Abstract
Background
Previously, we demonstrated that pollen chamber formation (PCF) in G. biloba ovules was a process of programmed cell death (PCD) within the nucellar cells at the micropylar end. However, the signal triggering the cascades of the programmed events in these nucellar cells remains unexplored.
Results
A transcriptomic strategy was employed to unravel the mechanism underlying the nucellar PCD via the comparative profiles of RNA-seq between pre-PCF and post-PCF ovules. A total of 5599 differentially expressed genes (DEGs) with significance was identified from G. biloba ovules and classified into three main categories of GO annotation, including 17 biological processes, 15 cellular components and 17 molecular functions. KEGG analysis showed that 72 DEGs were enriched in “Plant hormone signal transduction”. Furthermore, 99 DEGs were found to be associated with the PCD process, including the genes involved in ethylene signaling pathway, PCD initiation, and PCD execution. Moreover, calcium-cytochemical localization indicated that calcium could play a role in regulating PCD events within the nucellar cells during pollen chamber formation in G. biloba ovules.
Conclusions
A putative working model, consisting of three overlapping processes, is proposed for the nucellar PCD: at the stage of PCD preparation, ethylene signaling pathway is activated for transcriptional regulation of the downstream targets; subsequently, at the stage of PCD initiation, the upregulated expression of several transcription factors, i.e., NAC, bHLH, MADS-box, and MYB, further promotes the corresponding transcript levels of CYTOCHROME C and CALMODULINs, thereby, leads to the PCD initiation via the calcium-dependent signaling cascade; finally, at the stage of PCD execution, some proteases like metacaspases and vacuolar processing enzyme for hydrolysis, together with the process of autophagy, play roles in the clearance of cellular components. Afterwards, a pollen chamber is generated from the removal of specific nucellar cells in the developing ovule.
Electronic supplementary material
The online version of this article (10.1186/s12870-019-1671-8) contains supplementary material, which is available to authorized users.
Keywords: Ginkgo biloba L., Nucellus, Ovule, Pollen chamber, Programmed cell death (PCD), Transcriptomics
Background
During the reproductive development of Ginkgo biloba L., pollen chamber functions as a storage site for immature pollens pollinated onto the ovule [1]. Pollen chamber formation (PCF) is resulted from the degeneration of 5 ~ 7 layers of nucellar cells at the micropylar end of ovule [2]. Previous researches have demonstrated that the nucellar degeneration should involve programmed cell death (PCD), due to the occurrence of molecular and biochemical markers for PCD, including DNA ladder and terminal deoxynucleotidyl transferase-mediated dUTP nick end labeling (TUNEL) positive labeling on these nucellar cells, together with morphological characteristics, for instance, nuclear degradation, vacuole rupture, and the process of autophagy [2–4].
PCD represents a common mechanism underlying various developmental processes in both animals and plants [5–7]. In plants, developmental PCD (dPCD) has occurred concomitantly with reproductive and vegetative developments, for instance, cell death of the nucellar tissue, tapetum, sex determination, endosperm, embryonic suspensor, xylogenesis, organ senescence, and aerenchyma formation [8–10]. Other types of PCD have also been found to occur during hypersensitive response against invading pathogens [11], and in response to various abiotic stresses [12, 13].
Unlike the animal PCD, i.e., typically apoptosis which is under control by homologs of Bcl-2 proteins or caspases, the plant PCD is executed in a distinctive, plant-specific way, due to the lack of homologs of apoptotic regulators and executors [14]. A certain degree of common in the regulatory network of the diverse cases, which should be coordinated with the PCD preparation, initiation and execution, has been documented in the modulation of plant dPCD [8, 10]. Similar to many plant developmental processes, dPCD is frequently coordinated by hormone signaling through transcriptional control, most commonly ethylene [8, 15–17]. Ethylene and its signaling pathways promotes several types of dPCD, such as root aerenchyma formation and leaf senescence [17–19]. For other phytohormones, gibberellic acid (GA), abscisic acid (ABA), brassinosteroid (BR), jasmonic acid (JA), and salicylic acid (SA) are found to be regulators of leaf senescence-associated PCD [20]. Additionally, GA could promote tapetum PCD [21], and auxin, cytokinin and brassinosteroid promote the PCD during tracheary element (TE) differentiation [22].
Downstream of the hormone signaling, PCD initiation of particular cell types is triggered by diverse events like finetuning of cytoplasmic [Ca2+], cytoplasmic acidification, ROS accumulation, and cytoskeleton modification, respectively or redundantly to a certain extend [8, 13, 23, 24]. Several hydrolytic enzymes have been isolated as putative PCD executers including nucleases and various proteases. For instance, the nuclease BFN1and the aspartate protease PASPA3 from the xylem undergoing PCD, played roles in DNA fragmentation and cellular corpse clearance [23, 25, 26]. The cysteine protease CEP1 functions as an executor during Arabidopsis tapetal PCD [27], as well as XCP1 and XCP2 during TE PCD [28]. Another PCD executors are metacaspases, a type of arginine/lysinespecific cysteine proteases distantly related to animal caspases [29]. A type II metacaspase (McII-Pa) is required for suspensor PCD [30]. The metacaspases MC1 and MC9 are involved in pathogen-triggered PCD and TE PCD, respectively [31, 32]. Other plant proteases, including vacuolar processing enzymes (VPEs), are also shown to be frequently involved in plant PCD. In these cases, nucellain, a barley homologue of VPE, is involved in nucellar PCD during the developing barley grain [33], and δVPE is responsible for the PCD in Arabidopsis seed coats [34]. And the aspartic proteases OsAP25 and OsAP27 could promote tapetum dPCD in rice [35].
In the present study, we conducted a transcriptomic analysis to compare RNA-seq profiles between the ovules before PCF (i.e., pre-PCF) and those after PCF (post-PCF). The aims of this study are (i) to investigate patterns of enrichment for Gene Ontology (GO) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with the significantly differentially expressed genes (DEGs); (ii) to elucidate modulating pathways with respective to the nucellar PCD during pollen chamber formation in G. biloba ovules. Our results from GO and KEGG enrichments of DEGs between pre-PCF and post-PCF ovules presented possible candidates involved in the nucellar PCD and allowed us to identify components within regulatory and signaling pathways. These transcriptomic data will be useful for determining the molecular mechanism by which pollen chamber is formed in G. biloba ovules.
Results
Pollen chamber formation within G. biloba Ovule
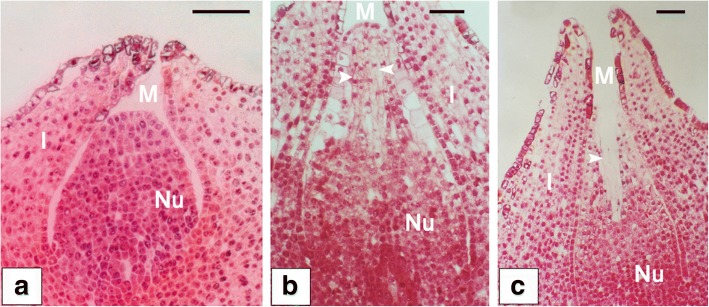
During the process of pollen chamber formation within G. biloba ovule, several stepwise developmental phases in morphology have been observed (Fig. 1). At the early stage of ovule development, nucellar cells at the micropylar end are morphologically similar in their size and shape with those at other parts of nucellus (Fig. 1a). At the later developmental stage, three to four layers of nucellar cells at the micropylar end are differentiated into longitudinally elongated-shape (Fig. 1b). Finally, these elongated nucellar cells are dead and removed for the formation of pollen chamber within an ovule (Fig. 1c).
Fig. 1.
Pollen chamber formation within G. biloba ovule observed under the microscope. (a) At the early stage of ovule development, nucellar cells are morphologically similar in their size and shape. (b) At the later developmental stage, nucellar cells at the micropylar end (indicated by arrows), are elongated longitudinally, and distinguishable from other nucellar cells. (c) With the developmental process, pollen chamber (indicated by an arrow) is formed after the death and clearance of nucellar cells at the micropylar end within an ovule. Bars = 250 μm. Abbreviations: I, integument; M, micropyle; Nu, nucellus
Sequencing data analysis
In the present study, two transcript libraries from G. biloba ovules at the stages of pre- and post- PCF were assayed by high throughout RNA-seq, resulting in 22,246,632 and 22,374,995 raw reads, respectively (Table 1). With the process of quality control for raw reads, clean reads were collected from the two libraries and mapped to the G. biloba genome, producing 90.94% (20,203,385 reads) and 91.42% (20,430,564 reads) matched reads in pre-PCF and post-PCF libraries, respectively. Further analysis revealed that 19,666,682 reads (88.52%) in pre-PCF library and 19,776,546 reads (88.50%) in post-PCF library were uniquely matched (Table 1).
Table 1.
Summary of read numbers based on the RNA-Seq data from G. biloba ovules
| Sample | pre-PCF | post-PCF |
|---|---|---|
| Raw reads | 22,246,632 (100%) | 22,374,995 (100%) |
| Total base pairs (Mb) | 50.99 | 50.97 |
| Clean reads | 22,216,542 (100%) | 22,347,120 (1000%) |
| Total mapped reads | 20,203,385 (90.94%) | 20,430,564 (91.42%) |
| Unique match reads | 19,666,682 (88.52%) | 19,776,546 (88.50%) |
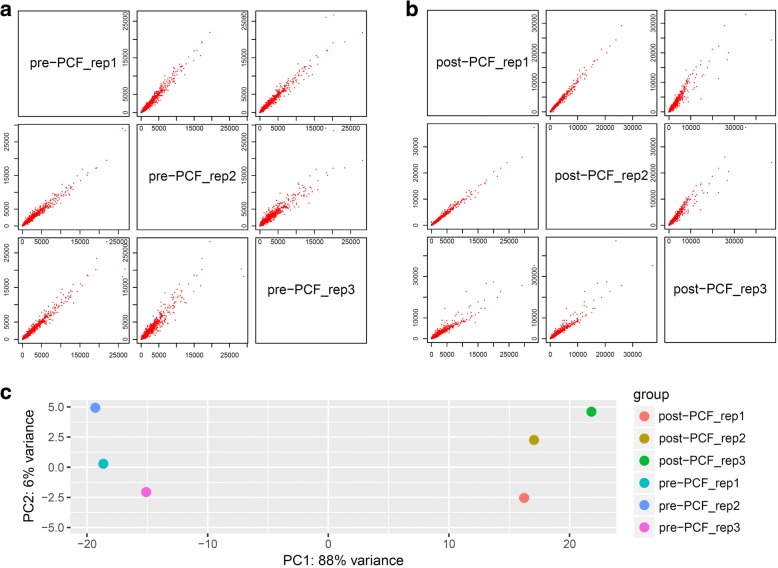
To assess the reliability of the tested samples, the varied degrees among intra- or inter-groups were performed with analysis of principal component (PCA) and replicate scatter (Fig. 2). Both read-counts from the intra-replicates of pre-PCF and post-PCF showed linear-type scatter and correlations, respectively (Fig. 2a and b). On the contrary, there were significant difference between inter-groups (pre-PCF vs. post-PCF) with 88% variance, compared to 6% variance within the intra-groups (Fig. 2c). These results suggested that sampling of G. biloba ovules in the present experiment were reliable and suitable for further analysis.
Fig. 2.
The assessment of correlations among the tested G. biloba ovules by analysis of replicate scatter (a and b) and PCA (c), with each sample group (pre-PCF or post-PCF) including three biological replicates (rep_1, 2, and 3), respectively
Identification and functional enrichment analysis of DEGs
Based on the FPKM method, the transcript abundance of each gene from pre-PCF and post-PCF data was analyzed. A total of 21,435 DEGs were identified between pre-PCF and post-PCF libraries (Fig. 3 and Additional file 1: Table S1). Out of these DEGs, 5599 genes exhibited a significant difference in their expression levels with the threshold of FDR ≤ 0.05, including 2533 up-regulated and 3066 down-regulated ones (Additional file 1: Table S1).
Fig. 3.
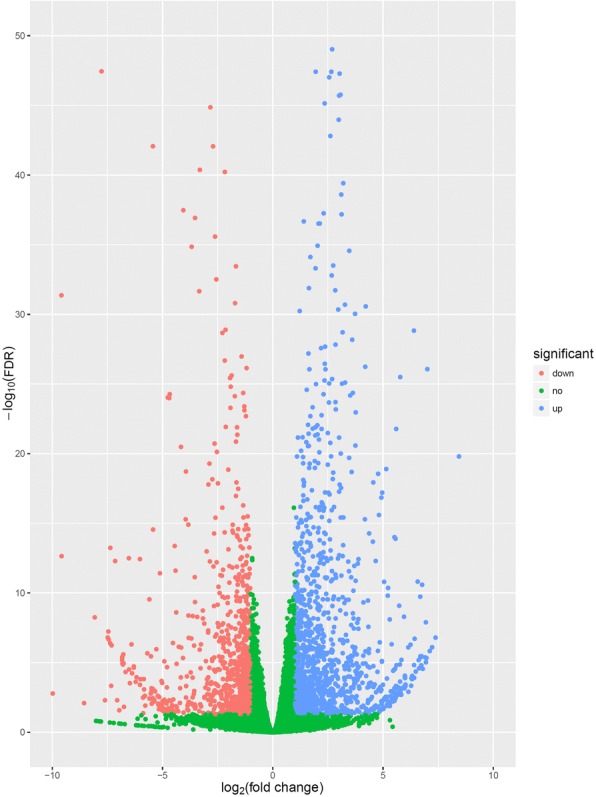
The expression profiles of the identified DEGs. Red and blue points represent the significant DEGs with FDR ≤ 0.05 and log2(fold change) > 1, and green ones show those without significance, respectively. Fold change refers to the values of FPKM change of post-PCF vs. pre-PCF libraries
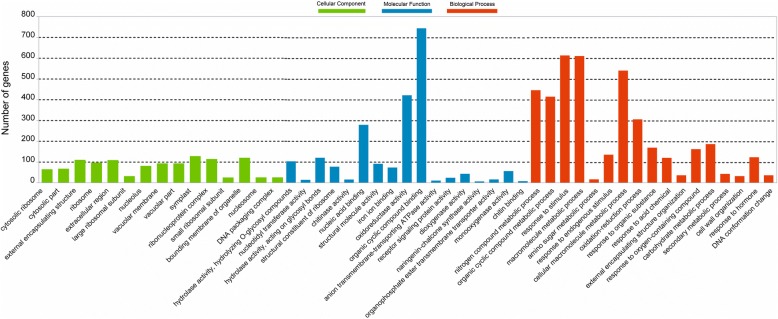
These identified DEGs were annotated with 17 biological processes, 15 cellular components and 17 molecular functions in GO categories (Fig. 4), and significantly enriched (P ≤ 0.001) into 106 GO terms (Additional file 2: Table S2). The terms of “binding” (GO:0005488), “organic cyclic compound binding” (GO:0097159), “heterocyclic compound binding” (GO:1901363), “oxidoreductase activity” (GO:0016491), “nucleic acid binding” (GO:0003676), and “hydrolase activity” (GO:0016798 and GO:0004553) were the dominant groups in the molecular functions; “primary metabolic process” (GO:0044238), “response to stimulus” (GO:0050896), “macromolecule metabolic process” (GO:0043170), “nitrogen compound metabolic process” (GO:0006807), “response to chemical” (GO:0042221), and “oxidation-reduction process” (GO:0055114) were of the representative groups in the biological processes. Among the cellular components, a great number of DEGs were focused on categories of “symplast” (GO:0055044), “cell-cell junction” (GO:0005911), “plasmodesma” (GO:0009506), “bounding membrane of organelle” (GO:0098588), “ribonucleoprotein complex” (GO:0030529), “external encapsulating structure” (GO:0030312), “cell wall” (GO:0005618), and “extracellular region” (GO:0005576).
Fig. 4.
The Gene Ontology (GO) classification of 5599 DEGs. GO terms are summarized in three main categories of cellular component, molecular function and biological process
Moreover, GO terms associated with PCD or autophagy, were found among some of the DEGs, for instance, “programmed cell death” (GO:0012501), “induction of programmed cell death” (GO:0012502), “regulation of programmed cell death” (GO:0043067), “positive of regulation of programmed cell death” (GO:0043068), “negative regulation of programmed cell death” (GO:0043069), “singlet oxygen-mediated programmed cell death” (GO:0010343), “autophagy” (GO:0006914), “process utilizing autophagic mechanism” (GO:0061919), and “ribonuclease activity” (GO:0004540).
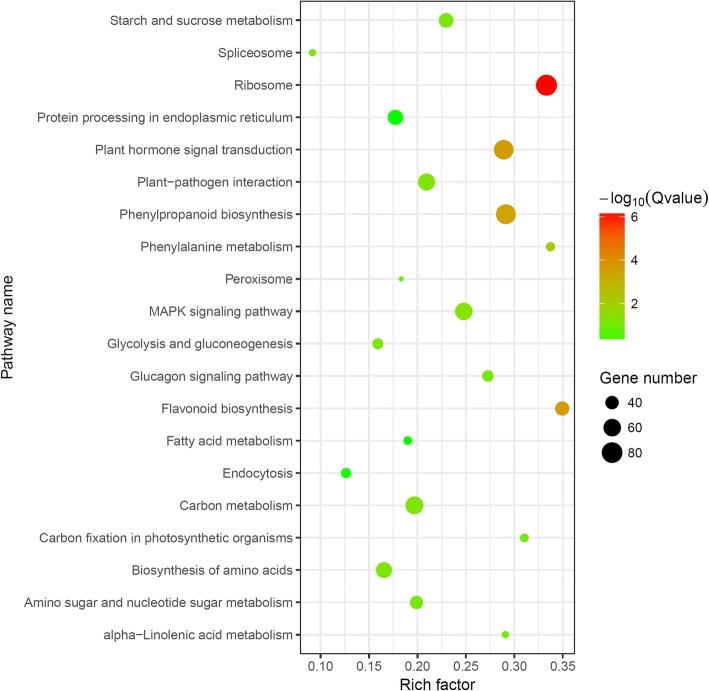
Furthermore, 5599 of the significantly DEGs were blasted to the KEGG database to analyze their biological pathways. Among 205 of the enriched KEGG pathways (Additional file 3: Table S3), the most significant ones (Q ≤ 0.05) consist of “Ribosome”, “Phenylpropanoid biosynthesis”, “Plant hormone signal transduction”, “Flavonoid biosynthesis”, and “Phenylalanine metabolism” (Table 2 and Fig. 5), followed by others, such as “Carbon metabolism” (ko01200), “MAPK signaling pathway” (ko04016), “Plant-pathogen interaction” (ko04626), “Glucagon signaling pathway” (ko04922), “Biosynthesis of amino acids” (ko01230), and “Starch and sucrose metabolism” (ko00500).
Table 2.
The significantly enriched pathways for DEGs in G. biloba ovules
| Pathway | Pathway ID | Number of DEGs | P value | Q value |
|---|---|---|---|---|
| Ribosome | ko03010 | 82 (1.09%) | 2.74E-09 | 7.43E-07 |
| Flavonoid biosynthesis | ko00941 | 43 (0.57%) | 2.69E-06 | 2.44E-04 |
| Plant hormone signal transduction | ko04075 | 72 (0.95%) | 2.70E-06 | 2.44E-04 |
| Phenylpropanoid biosynthesis | ko00940 | 74 (0.98%) | 6.07E-06 | 4.11E-04 |
| Phenylalanine metabolism | ko00360 | 28 (0.37%) | 2.13E-04 | 1.15E-02 |
Fig. 5.
Top 20 pathways of KEGG functional enrichment among DEGs. Coloring indicates -log10(Q value) with higher in red and lower in green. And the lower Q value indicates the more significantly enriched. Point size indicates DEG number
DEGs involved in hormone signal transduction pathway
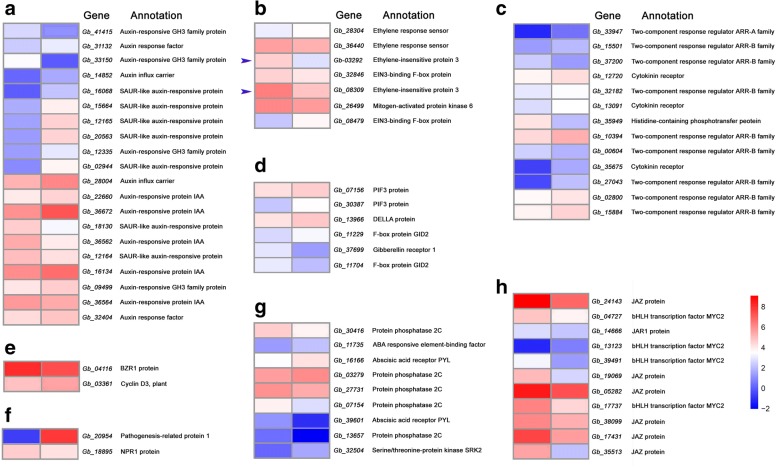
Within the pathway of “Plant hormone signal transduction” (ko04075), there enriched 72 of the significantly DEGs (Table 3 and Additional file 4: Table S4), most of which were involved in auxin, cytokinin, ethylene, JA, ABA, GA, SA, and BR signaling pathways (Fig. 6). In the ethylene pathway, ETR (Ethylene Receptor), MPK6 (Mitogen-Activated Protein Kinase 6), EIN3 (Ethylene-Insensitive Protein 3), and EBF1/2 (EIN3-Binding F-Box Protein), have been documented to participate in the regulation of fruit ripening, senescence, and developmental PCD. In the present study, two orthologs of EIN3 genes and one of MPK6 gene were up-regulated in pre-PCF, compared to those in post-PCF, while both of two ETR or EBF1/2 genes were inconsistent with each other in the transcript levels, respectively. In the auxin signaling pathway, both orthologs of two AUX1 encoding for auxin influx carrier, and ARF for auxin response factor, were up-regulated in post-PCF, compared to those in pre-PCF. Moreover, there were 6 up- and 3 down-regulated transcripts belonging to SAUR genes, and 3 up-and 2 down-regulated ones encoding for auxin-responsive proteins (IAA), respectively. In the cytokinin pathway, 3 CRE1 (encoding for cytokinin receptor), and 7 ARR (two-component response regulator), showed higher expression levels in post-PCF than in pre-PCF, while 1 AHP (histidine-containing phosphotransfer protein) was expressed in a reserve trend. In the pathway of JA signaling, a consistent down-regulation of 6 JAZ (encoding for jasmonate ZIM domain-containing protein), 4 MYC2 (bHLH transcription factors), and 1 JAR1 (jasmonic acid-amino synthetase), was presented. In the ABA pathway, ABF (encoding for ABA responsive element binding factor) and SnRK2 (serine/threonine protein kinase), were up-regulated in post-PCF, compared to those in pre-PCF, while 4 PP2C (protein phosphatase 2C) were down-regulated. In the GA pathway, GID1 (encoding for gibberellin receptor 1) was down-regulated, while both of DELLA and PIF3 (phytochrome-interacting factor 3) were up-regulated in post-PCF, in contrast to those in pre-PCF, respectively. In addition, some DEGs involved in SA and BR signaling pathways were expressed in up- (PR-1, CYCD3) or down-regulated patterns (NPR1, BZR1/2), respectively.
Table 3.
The DEGs involved in plant hormone signal transduction pathway in G. biloba ovules
| Gene ID | KEGG Orthology | Signaling Pathway | Annotation |
|---|---|---|---|
| Gb_28304 | K14509 | Ethylene | ETR, ethylene response sensor |
| Gb_36440 | K14509 | Ethylene | ETR, ethylene response sensor |
| Gb_26499 | K14512 | Ethylene | MPK6, Mitogen-activated protein kinase 6 |
| Gb_03292 | K14514 | Ethylene | EIN3, ethylene-insensitive protein 3 |
| Gb_08309 | K14514 | Ethylene | EIN3, ethylene-insensitive protein 3 |
| Gb_08479 | K14515 | Ethylene | EBF1/2, EIN3-binding F-box protein |
| Gb_32846 | K14515 | Ethylene | EBF1/2, EIN3-binding F-box protein |
| Gb_14852 | K13946 | Auxin | AUX1, auxin influx carrier |
| Gb_28004 | K13946 | Auxin | AUX1, auxin influx carrier |
| Gb_16134 | K14484 | Auxin | Auxin-responsive protein IAA |
| Gb_22660 | K14484 | Auxin | Auxin-responsive protein IAA |
| Gb_36562 | K14484 | Auxin | Auxin-responsive protein IAA |
| Gb_36564 | K14484 | Auxin | Auxin-responsive protein IAA |
| Gb_36672 | K14484 | Auxin | Auxin-responsive protein IAA |
| Gb_31132 | K14486 | Auxin | ARF, auxin response factor |
| Gb_32404 | K14486 | Auxin | ARF, auxin response factor |
| Gb_09499 | K14487 | Auxin | Auxin-responsive GH3 family protein |
| Gb_12335 | K14487 | Auxin | Auxin-responsive GH3 family protein |
| Gb_33150 | K14487 | Auxin | Auxin-responsive GH3 family protein |
| Gb_41415 | K14487 | Auxin | Auxin-responsive GH3 family protein |
| Gb_02944 | K14488 | Auxin | SAUR-like auxin-responsive protein |
| Gb_12164 | K14488 | Auxin | SAUR-like auxin-responsive protein |
| Gb_12165 | K14488 | Auxin | SAUR-like auxin-responsive protein |
| Gb_15664 | K14488 | Auxin | SAUR-like auxin-responsive protein |
| Gb_16068 | K14488 | Auxin | SAUR-like auxin-responsive protein |
| Gb_18130 | K14488 | Auxin | SAUR-like auxin-responsive protein |
| Gb_20563 | K14488 | Auxin | SAUR-like auxin-responsive protein |
| Gb_39828 | K14488 | Auxin | SAUR-like auxin-responsive protein |
| Gb_41374 | K14488 | Auxin | SAUR-like auxin-responsive protein |
| Gb_12720 | K14489 | Cytokinin | CRE1, cytokinin receptor |
| Gb_13091 | K14489 | Cytokinin | CRE1, cytokinin receptor |
| Gb_35675 | K14489 | Cytokinin | CRE1, cytokinin receptor |
| Gb_35949 | K14490 | Cytokinin | AHP, histidine-containing phosphotransfer protein |
| Gb_00604 | K14491 | Cytokinin | Two-component response regulator ARR-B family |
| Gb_02800 | K14491 | Cytokinin | Two-component response regulator ARR-B family |
| Gb_10394 | K14491 | Cytokinin | Two-component response regulator ARR-B family |
| Gb_15501 | K14491 | Cytokinin | Two-component response regulator ARR-B family |
| Gb_15884 | K14491 | Cytokinin | Two-component response regulator ARR-B family |
| Gb_27043 | K14491 | Cytokinin | Two-component response regulator ARR-B family |
| Gb_32182 | K14491 | Cytokinin | Two-component response regulator ARR-B family |
| Gb_37200 | K14491 | Cytokinin | Two-component response regulator ARR-B family |
| Gb_33947 | K14492 | Cytokinin | Two-component response regulator ARR-A family |
| Gb_04727 | K13422 | Jasmonic acid | bHLH transcription factor MYC2 |
| Gb_13123 | K13422 | Jasmonic acid | bHLH transcription factor MYC2 |
| Gb_17737 | K13422 | Jasmonic acid | bHLH transcription factor MYC2 |
| Gb_39491 | K13422 | Jasmonic acid | bHLH transcription factor MYC2 |
| Gb_14666 | K14506 | Jasmonic acid | JAR1, jasmonic acid-amino synthetase |
| Gb_05282 | K13464 | Jasmonic acid | JAZ, jasmonate ZIM domain-containing protein |
| Gb_17431 | K13464 | Jasmonic acid | JAZ, jasmonate ZIM domain-containing protein |
| Gb_19069 | K13464 | Jasmonic acid | JAZ, jasmonate ZIM domain-containing protein |
| Gb_24143 | K13464 | Jasmonic acid | JAZ, jasmonate ZIM domain-containing protein |
| Gb_35513 | K13464 | Jasmonic acid | JAZ, jasmonate ZIM domain-containing protein |
| Gb_38099 | K13464 | Jasmonic acid | JAZ, jasmonate ZIM domain-containing protein |
| Gb_11735 | K14432 | Abscisic acid | ABF, ABA responsive element-binding factor |
| Gb_16166 | K14496 | Abscisic acid | Abscisic acid receptor PYL |
| Gb_39601 | K14496 | Abscisic acid | Abscisic acid receptor PYL |
| Gb_03279 | K14497 | Abscisic acid | PP2C, protein phosphatase 2C |
| Gb_07154 | K14497 | Abscisic acid | PP2C, protein phosphatase 2C |
| Gb_13657 | K14497 | Abscisic acid | PP2C, protein phosphatase 2C |
| Gb_27731 | K14497 | Abscisic acid | PP2C, protein phosphatase 2C |
| Gb_30416 | K14497 | Abscisic acid | PP2C, protein phosphatase 2C |
| Gb_32504 | K14498 | Abscisic acid | SnRK2, serine/threonine-protein kinase SRK2 |
| Gb_07156 | K12126 | Gibberellin | PIF3, phytochrome-interacting factor 3 |
| Gb_30387 | K12126 | Gibberellin | PIF3, phytochrome-interacting factor 3 |
| Gb_37699 | K14493 | Gibberellin | GID1, gibberellin receptor 1 |
| Gb_13966 | K14494 | Gibberellin | DELLA protein |
| Gb_11229 | K14495 | Gibberellin | F-box protein GID2 |
| Gb_11704 | K14495 | Gibberellin | F-box protein GID2 |
| Gb_20954 | K13449 | Salicylic acid | PR1, pathogenesis-related protein 1 |
| Gb_18895 | K14508 | Salicylic acid | regulatory protein NPR1 |
| Gb_04116 | K14503 | Brassinosteroid | BZR1/2, brassinosteroid resistant 1/2 |
| Gb_03361 | K14505 | Brassinosteroid | CYCD3, cyclin D3, plant |
Fig. 6.
Heatmaps of expression patterns for DEGs involved in phytohormone signaling pathways, including auxin (a), ethylene (b), cytokinin (c), GA (d), BR (e), SA (f), ABA (g), and JA (h). Left and right box-columns represent pre-PCF and post-PCF libraries, respectively. Arrowheads indicate the significantly differentially expressed EIN3 genes
DEGs Involved in the Regulation of PCD in G. biloba Ovules.
To identify DEGs associated with the nucellar PCD during pollen chamber formation in G. biloba ovules, the functional annotations of DEGs were blasted against the Nr database. According to the resultant annotations, a total of 99 significantly DEGs related to PCD were identified and involved in different PCD processes including hormone signaling pathways, initiation, and execution (Additional file 5: Table S5). In the present study, ethylene signaling pathway was one of the significantly enriched KEGG pathways. Some genes were identified from the ethylene signaling pathway, including MPK6, EIN3, ethylene receptor gene ETR, and EBF1/2. Moreover, three DEGs involved in the process of ethylene biosynthesis were found out with significantly higher expression levels in the pre-PCF ovules than those in the post-PCF ones. Compared with their transcriptional amounts in the pre-PCF ovules, Gb_41401 (encoding for 1-aminocyclopropane-1-carboxylate synthase, ACS), Gb_08184 and Gb_31357 (encoding for 1-aminocyclopropane-1-carboxylate oxidase, ACO) were down-regulated by 0.95-, 0.79-, and 3.30-fold in the post-PCF ovules, respectively (Additional file 5: Table S5 and Additional file 6: Figure S1). Furthermore, ethylene contents in between the two developmental ovules were manifested by a concomitant elevation in the pre-PCF ovules and a significant decrease in the post-PCF ones (Additional file 5: Table S5 and Additional file 6: Figure S1).
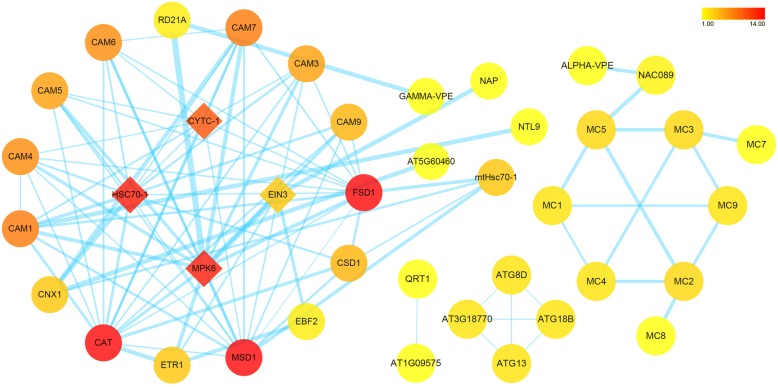
Downstream of ethylene signaling pathway, a distinct PCD-initiating and PCD-executing network is being coordinated to control the PCD process in a precise temporal and spatial pattern. In the study, some genes involved in the PCD initiation are mainly focused on the specific transcription factors (TFs), including MYB, MADS-boxes, bHLHs, and NACs. Other functional components, such as CYTOCHROME C (CYTC-1), CALMODULIN (CAM), MAPK, LESION SIMULATING DISEASE 1 (LSD1), and calcium uniporter protein MCU, were also identified. A total of 45 DEGs was found out to be associated to calcium signaling (Additional file 7: Table S6), consisting of 15 calcineurin B-like proteins (CBLs), 4 calcium channels, 15 calmodulins (CAMs), and 11 calcium-dependent protein kinases (CDPKs). Moreover, a variety of genes associated with the PCD execution were enriched with two categories: protease and autophagy. The identified protease genes consist of CYSTEINE PROTEINASE RD21A, metacaspase (MC) genes MC2 and MC6, ASPARTIC PROTEASE, SENESCENCE-ASSOCIATED PROTEIN, VACUOLAR PROCESSING ENZYME (VPE), serine-type Clp protease CLPP, ENDOGLUCANASEs 17 and 23-like, PECTINESTERASEs 8 and QUARTET 1 (QRT1), XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASEs 7 and 9, exosome complex component RRP45, and Werner Syndrome-like exonuclease WRNEXO. On the other hand, autophagy-related genes comprise AUTOPHAGY-RELATED PROTEINs 5 (ATG5), 8C and 16–1, syntaxin-related protein KNOLLE, protein-transport protein SEC61B, vesicle-transport protein SEC22, calnexin CNX1, calreticulin family protein CALR and CALR3. Additionally, some genes within the longevity regulating pathway were found out, including heat shock 70 kDa protein 1/2/6/8 (HSC70–1), Cu-Zn superoxide dismutase (CSD1), Fe-Mn superoxide dismutase (FSD1 and MSD1), catalase CAT, molecular chaperone DnaK, and chaperonin GroEL.
To further investigate regulatory pathways of the nucellar PCD, a protein-protein interaction network was constructed for some significantly DEGs, based on their homologs from A. thaliana (Fig. 7). Within the resultant network, there presented two discrete pathways, albeit overlapping at some nodes. One is the MPK6-EIN3 network, containing some members involved in “Plant hormone signal transduction” (ko04075) pathway (Additional file 8: Figure S2). Noticeable, MPK6 has a strong interaction with RD21A and GAMMA-VPE. Another type of VPE, ALPHA-VPE, forms a protein-protein interaction network with various types of metacaspases (Fig. 7). And EIN3 has a direct interaction with NAP, a transcription factor of the NAC family involved in senescence. The other is the CYTC-1/HSC70–1 network. Within this network, both CYTC-1 and HSC70–1 could interact with a number of CAMs, as well as CAT, FSD1 and MSD1 (Fig. 7). In addition, CNX1, a calcium-binding protein, has interaction with AT5G60460 protein (a transport protein SEC61B), and CAM1 has interaction with NTL9 (a NAC transcription factor). Moreover, four autophagy-related proteins (ATG8D, ATG13, ATG18B, and AT3G18770) show cross-over interactions with each other. And pectinesterase QRT1, has interaction with AT1G09575 protein, a mitochondrial inner membrane calcium uniporter (Fig. 7).
Fig. 7.
The protein-protein interaction network of the significantly DEGs involved in the nucellar PCD. The nodes represent target proteins shown by their gene names, respectively. The color band is illustrated from red to yellow in descending order of degree values. Edge size is mapped according to its corresponding parameter EdgeBetweenness with lower value to smaller size
Expression profile analysis by RT-qPCR
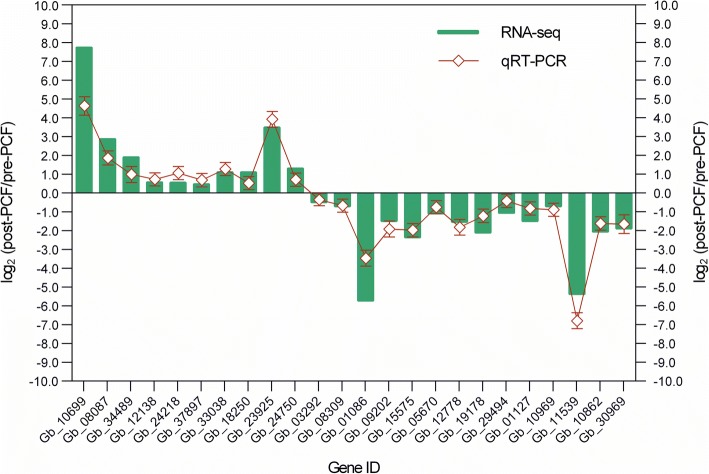
10 of the up-regulated and 14 of the down-regulated DEGs were randomly selected for real-time quantitative PCR (RT-qPCR), to validate their expression levels during the process of nucellar PCD during pollen chamber formation in G. biloba ovules. Based on RT-qPCR results, it was found that the transcriptional levels of the tested genes were in a correlated trend with the respective abundance estimated by RNA-seq (Fig. 8), suggesting a relative rationality and accuracy of the transcriptome analysis in the present study.
Fig. 8.
Expression level validation of DEG using qRT-PCR, in comparation to corresponding data detected in RNA-Seq. Relative expression ratio of each DEG is presented in a log2 value of post-PCF vs. pre-PCF libraries. The values are mean ± SE (n = 3)
Subcellular localization of calcium within Nucellar cells
Using in situ cytochemical method, dynamic changes in calcium concentration were visualized for the nucellar cells involved in pollen chamber formation (Fig. 9). At the early stage of ovule development, Ca2+ precipitation was aggregated in both of vacuoles and nucleus, in contrast to a scarce precipitation in the cytoplasm (Fig. 9a). Along with the developmental stage of ovule, an increased Ca2+ precipitation was found to be distributed in the elongated nucellar cell (Fig. 9c, d, f and g). In addition to their distribution in the vacuole (Fig. 9c and d), a large number of Ca2+ particles appeared in the cytoplasm (Fig. 9f and g). Some subcellular structures, such as endoplasmic reticula, were enclosed into the vacuole, showing a deformed shape and likely in a state of hydrolysis (Fig. 9g). For the negative control of corresponding nucellar cells, there were no visible Ca2+ particles (Fig. 9b and e). The amounts of detectable Ca2+ particles were significantly decreased in the dying nucellar cells. Within these nucellar cell, whose vacuole has been collapsed, few of Ca2+ particles were located along the cytoplasm debris and there presents no detectable aggregation of Ca2+ in the nucleus (Fig. 9h).
Fig. 9.
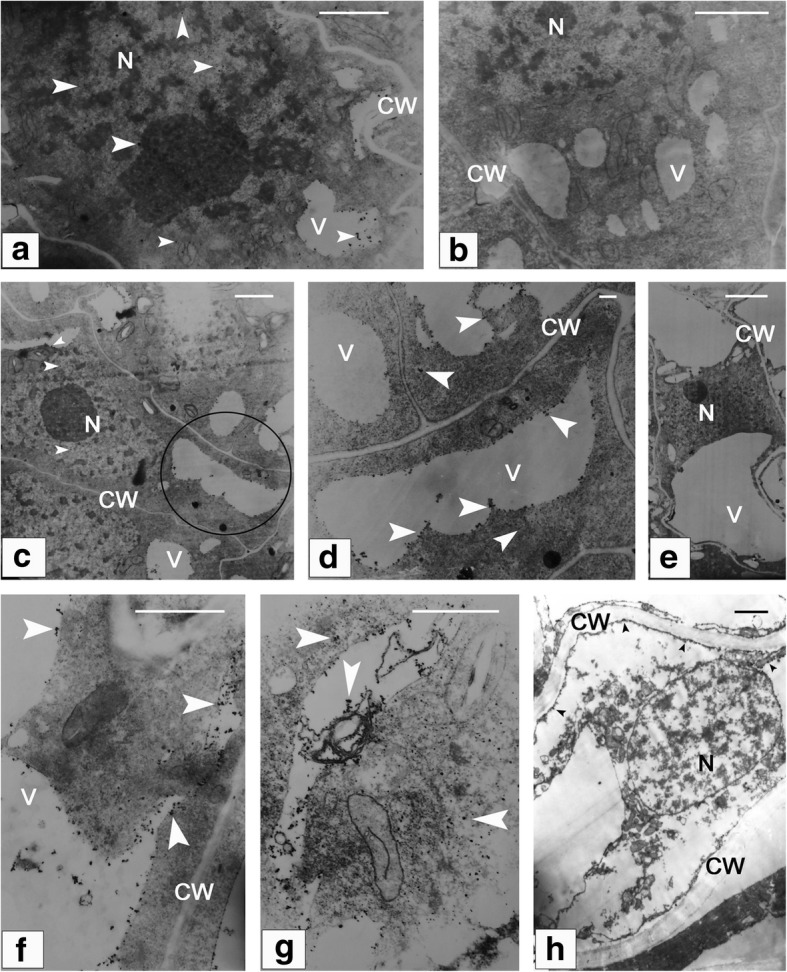
Subcellular localization of calcium within the nucellar cells involved in pollen chamber formation. (a) At the early stage of ovule development, a relatively higher Ca2+ precipitation is found in both of vacuoles and nucleus, compared with that in the cytoplasm. (b) A negative control of (a) without Ca2+ particles. (c), (d), (f), and (g) The nucellar cells at the micropylar end become elongated in shape, and distribution of Ca2+ precipitation is found to be increased in these nucellar cells. (d) Magnified view of the circled area in (c) shows numerous Ca2+ particles within the vacuole and cytoplasm. (e) A negative control of (c) without Ca2+ particles. (g) Deformed endoplasmic reticula are enclosed within the vacuole. (h) Few of Ca2+ particles are distributed along the cytoplasm debris, with no detectable aggregation of Ca2+ in the nucleus of the dying nucellar cell. Arrows indicate Ca2+ particles. Bars in (a), (b), (c), (e), and (h) = 2 μm, and in (d), (f), and (g) = 0.2 μm. CW, cell wall; N, nucleus; V, vacuole
Discussion
During the reproductive development, female G. biloba tree produces ovules, within which megaspores and female gametophyte are to be established for fertilization. Prior to the generation of female gametophyte, pollen grains have been pollinated onto the G. biloba ovules and stored in a cavity, namely pollen chamber, for around four months until completion of fertilization. Formation of pollen chamber is a natural process through the cell death of the specific nucellus at the early development of G. biloba ovules. Nucellar degeneration has been a developmental event widely existed in both gymnosperms and angiosperms [9]. Accumulated evidences have suggested that nucellar degeneration occurs by means of a developmental PCD [33, 36–38]. In the previous research, PCF concomitantly with the death of specific nucellar cells has been described as a timely and spatially regulated PCD process in G. biloba ovules [2–4]. However, little has been known about the molecular network controlling the nucellar PCD. Intensive genetic and genomic studies during the past decade have led to major advances in our understanding of animal and plant PCD at the molecular level. Base on the recent advances in regulatory mechanisms underlying diverse types of PCD, a comprehensive identification of DEGs and modulating pathways related to the nucellar PCD during PCF development was profiled using transcriptomic data via comparation between G. biloba ovules at the stages of pre- and post-PCF.
Role of ethylene signaling pathway in nucellar PCD during PCF in G. biloba ovules
Phytohormone signaling has been thought as the common mechanism underlying upstream regulation of various dPCD for occurrence in a precise spatial and temporal way [8], including ethylene in aerenchyma formation and leaf senescence [17–19]; GA in tapetum PCD; auxin, cytokinin and BR in TE PCD [21, 22], respectively. In this study, KEGG-based analysis on the pathway enrichment of the DEGs showed that the pathway “Plant hormone signal transduction” (ko04075) was significantly enriched, in addition to those concerning the basic and secondary metabolisms. Noticeably, transcriptional expression of some key components within the ethylene signaling pathway, including MPK6 and EIN3, were found to be significantly differential with their expression down-regulated in the post-PCF ovules compared to those in the pre-PCF ones. EIN3 has been shown to be involved in the trifurcate feed-forward pathway of age-dependent senescence and cell death [18]. According to the model for the EIN2–EIN3–NAC TFs regulatory cascade in the control of leaf senescence-associated PCD, EIN3 directly induces the expression of two key positive regulators of leaf senescence, i.e., NAC transcription factors ORE1 and AtNAP. Simultaneously, EIN3 directly suppresses the expression of miR164, which negatively regulates ORE1 at the post-transcriptional level [18, 19]. By analogy, the upregulated expression of EIN3 in the pre-PCF ovules were likely to promote the nucellar PCD in this study. These results indicated that ethylene signaling pathway was activated for transcriptional regulation of the downstream targets at the stage of nucellar PCD preparation in G. biloba ovules. To investigate the ethylene signal involved in the PCF process, ethylene contents from the pre- and post-PCF ovules were assayed, respectively (Additional file 6: Fig. S1). It was found out that ethylene contents from the pre-PCF ovules were significantly higher than those in the post-PCF ones. And genes encoding for ACS and ACO, which play key roles in promoting ethylene biosynthesis, were significantly differentially expressed with a trend in accordance with that in ethylene contents (Additional file 5: Table S5 and Additional file 6: Figure S1), suggesting the molecular cues for the activated ethylene signaling pathway. With regard to the roles of other phytohormone, it is proposed that they should function as integrated regulators for developmental processes of whole ovules, such as cell division, elongation, and enlargement, etc., based on the pathway “Plant hormone signal transduction” in KEGG database. A set of differentially expressed genes within various types of hormone signaling, has been demonstrated during the nucellar PCD in G. biloba ovules (Table 3 and Additional file 4: Table S4), indicating that there might existed an interaction effect of ethylene and other hormones on the nucellar PCD. During the process of aleurone cell death, it has been reported that GA and ABA are in an antagonistic way to regulate its degeneration [8]. The antagonistic effects have also been suggested during leaf senescence and xylogenesis, respectively [20, 22]. For instance, SA, ABA, JA and ethylene promote leaf senescence, while cytokinin, GA, and auxin delay its senescence [20]. As showed in Fig. 6, gens within ethylene signaling, such as ETR (Gb_36440), EIN3 (Gb_03292 and Gb_08309), and MPK6 (Gb_26499), were downregulated in their expression levels compared between post-PCF and pre-PCF. On the contrary, some genes involved in auxin pathway, such as AUX1 (Gb_14852 and Gb_28004), SAUR-like auxin-responsive protein (Gb_02944, Gb_12164, Gb_12165, Gb_15664, Gb_16068, and Gb_20563), Auxin-responsive protein (Gb_16134, Gb_22660, Gb_36672, and Gb_32404) or in cytokinin pathway, like CRE1 (Gb_12720 and Gb_13091), and two-component response regulator ARR-B family (Gb_15884, Gb_02800, and Gb_10394) were upregulated. Therefore, it is feasible that nucellar PCD process in G. biloba ovules should mainly be controlled by ethylene, antagonistically interacting with other acting hormones (auxin and cytokinin).
Initiation and execution in nucellar PCD during PCF in G. biloba ovules
Various key TFs have been described as bridges linking hormone signaling with PCD control [7, 8]. In the present study, several DEGs encoding for homologs of the specific TFs associated with particular PCD have been identified, including one MYB, two MADS-boxes, six bHLHs, and thirteen NACs. All of these TF genes have exhibited a similarity in the expression trend with significantly higher levels in the pre-PCF than those in the post-PCF. Homologs of MYB [39] and bHLH [35] have been known for involvement in promoting tapetum PCD, respectively. MADS-box was found to promote nucellar PCD during rice seed development [40]. And some NAC TFs function downstream of ethylene signaling pathway to modulate the cascade of leaf senescence-associated PCD [41]. Altogether, these results suggested that downstream of hormone signaling, several TFs should play important roles in ensuring nucellar PCD initiation and execution during PCF in G. biloba ovules.
Changes in cytoplasmic levels of calcium and cytochrome c have been proposed as parts of dPCD signaling network [42, 43]. During apoptosis in animals, once released from the mitochondria, cytochrome c associates with cytoplasmic Apaf1 to form the apoptosome, a large complex that processes initiator caspases [44]. Several plant PCD cases, such as the floral organ senescence in petunia and the embryonic suspensor cell death in runner bean (Phaseolus coccineus), are accompanied by release of mitochondrial cytochrome c [37, 42]. In the present case of nucellar PCD, the transcript amounts of genes encoding for cytochrome c and calmodulin were significantly higher in the pre-PCF ovules compared to those in the post-PCF ones. And within the constructed protein-protein interaction network, CYTC-1 showed a multiple interaction with various CAMs (Fig. 7). The activities of a large number of enzymes and other proteins are under the control of CAM-Ca2+ complex. Ca2+ signaling has been documented to be associated with various PCD cases, including regulating activities of Ca2+-dependent endonucleases and hydrolytic enzymes [8, 45–47]. Cytoplasmic Ca2+-influx from vacuoles and endoplasmic reticulum may be an early event in the PCD regulation pathway [48]. In the present research, 45 DEGs associated to calcium signaling were identified, including CBLs for calcium sensors, MCU for calcium channel, CAMs and CDPKs for calcium-binding proteins. Although some of them were not significant DGEs, these genes were actively expressed and thus were functional within calcium signaling pathway (Additional file 5: Table S5 and Additional file 7: Table S6). With regard to calcium-binding protein CAM, its upregulated expression, along with the differential expression of other calcium sensors and MCU for calcium uniporter protein, suggests that cascade events are likely to contribute to the calcium signaling. Moreover, elevated calcium levels been observed in the nucellar cells undergoing PCD in G. biloba ovules (Fig. 9). Therefore, it is reasonable to infer that cytochrome c and calcium should be related to modulating the nucellar PCD initiation in G. biloba ovules. Despite both the dynamic changes in calcium influx and significantly DEGs associated to Ca2+ signaling, including CAM, CBL, and CDPK, were identified during the nucellar PCD in G. biloba ovules, downstream cascades within Ca2+ regulation pathway in the PCD process are still unknown. A Ca2+/Mg2+-dependent nuclease was reported to be involved in both wheat aleurone and nucellus cells undergoing PCD [47]. The significantly DEGs encoding for various proteinases have been showed in the present study. Whether their biological functions or activities in the nucellus PCD are proceed through the Ca2+-mediated regulatory network, provides a point for future research.
During the PCD execution in this study, the significantly DEGs encoding for a variety of proteases have been identified, including cysteine proteinases RD21A, MC2 and MC6, ASPARTIC PROTEASE, VPE, SENESCENCE-ASSOCIATED PROTEIN, ENDOGLUCANASEs 17 and 23-like, PECTINESTERASEs 8 and QRT1, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASEs 7 and 9, RRP45 and WRNEXO. In addition to mediating ethylene signaling, MPK6 could strongly interact with RD21A and GAMMA-VPE (Fig. 7 and Additional file 8: Figure S2), both of which are members of cysteine proteinase, and thought to be associated with several types of cell death. And ALPHA-VPE has interaction with various types of MCs mediated by a NAC TF (Fig. 7). These results suggest that there exist regulatory networks among these components. The identified MCs, VPE, and other proteases, might function as their homologs known to have a principal role in proceeding to degrade many essential cellular targets [26, 31, 32, 34, 49]. Endoglucanases, pectinesterases, and xyloglucan endotransglucosylase/hydrolases, might be associated with recycling of carbohydrates in dying cells. And homologs of exosome complex component RRP45 and Werner Syndrome-like exonuclease WRNexo are likely to participate in DNA degradation [50].
Apart from the effects of proteases, autophagy occurs concomitantly with the execution of nucellar PCD in G. biloba ovules. Autophagy is a highly regulated process during which cytoplasmic materials are enclosed in double-membrane-bound vesicles that are then targeted to the vacuole or lysosome for degradation [51]. Unlike those of animals, vacuoles participate in the autolysis process of plant PCD [45, 52]. In the previous research on nucellar PCD in G. biloba ovules, ultrastructural observation clearly showed that some double-membrane bodies were being engulfed in the vacuole, as well as a convoluted membrane structure fusing with the vacuole [3]. In accordance with the observation, autophagy-related genes, including AUTOPHAGY-RELATED PROTEINs 5, 8C and 16–1, KNOLLE, SEC61B, SEC22, CNX1, CALR and CALR3, are found to be differentially expressed (Additional file 1: Table S1).
Conclusions
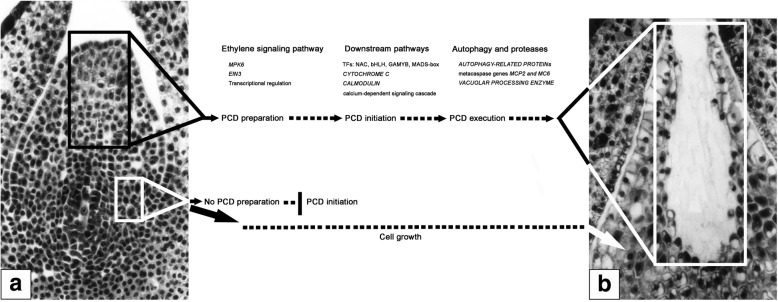
Transcriptomic profiling unravels the DEGs and modulating pathways with respective to the nucellar PCD during pollen chamber formation in G. biloba ovules. Based on these results, a putative working model, consisting of three overlapping processes, is proposed (Fig. 10): at the stage of PCD preparation, ethylene signaling pathway is activated for transcriptional regulation of the downstream targets; subsequently, at the stage of PCD initiation, the upregulated expression of several TFs, i.e., NAC, bHLH, MADS-box, and MYB, further promotes the corresponding transcript levels of CYTOCHROME C and CAMs, thereby, leads to the PCD initiation via the calcium-dependent signaling cascade; finally, at the stage of PCD execution, some proteases like MCs and VPE for hydrolysis, together with the process of autophagy, play roles in the clearance of cellular components. Afterwards, a pollen chamber is generated from the removal of specific nucellar cells in the developing ovule.
Fig. 10.
Tentative model for the nucellar PCD during pollen chamber formation in G. biloba ovules. Ovules at the stage of pre-PCF (a) and post-PCF (b) are sectioned longitudinally. The black-boxed area in (a) contains nucellar cells at the micropylar end of ovule, which are to undergo PCD to generated a pollen chamber, bounded by the white-box in (b). Other nucellar cells, represented by a white-boxed area in (a), keep growth throughout the stages of PCF
Methods
Plant materials
The ovules were collected from the same cultivar, Ginkgo biloba ‘Foshou’, at the campus of Anhui Agricultural University, Hefei, China. Sampling were performed every two days from 17th of March to 15th of April in 2016, respectively. The progressive development of pollen chamber in ovules were determined by micro-sections prepared in Epon 812. According to the ovule sections (Fig. 1 and Additional file 9: Figure S3), nucellar cells in the collected ovules on the 17th of March, were uniform in a rounded morphology and no occurrence of longitudinally elongation of nucellar cells (Fig. 1a and Additional file 9: Figure S3a and S3b), whereas nucellar cells at the micropylar end in those collected on the 1st of April, have been removed and a cavity of individual pollen chamber has been generated (Fig. 1c and Additional file 9: Figure S3e). Therefore, the ovules (collected on the 17th of March) at the stage of Fig. 1a (pre-PCF) and ones (on the 1st of April) at the stage of Fig. 1a (post-PCF), with three biological replicates, were used as samples for RNA-seq in this study (Fig. 11). In order to collect the right stage sample without microscope, the petiole length of megasporophyll where G. biloba ovules are situated were measured. Subsequently, a correlationship between the petiole length and the developmental stage of ovules was used as the criterion for sampling respective ovules. As showed in Additional file 9: Figure S3, ovules at pre-PCF, PCF in progress (Additional file 9: Fig. S3c and S3d), and post-PCF, had 0.60 ± 0.11 cm, 1.40 ± 0.16 cm, and 2.10 ± 0.17 cm on average in petiole length of megasporophyll, respectively. The climate data from March 10th to April 15th in 2016 were collected in Additional file 10: Table S7. As referred to the Additional file 10: Table S7, there is no occurrence of adverse weather condition during the period of sampling G. biloba ovules.
Fig. 11.
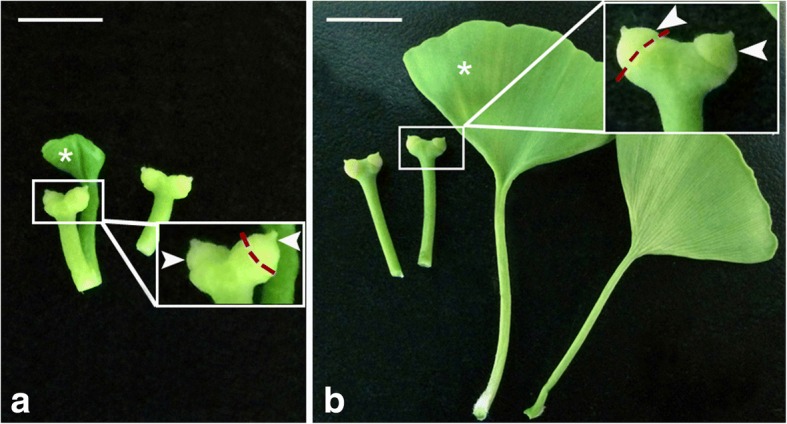
Ovule collection from G. biloba strobili at the stage of pre-PCF (a) or post-PCF (b). Magnified view of the boxed area shows ovules (arrowheads) collected from megasporophyll, and red-dash lines marking the junction region between ovules and megasporophyll. G. biloba leaves (asterisks) are positioned along with ovules. Bars = 1 cm
DGE library construction and Illumina sequencing
After grinding into powder in liquid nitrogen, ovules from each sample were applied to total RNA extraction, followed by isolating mRNAs. The individual cDNA was synthesized using the random hexamers as primers and mRNA templates. The resultant products were connected with adapters, followed by size selection and PCR amplification. The constructed library was analyzed by Illumina HiSeq™ 4000 sequencing platform (BGI, Shenzhen, China). The data of RNA-Seq have been assigned with the accession number SRP158368 in NCBI (http://www.ncbi.nlm.nih.gov/sra).
Analysis of differentially expressed gene and Functional annotation
Adaptor sequences and low-quality reads were initially filtered from the raw data. Then the remaining ones, called as clean reads, were aligned to the reference genome of G. biloba [53], using HISAT2 program [54]. Subsequently, unigene expression was calculated as the FPKM (fragments per kilo bases of exons for per million mapped reads) with the software package Cufflinks [55]. For both the pre-PCF and post-PCF libraries, the FPKM values were applied to measuring the log2 ratios (post-PCF/pre-PCF). And DEGs were filtered by a FDR (false discovery rate) value ≤0.05. Gene annotation were performed throughout blasting against the local Nr database (downloading from http://ftp-private.ncbi.nlm.nih.gov). Both annotations of GO (http://www.geneontology.org/) and KEGG (http://www.genome.jp/kegg/) were carried out to identify functional genes. Subsequently, functional enrichment of GO and KEGG pathway were analyzed using R package PHYPER, respectively. The terms were considered to be significantly enriched if FDR ≤ 0.001.
Assay for ethylene content in G. biloba ovules
G. biloba ovules (0.25 g) were collected as mentioned above. Ethylene contents from the pre- and post-PCF ovules were assayed on a Varian 3800 gas chromatograph (Walnut Creek, CA, USA), using the previous method by Wang et al. [56]. The method validation followed the control procedures for the prepared ethylene concentration (10 μL/L).
Construction and analysis of protein-protein interaction network
Protein-protein interactions were searched through the STRING (https://string-db.org/cgi). The proteins, putatively encoded by the significantly DEGs involved in the nucellar PCD, were mapped to the network according to information of their respective homologs from Arabidopsis thaliana. The protein interaction network was visualized using the software Cytoscape [57].
qRT-PCR analysis
qRT-PCR was performed using CFX96 Touch™ System (BIO-RAD, USA), with the parameters: 96 °C for 2 min, followed by cycling for 30 rounds (96 °C for 10 s, 58 °C for 10 s, and 68 °C for 30 s). To calculate the expression abundances of target genes, the method 2-ΔΔCt for statistics was applied with three biological replicates [58], and the G. biloba gene GAPDH was taken as an internal reference. All primer pairs for these qRT-PCR were deposited in Additional file 11: Table S8.
Microscope observation and calcium-Cytochemical localization
The sections (8 μm in thickness) of the collected G. biloba ovules were prepared using paraffin-cutting and stained with safranine O for light microscope observation. Calcium-cytochemical localization in G. biloba nucellar cells was visualized by producing precipitation with potassium pyroantimonate, according to the previous method [4]. Observation and photographs were taken under JEM-100CX transmission electron microscope.
Additional files
Table S1. All the identified DEGs between pre-PCF and post-PCF libraries. (XLSX 1565 kb)
Table S2. GO enrichment analysis of differentially expressed genes in G. biloba ovules. (XLSX 196 kb)
Table S3. KEGG pathway analysis of differentially expressed genes in G. biloba ovules. (XLSX 33 kb)
Table S4. The DEGs involved in plant hormone signal transduction pathway. (XLSX 20 kb)
Table S5.The significantly DEGs involved in the nucellar PCD during pollen chamber formation in G. biloba ovules. (XLSX 39 kb)
Figure S1. Transcriptional expression levels of the genes involved in ethylene biosynthesis (upper panel) and ethylene contents (lower panel) in pre- and post-PCF G. biloba ovules. Abundance of gene transcripts was presented by FPKM value resulted from the DEGs analysis in this study. Ethylene contents were normalized as nmol per gram fresh weight (Fw). (TIF 141 kb)
Table S6. The DEGs associated to calcium signaling in G. biloba ovules. (DOCX 16 kb)
Figure S2. The identified DEGs involved in plant hormone signal transduction by KEGG enrichment. (TIF 446 kb)
Figure S3. A correlationship between the petiole length of megasporophyll and the developmental stage of ovules. Ovules at the developmental stages of pre-PCF (a and b), PCF in progress (c and d), and post-PCF (e) were determined by micro-sections from the representative samples and observed under microscope. The petiole length was a mean value of 10 megasporophylls randomly selected from one set of ovule samples, prior to the preparation for their ovule sections. Bars = 250 μm. Abbreviations: I, integument; M, micropyle; Nu, nucellus. (TIF 527 kb)
Table S7. The climate data from March 10th to April 15th in 2016. (XLSX 28 kb)
Table S8. The primers of DEGs for RT-qPCR in G. biloba ovules. (XLSX 24 kb)
Acknowledgements
We would like to thank Dr. Guomin Han at Anhui Agricultural University for his helpful assistance during the transcriptomic analysis on this research.
Funding
The research was supported by the grants from the Key University Science Research Project of Anhui Province, China (No. KJ2016A225), the Natural Science Foundation of Anhui Province, China (No. 1808085MC58), the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry, China (No. 2015–1098), and the Provincial Quality Engineer Fund of Anhui Education Department, China (No. 2015GXK015).
Availability of data and materials
RNA-seq data generated is available with the SRA accession SRP158368 in NCBI (http://www.ncbi.nlm.nih.gov/sra).
Abbreviations
- ATGs
autophagy-related proteins
- CAMs
calmodulins
- DEGs
differentially expressed genes
- EBF1/2
EIN3-binding F-box protein
- EIN3
ethylene-insensitive protein 3
- ETR
ethylene receptor
- GO
Gene Ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MCs
metacaspases
- MPK6
mitogen-activated protein kinase 6
- PCD
programmed cell death
- PCF
pollen chamber formation
- VPE
vacuolar processing enzyme
Authors’ contributions
DHL and YPC designed the research project; DHL, DW, SZL, NG, JSG and XS carried out the experimental work; DHL and DW analyzed the data; DHL wrote the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Dahui Li, Email: dahui2@126.com.
Ning Guo, Email: guoning@ahau.edu.cn.
Junshan Gao, Email: gaojsh@ahau.edu.cn.
Yongping Cai, Email: swkx12@ahau.edu.cn.
References
- 1.Lee CL. Fertilization in Ginkgo biloba. Bot Gaz. 1955;117:79–100. doi: 10.1086/335894. [DOI] [Google Scholar]
- 2.Li DH, Yang X, Cui X, Cui KM, Li ZL. Early development of pollen chamber in Ginkgo biloba ovule. Acta Bot Sin. 2002;44:757–763. [Google Scholar]
- 3.Li DH, Yang X, Cui KM, Li ZL. Morphological changes in nucellar cells undergoing programmed cell death (PCD) during pollen chamber formation in Ginkgo biloba. Acta Bot Sin. 2003;45:53–63. [Google Scholar]
- 4.Li DH, Yang X, Cui KM. Formation of archegonium chamber is associated with nucellar-cell programmed cell death in Ginkgo biloba. Protoplasma. 2007;231:173–181. doi: 10.1007/s00709-007-0257-8. [DOI] [PubMed] [Google Scholar]
- 5.Greenberg JT. Programmed cell death: a way of life for plants. PNAS. 1996;93:12094–12097. doi: 10.1073/pnas.93.22.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pennel RI, Lamb C. Programmed cell death in plants. Plant Cell. 1997;9:1157–1168. doi: 10.1105/tpc.9.7.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huysmans M, Lema AS, Coll NS, Nowack MK. Dying two deaths - programmed cell death regulation in development and disease. Curr Opin Plant Biol. 2017;35:37–44. doi: 10.1016/j.pbi.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 8.Van Hautegem T, Waters AJ, Goodrich J, Nowack MK. Only in dying, life: programmed cell death during plant development. Trends Plant Sci. 2015;20:102–113. doi: 10.1016/j.tplants.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Daneva A, Gao Z, Van Durme M, Nowack MK. Functions and regulation of programmed cell death in plant development. Annu Rev Cell Dev Biol. 2016;32:441–468. doi: 10.1146/annurev-cellbio-111315-124915. [DOI] [PubMed] [Google Scholar]
- 10.Kabbage M, Kessens R, Bartholomay LC, Williams B. The life and death of a plant cell. Annu Rev Plant Biol. 2017;68:375–404. doi: 10.1146/annurev-arplant-043015-111655. [DOI] [PubMed] [Google Scholar]
- 11.Coll NS, Epple P, Dangl JL. Programmed cell death in the plant immune system. Cell Death Differ. 2011;18:1247–1256. doi: 10.1038/cdd.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gadjev I, Stone JM, Gechev TS. Programmed cell death in plants: new insights into redox regulation and the role of hydrogen peroxide. Int Rev Cell Mol Biol. 2008;270:87–144. doi: 10.1016/S1937-6448(08)01403-2. [DOI] [PubMed] [Google Scholar]
- 13.Petrov V, Hille J, Mueller-Roeber B, Gechev TS. ROS-mediated abiotic stress-induced programmed cell death in plants. Front Plant Sci. 2015;6:69. doi: 10.3389/fpls.2015.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Doorn WG. Classes of programmed cell death in plants, compared to those in animals. J Exp Bot. 2011;62:4749–4761. doi: 10.1093/jxb/err196. [DOI] [PubMed] [Google Scholar]
- 15.Gunawardena A, Pearce DM, Jackson MB, Hawes CR, Evans DE. Characterisation of programmed cell death during aerenchyma formation induced by ethylene or hypoxia in roots of maize (Zea mays L.) Planta. 2001;212:205–214. doi: 10.1007/s004250000381. [DOI] [PubMed] [Google Scholar]
- 16.Völz R, Heydlauff J, Ripper D, von Lyncker L, Groß-Hardt R. Ethylene signaling is required for synergid degeneration and the establishment of a pollen tube block. Dev Cell. 2013;25:310–316. doi: 10.1016/j.devcel.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 17.Yamauchi T, Watanabe K, Fukazawa A, Mori H, Abe F, Kawaguchi K, Oyanagi A, Nakazono M. Ethylene and reactive oxygen species are involved in root aerenchyma formation and adaptation of wheat seedlings to oxygen-deficient conditions. J Exp Bot. 2014;65:261–273. doi: 10.1093/jxb/ert371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Z, Peng J, Wen X, Guo H. Ethylene-insensitive3 is a senescence-associated gene that accelerates age-dependent leaf senescence by directly repressing miR164 transcription in Arabidopsis. Plant Cell. 2013;25:3311–3328. doi: 10.1105/tpc.113.113340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim HJ, Hong SH, Kim YW. Gene regulatory cascade of senescence-associated NAC transcription factors activated by ETHYLENE-INSENSITIVE2-mediated leaf senescence signalling in Arabidopsis. J Exp Bot. 2014;65:4023–4036. doi: 10.1093/jxb/eru112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarwat M, Naqvi AR, Ahmad P. Phytohormones and microRNAs as sensors and regulators of leaf senescence: assigning macro roles to small molecules. Biotechnol Adv. 2013;31:1153–1171. doi: 10.1016/j.biotechadv.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 21.Plackett AR, Thomas SG, Wilson ZA, Hedden P. Gibberellin control of stamen development: a fertile field. Trends Plant Sci. 2011;16:568–578. doi: 10.1016/j.tplants.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Milhinhos A, Miguel CM. Hormone interactions in xylem development: a matter of signals. Plant Cell Rep. 2013;32:867–883. doi: 10.1007/s00299-013-1420-7. [DOI] [PubMed] [Google Scholar]
- 23.Fendrych M, Van Hautegem T, Van Durme M, Olvera-Carrillo Y, Huysmans M, Karimi M, Lippens S, Guerin CJ, Krebs M, Schumacher K, Nowack MK. Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr Biol. 2014;24:931–940. doi: 10.1016/j.cub.2014.03.025. [DOI] [PubMed] [Google Scholar]
- 24.Bosch M, Poulter NS, Vatovec S, Franklin-Tong VE. Initiation of programmed cell death in selfincompatibility: role for cytoskeleton modifications and several caspase-like activities. Mol Plant. 2008;1:879–887. doi: 10.1093/mp/ssn053. [DOI] [PubMed] [Google Scholar]
- 25.Farage-Barhom S, Burd S, Sonego L, Perl-Treves R, Lers A. Expression analysis of the BFN1 nuclease gene promoter during senescence, abscission, and programmed cell death-related processes. J Exp Bot. 2008;59:3247–3258. doi: 10.1093/jxb/ern176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Pfeil JE, Gal S. The three typical aspartic proteinase genes of Arabidopsis thaliana are differentially expressed. Eur J Biochem. 2002;269:4675–4684. doi: 10.1046/j.1432-1033.2002.03168.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhang D, Liu D, Lv X, Wang Y, Xun Z, Liu Z, Li F, Lu H. The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell. 2014;26:2939–2961. doi: 10.1105/tpc.114.127282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Escamez S, Tuominen H. Programmes of cell death and autolysis in tracheary elements: when a suicidal cell arranges its own corpse removal. J Exp Bot. 2014;65:1313–1321. doi: 10.1093/jxb/eru057. [DOI] [PubMed] [Google Scholar]
- 29.Lam E, Zhang Y. Regulating the reapers: activating metacaspases for programmed cell death. Trends Plant Sci. 2012;17:487–494. doi: 10.1016/j.tplants.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 30.Sundström JF, Vaculova A, Smertenko AP, Savenkov EI, Golovko A, Minina E, Tiwari BS, Rodriguez-Nieto S, Zamyatnin AA Jr, Välineva T, Saarikettu J, Frilander MJ, Suarez MF, Zavialov A, Ståhl U, Hussey PJ, Silvennoinen O, Sundberg E, Zhivotovsky B, Bozhkov PV. Tudor staphylococcal nuclease is an evolutionarily conserved component of the programmed cell death degradome. Nat Cell Biol 2009;11:1347–1354. [DOI] [PubMed]
- 31.Coll NS, Smidler A, Puigvert M, Popa C, Valls M, Dangl JL. The plant metacaspase AtMC1 in pathogen triggered programmed cell death and aging: functional linkage with autophagy. Cell Death Differ. 2014;21:1399–1408. doi: 10.1038/cdd.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Escamez S, André D, Zhang B, Bollhöner B, Pesquet E, Tuominen H. METACASPASE9 modulates autophagy to confine cell death to the target cells during Arabidopsis vascular xylem differentiation. Biol Open. 2016;5:122–129. doi: 10.1242/bio.015529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linnestad C, Doan DNP, Brown RC, Lemmon BE, Meyer DJ, Jung R, Olsen O. Nucellain, a barley homolog of the dicot vacuolar-processing protease, is localized in nucellar cell walls. Plant Physiol. 1998;118:1169–1180. doi: 10.1104/pp.118.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakaune S, Yamada K, Kondo M, Kato T, Tabata S, Nishimura M. Hara-Nishimura I. A vacuolar processing enzyme, δVPE, is involved in seed coat formation at the early stage of seed development. Plant Cell. 2005;17:876–887. doi: 10.1105/tpc.104.026872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ko SS, Li MJ, Ku MSB, Ho YC, Lin YJ, Chuang MH, Hsing HX, Lien YC, Yang HT, Chang HC, Chan MT. The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant Cell. 2014;26:2486–2504. doi: 10.1105/tpc.114.126292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Domínguez F, Moreno J, Cejudo FJ. The nucellus degenerates by a process of programmed cell death during the early stages of wheat grain development. Planta. 2001;213:352–360. doi: 10.1007/s004250000517. [DOI] [PubMed] [Google Scholar]
- 37.Lombardi L, Casani S, Ceccarelli N, Galleschi L, Picciarelli P, Lorenzi R. Programmed cell death of the nucellus during Sechium edule Sw. Seed development is associated with activation of caspase-like proteases. J Exp Bot. 2007;58:2949–2958. doi: 10.1093/jxb/erm137. [DOI] [PubMed] [Google Scholar]
- 38.Radchuk V, Weier D, Radchuk R, Weschke W, Weber H. Development of maternal seed tissue in barley is mediated by regulated cell expansion and cell disintegration and coordinated with endosperm growth. J Exp Bot. 2011;62:1217–1227. doi: 10.1093/jxb/erq348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plackett AR, Ferguson AC, Powers SJ, Wanchoo-Kohli A, Phillips AL, Wilson ZA, Hedden P, Thomas SG. DELLA activity is required for successful pollen development in the Columbia ecotype of Arabidopsis. New Phytol. 2014;201:825–836. doi: 10.1111/nph.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yin LL, Xue HW. The MADS29 transcription factor regulates the degradation of the nucellus and the nucellar projection during rice seed development. Plant Cell. 2012;24:1049–1065. doi: 10.1105/tpc.111.094854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matallana-Ramirez LP, Rauf M, Farage-Barhom S, Dortay H, Xue GP, Dröge-Laser W, Lers A, Balazadeh S, Mueller-Roeber B. NAC transcription factor ORE1 and senescence-induced BIFUNCTIONAL NUCLEASE1 (BFN1) constitute a regulatory cascade in Arabidopsis. Mol Plant. 2013;6:1432–1452. doi: 10.1093/mp/sst012. [DOI] [PubMed] [Google Scholar]
- 42.Xu Y, Hanson MR. Programmed cell death during pollination-induced petal senescence in petunia. Plant Physiol. 2000;122:1323–1333. doi: 10.1104/pp.122.4.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Durme M, Nowack MK. Mechanisms of developmentally controlled cell death in plants. Curr Opin Plant Biol. 2016;29:29–37. doi: 10.1016/j.pbi.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 44.Zou H, Henzel WJ, Liu X, Lutschg A, Wang X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell. 1997;90:405–413. doi: 10.1016/S0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 45.Groover A, DeWitt N, Heidel A, Jones A. Programmed cell death of plant tracheary elements differentiating in vitro. Protoplasma. 1997;196:197–211. doi: 10.1007/BF01279568. [DOI] [Google Scholar]
- 46.Gupta PD, Pushkala K. Importance of the role of calcium in programmed cell death: a review. Cytobios. 1999;99:83–95. [PubMed] [Google Scholar]
- 47.Domínguez F, Cejudo FJ. Identification of a nuclear-localized nuclease from wheat cells undergoing programmed cell death that is able to trigger DNA fragmentation and apoptotic morphology on nuclei from human cells. Biochem J. 2006;397:529–536. doi: 10.1042/BJ20051809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nutt LK, Chandra J, McConkey DJ. Bax-mediated Ca2+ mobilization promotes cytochrome c release during apoptosis. J Biol Chem. 2002;277:20301–20308. doi: 10.1074/jbc.M201604200. [DOI] [PubMed] [Google Scholar]
- 49.Hara-Nishimura I, Hatsugai N, Nakaune S, Kuroyanagi M, Nishimura M. Vacuolar processing enzyme: an executor of plant cell death. Curr Opin Plant Biol. 2005;8:404–408. doi: 10.1016/j.pbi.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 50.Plchova H, Hartung F, Puchta H. Biochemical characterization of an exonuclease from Arabidopsis thaliana reveals similarities to the DNA exonuclease of the human Werner syndrome protein. J Biol Chem. 2003;278:44128–44138. doi: 10.1074/jbc.M303891200. [DOI] [PubMed] [Google Scholar]
- 51.Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 52.Obara K, Kuriyama H, Fukuda H. Direct evidence of active and rapid nuclear degradation triggered by vacuole rupture during programmed cell death in zinnia. Plant Physiol. 2001;125:615–626. doi: 10.1104/pp.125.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guan R, Zhao Y, Zhang H, Fan G, Liu X, Zhou W, Shi C, Wang J, Liu W, Liang X, Fu Y, Ma K, Zhao L, Zhang F, Lu Z, Lee SM, Xu X, Wang J, Yang H, Fu C, Ge S, Chen W. Draft genome of the living fossil Ginkgo biloba. GigaScience. 2016;5:49. doi: 10.1186/s13742-016-0154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang KLC, Yoshida H, Lurin C, Ecker JR. Regulation of ethylene gas biosynthesis by the Arabidopsis ET01 protein. Nature. 2004;428:945–950. doi: 10.1038/nature02516. [DOI] [PubMed] [Google Scholar]
- 57.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. All the identified DEGs between pre-PCF and post-PCF libraries. (XLSX 1565 kb)
Table S2. GO enrichment analysis of differentially expressed genes in G. biloba ovules. (XLSX 196 kb)
Table S3. KEGG pathway analysis of differentially expressed genes in G. biloba ovules. (XLSX 33 kb)
Table S4. The DEGs involved in plant hormone signal transduction pathway. (XLSX 20 kb)
Table S5.The significantly DEGs involved in the nucellar PCD during pollen chamber formation in G. biloba ovules. (XLSX 39 kb)
Figure S1. Transcriptional expression levels of the genes involved in ethylene biosynthesis (upper panel) and ethylene contents (lower panel) in pre- and post-PCF G. biloba ovules. Abundance of gene transcripts was presented by FPKM value resulted from the DEGs analysis in this study. Ethylene contents were normalized as nmol per gram fresh weight (Fw). (TIF 141 kb)
Table S6. The DEGs associated to calcium signaling in G. biloba ovules. (DOCX 16 kb)
Figure S2. The identified DEGs involved in plant hormone signal transduction by KEGG enrichment. (TIF 446 kb)
Figure S3. A correlationship between the petiole length of megasporophyll and the developmental stage of ovules. Ovules at the developmental stages of pre-PCF (a and b), PCF in progress (c and d), and post-PCF (e) were determined by micro-sections from the representative samples and observed under microscope. The petiole length was a mean value of 10 megasporophylls randomly selected from one set of ovule samples, prior to the preparation for their ovule sections. Bars = 250 μm. Abbreviations: I, integument; M, micropyle; Nu, nucellus. (TIF 527 kb)
Table S7. The climate data from March 10th to April 15th in 2016. (XLSX 28 kb)
Table S8. The primers of DEGs for RT-qPCR in G. biloba ovules. (XLSX 24 kb)
Data Availability Statement
RNA-seq data generated is available with the SRA accession SRP158368 in NCBI (http://www.ncbi.nlm.nih.gov/sra).