Sir,
Since the re-emergence of chikungunya virus (CHIKV) in India during 2005, several outbreaks have been reported in different parts of the country, including Maharashtra1. Muniaraj2 reported declining chikungunya incidence in India. However, outbreaks continue to occur in areas which were not affected previously, highlighting the need to investigate whether it is endemic in the country.
During the monsoon season (June-July) of 2016, there was a significant surge in suspected CHIKV and Dengue virus (DENV) fever cases in Pune city and the adjoining suburban areas of Maharashtra State, India. Both are RNA viruses and transmitted by the vectors Aedes aegypti and Ae. albopictus mosquitoes3. Co-circulation of CHIKV and DENV in Southeast Asia including India has been reported4. Since, the first record of CHIKV in West Bengal, India, during 19635, many sporadic cases have occurred in Maharashtra. DENV cases were also reported in many States including Maharashtra during 2010-20166. Overall in Maharashtra, a higher seropositivity for DENV was noted in urbanized villages as compared to rural villages7. Although CHIK and DEN fever display overlapping clinical manifestation and transmitted by the same vectors with similar epidemiology and geographic distribution, scanty data are available on CHIKV characterization.
Here we report the serological and molecular evidence of CHIKV and DENV infections among the suspected cases from the urbanized villages, namely Uttamnagar, Shivne, and Kondhwa Dhawde of Pune district, Maharashtra State, India in 2016.
Of the 282 suspected cases in these villages, 214 patients were admitted to the hospitals while 68 were outpatients. Blood samples (5 ml) were collected from both hospitalized and outpatients with symptoms of mild-to-high fever, severe headache, pain behind the eyes, muscle and joint pain and rash. The samples were transported on wet ice to the ICMR-National Institute of Virology, Pune and tested for immunoglobulin M (IgM) antibodies to CHIKV and DENV8. The study was approved by the Institutional Ethics Committee.
The suspected patients were aged between 6 and 75 years (144 females, aged between 9 and 75 yr and 138 males, aged between 6 and 75 yr). The median age was 34 years. The age-wise distribution of confirmed CHIKV and DENV cases did not vary significantly (i.e., 6-75 yr females 101 and males 97).
Of the 282 cases, 198 (70.2%) cases were positive for CHIKV IgM, DENV IgM or both. Among these, 32.6 per cent (n=92) patients were exclusively positive for CHIKV IgM while 14.5 per cent (n=41) were tested positive for DENV IgM alone. Twenty three per cent (n=65) patients tested positive both for CHIKV and DENV IgM. Of the 65 dual-positive cases, convalescent-phase serum samples were collected from 24 patients and tested for the presence of anti-DENV (Standard Diagnostics, Borahagal-ro, Republic of Korea)/CHIKV IgG capture ELISA kits (InBios International, Inc. Seattle, Washington, USA) as per manufacturer's instructions. All these patients were positive for anti-CHIKV and anti-DENV IgG antibodies. These samples were tested further for virus neutralization and a four-fold rise in antibody titre was observed in convalescent phase when compared with the acute phase (data not shown). In addition to fever, the predominant symptoms of serologically confirmed cases (n=198) were joint pain (98%), debilitating weakness (60%), severe myalgia (46%), rash (40%) and malaise (29%).
The possible reason of co-infection may be the abundance of Ae. aegypti and Ae. albopictus mosquitoes in this region. This might have resulted in a disease with overlapping signs and symptoms, making the diagnosis and treatment difficult8.
Fifteen acute serum samples available in sufficient quantity and positive for DENV (n=4) and CHIKV (n=11) by real-time polymerase chain reaction were subjected to virus isolation in Vero cells and yielded five CHIKV isolates. Only CHIKV could be isolated from the patient samples; this could be due to higher virus loads present in the acute samples.
CHIKV has been classified into three distinct genotypes: Asia; East/Central/South African (ECSA) and West African9. CHIK isolates identified in India during 1963, and 1973 belonged to Asian genotype; however, recent epidemics in Indian Ocean and Indian subcontinent were caused by virus of the Indian Ocean Lineage (IOL)9 which has emerged from ECSA genotype. The appearance of new genotypes of ECSA strains was resulted due to microevolutionary changes in the CHIKV genome10.
Full genome sequencing and phylogenetic analysis of CHIKV isolate were performed using MEGA5.1 (https://mega.software.informer.com/5.1) with the neighbour-joining method9. Comparative amino acid analysis was also performed using the CHIKV prototype (S27 East-Central South African) strain from Africa and the Karnataka 2006 strains (IOL group). This analysis revealed that CHIK 2016 Pune isolate GenBank accession number MG137428) belonged to the IOL of the ECSA genotype (Figure). Comparative amino acid analysis of this isolate with CHIKV prototype (S27 ECSA) strain from Africa and Karnataka 2006 strains (IOL group) indicated four mutations (E1: I317V, E1 K211E, E2: A76T, E2: V264A) in E protein. The changes in the amino acids are represented in the Table.
Figure.
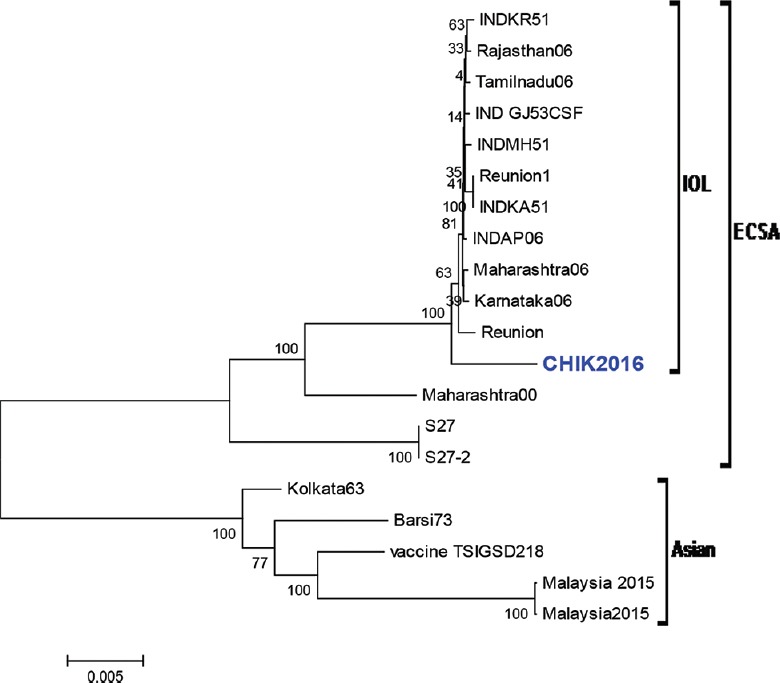
Phylogenetic analysis of CHIK 2016 isolate based on full genome sequence: tree depicting the genotypic status of Indian CHIK virus isolates, based on full-length genome sequences of 18 isolates. Numbers at nodes indicate bootstrap support (%). Isolates included are CHIK2016 (Pune isolate), Karnataka06 (EF027135), Andhra Pradesh06 (INDAP06), Maharashtra06 (EF027136), Tamilnadu06 (EF027138), Rajasthan06 (EF027137), INDKR51 (FJ000066), INDGJ53CSF (FJ000065), INDMH51 (FJ000067), INDKA51 (FJ000068), Reunion (DQ443544.2), Reunion1 (KT449801.1), Maharashtra (EF027139), Kolkatta63 (EF027140), Barsi73 (EF027141), S27 (AF369024), S27-2 (AF490259), Vaccine TSIGSD218 (EF452494), Malaysia2015 (KM923919), Malaysia2015 (KM923920). *All strains were obtained from humans except for Maharashtra00 (EF027139.1) which was obtained from a mosquito.
Table.
Amino acid changes in chikungunya virus Pune 2016 isolate compared to S27 prototype East/Central/South African strain from Africa and Karnataka 2006 strain
| Genomic region | Amino acid position in the protein | S27 (prototype ECSA strain) | Karnataka 2006 strain | CHIKV Pune 2016 isolate |
|---|---|---|---|---|
| NS1 | 171 | R | R | Q |
| NS2 | 130 | H | H | Y |
| NS2 | 374 | H | Y | H |
| NS2 | 676 | L | L | P |
| NS3 | 326 | - | S | P |
| NS4 | 85 | R | R | G |
| CAPSID | 79 | N | N | S |
| E2 | 76 | A | A | T |
| E2 | 264 | V | V | A |
| E1 | 211 | K | N | E |
| E1 | 317 | I | I | V |
ECSA, East/Central/South African; CHIKV, chikungunya virus
Notably, the previously reported single amino acid mutation (A226V) in the envelope protein 1 (E1), that determined the infectivity and vector specificity in isolates from Kerala, India11, was absent. The La Reunion island studies reported that a point mutation in E protein could increase its infectivity for Ae. albopictus12. Four mutations (E1:I317V, E1 K211E, E2:A76T, E2:V264A) were noted in the E protein of CHIK 2016 Pune isolate. The significance of these mutations is not known and needs to be studied.
The role of these mutations highlights the need to study their possible effects on virus neutralization and the occurrence of viral variants. The persistence and outbreaks of these infections in South Asia including India call for sustained monitoring of the viruses circulating in both the endemic and non-endemic areas.
Acknowledgment
The authors thank the services of the hospital staff, along with the health authorities of the study area of Pune district and Servshri Shivshankar Gaikwad and Ajay Koli for their support in specimen collection. The authors thank Dr D.T. Mourya, Director, NIV and Dr Kavita Lole for critical suggestions.
Footnotes
Financial support & sponsorship: The financial assistance for this study was provided by the Indian Council of Medical Research (ICMR), New Delhi, India.
Conflicts of Interest: None.
References
- 1.National Vector Borne Disease Control Programme India. [accessed on November 10, 2018]. Available from: http://www.nvbdcp.gov.in/index4.php?lang=1&level=0&linkid=407&lid=3683 .
- 2.Muniaraj M. Fading chikungunya fever from India: Beginning of the end of another episode? Indian J Med Res. 2014;139:468–70. [PMC free article] [PubMed] [Google Scholar]
- 3.Chahar HS, Bharaj P, Dar L, Guleria R, Kabra SK, Broor S, et al. Co-infections with chikungunya virus and dengue virus in Delhi, India. Emerg Infect Dis. 2009;15:1077–80. doi: 10.3201/eid1507.080638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nayar SK, Noridah O, Paranthaman V, Ranjit K, Norizah I, Chem YK, et al. Co-infection of dengue virus and chikungunya virus in two patients with acute febrile illness. Med J Malaysia. 2007;62:335–6. [PubMed] [Google Scholar]
- 5.Shah KV, Gibbs CJ, Jr, Banerjee G. Virological investigation of the epidemic of haemorrhagic fever in Calcutta: Isolation of three strains of chikungunya virus. Indian J Med Res. 1964;52:676–83. [PubMed] [Google Scholar]
- 6.Cecilia D. Current status of dengue and chikungunya in India. WHO South East Asia J Public Health. 2014;3:22–6. doi: 10.4103/2224-3151.206879. [DOI] [PubMed] [Google Scholar]
- 7.Yergolkar PN, Tandale BV, Arankalle VA, Sathe PS, Sudeep AB, Gandhe SS, et al. Chikungunya outbreaks caused by African genotype, India. Emerg Infect Dis. 2006;12:1580–3. doi: 10.3201/eid1210.060529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinkar A, Singh J, Prakash P, Das A, Nath G. Hidden burden of chikungunya in North India; a prospective study in a tertiary care centre. J Infect Public Health. 2018;11:586–91. doi: 10.1016/j.jiph.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Patil J, More A, Patil P, Jadhav S, Newase P, Agarwal M, et al. Genetic characterization of chikungunya viruses isolated during the 2015-2017 outbreaks in different states of India, based on their E1 and E2 genes. Arch Virol. 2018;163:3135–40. doi: 10.1007/s00705-018-3974-8. [DOI] [PubMed] [Google Scholar]
- 10.Shrinet J, Jain S, Sharma A, Singh SS, Mathur K, Rana V, et al. Genetic characterization of chikungunya virus from New Delhi reveal emergence of a new molecular signature in Indian isolates. Virol J. 2012;9:100. doi: 10.1186/1743-422X-9-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar NP, Joseph R, Kamaraj T, Jambulingam P. A226V mutation in virus during the 2007 chikungunya outbreak in Kerala, India. J Gen Virol. 2008;89:1945–8. doi: 10.1099/vir.0.83628-0. [DOI] [PubMed] [Google Scholar]
- 12.Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3:e201. doi: 10.1371/journal.ppat.0030201. [DOI] [PMC free article] [PubMed] [Google Scholar]


