Abstract
Background
Pancreatic cancer (PC) remains one of the most aggressive cancers worldwide. However, genetic factors underlying PC susceptibility remain largely unclear. Long noncoding RNA (lncRNA) HOX transcript antisense RNA (HOTAIR) acts as an oncogene and its genetic variation has been linked to many cancers. However, the associations between genetic variants in HOTAIR gene and PC risk has not yet been reported.
Methods
A two-stage, case–control study was conducted to investigate the associations between HOTAIR SNPs and the PC risk. Dual luciferase reporter assay and real-time -PCR (RT-PCR) was conducted to evaluate the potential regulatory function of HOTAIR rs4759314 and rs200349340.
Results
We found the minor alleles of rs4759314 (OR = 1.76; 95 CI 1.37–2.25; P = 0.001) and rs200349340 (OR = 1.32; 95 CI 1.12–1.56; P = 0.001) were significantly associated with PC susceptibility. In functional experiments, we found subjects carrying the minor alleles of rs4759314 and rs200349340 had significantly higher HOTAIR RNA levels (mean ± SD) than those carrying the major alleles in PC tissues. For rs4759314, cells transfected with rs4759314 -G allele construct showed higher relative luciferase activity; while for rs200349340, cells transfected with rs200349340 -G allele construct showed more sensitive change of the relative luciferase activity.
Conclusion
Our studies revealed that functional SNP rs4759314 and rs200349340 of HOTAIR had strong associations with PC susceptibility. These findings elucidate that functional genetic variants influencing lncRNA expression may explain a portion of PC genetic basis.
Electronic supplementary material
The online version of this article (10.1186/s12935-019-0761-x) contains supplementary material, which is available to authorized users.
Keywords: Pancreatic cancer, Variation, HOTAIR
Background
Pancreatic cancer (PC) remains one of the most aggressive cancers with a 5-year survival rate of less than 5%, and conventional treatment offers no hope of cure and has little palliative value [1, 2]. It was estimated that 55,440 new PC cases and 44,330 new PC deaths would occur in United States [3]. In contrast to the overall declining trends for the 4 major cancers (Lung, breast, prostate, and colorectum), death rates rose during 2011 through 2015 for PC in men by 0.3% per year [3]. In China, 90.1 thousands new PC cases and 79.4 thousands new PC deaths were estimated per year, according to the report of the National Office for Cancer Prevention and Control of China [4]. Although genome-wide association studies (GWASs) have identified several loci associated with PC risk; most of genetic factors remain unknown [5–7]. Therefore, identification and characterization of new genetic variations in predicting PC risk are essential.
Long non-coding RNAs (lncRNAs) are an important class of genes involved in various biological processes [8–10]. Among them, lncRNA HOX transcript antisense RNA (HOTAIR) was involved in pathogenesis and progress of multiple tumors, including PC [11–19]. Previous studies showed that HOTAIR could induce genome-wide retargeting of Polycomb repressive complex 2 (PRC2), contribute to altered H3 lysine 27 methylation, and further regulate various downstream genes and promote cancer cell metastasis [20, 21]. Significant associations between overexpression of HOTAIR and susceptibility, metastasis and/or poor prognosis of several human cancers such as liver, breast, pancreas, colon, lung, laryngeal, nasopharyngeal, and esophageal cancers have been reported [14, 22–24]. Meanwhile, studies showed that HOTAIR was significantly up-regulated in PC tissues, compared with normal pancreatic tissues [19, 25]. Furthermore, increasing publications are drawing attention to the associations between common polymorphisms in HOTAIR and risk of cancers, although meta-analysis showed these results had been controversial and inconsistent [26]. Even with the potential importance of HOTAIR in carcinogenesis, little has been investigated about the functional significance of HOTAIR locus with PC susceptibility.
Here, we hypothesized that genetic variants in the HOTAIR gene might affect HOTAIR expression and/or its function, which in turn, might influence consequential risk of developing PC. In current study, we conducted a two-stage, case–control study to investigate the associations between HOTAIR genotypes and PC risk.
Patients and methods
Study subjects
Totally 900 PC cases were collected and pathologically confirmed, while 900 matched controls were recruited in the same hospital. All the samples were continuously collected since Jan 2005. In stage 1, the samples were collected before Sep 2010. While the samples in stage 2 were collected from Oct 2010 to Jun 2016. The inclusion and exclusion criteria for PC patients were: pathologically confirmed and without treatment of any medications, while the inclusion and exclusion criteria for control group were: (1) without cancer or other serious illness; (2) matched with the PC patients by age (± 5 years), gender, and area of residence (urban and rural). Face-to-face interviews were conducted by trained interviewers using a structured questionnaire relating to demographic characteristics and lifestyle-related factors. Meanwhile, 10 mL peripheral blood from each individual was collected and stored in sodium citrate anticoagulant tubes at − 80 °C for DNA isolation. The study was performed in accordance with Declaration of Helsinki, and approved by the hospital’s Ethics Committee. All the participants provided signed informed consent before the study.
SNP selection and genotyping
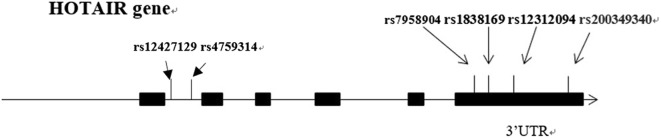
The tagSNPs covering the HOTAIR region were selected from 1000 genome CHB data (phase 3, minor allele frequency ≥ 5%, pairwise r2 ≥ 0.8) using the Haploview 4.2 software. We also considered the potential functional variants that were located in the binding site of miRNAs. Finally, six SNPs, including rs4759314, rs12427129, as well as four SNPs in 3′UTR of HOTAIR gene (rs200349340, rs12312094, rs1838169, and rs7958904), were finally selected as the candidates (Fig. 1). Genotyping was conducted using matrix assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) (the mass spectrum plots for three genotypes of rs200349340 were presented in Additional file 1: Figures S1–S3). Genotype calling was conducted using MassARRAY RT software version 3.0. Genotyping data was then analyzed by MassARRAY Typer software 4.0.3 (Sequenom, San Diego, CA, USA) in conjunction with Sequenom’s iPLEX™ Gold assay. Quality control was detected using Sanger sequencing of ~ 10% randomly selected samples, yielding a 100% concordance.
Fig. 1.
Relative location about the 6 candidate SNPs in HOTAIR gene
Real-time analyses of LncRNA HOTAIR
Total RNA was extracted from PC and adjacent normal tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Then cDNA was synthesized with M-MLV reverse transcriptase (Invitrogen). Real-time reverse transcription-polymerase chain reaction (RT-PCR) with SYBR Green assay (TaKaRa Biotechnology, Dalian, China) was performed to examine expression level of HOTAIR (forward-primer: GGTAGAAAAAGCAACCACGAAGC; reverse-primer: ACATAAACCTCTGTCTGTGAGTGCC). Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an internal control (forward-primer: CCGGGAAACTGTGGCGTGATGG; reverse-primer: AGGTGGAGGAGTGGGTGTCGCTGTT). The thermal cycling parameters were as follows: 95 °C for 5 min followed by 40 cycles of 95 °C for 30 s, 59 °C for 30 s, 72 °C for 30 s. The assay was conducted using the ABI 7300 system (Applied Biosystems). All reactions were done in triplicate and expression level of HOTAIR was calculated according to the equation 2−∆∆Ct.
Dual luciferase reporter assay
To evaluate the potential function of HOTAIR rs4759314 and rs200349340, two reporter constructs carrying two copies of rs4759314 -A allele/G allele, rs200349340 -T allele/G allele were created based on pGL3-plasmids (Fig. 2c, e presents the schematic line diagrams). First, about 200-bp DNA sequences centered at rs4759314 (A or G allele) and rs200349340 (T or G allele) were synthesized; second, 2 tandem copies of these sequences were cloned into pGL-3 basic, pGL-3 promoter vectors (Promega, Madison, WI, USA), respectively. Third, cultured Sw1990 cells were transfected with 0.8 μg pGL-3 vectors (inserted with rs4759314 -A allele/G allele or rs200349340 -T allele/G allele), while tThe pRL-TK Renilla control vector (0.1 μg) (Promega, Madison, WI, USA) was also co-transfected as an internal control for transfection efficiency. Furthermore, for pGL-3 promoter vectors, Sw1990 Cells were transfected with mimic control, antagomir control, miR-29a mimic or antagomir miR-29a (GenePharma, Shanghai) in different groups. Finally, firefly and renilla luciferase activity was measured on Synergy H1 microplate reader (BioTek Instruments, Winooski, VT, USA) using Dual-luciferase Reporter Assay System (Promega, Madison, WI, USA) according to the instructions of manufactures. Luciferase activity was normalized to renilla activity to correct for differences in transfection efficiency. For each reporter construct, three independent transfection experiments were performed, and each was done in triplicate.
Fig. 2.
HOTAIR rs4759314 and rs200349340 affect luciferase activities in Sw1990 Cells. a, b: The expression of HOTAIR in carriers of different genotypes of rs4759314 and rs200349340 was calculated relative to expression of GAPDH using the 2−∆∆Ct method.c: the schematic line diagram of the vector construct for rs4759314. d: relative luciferase activities (vs. Renila luciferase) of rs4759314 were measured in sw1990 cells transfected with A allele, or G allele. Three replicates for each group and the experiment were repeated three times; e: the schematic line diagram of the vector construct for rs200349340.f: relative luciferase activities (vs. Renila luciferase) of rs200349340 were measured in sw1990 cells transfected with G allele, or T allele. Cells in different groups were treated with control miR, miR-29a mimic or antagomir-miR-29a. Three replicates for each group and the experiment were repeated three times
Statistical analysis
Differences in the distribution of selected demographic variables and genotypes of SNPs were evaluated by Pearson’s χ2 test. Hardy–Weinberg equilibrium (HWE) for each SNP among controls was tested using a goodness-of-fit χ2-test. The associations of each SNP and PC susceptibility were estimated by using unconditional logistic regression analyses with odds ratios (ORs) and 95% confidence intervals (CIs). All analyses were conducted with SAS 9.3 software (SAS Institute, Inc., Cary, NC, USA), and a P value of less 0.05 for two-side was considered statistically significant.
Results
Characteristics of study subjects
In current study, a total of 900 PC cases and 900 controls were recruited in two stages, and there were no significant differences between PC cases and controls for each stage regarding to age, gender, drinking and smoking status (all P > 0.05, Table 1). However, we found PC cases are more likely to be diabetes patients in both stages (P < 0.001).
Table 1.
Distribution of selected characteristics among PC cases and controls
| Category | Stage 1 | Stage 2 | ||||
|---|---|---|---|---|---|---|
| Cases (N = 400) | Controls (N = 400) | P value | Cases (N = 500) | Controls (N = 500) | P value | |
| Age group | ||||||
| < 50 | 200 | 192 | 0.572 | 245 | 239 | 0.704 |
| ≥ 50 | 200 | 208 | 255 | 261 | ||
| Gender | ||||||
| Male | 240 | 245 | 0.717 | 304 | 299 | 0.747 |
| Female | 160 | 155 | 196 | 201 | ||
| Drinking | ||||||
| Never | 271 | 279 | 0.542 | 333 | 350 | 0.248 |
| Ever | 129 | 121 | 167 | 150 | ||
| Smoking | ||||||
| Never | 291 | 300 | 0.469 | 351 | 369 | 0.205 |
| Ever | 109 | 100 | 149 | 131 | ||
| Diabetes | ||||||
| Yes | 102 | 50 | < 0.001 | 137 | 66 | < 0.001 |
| No | 298 | 350 | 363 | 434 | ||
Categorical variables: numbers, and P-values from x2 test
Italic values indicate significant of p value (P<0.05)
Association of SNPs with PC susceptibility
Six SNPs, rs4759314, rs12427129, rs200349340, rs12312094, rs1838169, and rs7958904, were in accordance with Hardy–Weinberg equilibrium (HWE) in the control subjects. As shown in Table 2 and Table 3, we found that both SNP rs4759314 (OR = 1.84; 95 CI 1.26–2.67; P = 0.018) and rs200349340 (OR = 1.36; 95 CI 1.06–1.73; P = 0.014) were significantly associated with increased PC susceptibility, after adjusted for age, gender, Diabetes, drinking and smoking. Subsequently, we performed an independent validation set for the significant results and identified that rs4759314 and rs200349340 still had significant effects on PC risk (P < 0.05). When combined data of these two stages together, the minor alleles of rs4759314 (OR = 1.76; 95 CI 1.37–2.25; P = 0.001) and rs200349340 (OR = 1.32; 95 CI 1.12–1.56; P = 0.001) were significantly associated with PC susceptibility.
Table 2.
Genetic variants of HOTAIR and PC risk in Stage 1
| Genotype | Cases | Controls | Crude OR (95% CI) | Adjusted OR (95% CI)a |
|---|---|---|---|---|
| rs4759314 | ||||
| AA | 327 | 354 | 1.00 (reference) | 1.00 (reference) |
| AG | 65 | 44 | 1.60 (1.06–2.41) | 1.66 (1.09–2.54) |
| GG | 8 | 2 | 4.33 (1.04–18.1) | 4.50 (1.11–18.3) |
| G vs A | 1.76 (1.22–2.55) | 1.84 (1.26–2.67) | ||
| P trend | 0.024 | 0.018 | ||
| rs12427129 | ||||
| GG | 359 | 356 | 1.00 (reference) | 1.00 (reference) |
| AG | 36 | 41 | 0.87 (0.54–1.39) | 0.91 (0.66–1.25) |
| AA | 5 | 3 | 1.65 (0.4–6.87) | 1.72 (0.4–7.43) |
| A vs G | 0.98 (0.64–1.49) | 1.02 (0.76–1.35) | ||
| P trend | 0.564 | 0.546 | ||
| rs200349340 | ||||
| GG | 205 | 236 | 1.00 (reference) | 1.00 (reference) |
| TG | 161 | 140 | 1.32 (0.99–1.78) | 1.38 (1.00–1.89) |
| TT | 34 | 24 | 1.63 (0.94–2.83) | 1.70 (0.96–2.99) |
| T vs G | 1.31 (1.04–1.63) | 1.36 (1.06–1.73) | ||
| P trend | 0.061 | 0.014 | ||
| rs12312094 | ||||
| CC | 260 | 278 | 1.00 (reference) | 1.00 (reference) |
| CG | 128 | 116 | 1.18 (0.87–1.6) | 1.18 (0.87–1.60) |
| GG | 12 | 6 | 2.14 (0.81–5.66) | 2.13 (0.81–5.65) |
| G vs C | 1.23 (0.95–1.59) | 1.23 (0.89–1.70) | ||
| P trend | 0.284 | 0.214 | ||
| rs1838169 | ||||
| CC | 269 | 275 | 1.00 (reference) | 1.00 (reference) |
| CG | 122 | 118 | 1.06 (0.78–1.43) | 1.10 (0.67–1.80) |
| GG | 9 | 7 | 1.31 (0.48–3.57) | 1.37 (0.46–4.06) |
| G vs C | 1.07 (0.83–1.39) | 1.12 (0.76–1.64) | ||
| P trend | 0.721 | 0.577 | ||
| rs7958904 | ||||
| GG | 215 | 237 | 1.00 (reference) | 1.00 (reference) |
| CG | 156 | 136 | 1.26 (0.94–1.7) | 1.31 (0.94–1.82) |
| CC | 29 | 27 | 1.18 (0.68–2.06) | 1.23 (0.64–2.36) |
| C vs G | 1.17 (0.94–1.47) | 1.22 (0.93–1.59) | ||
| P trend | 0.119 | 0.147 | ||
aAdjusting for age, gender, diabetes, drinking and smoking
Italic values indicate significant of p value (P<0.05)
Table 3.
Associations of genetic variants of HOTAIR with PC risk in Stage 2 and combined results
| Genotype | Cases | Controls | Crude OR (95% CI) | Adjusted OR (95% CI)a |
|---|---|---|---|---|
| rs4759314 | ||||
| Stage 2 | ||||
| AA | 406 | 437 | 1.00 (reference) | 1.00 (reference) |
| AG | 83 | 59 | 1.51 (1.06–2.17) | 1.57 (1.08–2.29) |
| GG | 11 | 4 | 2.96 (0.99–8.89) | 3.08 (1.04–9.13) |
| G vs A | 1.63 (1.19–2.24) | 1.7 (1.23–2.36) | ||
| P trend | 0.023 | 0.017 | ||
| Combined | ||||
| AA | 733 | 791 | 1.00 (reference) | 1.00 (reference) |
| AG | 148 | 103 | 1.55 (1.18–2.03) | 1.61 (1.22–2.13) |
| GG | 19 | 6 | 3.42 (1.43–8.16) | 3.55 (1.51–8.36) |
| G vs A | 1.69 (1.33–2.15) | 1.76 (1.37–2.25) | ||
| P trend | 0.001 | 0.001 | ||
| rs200349340 | ||||
| Stage 2 | ||||
| GG | 264 | 295 | 1.00 (reference) | 1.00 (reference) |
| TG | 196 | 175 | 1.25 (0.96–1.63) | 1.30 (0.97–1.74) |
| TT | 40 | 30 | 1.49 (0.9–2.45) | 1.54 (0.92–2.61) |
| T vs G | 1.24 (1.01–1.52) | 1.29 (1.03–1.62) | ||
| P trend | 0.094 | 0.027 | ||
| Combined | ||||
| GG | 469 | 531 | 1.00 (reference) | 1.00 (reference) |
| TG | 357 | 315 | 1.28 (1.06–1.56) | 1.33 (1.07–1.65) |
| TT | 74 | 54 | 1.55 (1.07–2.25) | 1.61 (1.10–2.37) |
| T vs G | 1.27 (1.09–1.47) | 1.32 (1.12–1.56) | ||
| P trend | 0.013 | 0.001 | ||
aAdjusting for age, gender, diabetes, drinking and smoking
Italic values indicate significant of p value (P<0.05)
Functional relevance of rs4759314 and rs200349340 on HOTAIR expression
We found that subjects with the minor alleles of rs4759314 and rs200349340 had significantly higher HOTAIR RNA levels (mean ± SD) than those with the major alleles in PC tissues (Fig. 2a, b, P < 0.01). Then dual luciferase reporter assay was conducted to evaluate the potential function of HOTAIR rs4759314 and rs200349340. For rs4759314, cells transfected with rs4759314 -G allele construct showed higher relative luciferase activity (Fig. 2c, d, P < 0.01); while for rs200349340, cells transfected with rs200349340 -G allele construct, miR-29a mimic showed more sensitive change of the relative luciferase activity (Fig. 2e, f, P < 0.01).
Discussion
The current study explored the association between four functional SNPs located in 3′ UTR region of the HOTAIR gene and PC risk using a two-stage case–control study design. We found rs4759314 and rs200349340 were significantly associated with increased PC susceptibility. Functional studies revealed that minor alleles of rs4759314 and rs200349340 increased HOTAIR expression.
HOTAIR was one of the most studied lncRNAs in cancers [27–29]. LncRNA HOTAIR was first identified to impact H3K27me3 and breast cancer metastasis by Gupta et al. [30]. Since then, many studies have reported the function of HOTAIR regarding the pro-oncogenic and prognostic activities, radiosensitivity, biomarkers, cell proliferation, and metastasis of PC [18, 19, 25, 31–36]. Our study identified that expression of HOTAIR was significantly higher in PC tissues than that in adjacent normal tissues, which is consistent with the results of ONCOMINE (https://www.oncomine.org).
Genetic variants of lncRNA HOTAIR has been linked to many diseases, including cancers, congenital heart disease, lower extremity malformations, benign prostate hyperplasia, noise-induced hearing loss, lead poisoning, and so on [26, 37–47]. In 2014, Zhang et al. [37] first evaluated variants of HOTAIR in esophageal squamous cell carcinoma, and found SNP rs920778 that regulates the expression of lncRNA HOTAIR via a novel intronic enhancer. Following, it was linked to breast cancer, gastric cancer, colorectal cancer, lung cancer, cervical cancer, ovarian cancer, osteosarcoma, papillary thyroid carcinoma, hepatocellular carcinoma, glioma, prostate cancer, oral cancer, neuroblastoma [26, 42–44, 47, 48]. Li found rs4759314 polymorphism was associated with the risk of congenital heart disease by alternating downstream signaling via reducing its expression [38]. A significant association between polymorphism rs4759314 and susceptibility to gastric cancer in allele contrast was identified by an epidemiological investigation and a meta-analysis [21, 49]. This was similar to our results, that rs4759314 was significantly associated with increased PC susceptibility. Another interesting finding was that SNP rs200349340, resided in 3′UTR region of the HOTAIR gene, contributing to a genotype specific effect on the expression of lncRNA HOTAIR via interfering the binding of miR-29a. The miRNA-29 family, including miR-29a, miR-29b and miR-29c, was recently reported to be aberrantly expressed in multiple cancers [50]. Previously, miR-29a-3p were identified to be associated with prognosis of hepatocellular carcinoma [51]. Meanwhile, miR-29a was found to function as tumor suppressors by targeting the MUC1 mucin in pancreatic cancer cells [52]. To the best of our knowledge, the functional polymorphism- rs200349340 has not been reported before.
One of the major strengths of the current study is the large sample size, since with a total of 1800 subjects were included in this study. Our statistical power reached 99.5% for detecting an association for rs4759314, 95.5% for detecting an association for rs200349340. The second strength should be two-stage case–control study design, which makes the positive findings more reliable. Finally, the functional validation of the variant rs200349340 further convinced the results. Several limitations may also exist in the current study. For example, there might be inherent selection bias for the hospital-based study. Thus, the findings of our study are warranted to be validated in different populations. Also, we only obtained information on smoking, drinking and diabetes in the present study. Further studies should focus on more environmental risk factors, such as dietary habits, life style of the subjects, to make the result more informative, and to perform potential interaction analyses.
Conclusions
In conclusion, we identified that functional SNP rs4759314 and rs200349340 of HOTAIR had strong associations with PC susceptibility. These indicated that lncRNA HOTAIR and its genetic variations may be a potential target for risk evaluation, early detection and therapeutic target of PC; and larger prospective and experimental studies were warranted to further validation.
Additional file
Additional file 1: Figure S1. The mass spectrum plot for rs200349340 TT genotype. Figure S2. The mass spectrum plot for rs200349340 GT genotype. Figure S3. The mass spectrum plot for rs200349340 GG genotype.
Authors’ contributions
LX, DJ: study design, data analysis, writing of the article, and final approval of the article. DJ, JN, MC, LS: data collection, data analysis, writing of the article and final approval of the article. All authors read and approved the final manuscript.
Acknowledgements
The authors would like to thank the patients and the supporting staff in this study.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during the present study are included in this published article.
Consent for publication
All the participants provided signed informed consent before the study.
Ethics approval and consent to participate
Informed consent was obtained from each participant and all procedures and study guidelines were approved by the ethics Committee.
Funding
We acknowledge financial support from Zhejiang Medical and Health Science and Technology Plan Project (2019KY691).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- PC
pancreatic cancer
- lncRNA
long noncoding RNA
- HOTAIR
HOX transcript antisense RNA
- RT-PCR
real-time reverse transcription-polymerase chain reaction
- PRC2
polycomb repressive complex 2
- MALDI-TOF MS
matrix assisted laser desorption ionization time-of-flight mass spectrometry
Contributor Information
Dawei Jiang, Email: jiandawei@jiaxing.com.
Liu Xu, Phone: +86 0573 82519768, Email: xuliuzj@aliyun.com.
Jianqi Ni, Email: nijianqi@jiaxing.com.
Jie Zhang, Email: zhangjie@jiaxing.com.
Min Cai, Email: caimin@jiaxing.com.
Lan Shen, Email: shenlan@jiaxing.com.
References
- 1.Michl P, Gress TM. Current concepts and novel targets in advanced pancreatic cancer. Gut. 2013;62(2):317–326. doi: 10.1136/gutjnl-2012-303588. [DOI] [PubMed] [Google Scholar]
- 2.Poston GJ, Gillespie J, Guillou PJ. Biology of pancreatic cancer. Gut. 1991;32(7):800–812. doi: 10.1136/gut.32.7.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 4.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 5.Zheng J, Huang X, Tan W, Yu D, Du Z, Chang J, Wei L, Han Y, Wang C, Che X, et al. Pancreatic cancer risk variant in LINC00673 creates a miR-1231 binding site and interferes with PTPN11 degradation. Nat Genet. 2016;48(7):747–757. doi: 10.1038/ng.3568. [DOI] [PubMed] [Google Scholar]
- 6.Seufferlein T, Mayerle J. Pancreatic cancer in 2015: precision medicine in pancreatic cancer–fact or fiction? Nat Rev Gastroenterol Hepatol. 2016;13(2):74–75. doi: 10.1038/nrgastro.2015.215. [DOI] [PubMed] [Google Scholar]
- 7.Wolpin BM, Rizzato C, Kraft P, Kooperberg C, Petersen GM, Wang Z, Arslan AA, Beane-Freeman L, Bracci PM, Buring J, et al. Genome-wide association study identifies multiple susceptibility loci for pancreatic cancer. Nat Genet. 2014;46(9):994–1000. doi: 10.1038/ng.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leveille N, Melo CA, Rooijers K, Diaz-Lagares A, Melo SA, Korkmaz G, Lopes R, Akbari Moqadam F, Maia AR, Wijchers PJ, et al. Genome-wide profiling of p53-regulated enhancer RNAs uncovers a subset of enhancers controlled by a lncRNA. Nat Commun. 2015;6:6520. doi: 10.1038/ncomms7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang Q, Wang J, Wu X, Ma R, Zhang T, Jin S, Han Z, Tan R, Peng J, Liu G, et al. LncRNA2Target: a database for differentially expressed genes after lncRNA knockdown or overexpression. Nucleic Acids Res. 2015;43(Database issue):D193–D196. doi: 10.1093/nar/gku1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang W, Thomas B, Flynn RA, Gavzy SJ, Wu L, Kim SV, Hall JA, Miraldi ER, Ng CP, Rigo FW, et al. DDX5 and its associated lncRNA Rmrp modulate TH17 cell effector functions. Nature. 2015;528(7583):517–522. doi: 10.1038/nature16193. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Ding J, Yeh CR, Sun Y, Lin C, Chou J, Ou Z, Chang C, Qi J, Yeh S. Estrogen receptor beta promotes renal cell carcinoma progression via regulating LncRNA HOTAIR-miR-138/200c/204/217 associated CeRNA network. Oncogene. 2018;37:5037–5053. doi: 10.1038/s41388-018-0175-6. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Wang W, Zhang Q, Gu D, Zhang K, Ge Y, Chu H, Du M, Xu B, Wang M, et al. Tagging SNPs in the HOTAIR gene are associated with bladder cancer risk in a Chinese population. Gene. 2018;664:22–26. doi: 10.1016/j.gene.2018.04.039. [DOI] [PubMed] [Google Scholar]
- 13.Lv R, Zhang J, Zhang W, Huang Y, Wang N, Zhang Q, Qu S. Circulating HOTAIR expression predicts the clinical response to neoadjuvant chemotherapy in patients with breast cancer. Cancer Biomark. 2018;22:249–256. doi: 10.3233/CBM-171019. [DOI] [PubMed] [Google Scholar]
- 14.Tan SK, Pastori C, Penas C, Komotar RJ, Ivan ME, Wahlestedt C, Ayad NG. Serum long noncoding RNA HOTAIR as a novel diagnostic and prognostic biomarker in glioblastoma multiforme. Mol Cancer. 2018;17(1):74. doi: 10.1186/s12943-018-0822-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y, Fang Z, Lin Z, Li Z, Zhao J, Luo Y, Xu B. The prognostic impact of long noncoding RNA HOTAIR in leukemia and lymphoma: a meta-analysis. Hematology. 2018;23:1–8. doi: 10.1080/10245332.2018.1446572. [DOI] [PubMed] [Google Scholar]
- 16.Zhao W, Geng D, Li S, Chen Z, Sun M. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018;7(3):842–855. doi: 10.1002/cam4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Dasgupta P, Kulkarni P, Majid S, Shahryari V, Hashimoto Y, Bhat NS, Shiina M, Deng G, Saini S, Tabatabai ZL, et al. MicroRNA-203 inhibits long noncoding RNA HOTAIR and regulates tumorigenesis through epithelial-to-mesenchymal transition pathway in renal cell carcinoma. Mol Cancer Ther. 2018;17(5):1061–1069. doi: 10.1158/1535-7163.MCT-17-0925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li CH, Xiao Z, Tong JH, To KF, Fang X, Cheng AS, Chen Y. EZH2 coupled with HOTAIR to silence MicroRNA-34a by the induction of heterochromatin formation in human pancreatic ductal adenocarcinoma. Int J Cancer. 2017;140(1):120–129. doi: 10.1002/ijc.30414. [DOI] [PubMed] [Google Scholar]
- 19.Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, Kim S, Safe S. HOTAIR is a negative prognostic factor and exhibits pro-oncogenic activity in pancreatic cancer. Oncogene. 2013;32(13):1616–1625. doi: 10.1038/onc.2012.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan W, Liu L, Wei J, Ge Y, Zhang J, Chen H, Zhou L, Yuan Q, Zhou C, Yang M. A functional lncRNA HOTAIR genetic variant contributes to gastric cancer susceptibility. Mol Carcinog. 2015;55:90–96. doi: 10.1002/mc.22261. [DOI] [PubMed] [Google Scholar]
- 21.Du M, Wang W, Jin H, Wang Q, Ge Y, Lu J, Ma G, Chu H, Tong N, Zhu H, et al. The association analysis of lncRNA HOTAIR genetic variants and gastric cancer risk in a Chinese population. Oncotarget. 2015;6(31):31255–31262. doi: 10.18632/oncotarget.5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao Z, Qu Z, Chen Z, Fang Z, Zhou K, Huang Z, Guo X, Zhang Y. LncRNA HOTAIR is a prognostic biomarker for the proliferation and chemoresistance of colorectal cancer via MiR-203a-3p-Mediated Wnt/ss-catenin signaling pathway. Cell Physiol Biochem. 2018;46(3):1275–1285. doi: 10.1159/000489110. [DOI] [PubMed] [Google Scholar]
- 23.Qiu JJ, Lin YY, Ye LC, Ding JX, Feng WW, Jin HY, Zhang Y, Li Q, Hua KQ. Overexpression of long non-coding RNA HOTAIR predicts poor patient prognosis and promotes tumor metastasis in epithelial ovarian cancer. Gynecol Oncol. 2014;134(1):121–128. doi: 10.1016/j.ygyno.2014.03.556. [DOI] [PubMed] [Google Scholar]
- 24.Loewen G, Jayawickramarajah J, Zhuo Y, Shan B. Functions of lncRNA HOTAIR in lung cancer. J Hematol Oncol. 2014;7:90. doi: 10.1186/s13045-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Z, Chen X, Li J, Guo Y, Li H, Pan X, Jiang J, Liu H, Wu B. Salivary HOTAIR and PVT1 as novel biomarkers for early pancreatic cancer. Oncotarget. 2016;7(18):25408–25419. doi: 10.18632/oncotarget.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Cui Z, Li H, Lv X, Gao M, Yang Z, Bi Y, Zhou B, Yin Z. Long non-coding RNA HOTAIR polymorphism and susceptibility to cancer: an updated meta-analysis. Environ Health Prev Med. 2018;23(1):8. doi: 10.1186/s12199-018-0697-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fagoonee S, Durazzo M. HOTAIR and gastric cancer: a lesson from two meta-analyses. Panminerva Med. 2017;59(3):201–202. doi: 10.23736/S0031-0808.17.03333-X. [DOI] [PubMed] [Google Scholar]
- 28.Avazpour N, Hajjari M, Tahmasebi Birgani M. HOTAIR: a promising long non-coding RNA with potential role in breast invasive carcinoma. Front Genet. 2017;8:170. doi: 10.3389/fgene.2017.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhan A, Mandal SS. LncRNA HOTAIR: a master regulator of chromatin dynamics and cancer. Biochim Biophys Acta. 2015;1856(1):151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu C, Yang L, Qi X, Wang T, Li M, Xu K. Inhibition of long non-coding RNA HOTAIR enhances radiosensitivity via regulating autophagy in pancreatic cancer. Cancer Manag Res. 2018;10:5261–5271. doi: 10.2147/CMAR.S174066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Dong P, Wang W, Huang M, Tian B. Gemcitabine treatment causes resistance and malignancy of pancreatic cancer stem-like cells via induction of lncRNA HOTAIR. Exp Ther Med. 2017;14(5):4773–4780. doi: 10.3892/etm.2017.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang SZ, Xu F, Zhou T, Zhao X, McDonald JM, Chen Y. The long non-coding RNA HOTAIR enhances pancreatic cancer resistance to TNF-related apoptosis-inducing ligand. J Biol Chem. 2017;292(25):10390–10397. doi: 10.1074/jbc.M117.786830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai H, Yao J, An Y, Chen X, Chen W, Wu D, Luo B, Yang Y, Jiang Y, Sun D, et al. LncRNA HOTAIR acts a competing endogenous RNA to control the expression of notch3 via sponging miR-613 in pancreatic cancer. Oncotarget. 2017;8(20):32905–32917. doi: 10.18632/oncotarget.16462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai H, An Y, Chen X, Sun D, Chen T, Peng Y, Zhu F, Jiang Y, He X. Epigenetic inhibition of miR-663b by long non-coding RNA HOTAIR promotes pancreatic cancer cell proliferation via up-regulation of insulin-like growth factor 2. Oncotarget. 2016;7(52):86857–86870. doi: 10.18632/oncotarget.13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang Y, Li Z, Zheng S, Chen H, Zhao X, Gao W, Bi Z, You K, Wang Y, Li W, et al. The long non-coding RNA HOTAIR affects the radiosensitivity of pancreatic ductal adenocarcinoma by regulating the expression of Wnt inhibitory factor 1. Tumour Biol. 2016;37(3):3957–3967. doi: 10.1007/s13277-015-4234-0. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, Zhou L, Fu G, Sun F, Shi J, Wei J, Lu C, Zhou C, Yuan Q, Yang M. The identification of an ESCC susceptibility SNP rs920778 that regulates the expression of lncRNA HOTAIR via a novel intronic enhancer. Carcinogenesis. 2014;35(9):2062–2067. doi: 10.1093/carcin/bgu103. [DOI] [PubMed] [Google Scholar]
- 38.Li Y, Zhao W, Shi R, Jia J, Li X, Cheng J. Rs4759314 polymorphism located in HOTAIR is associated with the risk of congenital heart disease by alternating downstream signaling via reducing its expression. J Cell Biochem. 2018;119(10):8112–8122. doi: 10.1002/jcb.26736. [DOI] [PubMed] [Google Scholar]
- 39.Alvarado DM, McCall K, Hecht JT, Dobbs MB, Gurnett CA. Deletions of 5′ HOXC genes are associated with lower extremity malformations, including clubfoot and vertical talus. J Med Genet. 2016;53(4):250–255. doi: 10.1136/jmedgenet-2015-103505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taheri M, Habibi M, Noroozi R, Rakhshan A, Sarrafzadeh S, Sayad A, Omrani MD, Ghafouri-Fard S. HOTAIR genetic variants are associated with prostate cancer and benign prostate hyperplasia in an Iranian population. Gene. 2017;613:20–24. doi: 10.1016/j.gene.2017.02.031. [DOI] [PubMed] [Google Scholar]
- 41.Chen L, Zhao QN, Qian XR, Zhu BL, Ding EM, Wang BS, Zhang HD, Yang H. Association between the HOTAIR Polymorphism and Susceptibility to Lead Poisoning in a Chinese Population. Biomed Environ Sci BES. 2018;31(6):473–478. doi: 10.3967/bes2018.063. [DOI] [PubMed] [Google Scholar]
- 42.Tian T, Li C, Xiao J, Shen Y, Lu Y, Jiang L, Zhuang X, Chu M. Quantitative assessment of the polymorphisms in the HOTAIR lncRNA and cancer risk: a meta-analysis of 8 case-control studies. PLoS ONE. 2016;11(3):e0152296. doi: 10.1371/journal.pone.0152296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang J, Liu X, You LH, Zhou RZ. Significant association between long non-coding RNA HOTAIR polymorphisms and cancer susceptibility: a meta-analysis. OncoTargets Ther. 2016;9:3335–3343. doi: 10.2147/OTT.S107190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chu H, Chen Y, Yuan Q, Hua Q, Zhang X, Wang M, Tong N, Zhang W, Chen J, Zhang Z. The HOTAIR, PRNCR1 and POLR2E polymorphisms are associated with cancer risk: a meta-analysis. Oncotarget. 2017;8(26):43271–43283. doi: 10.18632/oncotarget.14920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X, Duan Q, Zhang J. Quantitative assessment of lncRNA HOTAIR polymorphisms and cancer risk in Chinese population: a meta-analysis based on 26,810 subjects. Oncotarget. 2017;8(35):59698–59708. doi: 10.18632/oncotarget.19776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang ZX, Tong X, Zhang WN, Fu WN. Association between the HOTAIR polymorphisms and cancer risk: an updated meta-analysis. Oncotarget. 2017;8(3):4460–4470. doi: 10.18632/oncotarget.13880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Min L, Mu X, Tong A, Qian Y, Ling C, Yi T, Zhao X. The association between HOTAIR polymorphisms and cancer susceptibility: an updated systemic review and meta-analysis. OncoTargets Ther. 2018;11:791–800. doi: 10.2147/OTT.S151454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu FT, Zhou L, Qiu C, Xia G, Zhu PQ, Luo HL. Association of the HOTAIR rs4759314 polymorphism with cancer risk: a meta-analysis. J BUON. 2016;21(4):1016–1023. [PubMed] [Google Scholar]
- 49.Qi Q, Wang J, Huang B, Chen A, Li G, Li X, Wang J. Association of HOTAIR polymorphisms rs4759314 and rs920778 with cancer susceptibility on the basis of ethnicity and cancer type. Oncotarget. 2016;7(25):38775–38784. doi: 10.18632/oncotarget.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y, Zhang X, Li H, Yu J, Ren X. The role of miRNA-29 family in cancer. Eur J Cell Biol. 2013;92(3):123–128. doi: 10.1016/j.ejcb.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 51.Zhu HT, Hasan AM, Liu RB, Zhang ZC, Zhang X, Wang J, Wang HY, Wang F, Shao JY. Serum microRNA profiles as prognostic biomarkers for HBV-positive hepatocellular carcinoma. Oncotarget. 2016;7:45637. doi: 10.18632/oncotarget.10082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trehoux S, Lahdaoui F, Delpu Y, Renaud F, Leteurtre E, Torrisani J, Jonckheere N, Van Seuningen I. Micro-RNAs miR-29a and miR-330-5p function as tumor suppressors by targeting the MUC1 mucin in pancreatic cancer cells. Biochim Biophys Acta. 2015;1853(10 Pt A):2392–2403. doi: 10.1016/j.bbamcr.2015.05.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. The mass spectrum plot for rs200349340 TT genotype. Figure S2. The mass spectrum plot for rs200349340 GT genotype. Figure S3. The mass spectrum plot for rs200349340 GG genotype.
Data Availability Statement
All data generated or analyzed during the present study are included in this published article.