Abstract
Zika virus (ZIKV) is an arbovirus belonging to the Flaviviridae family and the genus Flavivirus. Infection with ZIKV causes a mild, self-limiting febrile illness called Zika fever. However, ZIKV infection has been recently associated with microcephaly and Guillain-Barré syndrome. Vaccines for the disease are a high priority of World Health Organization. Several studies are currently being conducted to develop a vaccine against ZIKV, but until now there is no licensed ZIKV vaccine. This study used a novel immunoinformatics approach to identify potential T-cell immunogenic epitopes present in the structural and nonstructural proteins of ZIKV. Fourteen T-cell candidate epitopes were identified on ZIKV structural and nonstructural proteins: pr36−50; C61−75; C103−117; E374−382; E477−491; NS2a90−104; NS2a174−188; NS2a179−193; NS2a190−204; NS2a195−209; NS2a200−214; NS3175−189; and NS4a82−96; NS4a99−113. Among these epitopes, only E374−382 is a human leukocyte antigen (HLA) type I restricted epitope. All identified epitopes showed a low similarity with other important flaviviruses but had a high conservation rate among the ZIKV strains and a high population coverage rate. Therefore, these predicted T-cell epitopes are potential candidates targets for development of vaccines to prevent ZIKV infection.
Keywords: Diagnostic test, Immunoinformatic, T-cell epitope, vaccine, Zika virus
Introduction
Zika virus (ZIKV) is an arbovirus transmitted in urban cycles to humans by the bite of infected female mosquitoes of the Aedes genus, mainly Aedes aegypti. ZIKV is a positive-sense, single-strand RNA virus classified in the genus Flavivirus, family Flaviviridae [1], [2], [3]. The virus genome contains one open reading frame, which is translated into a large polyprotein that should be cleaved by viral and cellular proteases in order to generate three structural proteins (capsid, membrane and envelope proteins) and seven nonstructural proteins (NS1, NS2a, NS2b, NS3, NS4a, NS4b and NS5) [4].
ZIKV was first isolated in 1947 in Uganda [5], and six decades after its discovery, ZIKV caused an epidemic in the Federated States of Micronesia [6]. In 2013 the virus was introduced to Brazil, and after this event, a major pandemic in the Americas was observed [7], [8], [9]. The spread of ZIKV in the Americas was associated with the appearance of severe adverse outcomes, such as congenital Zika syndrome and Guillain-Barré syndrome [10], [11], [12]. Additionally the possibility of sexual transmission has intensified efforts to develop a vaccine for ZIKV [13], [14]. Until now several approaches have been used to develop ZIKV vaccine, such as DNA and messenger RNA vaccines, inactivated ZIKV strains and recombinant proteins [15], [16], [17]. In general these vaccines are immunogenic and able to induce protection in mice and nonhuman primates after challenge. However, vaccines developed using these approaches have yet to complete phase 2 clinical trials [18], [19]. In addition, the success of preclinical test of these vaccines—the development of an effective ZIKV vaccine—could be complicated by previous immunity against other flaviviruses such as Dengue virus (DENV) [20], [21], [22]. Additionally, the antibody-dependent enhancement (ADE) phenomenon could confound the development of any flavivirus vaccine [23].
Traditionally vaccines have been developed by isolating one or more antigenic components from a given pathogen and checking whether these components are able of inducing a protective immune response. Although this approach has presented a series of successes throughout the history of vaccinology, the knowledge generated with the new genomic, transcriptomic and proteomic technologies has contributed to the advancement of vaccinology and the production of more effective and safe vaccines [24], [25], [26]. One of the most promising approaches is the use of computational tools that allow the identification of genes in the genome of different pathogens encoding proteins with antigenic potential. This approach, called reverse vaccinology (Rvac), is a direct consequence of new ‘omic’ technologies and is currently recognized as a promising technique for the theoretical determination of proteins or peptides that have a potential for induction of an immune response. The use of Rvac could reduce time and cost related to development of new vaccines [27], [28], [29], [30], [31], [32], [33]. Therefore, Rvac can find important peptide sequences that can be used in the development of new vaccines [33].
To overcome the confounding effects of cross-reactivity between DENV and ZIKV, the potential influence of a previous antiflavivirus immunity on the outcome of ZIKV infection, the difficulty that this cross-reactivity implies in the development of vaccines and diagnostic tests of high sensitivity and specificity, we used the strategy of Rvac to find conserved, exclusive and potential immunogenic regions present in the structural and nonstructural proteins of ZIKV. These regions could be used as a guide to develop new vaccines and diagnostic technologies for the disease.
Materials and methods
T-cell epitope prediction
T-cell epitopes of all structural and nonstructural protein sequences of a Brazilian ZIKV strain (PE243/2015; GenBank accession no. ANC90426.1) were predicted by using the TepiTool tool available in the Immune Epitope Database (IEDB) Analysis Resource (http://tools.iedb.org/tepitool) [34]. MHC-I and MHC-II are highly polymorphic molecules with very different allele specificity. Therefore, all predictions were performed to cover polymorphic loci using the most representative alleles in each class. The NetMCHpan method was used for MHC-I binding predictions and the NetMHCpanII for MHC-II binding predictions. A binding affinity threshold of 500 nM was selected as the cutoff for MHC-I binding and a 1000 nM cutoff for MHC-II binding, according to parameters recommended by TepiTool. The number of binding human leukocyte antigen (HLA) I and HLA-II alleles was recorded for each epitope.
Epitope specificity analysis
The selected epitopes for ZIKV were compared for similarity with seven viruses belonging to family Flaviviridae, genus Flavivirus (Table 1). The full polyprotein sequences of these viruses were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/genbank) and submitted to epitope conservation analysis using the Epitope Conservancy Analysis tool available online at the IEDB Analysis Resource (http://tools.iedb.org/conservancy) [35]. Data from previous studies have shown that epitopes with a high degree of similarity in amino acid sequences and three-dimensional structures could be recognized by cross-reaction antibodies [36]. Structural bioinformatics analysis showed cross-reactivity between allergenic proteins available with sequence identity >60% [37]. Therefore, only epitopes that showed a similarity of <60% with all other flaviviruses were considered to be specific for ZIKV and used for downstream analysis. Therefore, epitopes with similarity ≥60% with one or more virus were excluded from downstream analysis.
Table 1.
Genomic sequences of flaviviruses included in this study
| Vector | Name | GenBank accession no. |
|---|---|---|
| Tick | Omsk hemorrhagic fever virus | NP_878909.1 |
| Tick-borne encephalitis virus | ANN44512.1 | |
| Mosquito | Dengue virus serotype 1 | AGN94865.1 |
| Dengue virus serotype 2 | AGE89225.1 | |
| Dengue virus serotype 3 | AHG23238.1 | |
| Dengue virus serotype 4 | ANK35834.1 | |
| Japanese encephalitis virus | ANH21067.1 | |
| St Louis encephalitis virus | AIW82235.1 | |
| West Nile virus | ALA10710.1 | |
| Yellow fever virus | AFH35033.1 | |
| Zika virus | ANC90426.1 |
Selection of immunogenic epitopes
The epitopes with low identity to other flaviviruses and a potential to interact with four or more HLA-I or HLA-II alleles were assessed for their antigenicity through the VaxiJen server, version 2.0 (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html). For antigenicity analysis, a threshold of ≥0.5 was adopted, which corresponds to an accuracy of 87% for viruses [38]. The epitopes with values of ≥0.5 were submitted to a conservation analysis of amino acid sequences among South American and African ZIKV isolates. Nucleotide and amino acid sequences of 684 ZIKV isolates were obtained from GenBank (https://www.ncbi.nlm.nih.gov/genbank) for comparative analyses using Epitope Conservancy Analysis tool available at the IEDB Analysis Resource (http://tools.iedb.org/conservancy). All epitopes presenting antigenic potential were also evaluated for theoretical determination of toxicity by using ToxinPred. This tool has an accuracy of 90% (http://crdd.osdd.net/raghava/toxinpred) [39] and is widely used in the context of Rvac [40], [41], [42].
The selected epitopes were submitted to population coverage determination based on the group of alleles with which each epitope can interact according to the initial prediction using TepiTool. The calculation was performed using the Population Coverage Analysis tool of the IEDB [43]. Finally, the predicted epitopes on the M, E and NS1 proteins were evaluated for the solvent accessible surface (SAS) in the corresponding protein using UCSF Chimera software version 1.12 and the 3-D cryostructure of E and M protein trimer (5IRE) and NS1 dimer protein (5GS6) deposited on the Protein Data Bank server.
Results
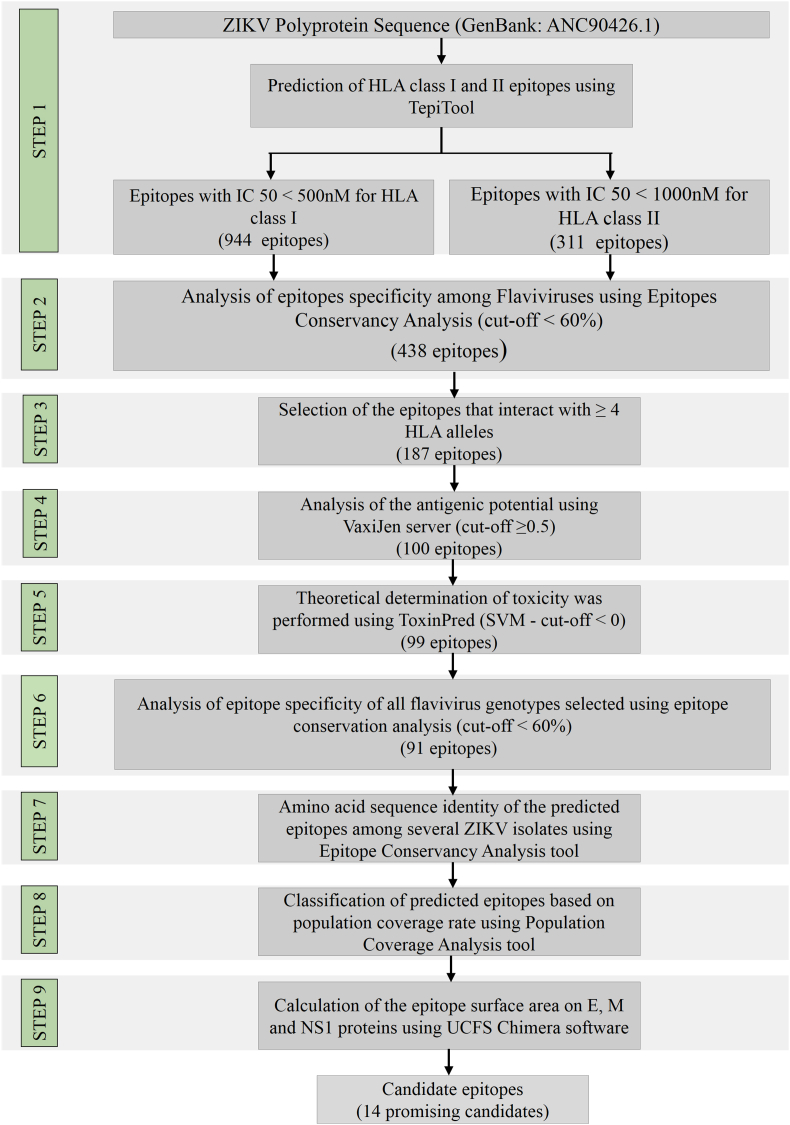
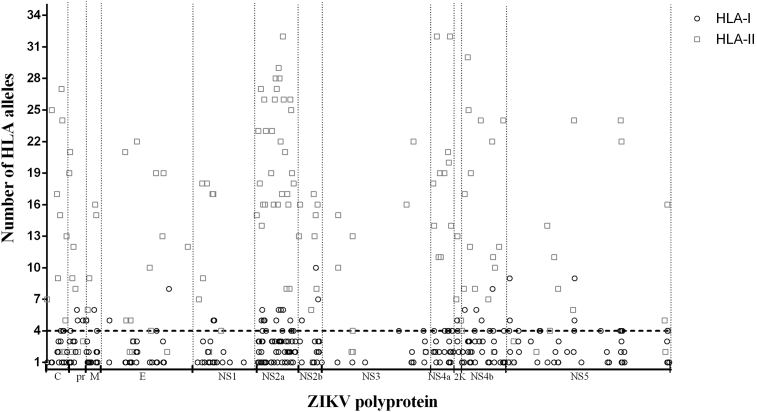
As illustrated in Fig. 1, a nine-step immunoinformatic approach was used to identify T-cell epitopes with the potential to contribute to the development of vaccine candidates and diagnostic tests for ZIKV. Using TepiTool, 1457 epitopes were predicted within ZIKV polyprotein. Among these predicted epitopes, 944 (64.79%) were predicted for binding to HLA-I and 513 for HLA-II (31.21%). A total of 311 epitopes were identified from structural proteins and 1146 epitopes from nonstructural proteins (Table 2). Conservation analysis of predicted epitopes showed that 1019 (88.92%) had a higher similarity degree with other flaviviruses (≥60% similarity) (Supplementary Table S1). Therefore, only 438 predicted epitopes (11.08%) had a similarity of <60% with structural and nonstructural proteins from other flaviviruses. Among these epitopes, only 187 (42.69%) showed a potential to interact with four or more HLA-I or HLA-II alleles (Fig. 2 and Supplementary Table S2). The data showed that HLA-II predicted epitopes are probably able to bind more alleles than the HLA-I predicted epitopes.
Fig. 1.
Schematic representation of entire in silico approach for predictions and selection of T-cell epitopes on structural and nonstructural Zika virus proteins.
Table 2.
T-cell epitopes predicted from Zika virus structural and nonstructural proteins by TepiTool HLA-I and HLA-II prediction programme
| Protein | No. of predicted epitopes |
|
|---|---|---|
| HLA-I | HLA-II | |
| C | 40 | 19 |
| PR | 21 | 74 |
| M | 31 | 12 |
| E | 104 | 10 |
| NS1 | 75 | 49 |
| NS2a | 96 | 35 |
| NS2b | 35 | 18 |
| NS3 | 151 | 93 |
| NS4a | 43 | 20 |
| 2k | 8 | 2 |
| NS4b | 96 | 41 |
| NS5 | 244 | 140 |
| Total | 944 | 513 |
HLA, human leukocyte antigen.
Fig. 2.
Number of HLA-I and HLA-II binding alleles by each predicted epitopes. Predicted epitopes with similarity less than 60% with structural and nonstructural proteins from other flaviviruses and theoretical number of binding HLA-I or HLA-II for each epitope are shown. Dotted line represents cutoff value (four or more binding HLA-I or HLA-II alleles). Only epitopes above cutoff were submitted to antigenicity calculation through VaxiJen server version 2.0 (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html). HLA, human leukocyte antigen.
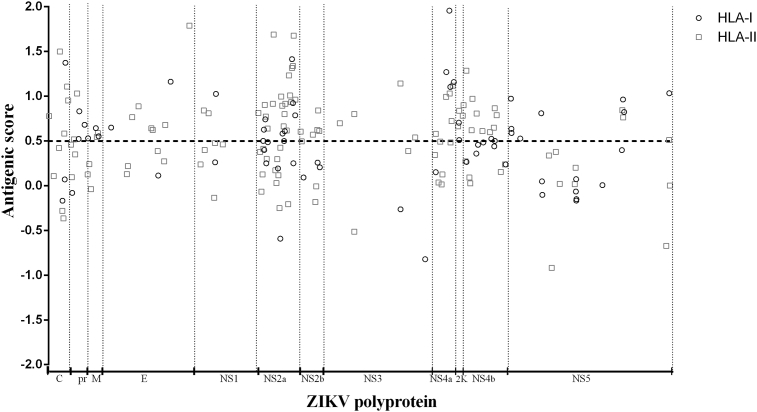
The immunogenicity potential of all 187 epitopes was evaluated using the VaxiJen server. The results showed that 87 epitopes (46.52%) were not predicted to be immunogenic. Thus, the number of candidates was reduced to 100 peptides with a range of antigenic potential score of between 0.5011 and 1.9567 (Fig. 3). The theoretical determination of toxicity was performed using ToxinPred, and only the epitope pr43−51 showed a potential to trigger toxicity in vivo (Supplementary Table S3). Therefore, this epitope was excluded from the next analysis.
Fig. 3.
Antigenicity prediction score computed by VaxiJen server. Antigenicity score was calculated by Vaxijen server version 2.0 (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html). Dotted line represents cutoff value (> 0.5). Only epitopes above cutoff were submitted to conservation analysis of amino acid sequence between different South American and African Zika virus strains.
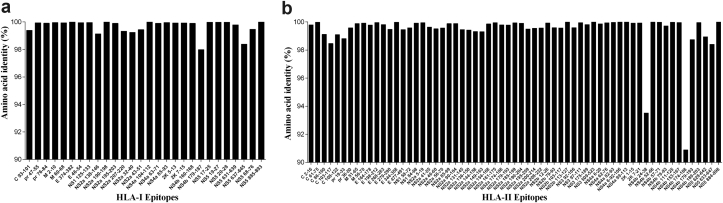
The 99 selected epitopes were submitted to a new conservation analysis using ten more sequences of DENV serotype 1, ten of DENV serotype 2, ten of DENV serotype 3, ten of DENV serotype 4, ten of Japanese encephalitis virus, four of Omsk hemorrhagic fever virus, nine of tick-borne encephalitis virus, four of West Nile virus, 16 of St Louis encephalitis virus and eight of yellow fever virus (Supplementary Table S4). Only eight epitopes presented a similarity of ≥60% with other flavivirus sequences; these were excluded from downstream analysis. These immunogenic epitopes were submitted to an analysis of conservation using 684 strains of ZIKV (Supplementary Table S5). The analysis showed a high degree of identity of all 100 epitopes among the ZIKV strains (90.89–100%) (Fig. 4). The evaluation of the population coverage based on the HLA alleles that each epitope can theoretically interact with varied from 17.68% and 59.52% for the epitopes predicted for HLA-I and from 59.39% and 99.97% for the epitopes for HLA-II. The HLA-II–predicted epitopes showed the highest coverage (≥50%) if compared to HLA-I–predicted epitopes (≤50%) (Fig. 5). The 91 selected epitopes and their properties are listed in Supplementary Table S6.
Fig. 4.
Amino acid sequence identity of predicted epitopes among Zika virus (ZIKV) isolates. Peptide conservation analysis was performed by Epitope Conservancy Analysis tool at Immune Epitope Database (IEDB) (http://tools.iedb.org/conservancy). Six hundred eighty-four ZIKV isolates were included in this analysis. (A) Amino acid sequence identity of predicted epitopes for HLA-I alleles. (B) Amino acid sequence identity of predicted epitopes for HLA-II alleles. HLA, human leukocyte antigen.
Fig. 5.
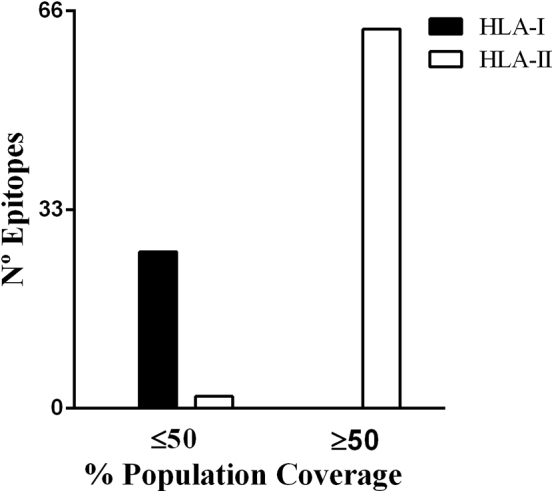
Classification of predicted epitopes based on population coverage rate. Selected epitopes were submitted to population coverage determination based on group of alleles that each epitope can interact according to initial prediction using TepiTool. Calculation was performed using Population Coverage Analysis tool of Immune Epitope Database (IEDB).
To gain insight into the spatial localization of the selected epitopes on E, M and NS1 proteins, these epitopes were located in the cryostructure of their respective proteins. The localization showed that all epitopes were exposed on the protein structures (Supplementary Fig. S1). The SAS calculation showed a high value of all epitopes (1471.71 ± 309.2) (Table 3).
Table 3.
Predicted epitopes for proteins E, M and NS1 and calculation of SAS
| Protein | Position | Epitope | HLA class | SAS (Å2) |
|---|---|---|---|---|
| M | 2 | VTLPSHSTR | I | 1014.30 |
| 60 | KVIYLVMIL | I | 968.19 | |
| 51 | WLLGSSTSQKVIYLV | II | 1430.80 | |
| 56 | STSQKVIYLVMILLI | II | 1386.61 | |
| E | 46 | TTVSNMAEV | I | 1187.39 |
| 164 | RAKVEITPNSPRAEA | II | 1886.30 | |
| 198 | FSDLYYLTMNNKHWL | II | 1742.05 | |
| 269 | LAGALEAEMDGAKGR | II | 1569.30 | |
| 276 | EMDGAKGRLSSGHLK | II | 1776.53 | |
| 344 | QMAVDMQTLTPVGRL | II | 1376.53 | |
| 374 | MMLELDPPF | I | 1217.23 | |
| 477 | LNTKNGSISLMCLAL | II | 1252.47 | |
| NS1 | 58 | SSVSRMENIMWRSVE | II | 1794.59 |
| 84 | VQLTVVVGSVKNPMW | II | 2026.50 | |
| 125 | RAAKTNNSF | I | 1446.91 |
HLA, human leukocyte antigen; SAS, solvent-accessible surface.
In order to identify the most promising candidates for the development of future vaccines and/or to create diagnostic tests to identify ZIKV, the selected epitopes were ranked. Further selection of epitopes with conservation rate ≥95% among the ZIKV strains, population coverage >50%, antigenic score ≥1.0 and SAS ≥1050 Å2 (in this case, epitopes for M, E and NS1 proteins) resulted in a final list of 14 candidates epitopes (Table 4). These epitopes were located on C (2/14.3%), E (2/14.3%), prM (1/7.1%), NS2a (6/42.9%), NS3 (1/7.1%) and NS4a (2/14.3%) proteins. Among these epitopes, only epitope E374−382 was a HLA-I–predicted epitope.
Table 4.
List of candidate T-cell epitopes predicted from ZIKV structural and nonstructural proteins
| Protein | Position | Epitope | HLA class | Antigenic score | Conservation among ZIKV strains (%) | Population coverage (%) | SAS (Å2) | Reference |
|---|---|---|---|---|---|---|---|---|
| E | 374 | MMLELDPPF | I | 1.1639 | 100 | 59.52 | 1.217.23 | [40], [44] |
| 477 | LNTKNGSISLMCLAL | II | 1.7899 | 99.46 | 92.7 | 1.252.47 | This study | |
| C | 61 | PSLGLINRWGSVGKK | II | 1.4993 | 99.98 | 74.37 | — | This study |
| 103 | RRGADTSVGIVGLLL | II | 1.1073 | 98.47 | 93.45 | — | [45] | |
| pr | 36 | IQIMDLGHMCDATMS | II | 1.0307 | 99.59 | 79.11 | — | This study |
| NS2a | 90 | LVSFIFRANWTPRES | II | 1.6900 | 99.88 | 99.5 | — | [45] |
| 174 | LATCGGFMLLSLKGK | II | 1.2330 | 99.80 | 99.3 | — | This study | |
| 179 | GFMLLSLKGKGSVKK | II | 1.0070 | 99.78 | 62.28 | — | This study | |
| 190 | SVKKNLPFVMALGLT | II | 1.3171 | 99.91 | 99.78 | — | This study | |
| 195 | LPFVMALGLTAVRLV | II | 1.3379 | 99.50 | 99.69 | — | [45] | |
| 200 | ALGLTAVRLVDPINV | II | 1.6769 | 99.55 | 97.2 | — | [45] | |
| NS3 | 175 | PVECFEPSMLKKKQL | II | 1.1434 | 99.82 | 98.09 | — | This study |
| NS4a | 82 | GFGMVTLGASAWLMW | II | 1.0313 | 99.97 | 99.6 | — | This study |
| 99 | EIEPARIACVLIVVF | II | 1.1164 | 100 | 94.92 | — | [45] |
HLA, human leukocyte antigen; SAS, solvent-accessible surface; ZIKV, Zika virus.
Discussion
ZIKV is a mosquito-borne Flavivirus belonging to the Flaviviridae family [5]. It has been classified as an emerging pathogen because of its fast spread across the Americas as well as its association with cases of Guillain-Barré syndrome and prenatal microcephaly in ZIKV-endemic regions [46], [47], [48]. Therefore, there is an urgent need for vaccines against ZIKV infection [49], [50], [51], [52]. Different immunoinformatics approaches aiming to identify the immunogenic peptides in the context of Rvac have been reported [40], [44], [53], [54], [55], [56], [57]. However, information about the prediction and selection of exclusive epitopes in ZIKV structural and nonstructural proteins has not yet been published. In this study, we used several bioinformatic tools to identify immunoreactive epitopes within ZIKV structural and nonstructural proteins and to see that they have no similarity with epitopes from other important flaviviruses. Therefore, these epitopes could be used to develop new vaccines and diagnostic tests for ZIKV.
The cross-reactivity of some antibodies is related to the high similarity between the amino acids and the tridimensional structure between epitopes [36]. The identification of exclusive epitopes for the development of ZIKV vaccines may contribute to avoid the ADE observed in some flaviviruses infections. The ADE predicts that previous infection with a flaviviruses could contribute to virus propagation in a subsequent infection by another related flaviviruses [58], [59], [60]. Although there are no definitive data that indicate that the ADE in humans, the overlap of Flavivirus-endemic areas [61] makes the ADE an important challenge in the elaboration of a vaccine candidate against ZIKV [62]. In this sense, it is reasonable to propose a vaccine platform based on epitopes to ZIKV that could induce an effective immunologic response without any cross-reacting activity against other flaviviruses.
Using in silico analysis, several groups have predicted ZIKV-specific mouse or human T-cell epitopes within E [40], [44], [54], [63], NS2a [63], NS5 proteins [56] or the entire ZIKV polyprotein [45], [55], [64], [65], [66], [67]. In this study, we found 91 epitopes with immunogenic properties on structural and nonstructural ZIKV proteins. Twenty-four epitopes had previously been identified (Supplementary Table S7). These studies showed that some epitopes (pr47−55, M56−70, E46−54, NS2a43−51 and NS519−27) were able to induce T-helper 1 type cytokines, including interferon gamma and/or tumor necrosis factor alpha secretion by CD4+ and CD8+ T cells in mice [63], [64], [65], [67].
The data we present here show that the predicted epitopes for HLA-II binding are likely to bind to more alleles than the predicted epitopes for HLA-I binding. This could be related to the structure of the HLA-I and HLA-II molecules. The open structure of the binding groove and the longer length of HLA-II–bound peptides allow greater flexibility in binding and association with more HLA-II alleles [68]. Therefore, this could be responsible for the greater population coverage showed by predicted binding HLA-II epitopes (Fig. 5). It also identified a high number of predicted epitopes on the NS5 protein (384/26.36%). This result is probably the result of the length and immunogenicity of this protein [69], [70], [71]. The NS5 protein is the most conserved nonstructural protein of flaviviruses [72], [73]. Therefore, because of the high conservation rate between the NS5 sequences of flaviviruses, only ten unique epitopes were identified on ZIKV NS5 protein (Supplementary Table S6).
Determination of the SAS is important to identify candidate epitopes exposed in the structural proteins and the NS1 nonstructural protein. Exposed epitopes are more readily recognized by antibodies, and they are probably potential candidates for vaccine and diagnostic test development [74], [75]. Previous studies showed that the interaction area of broadly neutralizing antibodies to DENV epitopes on protein E ranges from 1050 to 1400 Å2 [76]. Most of the epitopes identified in M, E and NS1 proteins have a larger exposed surface area (Table 3). Therefore, it is possible that antibodies produced after ZIKV infection recognize these epitopes.
An ideal candidate ZIKV vaccine will be able to induce virus-specific immune response to all ZIKV strains [77]. Our analysis showed 91 potentially immunogenic epitopes present in the structural and nonstructural ZIKV proteins. Thus, considering these peptides are highly conserved among the 684 strains analysed in this study (90.89–100%), they are likely to be used in a vaccine formulation to induce protective immunity against any ZIKV strain. In order to identify the most promising candidates, we listed the epitopes that have a greater number of desired characteristics (conservation rate ≥95% among the ZIKV strains; population coverage >50%; antigenic score ≥1.0 and SAS ≥1050 Å2). The threshold for this classification was defined on the basis of the highest scores obtained in this study and prioritizing epitopes with ≥95% conservation because strains with a similarity of >95% were previously able to produce neutralizing antibodies for infections by different strains [78]. Among the epitopes that are able to bind HLA-I, E374−382 is the only one that meets all the predetermined requirements, confirming the immunogenic potential of this epitope previously reported [40], [44]. Thirteen HLA-II epitopes met all the requirements, including four epitopes present in structural proteins (pr36−50, C61−75, C103−117, E477−491) and 9 in nonstructural proteins (NS2a90−104, NS2a174−188, NS2a179−193, NS2a190−204, NS2a195−209, NS2a200−214, NS3175−189, NS4a82−96, NS4a99−113). Among the epitopes present in structural proteins, E477−491 has the highest antigenic score (1.7899). This epitope is located in domain III of the E protein, which is the main domain associated with the induction of neutralizing antibodies [79], [80]. Of the nine HLA-II epitopes present in the nonstructural proteins, six (66.66%) were identified on NS2a protein (Table 4). The NS2a protein is involved in viral RNA replication, virus assembly and secretion in flaviviruses [81]. It was also demonstrated that ZIKV NS2a protein disrupts mammalian cortical neurogenesis by degrading adherens junction protein [82]. Therefore, epitopes predicted in this protein appear as potential candidates for the development of new vaccine formulations. Epitopes in NS2a have already been identified [45], [63]. However, here we identify new and exclusive epitopes on the ZIKV NS2a protein (Table 4).
The 14 predicted T-cell epitopes identified in this study, taken together, are of special interest as potential candidate regions for inclusion in developing epitope-driven vaccines against ZIKV or diagnostic tests for the disease. Therefore, these epitopes have a biotechnologic relevance to the rational design of vaccine and diagnostic tests for ZIKV. However, in the next years, future in vitro and in vivo studies should be carried out to best characterize the immunogenicity of these epitopes and to establish the role of these epitopes as new vaccines or diagnostic tests.
Acknowledgements
We thank S. Weaver (University of Texas Medical Branch) for his careful review. Supported in part by the FAPEMIG (grants APQ-01165–16 and PPM-00399–18). EAS received fellowships from FAPEMIG (PIBICT) and CNPq (PIBIC). GAPS received fellowships from FAPEMIG. LFLC received PQ fellowship from CNPq. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasil (CAPES), Finance Code 001.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nmni.2019.01.002.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Göertz G.P., Abbo S.R., Fros J.J., Pijlman G.P. Functional RNA during Zika virus infection. Virus Res. 2018;254:41–53. doi: 10.1016/j.virusres.2017.08.015. [DOI] [PubMed] [Google Scholar]
- 2.Rückert C., Weger-Lucarelli J., Garcia-Luna S.M., Young M.C., Byas A.D., Murrieta R.A. Impact of simultaneous exposure to arboviruses on infection and transmission by Aedes aegypti mosquitoes. Nat Commun. 2017;8:15412. doi: 10.1038/ncomms15412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simmonds P., Becher P., Bukh J., Gould E.A., Meyers G., Monath T. ICTV virus taxonomy profile: Flaviviridae. J Gen Virol. 2017;98:2–3. doi: 10.1099/jgv.0.000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hou W., Cruz-cosme R., Armstrong N., Obwolo L.A., Wen F., Hu W. Molecular cloning and characterization of the genes encoding the proteins of Zika virus. Gene. 2017;628:117–128. doi: 10.1016/j.gene.2017.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dick G.W., Kitchen S.F., Haddow A.J. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46:509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 6.Duffy M.R., Chen T.H., Hancock W.T., Powers A.M., Kool J.L., Lanciotti R.S. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med. 2009;360:2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- 7.Zinszer K., Morrison K., Brownstein J.S., Marinho F., Santos A.F., Nsoesie E.O. Reconstruction of Zika virus introduction in Brazil. Emerg Infect Dis. 2017;23:91–94. doi: 10.3201/eid2301.161274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Passos S.R.L., Borges dos Santos M.A., Cerbino-Neto J., Buonora S.N., Souza T.M.L., de Oliveira R.V.C. Detection of Zika virus in April 2013 patient samples, Rio de Janeiro, Brazil. Emerg Infect Dis. 2017;23:2120–2121. doi: 10.3201/eid2312.171375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musso D. Zika virus transmission from French Polynesia to Brazil. Emerg Infect Dis. 2015;21:1887–1889. doi: 10.3201/eid2110.151125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oehler E., Watrin L., Larre P., Leparc-Goffart I., Lastere S., Valour F. Zika virus infection complicated by Guillain-Barre syndrome—case report, French Polynesia, December 2013. Euro Surveill. 2014;19:20720. doi: 10.2807/1560-7917.es2014.19.9.20720. [DOI] [PubMed] [Google Scholar]
- 11.Edge L. Zika virus on the move. Cell. 2016;164:585–587. [Google Scholar]
- 12.Malkki H. Zika virus infection could trigger Guillain-Barré syndrome. Nat Rev Neurol. 2016;12:187. doi: 10.1038/nrneurol.2016.30. [DOI] [PubMed] [Google Scholar]
- 13.Musso D., Roche C., Robin E., Nhan T., Teissier A., Cao-Lormeau V.M. Potential sexual transmission of Zika virus. Emerg Infect Dis. 2015;21:359–361. doi: 10.3201/eid2102.141363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zamani M., Zamani V. Sexual transmission of Zika virus: an assessment of the evidence. Iran J Public Health. 2017;46:1305–1306. [PMC free article] [PubMed] [Google Scholar]
- 15.Tebas P., Roberts C.C., Muthumani K., Reuschel E.L., Kudchodkar S.B., Zaidi F.I. Safety and immunogenicity of an anti-Zika virus DNA vaccine – preliminary report. N Engl J Med. 2017 doi: 10.1056/NEJMoa1708120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaudinski M.R., Houser K.V., Morabito K.M., Hu Z., Yamshchikov G., Rothwell R.S. Safety, tolerability, and immunogenicity of two Zika virus DNA vaccine candidates in healthy adults: randomised, open-label, phase 1 clinical trials. Lancet. 2018;391(10120):552–562. doi: 10.1016/S0140-6736(17)33105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richner J.M., Himansu S., Dowd K.A., Pierson T.C., Ciaramella G., Diamond M.S. Modified mRNA vaccines protect against Zika virus infection. Cell. 2017;168 doi: 10.1016/j.cell.2017.02.017. 1114–25.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barouch D.H., Thomas S.J., Michael N.L. Perspective prospects for a Zika virus vaccine. Immunity. 2017;46:176–182. doi: 10.1016/j.immuni.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barrett A.D.T. Current status of Zika vaccine development: Zika vaccines advance into clinical evaluation. NPJ Vaccines. 2018;3:24. doi: 10.1038/s41541-018-0061-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Culshaw A., Mongkolsapaya J., Screaton G. The immunology of Zika virus. F1000Res. 2018;7:203. doi: 10.12688/f1000research.12271.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawiecki A.B., Christofferson R.C. Zika virus–induced antibody response enhances dengue virus serotype 2 replication in vitro. J Infect Dis. 2016;214:1357–1360. doi: 10.1093/infdis/jiw377. [DOI] [PubMed] [Google Scholar]
- 22.Priyamvada L., Quicke K.M., Hudson W.H., Onlamoon N., Sewatanon J., Edupuganti S. Human antibody responses after dengue virus infection are highly cross-reactive to Zika virus. Proc Natl Acad Sci U S A. 2016;113:7852–7857. doi: 10.1073/pnas.1607931113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rey F.A., Stiasny K., Vaney M., Dellarole M., Heinz F.X. The bright and the dark side of human antibody responses to flaviviruses: lessons for vaccine design. EMBO Rep. 2018;19:206–224. doi: 10.15252/embr.201745302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiang M.H., Sung W.C., Lien S.P., Chen Y.Z., Lo A.F., Huang J.H. Identification of novel vaccine candidates against Acinetobacter baumannii using reverse vaccinology. Hum Vaccin Immunother. 2015;11:1065–1073. doi: 10.1080/21645515.2015.1010910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Contreras M., Villar M., Artigas-Jerónimo S., Kornieieva L., Mуtrofanov S., de la Fuente J. A reverse vaccinology approach to the identification and characterization of Ctenocephalides felis candidate protective antigens for the control of cat flea infestations. Parasit Vectors. 2018;11:43. doi: 10.1186/s13071-018-2618-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Backert L., Kohlbacher O. Immunoinformatics and epitope prediction in the age of genomic medicine. Genome Med. 2015;7:119. doi: 10.1186/s13073-015-0245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bruno L., Cortese M., Rappuoli R., Merola M. Lessons from reverse vaccinology for viral vaccine design. Curr Opin Virol. 2015;11:89–97. doi: 10.1016/j.coviro.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Heinson A.I., Woelk C.H., Newell M.L. The promise of reverse vaccinology. Int Health. 2015;7:85–89. doi: 10.1093/inthealth/ihv002. [DOI] [PubMed] [Google Scholar]
- 29.Liljeroos L., Malito E., Ferlenghi I., Bottomley M.J. Structural and computational biology in the design of immunogenic vaccine antigens. J Immunol Res. 2015;2015:1–17. doi: 10.1155/2015/156241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schussek S., Trieu A., Doolan D.L. Genome- and proteome-wide screening strategies for antigen discovery and immunogen design. Biotechnol Adv. 2014;32:403–414. doi: 10.1016/j.biotechadv.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 31.Huber S.R., van Beek J., de Jonge J., Luytjes W., van Baarle D. T cell responses to viral infections—opportunities for peptide vaccination. Front Immunol. 2014;5(APR):1–12. doi: 10.3389/fimmu.2014.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nandy A., Basak S.C. A brief review of computer-assisted approaches to rational design of peptide vaccines. Int J Mol Sci. 2016;17:666. doi: 10.3390/ijms17050666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patronov A., Doytchinova I. T-cell epitope vaccine design by immunoinformatics. Open Biol. 2013;3:120139. doi: 10.1098/rsob.120139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paul S., Sidney J., Sette A., Peters B. TepiTool: a pipeline for computational prediction of T cell epitope candidates. Curr Protoc Immunol. 2016:18.19.1–18.19.24. doi: 10.1002/cpim.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bui H.H., Sidney J., Li W., Fusseder N., Sette A. Development of an epitope conservancy analysis tool to facilitate the design of epitope-based diagnostics and vaccines. BMC Bioinform. 2007;8:361. doi: 10.1186/1471-2105-8-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Breiteneder H., Mills C. Structural bioinformatic approaches to understand cross-reactivity. Mol Nutr Food Res. 2006;50:628–632. doi: 10.1002/mnfr.200500274. [DOI] [PubMed] [Google Scholar]
- 37.Negi S.S., Braun W. Cross-React: a new structural bioinformatics method for predicting allergen cross-reactivity. Bioinformatics. 2017;33:1014–1020. doi: 10.1093/bioinformatics/btw767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doytchinova I.A., Flower D.R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 2007;8:4. doi: 10.1186/1471-2105-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gupta S., Kapoor P., Chaudhary K., Gautam A., Kumar R. Open Source Drug Discovery Consortium, et al. In silico approach for predicting toxicity of peptides and proteins. PLoS One. 2013;8:e73957. doi: 10.1371/journal.pone.0073957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janahi E.M., Dhasmana A., Srivastava V., Sarangi A.N., Raza S., Arif J.M. In silico CD4+, CD8+ T-cell and B-cell immunity associated immunogenic epitope prediction and HLA distribution analysis of Zika virus. EXCLI J. 2017;16:63–72. doi: 10.17179/excli2016-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Solanki V., Tiwari V. Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Sci Rep. 2018;8:9044. doi: 10.1038/s41598-018-26689-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng J., Lin X., Wang X., Zheng L., Lan S., Jin S. In silico analysis of epitope-based vaccine candidates against hepatitis B virus polymerase protein. Viruses. 2017;9:112. doi: 10.3390/v9050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bui H.H., Sidney J., Dinh K., Southwood S., Newman M.J., Sette A. Predicting population coverage of T-cell epitope–based diagnostics and vaccines. BMC Bioinform. 2006;7:153. doi: 10.1186/1471-2105-7-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alam A., Ali S., Ahamad S., Malik M.Z., Ishrat R. From ZikV genome to vaccine: in silico approach for the epitope-based peptide vaccine against Zika virus envelope glycoprotein. Immunology. 2016;149:386–399. doi: 10.1111/imm.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramaiah A., Dai L., Contreras D., Sinha S., Sun R., Arumugaswami V. Comparative analysis of protein evolution in the genome of pre-epidemic and epidemic Zika virus. Infect Genet Evol. 2017;51:74–85. doi: 10.1016/j.meegid.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 46.Marano G., Pupella S., Vaglio S., Liumbruno G.M., Grazzini G. Zika virus and the never-ending story of emerging pathogens and transfusion medicine. Blood Transfus. 2016;14:95–100. doi: 10.2450/2015.0066-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beckham J.D., Pastula D.M., Massey A., Tyler K.L. Zika virus as an emerging global pathogen: neurological complications of Zika virus. JAMA Neurol. 2016;73:875–879. doi: 10.1001/jamaneurol.2016.0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basarab M., Bowman C., Aarons E.J., Cropley I. Zika virus. BMJ. 2016;352:1–7. doi: 10.1136/bmj.i1049. [DOI] [PubMed] [Google Scholar]
- 49.Makhluf H., Shresta S. Development of Zika virus vaccines. Vaccines. 2018;6:7. doi: 10.3390/vaccines6010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du L., Zhou Y., Jiang S. The latest advancements in Zika virus vaccine development. Expert Rev Vaccines. 2017;16:951–954. doi: 10.1080/14760584.2017.1363648. [DOI] [PubMed] [Google Scholar]
- 51.Usman Mirza M., Rafique S., Ali A., Munir M., Ikram N., Manan A. Towards peptide vaccines against Zika virus: immunoinformatics combined with molecular dynamics simulations to predict antigenic epitopes of Zika viral proteins. Sci Rep. 2016;6:37313. doi: 10.1038/srep37313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noor R., Ahmed T. Zika virus: epidemiological study and its association with public health risk. J Infect Public Health. 2018;11:611–616. doi: 10.1016/j.jiph.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 53.Yasmin T., Akter S., Debnath M., Ebihara A., Nakagawa T., Nabi A.H.M.N. In silico proposition to predict cluster of B- and T-cell epitopes for the usefulness of vaccine design from invasive, virulent and membrane associated proteins of C. jejuni. Silico Pharmacol. 2016;4:5. doi: 10.1186/s40203-016-0020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ashfaq U.A., Ahmed B. De novo structural modeling and conserved epitopes prediction of Zika virus envelop protein for vaccine development. Viral Immunol. 2016;29:436–443. doi: 10.1089/vim.2016.0033. [DOI] [PubMed] [Google Scholar]
- 55.Dar H., Zaheer T., Rehman M.T., Ali A., Javed A., Khan G.A. Prediction of promiscuous T-cell epitopes in the Zika virus polyprotein: an in silico approach. Asian Pac J Trop Med. 2016;9:844–850. doi: 10.1016/j.apjtm.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 56.dos Santos Franco L., Oliveira Vidal P., Amorim J.H. In silico design of a Zika virus non-structural protein 5 aiming vaccine protection against Zika and dengue in different human populations. J Biomed Sci. 2017;24:88. doi: 10.1186/s12929-017-0395-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ebrahimi S., Mohabatkar H. Prediction of T-cell epitopes for designing a reverse vaccine against streptococcal bacteria. Mol Biol Res Commun. 2018;7:35–41. doi: 10.22099/mbrc.2018.28775.1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dejnirattisai W., Supasa P., Wongwiwat W., Rouvinski A., Barba-Spaeth G., Duangchinda T. Dengue virus sero-cross-reactivity drives antibody-dependent enhancement of infection with Zika virus. Nat Immunol. 2016;17:1102–1108. doi: 10.1038/ni.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kam Y.W., Lee C.Y.P., Teo T.H., Howland S.W., Amrun S.N., Lum F.M. Cross-reactive dengue human monoclonal antibody prevents severe pathologies and death from Zika virus infections. JCI Insight. 2017;2:e92428. doi: 10.1172/jci.insight.92428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Paul L.M., Carlin E.R., Jenkins M.M., Tan A.L., Barcellona C.M., Nicholson C.O. Dengue virus antibodies enhance Zika virus infection. Clin Transl Immunol. 2016;5:e117. doi: 10.1038/cti.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Azeredo E.L., Dos Santos F.B., Barbosa L.S., Souza T.M.A., Badolato-Correa J., Sanchez-Arcila J.C. Clinical and laboratory profile of zika and dengue infected patients: lessons learned from the co-circulation of dengue, zika and chikungunya in Brazil. PLoS Curr. 2018;10 doi: 10.1371/currents.outbreaks.0bf6aeb4d30824de63c4d5d745b217f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fernandez E., Diamond M.S. Vaccination strategies against Zika virus. Curr Opin Virol. 2017;23:59–67. doi: 10.1016/j.coviro.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang H., Li S., Zhang Y., Han X., Jia B., Liu H. CD8(+) T cell immune response in immunocompetent mice during Zika virus infection. J Virol. 2017;91:e00900–e00917. doi: 10.1128/JVI.00900-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hassert M., Wolf K.J., Schwetye K.E., DiPaolo R.J., Brien J.D., Pinto A.K. CD4+ T cells mediate protection against Zika associated severe disease in a mouse model of infection. PLoS Pathog. 2018;14:e1007237. doi: 10.1371/journal.ppat.1007237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ngono A.E., Vizcarra E.A., Tang W.W., Sheets N., Joo Y., Kim K. Mapping and role of the CD8(+) T cell response during primary Zika virus infection in mice. Cell Host Microbe. 2017;21:35–46. doi: 10.1016/j.chom.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dikhit M.R., Ansari Y., Mansuri R., Sahoo B.R., Dehury B., Amit A. Infection, genetics and evolution computational prediction and analysis of potential antigenic CTL epitopes in Zika virus: a first step towards vaccine development. MEEGID. 2016;45:187–197. doi: 10.1016/j.meegid.2016.08.037. [DOI] [PubMed] [Google Scholar]
- 67.Wen J., Tang W.W., Sheets N., Ellison J., Sette A., Kim K. Identification of Zika virus epitopes reveals immunodominant and protective roles for dengue virus cross-reactive CD8+ T cells. Nat Microbiol. 2017;2:17036. doi: 10.1038/nmicrobiol.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wieczorek M., Abualrous E.T., Sticht J., Álvaro-Benito M., Stolzenberg S., Noé F. Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front Immunol. 2017;8:292. doi: 10.3389/fimmu.2017.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Laurent-Rolle M., Morrison J., Rajsbaum R., Macleod J.M.L., Pisanelli G., Pham A. The interferon signaling antagonist function of yellow fever virus NS5 protein is activated by type I interferon. Cell Host Microbe. 2014;16:314–327. doi: 10.1016/j.chom.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Morrison J., Laurent-Rolle M., Maestre A.M., Rajsbaum R., Pisanelli G., Simon V. Dengue virus co-opts UBR4 to degrade STAT2 and antagonize type I interferon signaling. PLoS Pathog. 2013;9:e1003265. doi: 10.1371/journal.ppat.1003265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lubick K.J., Robertson S.J., Mcnally K.L., Boehm M., Yoshii K., Best S.M. Flavivirus antagonism of type I interferon signaling reveals prolidase as a regulator of IFNAR1 surface article Flavivirus antagonism of type I interferon signaling reveals prolidase as a regulator of IFNAR1 surface expression. Cell Host Microbe. 2015;18:61–74. doi: 10.1016/j.chom.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grant A., Ponia S.S., Tripathi S., Balasubramaniam V., Miorin L., Sourisseau M. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe. 2016;19:882–890. doi: 10.1016/j.chom.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Iacono-Connors L.C., Schmaljohn C.S. Cloning and sequence analysis of the genes encoding the nonstructural proteins of langat virus and comparative analysis with other flaviviruses. Virology. 1992;188:875–880. doi: 10.1016/0042-6822(92)90545-z. [DOI] [PubMed] [Google Scholar]
- 74.Dowd K.A., Mukherjee S., Kuhn R.J., Pierson T.C. Combined effects of the structural heterogeneity and dynamics of flaviviruses on antibody recognition. J Virol. 2014;88:11726–11737. doi: 10.1128/JVI.01140-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Heinz F.X., Stiasny K. Flaviviruses and their antigenic structure. J Clin Virol. 2012;55:289–295. doi: 10.1016/j.jcv.2012.08.024. [DOI] [PubMed] [Google Scholar]
- 76.Rouvinski A., Guardado-Calvo P., Barba-Spaeth G., Duquerroy S., Vaney M.C., Kikuti C.M. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature. 2015;520(7545):109–113. doi: 10.1038/nature14130. [DOI] [PubMed] [Google Scholar]
- 77.Beaver J.T., Lelutiu N., Habib R., Skountzou I. Evolution of two major Zika virus lineages: implications for pathology, immune response, and vaccine development. Front Immunol. 2018;9:1640. doi: 10.3389/fimmu.2018.01640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dowd K.A., DeMaso C.R., Pelc R.S., Speer S.D., Smith A.R.Y., Goo L. Broadly neutralizing activity of Zika virus–immune sera identifies a single viral serotype. Cell Rep. 2016;16:1485–1491. doi: 10.1016/j.celrep.2016.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Deng Y.Q., Dai J.X., Ji G.H., Jiang T., Wang H.J., Yang H. A broadly Flavivirus cross-neutralizing monoclonal antibody that recognizes a novel epitope within the fusion loop of E protein. PLoS One. 2011;6:e16059. doi: 10.1371/journal.pone.0016059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chávez J.H., Silva J.R., Amarilla A.A., Moraes Figueiredo L.T. Domain III peptides from Flavivirus envelope protein are useful antigens for serologic diagnosis and targets for immunization. Biologicals. 2010;38:613–618. doi: 10.1016/j.biologicals.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 81.Leung J.Y., Pijlman G.P., Kondratieva N., Hyde J., Mackenzie J.M., Khromykh A.A. Role of nonstructural protein NS2A in Flavivirus assembly. J Virol. 2008;15(82):4731–4741. doi: 10.1128/JVI.00002-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yoon K.J., Song G., Qian X., Pan J., Xu D., Rho H.S. Zika virus–encoded NS2A disrupts mammalian cortical neurogenesis by degrading adherens junction proteins. Cell Stem Cell. 2017;21:349–358.e6. doi: 10.1016/j.stem.2017.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.