsc8 isw1Δ and the rsc8 chd1Δ double mutants both show the weaker phasing typical of the isw1Δ and chd1Δ single mutants, whereas the rsc8 isw2Δ double mutant is similar to the rsc8 single mutant, indicating little or no contribution of ISW2. Thus, chromatin organization in the rsc8 double mutants is the combination of the effects observed in the single mutants. This conclusion is supported by analysis of the triple mutants. The rsc8 chd1Δ isw2Δ and the rsc8 isw1Δ isw2Δ triple mutants resemble their respective double mutants without isw2Δ (Fig. 1B, cf. upper and lower rows). The rsc8 isw1Δ chd1Δ triple mutant is essentially a combination of the array shifts typical of rsc8 and the very poor phasing observed when isw1Δ and chd1Δ are combined. Finally, the quadruple mutant (rsc8 isw1Δ chd1Δ isw2Δ) resembles the rsc8 isw1Δ chd1Δ triple mutant, as expected given that isw2Δ has no discernible effect in the other mutants. In conclusion, to a first approximation, the global chromatin organization of the quadruple mutant is the sum of the separate contributions of the four remodelers.
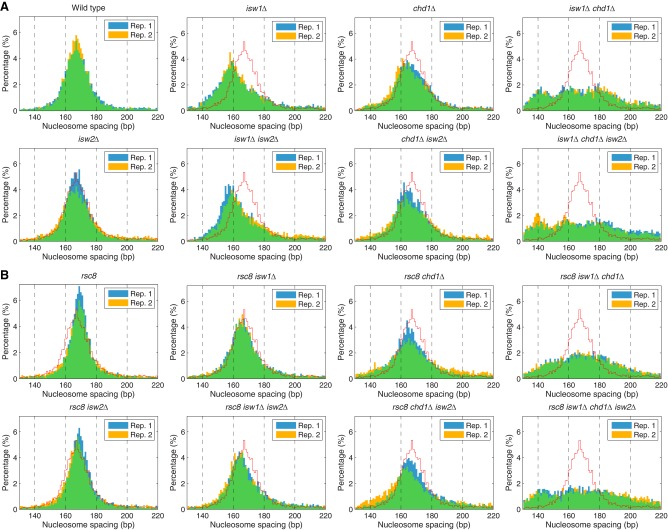
A quantitative analysis of the average nucleosome spacing reveals that spacing changes in the rsc8 isw1Δ and rsc8 chd1Δ double mutants do not follow the combinatorial rule, although the deviations are small (Fig. 1). The rsc8 isw1Δ double mutant shows a smaller reduction in spacing (163 bp) than expected from the combination of the rsc8 (167 bp; same as wild type) and isw1Δ (159 bp) mutations (Fig. 1). Conversely, the spacing in the rsc8 chd1Δ double mutant (160 bp) is significantly less than in the chd1Δ mutant (164 bp). Thus, rsc8 partially suppresses the effect of isw1Δ on spacing, but enhances the effect of chd1Δ. However, the global average spacing hides the fact that different genes have different spacing (Ocampo et al. 2016). Data for all of the mutants are presented as histograms of the percentage of genes with a given spacing (Fig. 2). In wild-type cells, genes exhibit a broad spacing distribution, ranging from 140 bp to >200 bp, although most genes are in the 155–180 bp range, with a single peak at ∼166 bp, as expected from the average (Fig. 1). The spacing distribution in rsc8 cells is somewhat narrower than in wild-type cells even though the average spacing is the same as in wild type (Fig. 2). This effect is consistent with reduced levels of transcription in rsc8 cells (Parnell et al. 2008), because very active genes have disrupted chromatin with extremely short or extremely long spacing (for some examples, see Supplemental Fig. S2; Ocampo et al. 2016). Although the narrowing effect is also observed in the rsc8 isw2Δ double mutant, it is not observed in any of the other rsc8 mutants, indicating that both ISW1 and CHD1 are important for maintaining the narrower range of nucleosome spacing on individual genes in rsc8 cells.
Figure 2.
Nucleosome spacing distributions in the chromatin remodeler mutants. Histograms of the percentage of genes having a given average spacing (1-bp bins). Data from two biological replicates are shown, indicated by blue and yellow bars; overlap is indicated by green bars. For comparison, the wild-type histogram is indicated by the red line. (A) Spacing distributions in wild type, isw1Δ, chd1Δ, and isw2Δ mutants (from Ocampo et al. 2016). (B) Spacing distributions in rsc8 mutants.
Loss of remodelers generally increases chromatin disruption
Nucleosome phasing occurs when regularly spaced nucleosomes are positioned relative to a fixed point in the DNA sequence, such as the TSS. In an array of perfectly positioned nucleosomes on a specific gene, the +1, +2, +3 nucleosomes, etc., would adopt exactly the same positions in all cells with a fixed length of linker DNA. In reality, these arrays are far from perfect, as is clear from the width of each nucleosome dyad peak in phasing plots (Fig. 1). We used the width of each peak to quantify the degree of phasing in the mutants, as described previously (Ocampo et al. 2016). The phasing parameter (σ) is related to the width of the Gaussian curve fit to each nucleosome peak on the average gene, such that higher values of σ indicate weaker phasing (σ = 0 indicates a perfectly positioned array). Physically, the phasing value reflects both cell-to-cell heterogeneity in the +1 nucleosome position (rotational positioning) and in the linker length, which follows the (10n + 5) rule, where n is an integer and predicts linker lengths of 5, 15, 25 bp, etc. (Lohr and Van Holde 1979; Brogaard et al. 2012; Chereji et al. 2018). Thus, wild-type and isw2Δ cells have σ values of ∼15 and ∼16, respectively, whereas rsc8, isw1Δ, and chd1Δ have weaker phasing (∼18, ∼19, and ∼20, respectively) and isw1Δ chd1Δ cells have much worse phasing (σ = ∼28) (Supplemental Fig. S3). The slightly weaker phasing in the rsc8 single mutant reflects the fact that the upstream shift of the nucleosomal array toward the promoter does not occur on all genes (Ganguli et al. 2014). The rsc8 double mutants have worse phasing than the single mutants: rsc8 isw2Δ, rsc8 isw1Δ, and rsc8 chd1Δ have σ = ∼19, ∼22, and ∼25, respectively. In general, the loss of each remodeler results in weaker phasing, indicating increasing chromatin disorganization.
Rsc8-depleted cells exhibit altered gene expression patterns
Cells lacking RSC activity have much lower transcriptional activity than wild-type cells (Parnell et al. 2008). We compared the Pol II distributions in 10 of our 16 strains by ChIP-seq for the Rpb3 subunit (Supplemental Fig. S4A). The Pol II occupancy patterns of the isw1Δ, chd1Δ, and isw2Δ single mutants are very similar to wild type (Spearman's rank correlation ∼0.9); very few genes show changes greater than twofold relative to wild type, although smaller differences could be physiologically important. Even the isw1Δ chd1Δ double mutant, which shows very poor global phasing, is similar to wild type (R > 0.9), as shown previously (Gkikopoulos et al. 2011; Ocampo et al. 2016). The gene expression pattern in rsc8 cells is less similar to that of wild type and the other single mutants (R < 0.8) (Supplemental Fig. S4A). Hierarchical clustering analysis shows that the gene expression patterns of all three rsc8 double mutants are relatively poorly correlated with wild type but correlate well with the rsc8 single mutant; the R values appear to define two sets of closely correlated strains (R > 0.9): those like wild type and those like rsc8 strains (Supplemental Fig. S4A).
Because RSC-depleted cells arrest in the G2/M phase of the cell cycle (Tsuchiya et al. 1992; Treich and Carlson 1997), we considered the possibility that the changes in gene expression in rsc8 cells are primarily due to growth arrest. However, we found that nocodazole-induced arrest of wild-type cells in G2/M has only minor effects on gene expression (R ∼0.9 compared with no nocodazole) (Supplemental Fig. S4B). In contrast, the gene expression patterns of cells arrested in G2/M by nocodazole or by Rsc8 depletion are quite different (R ∼0.75) (Supplemental Fig. S4B), indicating that Rsc8 depletion substantially alters gene expression patterns independently of cell cycle arrest.
RSC and ISW1/CHD1 have opposite effects on the levels of elongating and terminating Pol II
It is surprising that genes in rsc8 cells are associated with significant amounts of Pol II because transcriptional activity, as measured by RNA levels in RSC-depleted cells, is low (Parnell et al. 2008). We hypothesized that Pol II might be paused in rsc8 cells, such that it is engaged but not actively transcribing. Promoter-proximal pausing of Pol II is common in higher eukaryotes, but it is not generally observed in yeast (Mayer et al. 2017). In wild-type cells, Pol II levels are low at the promoter and high on the gene (elongating Pol II), with a well-resolved dip at the transcript termination site (TTS), and a peak just downstream from the TTS that sometimes trails into the next gene, corresponding to terminating Pol II (Cole et al. 2014; Rawal et al. 2018b). Pol II transcribes through the TTS, which refers to the mRNA cleavage site, where poly(A) is added and the mRNA is released. Pol II is still engaged downstream from the TTS and continues to transcribe before it is forced to dissociate from the DNA by Pol II termination factors (Porrua and Libri 2015).
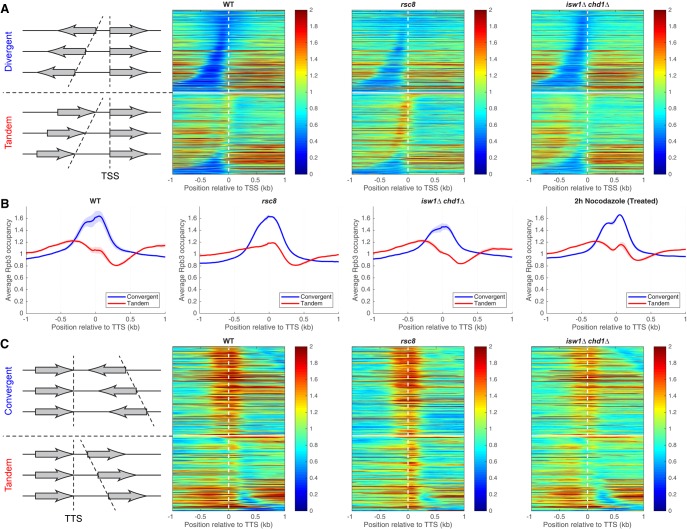
To determine whether the genic distribution of Pol II is altered in the mutants, we divided genes into divergent, tandem, and convergent pairs to resolve promoters from termination sites. We constructed heat maps of Pol II occupancy for divergent gene pairs aligned on the TSS and sorted by the distance between their TSSs (Fig. 3A; Supplemental Fig. S5). Divergent gene pairs in wild type and in all of the mutants have relatively low levels of Pol II at promoters and higher levels on the gene, with little evidence for promoter-proximal pausing. Tandem gene pairs were sorted by the distance between the TTS of the upstream gene and the TSS of the downstream gene (Fig. 3A). In rsc8 cells, a peak of Pol II is apparent in the termination regions of the upstream genes of tandem pairs, sometimes reaching as far as the TSS of the downstream gene (Fig. 3A, upper rows in the lower panel). A weak peak corresponding to terminating Pol II is apparent in wild-type cells but almost absent in the isw1Δ chd1Δ mutant (Fig. 3A). Thus, Pol II accumulates at the 3′ ends of genes in rsc8 cells, but its levels are relatively low at 3′ ends in isw1Δ chd1Δ cells, suggesting that these mutants have opposite termination defects.
Figure 3.
RSC and ISW1/CHD1 have opposite effects on the levels of elongating and terminating Pol II. (A) Pol II occupancy heat maps. Genes were divided into divergent (top) and tandem (bottom) pairs (separated by the white horizontal line), which were then sorted by intergenic distance and aligned on the TSS of the downstream gene (dashed vertical line). Data for wild type, rsc8, and isw1Δ chd1Δ double mutant cells. Rpb3 ChIP-seq data for DNA fragments of 300 bp or less normalized to the genomic average (set at 1). Shaded areas indicate the range of biological replicate data. (B) Aggregate plots for Pol II occupancy (IP/Input) relative to the TTS of the upstream gene for convergent and tandem gene pairs in wild type, rsc8, isw1Δ chd1Δ double mutant, and nocodazole-treated wild-type cells. (C) Heat maps for the data in B, aligned on the TTS of the upstream gene and sorted by the distance to the TSS of the downstream gene. The white horizontal line separates convergent (top) from tandem gene pairs (bottom). Equivalent plots for the other strains are shown in Supplemental Figures S5, S6.
We confirmed these observations by examining tandem and convergent gene pairs (Fig. 3B,C; Supplemental Fig. S6). The average Pol II distributions on tandem gene pairs relative to the TTS of the upstream gene show that the terminating Pol II peak is more prominent relative to the level of elongating Pol II in rsc8 cells than in wild-type cells and less prominent in isw1Δ chd1Δ cells (Fig. 3B). The level of terminating Pol II in the chd1Δ single mutant is slightly lower than in wild type, suggesting that CHD1 contributes more to the larger effect observed in the isw1Δ chd1Δ double mutant than ISW1 (Supplemental Fig. S6A). Convergent gene pairs exhibit two poorly separated peaks flanking the TTS in all strains; these peaks represent terminating Pol II on both genes in the pair, such that the upstream peak belongs to the downstream gene and vice versa (Fig. 3B,C; Supplemental Fig. S6).
The accumulation of Pol II downstream from the TTS in rsc8 cells suggests that termination is inhibited (i.e., that RSC facilitates Pol II termination), and the low levels of mRNA in RSC-depleted cells (Parnell et al. 2008) may at least partly reflect a termination defect. This Pol II termination defect cannot be attributed to the arrest of these cells in G2/M, because nocodazole-induced arrest of wild-type cells at the same stage of the cell cycle has little effect on the genic distribution of Pol II (Fig. 3B; Supplemental Fig. S6).
For the isw1Δ chd1Δ double mutant, the simplest interpretation is that Pol II dissociation is facilitated, implying that ISW1 and CHD1 normally inhibit termination and/or dissociation. It is unlikely that Pol II transcribes farther downstream from the TTS before dissociating, because a lower, extended peak is expected but not observed (Fig. 3). Alternatively, Pol II elongation may be inhibited in isw1Δ chd1Δ cells, resulting in higher levels of Pol II on the gene than downstream from the TTS (see below). The effect of RSC on termination is epistatic to the separate effects of ISW1 and CHD1, because the rsc8 isw1Δ and rsc8 chd1Δ double mutants and the quadruple mutant all have higher levels of terminating Pol II than the isw1Δ chd1Δ double mutant (Supplemental Figs. S5, S6).
Chromatin organization around the TTS in rsc8 and isw1Δ chd1Δ cells
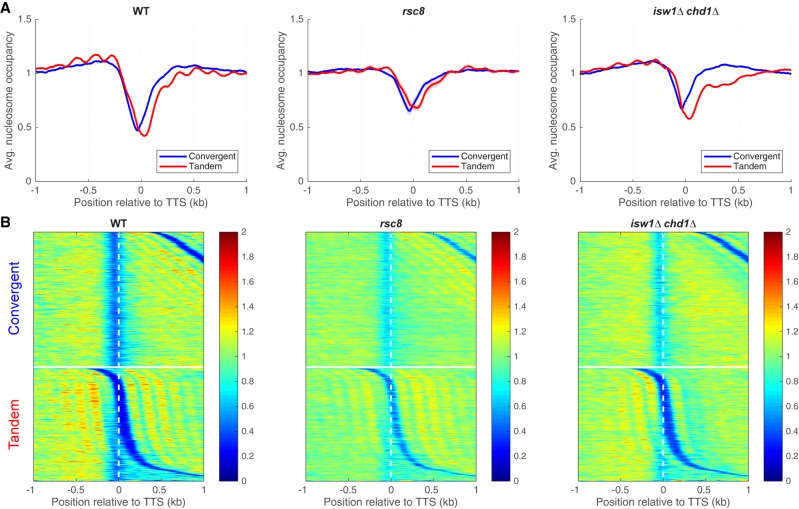
We examined nucleosome occupancy at the TTS to determine whether altered chromatin structure can account for the termination defect in rsc8 cells (Fig. 4; Supplemental Fig. S7). In wild-type cells, there is an NDR at the TTS between tandem genes, which is reproducibly shallower in rsc8 and isw1Δ chd1Δ cells (Fig. 4A; Supplemental Fig. S7). However, the depth of the 3′ NDR is strongly dependent on the extent of MNase digestion, because 3′ UTRs tend to be AT-rich and therefore nucleosomes covering 3′ UTRs are digested faster than nucleosomes with normal AT-content (Schwabish and Struhl 2004; Chereji et al. 2017). Consequently, it is difficult to rule out the possibility that these differences in 3′-NDR depth are due to small differences in the extent of MNase digestion. In any case, higher nucleosome occupancy is observed at the TTSs of convergent genes in both rsc8 and isw1Δ chd1Δ cells (Fig. 4), even though these mutants have quite different levels of terminating Pol II, suggesting that higher nucleosome occupancy does not necessarily inhibit termination.
Figure 4.
Chromatin organization in the vicinity of the TTS in rsc8 and isw1Δ chd1Δ cells. (A) Nucleosome occupancy aggregate plots for convergent and tandem gene pairs aligned on the TTS of the upstream gene. MNase-seq data (for fragments of 120–160 bp) normalized to the genomic average (set at 1). (B) Heat maps for the data in A, aligned on the TTS of the upstream gene and sorted by the distance to the TSS of the downstream gene. The white horizontal line separates convergent from tandem gene pairs. Equivalent plots for the other strains are shown in Supplemental Figure S7.
High levels of closely packed dinucleosomes involving the +2 nucleosome are present in isw1Δ chd1Δ cells
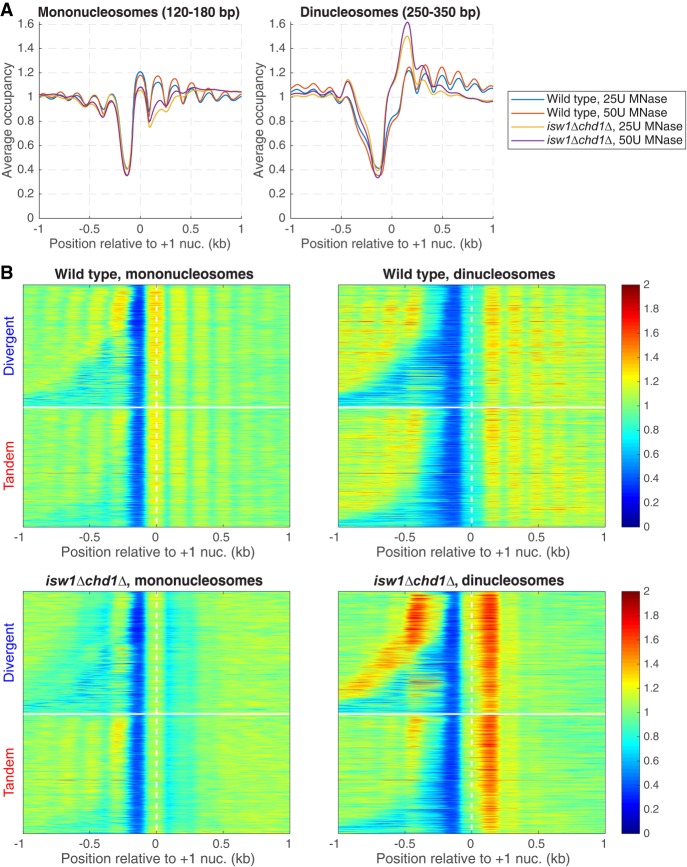
There is an apparent reduction in nucleosome occupancy on the downstream gene of tandem gene pairs in wild-type cells (Fig. 4A). This is not actually the case, because the NDR at the promoter belonging to the downstream gene is located at a variable distance from the TTS, resulting in reduced average occupancy (see heat maps, Fig. 4B). However, this effect is extreme for tandem genes in the isw1Δ chd1Δ double mutant: Nucleosome occupancy at the 5′ ends of downstream genes is much reduced (Fig. 4A), with a disproportionately greater effect on the +2 nucleosome that is obvious in the heat maps (Fig. 4B). This observation suggests that nucleosomes are missing from the 5′ ends of genes in the isw1Δ chd1Δ double mutant. However, an alternative explanation for the reduced nucleosome occupancy is suggested by the close proximity of two nucleosomes at the 5′ end of MET16 in an isw1Δ mutant (Morillon et al. 2003), which prevents MNase from cutting between them. We considered the possibility that nucleosome occupancy is normal in the isw1Δ chd1Δ double mutant, and that nucleosomes are sometimes pushed together, eliminating the linker DNA. This model predicts that the missing nucleosomes are actually present as MNase-resistant dinucleosomes. Such dinucleosomes would have been eliminated from our data when the mononucleosome band was excised from the gel.
To test the model, we sequenced MNase-digested DNA that had not been gel-purified. Wild-type mononucleosomes show a normal phasing pattern relative to the +1 nucleosome, whereas mononucleosomes from the isw1Δ chd1Δ double mutant show weak phasing and reduced occupancy of the +1 nucleosome and, in particular, of the +2 nucleosome (Fig. 5A), as observed for gel-purified mononucleosomes (as expected from Fig. 4). Wild-type dinucleosomes show a fairly even occupancy similar to that of mononucleosomes, except for the +1 nucleosome, which is reduced, probably because the probability of MNase cutting upstream in the NDR is always high, increasing the probability of releasing the +1 nucleosome as a mononucleosome (Fig. 5A). This could account for the slightly higher occupancy of the +1 nucleosome in the mononucleosome fraction (Fig. 5A). In contrast, the isw1Δ chd1Δ double mutant shows a strong enrichment of dinucleosomes that include the +2 nucleosome (i.e., the +1/+2 and +2/+3 dinucleosomes) (Fig. 5A). We separated divergent and tandem gene pairs in heat maps aligned on the +1 nucleosome dyad and sorted by intergenic distance (Fig. 5B). In wild-type cells, phasing with respect to the NDR is obvious for both mononucleosomes and dinucleosomes. In the isw1Δ chd1Δ double mutant, the +2 nucleosome is clearly depleted from the mononucleosome fraction and enhanced in the dinucleosomes. In the tandem gene heat maps, nucleosome phasing at the 3′ end of the upstream gene is only mildly affected in the isw1Δ chd1Δ double mutant, even though phasing on the downstream gene is heavily disrupted. In conclusion, the +2 nucleosome is often located immediately adjacent to the +1 or the +3 nucleosome in isw1Δ chd1Δ cells. Thus, ISW1 and CHD1 together prevent close packing of the +2 nucleosome by maintaining regular spacing.
Figure 5.
Closely packed dinucleosomes at the 5′ ends of genes in isw1Δ chd1Δ cells. MNase-seq data for wild-type and isw1Δ chd1Δ cells obtained without gel-purification of mononucleosomal DNA (two levels of digestion: 25 and 50 units MNase). (A) Nucleosome occupancy aggregate plots for mononucleosomes (120–180 bp) and dinucleosomes (250–350 bp) for all genes, aligned on the +1 nucleosome. Normalized to the genomic average (set at 1). (B) Heat maps for the data in A, separated into divergent and tandem gene pairs, sorted by intergenic distance and aligned on the +1 nucleosome of the downstream gene. The white horizontal line separates divergent from tandem gene pairs.
Discussion
Combinatorial contributions to chromatin organization by RSC, ISW1, CHD1, and ISW2
We report a systematic analysis of chromatin organization and gene expression in isogenic strains representing all possible combinations of RSC depletion with null mutations in three nucleosome spacing enzymes. We confirm that, in RSC-depleted cells, the upstream and downstream nucleosomal arrays shift toward the promoter with narrowing and filling of the NDR (Hartley and Madhani 2009; Ganguli et al. 2014). These effects persist when RSC depletion is combined with any of the spacing enzyme null mutations. RSC-depleted cells also have a narrower nucleosome spacing distribution, indicating that there are fewer genes with extreme spacing. Because extreme spacing correlates with heavy transcription (Ocampo et al. 2016), we attribute the narrower spacing distribution in RSC-depleted cells to reduced transcription (Parnell et al. 2008). Generally speaking, the chromatin organization in the double, triple, and quadruple mutants can be predicted by combining the effects of the single mutations. Thus, RSC depletion results in nucleosome array shifts into the promoter, loss of ISW1 or CHD1 results in weaker phasing, whereas loss of ISW2 has only minor global effects. The exception to this rule concerns nucleosome spacing, since the global average spacings are not as predicted from a simple combination of the single mutants. That is, all isw1Δ mutants should have short spacing and all chd1Δ mutants should have longer spacing than wild type. Our previous proposal that ISW1 activity may depend partly on CHD1 activity may explain this effect (Ocampo et al. 2016). The INO80 remodeling complex also contributes to spacing, since ino80Δ cells have shorter spacing than wild type (Udugama et al. 2011; Yen et al. 2012; van Bakel et al. 2013; Krietenstein et al. 2016).
In summary, we envisage that RSC determines the width of the NDR, either directly by setting the positions of the −1 and +1 nucleosomes (Krietenstein et al. 2016), or indirectly by determining the size of the barrier complex (Mavrich et al. 2008; Jiang and Pugh 2009; Chereji et al. 2017). Then ISW1, CHD1, and, to a much lesser extent, ISW2, compete to space nucleosomes on the gene (Ocampo et al. 2016) using the +1 nucleosome as a reference position. The outcome of the competition on each gene determines its spacing and depends on its transcriptional activity, since CHD1 (the short-spacing enzyme) is associated with active transcription (Simic et al. 2003) and ISW2 (the long-spacing enzyme) affects only relatively inactive genes (Ocampo et al. 2016).
RSC and gene expression
The gene expression pattern in RSC-depleted cells is quite different from that of wild-type cells, which may reflect both direct and indirect effects on chromatin organization. Indirect effects may be expected as a result of growth arrest, which occurs primarily in the G2/M phase of the cell cycle (Tsuchiya et al. 1992; Treich and Carlson 1997). However, our nocodazole experiments indicate that cell cycle arrest in G2/M has only minor effects on transcription, unlike RSC depletion, which also arrests cells at the spindle assembly checkpoint (Tsuchiya et al. 1992). We note that growth arrest as a result of Rsc8 depletion may also affect nucleosome positioning. Direct effects of RSC are more difficult to predict because RSC localization data are somewhat contradictory. The major contribution of RSC to chromatin organization is at the NDR, as discussed above, predicting that RSC binds at or near promoters in vivo. MNase-ChIP-seq experiments are consistent with this expectation: RSC is bound to the promoter-flanking +1 and −1 nucleosomes, although it is also enriched at the +2 and +3 nucleosomes (Yen et al. 2012). On the other hand, conventional ChIP-seq experiments indicate that RSC is not generally enriched at promoters, but modestly enriched on active genes (Ng et al. 2002; Floer et al. 2010; Ganguli et al. 2014; Spain et al. 2014; Rawal et al. 2018a), suggesting that RSC affects elongation rather than initiation. RSC may affect both stages of transcription by adjusting the distance between the +1 and −1 nucleosomes and evicting nucleosomes from NDRs (Rawal et al. 2018a) to facilitate transcription complex formation and by moving or removing nucleosomes on active genes to regulate elongation.
RSC stimulates Pol II termination/dissociation
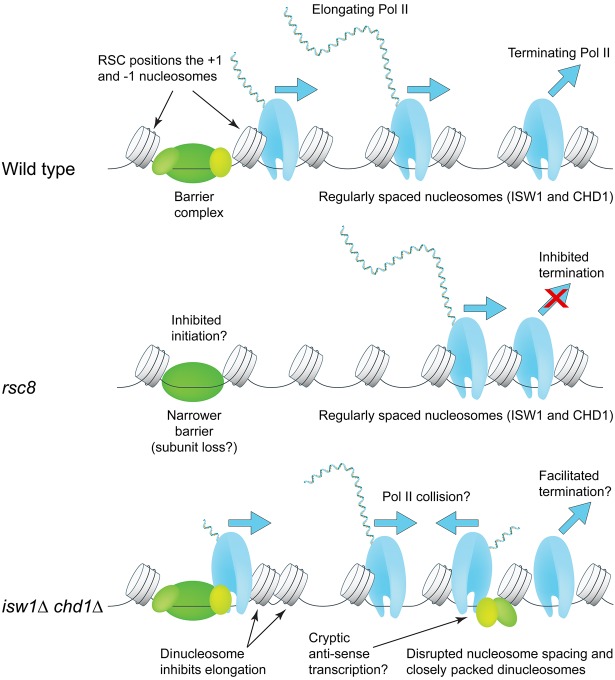
We find that RSC has an unexpected stimulatory effect on Pol II termination and/or dissociation after transcription. Dissociation of Pol II appears to be slower in rsc8 cells than in wild-type cells, since ChIP-seq experiments measure bound Pol II. However, the actual defect could be slow termination of transcription rather than slow dissociation. Slow dissociation seems less likely than a termination defect, because the “closed” Pol II complex is much less stable than the transcribing “open” complex. Inhibition of termination alone cannot account for the observed Pol II distribution in RSC-depleted cells, because a pileup of elongating polymerases behind the terminating polymerase would be expected if dissociation is prevented, leading to a higher level of elongating polymerase, which is not observed. A plausible scenario is that transcript initiation is also inhibited in RSC-depleted cells, perhaps due to partial disassembly of the barrier complex (Ganguli et al. 2014; Chereji et al. 2017) and defects in nucleosome eviction or sliding at promoters (Rawal et al. 2018a). An initiation defect is consistent with the fact that RSC is not enriched on terminal nucleosomes (Yen et al. 2012). We speculate that, in wild-type cells, Pol II termination or dissociation might be slow in order to facilitate reinitiation by Pol II (i.e., transfer of Pol II from just downstream from the TTS back to the promoter without release into the nucleoplasm) (Shandilya and Roberts 2012; Cole et al. 2014). Such a mechanism has been proposed to explain fast reinitiation by Pol III (Arimbasseri et al. 2013). In this speculative model, loss of RSC activity would result in the formation of a defective transcription complex in the NDR, reducing initiation and increasing the residence time of Pol II just downstream from the TTS as it waits for transfer back to the promoter.
ISW1 and CHD1 resolve closely packed dinucleosomes
A role for all three nucleosome spacing enzymes in Pol II termination is suggested by single-gene studies (Alén et al. 2002; Morillon et al. 2003). Transcription run-on assays indicate that Pol II fails to terminate correctly in chd1, isw1, and isw2 mutants, although the strength of the effect depends on the gene. Instead, Pol II continues transcription into the downstream promoter, interfering with its expression (Alén et al. 2002). Pol II ChIP data for a reporter gene (Morillon et al. 2003) indicate that, unlike in wild-type cells, there is a very strong Pol II peak at the 3′ end in isw1 cells, suggesting a termination defect. However, our data for the average gene in isw1Δ cells show the opposite effect: a decrease in Pol II just downstream from the TTS, not the large increase observed by Morillon et al. (2003), although the small number of genes examined by Morillon et al. (2003) may be atypical.
We show here that chromatin disruption in isw1Δ chd1Δ cells involves the close packing of nucleosomes on genes, particularly of the +2 nucleosome, such that it is positioned immediately adjacent to the +1 nucleosome or to the +3 nucleosome. This observation implies that an important function of ISW1 and CHD1 is to prevent close packing by spacing the nucleosomes out. An interesting question is whether these closely packed dinucleosomes are formed by spontaneous sliding or by other remodelers. Support for the latter model comes from observations that, in vitro, human SWI/SNF can generate “altosomes” (Ulyanova and Schnitzler 2005), and yeast RSC can form “overlapping dinucleosomes” lacking one H2A-H2B dimer (Engeholm et al. 2009; Kato et al. 2017). If RSC forms the dinucleosomes we have observed, we would expect chromatin organization in the rsc8 isw1Δ chd1Δ triple mutant to be closer to wild type than to the isw1Δ chd1Δ double mutant, but this is not the case, suggesting that RSC is not important for dinucleosome formation, although it may be redundant with SWI/SNF.
Although our data are consistent with a termination defect in isw1Δ chd1Δ cells, it is unclear how closely packed dinucleosomes near the 5′ end of a gene could facilitate Pol II termination or dissociation at the 3′ end. Instead, we propose that Pol II elongation through closely packed nucleosomes at the 5′ end of the gene is slow, eventually resulting in normal termination (Fig. 6). Other important factors to consider are the high levels of histone exchange and cryptic initiation in the isw1Δ chd1Δ double mutant (Smolle et al. 2012). Close packing of nucleosomes may be linked to histone exchange if one of the nucleosomes loses an H2A-H2B dimer, resulting in octasome-hexasome dimers (Engeholm et al. 2009; Kato et al. 2017). Cryptic antisense transcription may result in collisions with elongating sense polymerases, such that fewer polymerases reach the TTS, which would also increase the amount of Pol II on the gene relative to the region just downstream from the TTS (Fig. 6). Thus, ISW1 and CHD1 maintain a chromatin structure conducive to efficient elongation and termination by resolving dinucleosomes and creating regularly spaced nucleosomal arrays.
Figure 6.
Roles of the RSC, ISW1, and CHD1 remodelers in Pol II initiation, elongation, and termination. RSC widens the promoter NDR by positioning the +1 and −1 nucleosomes farther apart to accommodate a complete barrier complex. CHD1 (short spacing) and ISW1 (longer spacing) compete to set nucleosome spacing on the gene using the +1 nucleosome as a reference. In rsc8 cells, nucleosomes shift toward the promoter but the spacing is the same as wild type; Pol II remains 3′ of the TTS. In isw1Δ chd1Δ cells, nucleosome spacing is disrupted, with the +2 nucleosome in particular being pushed against the +1 or +3 nucleosome. High cryptic initiation in these cells may result in collisions between transcribing cryptic antisense Pol II and promoter-initiated Pol II (elongation defect). Termination and/or dissociation in these cells may be faster.
Methods
Yeast strains
All yeast strains used in this work are listed in Supplemental Table S1. In our previous study of RSC (Ganguli et al. 2014), we used a strain with a GAL promoter driving the essential RSC8 gene on a plasmid (Treich and Carlson 1997), such that the cells grow in galactose medium but eventually arrest when switched to glucose. However, this strain derives from S288C, whereas our isw1Δ, isw2Δ, and chd1Δ mutants are all derived from W303. To maintain isogenicity, we constructed a new GAL-RSC8 strain in the same W303 background. A GAL-RSC8 cassette with three N-terminal HA epitope tags was integrated into the genome instead of using a plasmid. We did not observe any differences in chromatin organization between the two GAL-RSC8 strains. GAL-RSC8 strains were grown in synthetic complete (SC) medium containing 2% galactose and switched to glucose as described (Ganguli et al. 2014); cells were harvested when growth reached a plateau (after ∼7.5 h in glucose). Rsc8 depletion was verified by immunoblotting using anti-HA-peroxidase antibody clone 3F10 (Roche 12013819001) (Supplemental Fig. S8).
MNase-seq and Pol II ChIP-seq
Nucleosomal DNA was prepared by MNase digestion of isolated nuclei and subjected to paired-end sequencing (Cole et al. 2011, 2012). These experiments, including biological replicates, were performed as described (Ocampo et al. 2016). For dinucleosome analysis, paired-end libraries were prepared from MNase-digested samples without gel-purification of the DNA (both mono- and dinucleosomes were sequenced).
Nocodazole arrest
Wild-type strain YDC111 (Kim et al. 2006) was grown in SC medium to A600 ∼ 0.25 and nocodazole (Abcam ab120630) was added to 6 µg/mL. Cells were monitored by microscopy for 2 h, by which time the large majority displayed the dumbbell shape characteristic of G2/M arrest. Arrest was confirmed using fluorescence microscopy after staining for DNA using propidium iodide. Cells were fixed with formaldehyde and processed for Pol II ChIP-seq as described (Ocampo et al. 2016).
Bioinformatic analysis
For phasing analysis, nucleosome sequences in the range of 120–160 bp were used. To facilitate comparisons between different samples, we normalized the sequencing depths of all samples to one read per bp. Details and scripts for estimation of the phasing parameter and nucleosome spacing on individual genes described by Ocampo et al. (2016) and in Supplemental Methods are available as Supplemental Code and at https://github.com/rchereji/Nucleosome_spacing_estimation/tree/GenomeRes_2019.
Data access
The sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GSE73428 and GSE117514.
Supplementary Material
Acknowledgments
We thank Alan Hinnebusch for comments on the manuscript. This research was supported by the Intramural Research Program of the NIH (NICHD). We thank the NHLBI Core Facility (Yan Luo, Poching Liu, and Jun Zhu) for paired-end sequencing. This study utilized the high performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.242032.118.
References
- Alén C, Kent NA, Jones HS, O'Sullivan J, Aranda A, Proudfoot NJ. 2002. A role for chromatin remodeling in transcriptional termination by RNA polymerase II. Mol Cell 10: 1441–1452. 10.1016/S1097-2765(02)00778-5 [DOI] [PubMed] [Google Scholar]
- Arimbasseri AG, Rijal K, Maraia RJ. 2013. Transcription termination by the eukaryotic RNA polymerase III. Biochim Biophys Acta 1829: 318–330. 10.1016/j.bbagrm.2012.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B. 2014. Regulating the chromatin landscape: structural and mechanistic perspectives. Annu Rev Biochem 83: 671–696. 10.1146/annurev-biochem-051810-093157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogaard K, Xi L, Wang JP, Widom J. 2012. A map of nucleosome positions in yeast at base-pair resolution. Nature 486: 496–501. 10.1038/nature11142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereji RV, Ocampo J, Clark DJ. 2017. MNase-sensitive complexes in yeast: nucleosomes and non-histone barriers. Mol Cell 65: 565–577.e3. 10.1016/j.molcel.2016.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereji RV, Ramachandran S, Bryson TD, Henikoff S. 2018. Precise genome-wide mapping of single nucleosomes and linkers in vivo. Genome Biol 19: 19 10.1186/s13059-018-1398-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung V, Chua G, Batada NN, Landry CR, Michnick SW, Hughes TR, Winston F. 2008. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol 6: e277 10.1371/journal.pbio.0060277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. 2009. The biology of chromatin remodeling complexes. Annu Rev Biochem 78: 273–304. 10.1146/annurev.biochem.77.062706.153223 [DOI] [PubMed] [Google Scholar]
- Cole HA, Howard BH, Clark DJ. 2011. Activation-induced disruption of nucleosome position clusters on the coding regions of Gcn4-dependent genes extends into neighbouring genes. Nucleic Acids Res 39: 9521–9535. 10.1093/nar/gkr643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole HA, Howard BH, Clark DJ. 2012. Genome-wide mapping of nucleosomes in yeast using paired-end sequencing. Meth Enzymol 513: 145–168. 10.1016/B978-0-12-391938-0.00006-9 [DOI] [PubMed] [Google Scholar]
- Cole HA, Ocampo J, Iben JR, Chereji RV, Clark DJ. 2014. Heavy transcription of yeast genes correlates with differential loss of histone H2B relative to H4 and queued RNA polymerases. Nucleic Acids Res 42: 12512–12522. 10.1093/nar/gku1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engeholm M, de Jager M, Flaus A, Brenk R, van Noort J, Owen-Hughes T. 2009. Nucleosomes can invade DNA territories occupied by their neighbors. Nat Struct Mol Biol 16: 151–158. 10.1038/nsmb.1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, et al. 2010. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell 141: 407–418. 10.1016/j.cell.2010.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguli D, Chereji RV, Iben JR, Cole HA, Clark DJ. 2014. RSC-dependent constructive and destructive interference between opposing arrays of phased nucleosomes in yeast. Genome Res 24: 1637–1649. 10.1101/gr.177014.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkikopoulos T, Schofield P, Singh V, Pinskaya M, Mellor J, Smolle M, Workman JL, Barton GJ, Owen-Hughes T. 2011. A role for Snf2-related nucleosome-spacing enzymes in genome-wide nucleosome organization. Science 333: 1758–1760. 10.1126/science.1206097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark JP, Fazzio TG, Estep PW, Church GM, Tsukiyama T. 2000. The Isw2 chromatin remodeling complex represses early meiotic genes upon recruitment by Ume6p. Cell 103: 423–433. 10.1016/S0092-8674(00)00134-3 [DOI] [PubMed] [Google Scholar]
- Hartley PD, Madhani HD. 2009. Mechanisms that specify promoter nucleosome location and identity. Cell 137: 445–458. 10.1016/j.cell.2009.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. 1997. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90: 145–155. 10.1016/S0092-8674(00)80321-9 [DOI] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. 2009. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet 10: 161–172. 10.1038/nrg2522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato D, Osakabe A, Arimura Y, Mizukami Y, Horikoshi N, Saikusa K, Akashi S, Nishimura Y, Park SY, Nogami J, et al. 2017. Crystal structure of the overlapping dinucleosome composed of hexasome and octasome. Science 356: 205–208. 10.1126/science.aak9867 [DOI] [PubMed] [Google Scholar]
- Kent NA, Karabetsou N, Politis PK, Mellor J. 2001. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev 15: 619–626. 10.1101/gad.190301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, McLaughlin N, Lindstrom K, Tsukiyama T, Clark DJ. 2006. Activation of Saccharomyces cerevisiae HIS3 results in Gcn4p-dependent, SWI/SNF-dependent mobilization of nucleosomes over the entire gene. Mol Cell Biol 26: 8607–8622. 10.1128/MCB.00678-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krietenstein N, Wal M, Watanabe S, Park B, Peterson CL, Pugh BF, Korber P. 2016. Genomic nucleosome organization reconstituted with pure proteins. Cell 167: 709–721.e12. 10.1016/j.cell.2016.09.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet 36: 900–905. 10.1038/ng1400 [DOI] [PubMed] [Google Scholar]
- Lohr D, Van Holde KE. 1979. Organization of spacer DNA in chromatin. Proc Natl Acad Sci 76: 6326–6330. 10.1073/pnas.76.12.6326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorch Y, Zhang M, Kornberg RD. 2001. RSC unravels the nucleosome. Mol Cell 7: 89–95. 10.1016/S1097-2765(01)00157-5 [DOI] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389: 251–260. 10.1038/38444 [DOI] [PubMed] [Google Scholar]
- Lusser A, Urwin DL, Kadonaga JT. 2005. Distinct activities of CHD1 and ACF in ATP-dependent chromatin assembly. Nat Struct Mol Biol 12: 160–166. 10.1038/nsmb884 [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. 2008. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res 18: 1073–1083. 10.1101/gr.078261.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Landry HM, Churchman LS. 2017. Pause & go: from the discovery of RNA polymerase pausing to its functional implications. Curr Opin Cell Biol 46: 72–80. 10.1016/j.ceb.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillon A, Karabetsou N, O'Sullivan J, Kent N, Proudfoot N, Mellor J. 2003. Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell 115: 425–435. 10.1016/S0092-8674(03)00880-8 [DOI] [PubMed] [Google Scholar]
- Musladin S, Krietenstein N, Korber P, Barbaric S. 2014. The RSC chromatin remodeling complex has a crucial role in the complete remodeler set for yeast PHO5 promoter opening. Nucleic Acids Res 42: 4270–4282. 10.1093/nar/gkt1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. 2013. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 154: 490–503. 10.1016/j.cell.2013.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. 2002. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev 16: 806–819. 10.1101/gad.978902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo J, Chereji RV, Eriksson PR, Clark DJ. 2016. The ISW1 and CHD1 ATP-dependent chromatin remodelers compete to set nucleosome spacing in vivo. Nucleic Acids Res 44: 4625–4635. 10.1093/nar/gkw068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell TJ, Huff JT, Cairns BR. 2008. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J 27: 100–110. 10.1038/sj.emboj.7601946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parnell TJ, Schlichter A, Wilson BG, Cairns BR. 2015. The chromatin remodelers RSC and ISW1 display functional and chromatin-based promoter antagonism. eLife 4: e06073 10.7554/eLife.06073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pointner J, Persson J, Prasad P, Norman-Axelsson U, Strålfors A, Khorosjutina O, Krietenstein N, Svensson JP, Ekwall K, Korber P. 2012. CHD1 remodelers regulate nucleosome spacing in vitro and align nucleosomal arrays over gene coding regions in S. pombe. EMBO J 31: 4388–4403. 10.1038/emboj.2012.289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrua O, Libri D. 2015. Transcription termination and the control of the transcriptome: why, where and how to stop. Nat Rev Mol Cell Biol 16: 190–202. 10.1038/nrm3943 [DOI] [PubMed] [Google Scholar]
- Quan TK, Hartzog GA. 2010. Histone H3K4 and K36 methylation, Chd1 and Rpd3S oppose the functions of Saccharomyces cerevisiae Spt4–Spt5 in transcription. Genetics 184: 321–334. 10.1534/genetics.109.111526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman-Livaja M, Quan TK, Valenzuela L, Armstrong JA, van Welsem T, Kim T, Lee LJ, Buratowski S, van Leeuwen F, Rando OJ, et al. 2012. A key role for Chd1 in histone H3 dynamics at the 3′ ends of long genes in yeast. PLoS Genet 8: e1002811 10.1371/journal.pgen.1002811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawal Y, Chereji RV, Qiu H, Ananthakrishnan S, Govind CK, Clark DJ, Hinnebusch AG. 2018a. SWI/SNF and RSC cooperate to reposition and evict promoter nucleosomes at highly expressed genes in yeast. Genes Dev 32: 695–710. 10.1101/gad.312850.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawal Y, Chereji RV, Valabhoju V, Qiu H, Ocampo J, Clark DJ, Hinnebusch AG. 2018b. Gcn4 binding in coding regions can activate internal and canonical 5′ promoters in yeast. Mol Cell 70: 297–311.e4. 10.1016/j.molcel.2018.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A, Wittmeyer J, Cairns BR. 2002. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev 16: 2120–2134. 10.1101/gad.995002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwabish MA, Struhl K. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol Cell Biol 24: 10111–10117. 10.1128/MCB.24.23.10111-10117.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shandilya J, Roberts SGE. 2012. The transcription cycle in eukaryotes: from productive initiation to RNA polymerase II recycling. Biochim Biophys Acta 1819: 391–400. 10.1016/j.bbagrm.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Shukla MS, Syed SH, Montel F, Faivre-Moskalenko C, Bednar J, Travers A, Angelov D, Dimitrov S. 2010. Remosomes: RSC generated non-mobilized particles with approximately 180 bp DNA loosely associated with the histone octamer. Proc Natl Acad Sci 107: 1936–1941. 10.1073/pnas.0904497107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J 22: 1846–1856. 10.1093/emboj/cdg179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, Washburn MP, Workman JL. 2012. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat Struct Mol Biol 19: 884–892. 10.1038/nsmb.2312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spain MM, Ansari SA, Pathak R, Palumbo MJ, Morse RH, Govind CK. 2014. The RSC complex localizes to coding sequences to regulate Pol II and histone occupancy. Mol Cell 56: 653–666. 10.1016/j.molcel.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. 2006. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J Biol Chem 281: 16279–16288. 10.1074/jbc.M600682200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swygert SG, Peterson CL. 2014. Chromatin dynamics: interplay between remodeling enzymes and histone modifications. Biochim Biophys Acta 1839: 728–736. 10.1016/j.bbagrm.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Sigal N, Barkai N. 2010. Widespread remodeling of mid-coding sequence nucleosomes by Isw1. Genome Biol 11: R49 10.1186/gb-2010-11-5-r49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treich I, Carlson M. 1997. Interaction of a Swi3 homolog with Sth1 provides evidence for a Swi/Snf-related complex with an essential function in Saccharomyces cerevisiae. Mol Cell Biol 17: 1768–1775. 10.1128/MCB.17.4.1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya E, Uno M, Kiguchi A, Masuoka K, Kanemori Y, Okabe S, Mikayawa T. 1992. The Saccharomyces cerevisiae NPS1 gene, a novel CDC gene which encodes a 160 kDa nuclear protein involved in G2 phase control. EMBO J 11: 4017–4026. 10.1002/j.1460-2075.1992.tb05495.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. 1999. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev 13: 686–697. 10.1101/gad.13.6.686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udugama M, Sabri A, Bartholomew B. 2011. The INO80 ATP-dependent chromatin remodeling complex is a nucleosome spacing factor. Mol Cell Biol 31: 662–673. 10.1128/MCB.01035-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulyanova NP, Schnitzler GR. 2005. Human SWI/SNF generates abundant, structurally altered dinucleosomes on polynucleosomal templates. Mol Cell Biol 25: 11156–11170. 10.1128/MCB.25.24.11156-11170.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Tsui K, Gebbia M, Mnaimneh S, Hughes TR, Nislow C. 2013. A compendium of nucleosome and transcript profiles reveals determinants of chromatin architecture and transcription. PLoS Genet 9: e1003479 10.1371/journal.pgen.1003479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vary JC, Gangaraju VK, Qin J, Landel CC, Kooperberg C, Bartholomew B, Tsukiyama T. 2003. Yeast Isw1p forms two separable complexes in vivo. Mol Cell Biol 23: 80–91. 10.1128/MCB.23.1.80-91.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. 2007. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450: 1031–1035. 10.1038/nature06391 [DOI] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. 2012. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell 149: 1461–1473. 10.1016/j.cell.2012.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. 2005. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309: 626–630. 10.1126/science.1112178 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.